Summary
The vascular system is essential for embryonic development and adult life. Aberrant vascularization is associated with numerous diseases, including cancer, atherosclerosis, retinopathy, and stroke. Vascular development begins when mesodermal cells differentiate into endothelial cells, which then form primitive vessels. It has been hypothesized that endothelial-specific gene expression may be regulated combinatorially, but the transcriptional mechanisms governing vascular gene expression remain incompletely understood. Here, we identify a transcriptional code, consisting of Forkhead and Ets factors, which is required and sufficient for vascular development and endothelial gene expression through combinatorial activation of a composite cis-acting element. We show that the presence of this FOX:ETS motif is an effective predictor of endothelial-specific enhancers. These studies establish a paradigm in which two broadly expressed classes of transcription factors regulate tissue specific expression combinatorially through a single composite cis-acting element. This mechanism has broad implications for understanding differentiation and gene expression in many tissues.
Introduction
The establishment of the vascular system begins prior to the beating of the heart and initially forms through a process referred to as vasculogenesis. Mesodermal cells differentiate into endothelial cell precursors and form primitive vessels, which are then rapidly remodeled through endothelial sprouting, branching, and intussusception from existing blood vessels (Flamme et al., 1997; Patan, 2004). This highly organized developmental program requires the correct spatial and temporal expression of a large number of genes; yet despite the importance of the vasculature in development and disease, the transcriptional mechanisms governing gene expression in these processes remain incompletely understood.
The Ets family of winged helix proteins plays a clear role in the transcriptional control of genes involved in vascular development (Dejana et al., 2007; Sato, 2001). All Ets factors share a highly conserved DNA binding domain and bind to the core DNA sequence GGA(A/T), and nearly every endothelial cell enhancer or promoter characterized to date contains multiple essential Ets binding sites (Dejana et al., 2007; Sato, 2001). Of the nearly 30 different members of the mammalian Ets family, at least 19 are expressed in endothelial cells, and several have been shown to play essential roles in vascular development (Hollenhorst et al., 2004). However, no Ets factor is unique to the vasculature, and Ets binding sites are not specific to endothelial-expressed genes (Hollenhorst et al., 2004; Maroulakou and Bowe, 2000). Thus, it is unclear exactly how Ets factors contribute to the specificity of endothelial gene regulation. It has been hypothesized that Ets proteins may achieve tissue specific activation through binding to lower affinity sites in cooperation with other proteins (Hollenhorst et al., 2007), but Ets partners in endothelial cells have yet to be identified.
Members of the Forkhead (Fox) transcription factor family also play important roles in vascular endothelial development. Forkhead transcription factors are helix-turn-helix proteins that typically bind asymmetric cis-acting elements of 15-17 bp, containing the core Fox protein consensus of RYMAAYA (Carlsson and Mahlapuu, 2002). FoxC1 and FoxC2 are expressed in the developing vasculature, although not exclusively, and Foxc1/Foxc2 compound null embryos die during embryonic development with profound vascular defects (Hosaka et al., 2004; Seo et al., 2006). However, the mechanisms by which Forkhead transcription factors control endothelial gene expression are not clear. It has been hypothesized that gene expression in the endothelium may be regulated via the combined action of multiple transcription factors, but direct evidence for such a putative combinatorial code has been lacking.
In the present study, we identified a 44-bp transcriptional enhancer that is sufficient to direct expression specifically and exclusively to the developing vascular endothelium. This enhancer is regulated by a composite cis-acting element, the FOX:ETS motif, which is bound and synergistically activated by Forkhead and Ets transcription factors. We demonstrate that coexpression of FoxC2 and the Ets protein Etv2 (Etsrp71, ER71) is sufficient to induce ectopic expression of vascular genes in Xenopus embryos, and that combinatorial knockdown of the orthologous genes in zebrafish embryos disrupts vascular development. Finally, we show that FOX:ETS motifs are present in many known endothelial specific enhancers and that this motif is an efficient predictor of endothelial enhancers in the human genome. Thus, these studies establish a novel transcriptional code for vascular development based on the presence of the FOX:ETS motif and the binding of the cognate transcription factors to this composite element. More generally, these results establish a paradigm for the regulation of tissue specific gene expression by the combinatorial activities of two widely expressed transcription factors on a single composite cis-acting element.
Results
Identification of a 44-bp enhancer sufficient to direct expression exclusively to endothelial cells in the developing embryo
The MEF2C transcription factor is expressed in endothelial cells soon after their initial specification, and is essential for vascular development in mice (Lin et al., 1998; Supplemental Material, Fig. S1). Based on evolutionary conservation, we identified a 5.6-kb region of the Mef2c locus (F10) that contained two separate enhancers that each direct expression to a single lineage in the developing mouse embryo (Fig. 1A). The activity of one enhancer was specific to the developing vascular endothelium (F10E), and the activity of the other was restricted to the neural crest and its derivatives (F10N) at E9.5 (Fig. 1A-D). Mef2c F10E is a distinct regulatory element from a previously identified enhancer, termed Mef2c F7 (Fig. 1A), which also directs vascular expression in vivo, although not as early in endothelial development as F10E (De Val et al., 2004). Mef2c F10E contains a highly conserved 44-bp region that shares 86% sequence homology with zebrafish (Danio rerio) (Fig. 2A). Deletion of this deeply conserved 44-bp fragment in the context of F10E or in the context of the larger 5.6-kb F10 construct resulted in the complete loss of endothelial expression (Fig. 1F, G). Remarkably, the 44-bp deeply conserved region of F10 (F10-44) was sufficient to direct endothelial cell-specific expression from the blood island stage at E7.5 through angiogenesis and remodeling at E9.5 (Fig. 1E, H-M). These results indicate that this minimal 44-bp region contains all the cis-regulatory information necessary for endothelial-specific gene activation and expression, and thus presented the opportunity to identify a minimal set of transcription factors sufficient to regulate endothelial-specific gene expression.
Figure 1. Identification of a 44-bp Mef2c endothelial-specific enhancer.
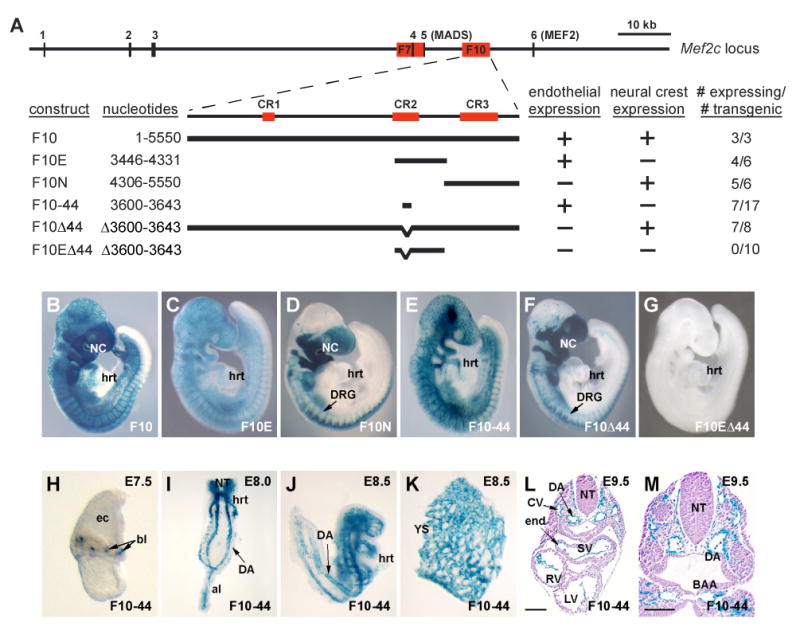
(A) A schematic representation of the mouse Mef2c locus is shown on the top line with exons depicted as vertical lines. The red boxes denotes the sizes and positions of the F7 and F10 fragments. F10 contains three evolutionarily conserved regions, denoted CR1-3. The lower portion of (A) depicts the deletion constructs of Mef2c F10. CR3 contains a neural crest specific enhancer. CR2 contains an endothelial specific enhancer, which encompasses a 44-bp deeply conserved region that is sufficient for endothelial enhancer activity in vivo. Endothelial and neural crest activity of each of the deletion constructs is denoted at the right as a + or -. The total number of transgenic embryos and the number that directed β-galactosidase expression to either the neural crest or endothelium are denoted at the far right of (A).
(B-G) Representative X-gal stained transgenic embryos for each of the Mef2c F10 transgene deletion constructs depicted in (A).
(H-M) Expression of the Mef2c F10-44-lacZ construct is specific to endothelial cells from blood island (bl) stage at E7.5 (H) throughout early endothelial development at E8.0 (I) and E8.5 (J, K). Transverse sections through an X-gal stained E9.5 transgenic embryo (L, M) demonstrate that transgene expression is restricted to endothelial cells throughout the vasculature, including the endocardium (end). al, allantois; BAA, branchial arch artery; CV, cardinal vein; DA, dorsal aorta; DRG, dorsal root ganglia; ec, ectoplacental cone; hrt, heart; LV, left ventricle; NC, neural crest; NT, neural tube; RV, right ventricle; SV, sinus venosus; YS, yolk sac.
Figure 2. Identification of a novel FOX:ETS motif simultaneously bound and synergistically activated by FoxC2 and Etv2.
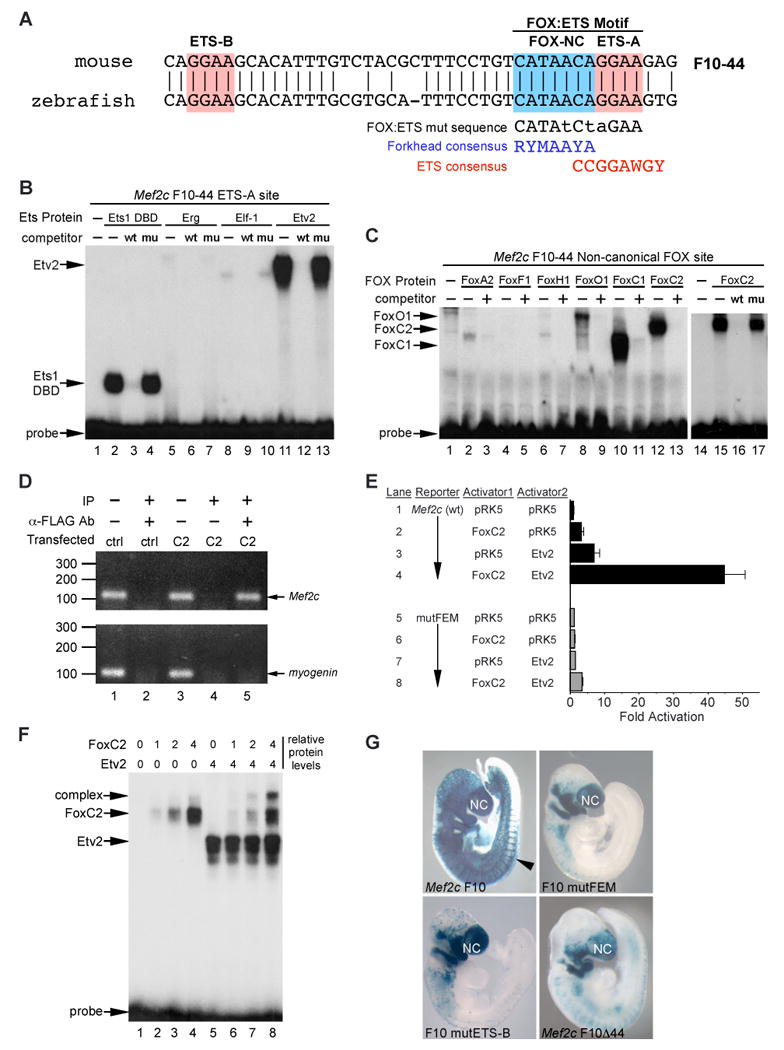
(A) Alignment of the mouse and zebrafish Mef2c F10-44 sequences. Red boxes denote core ETS binding sites, and the blue box denotes a non-consensus Forkhead binding element (FOX-NC). The novel, composite FOX:ETS motif is indicated above. Consensus Forkhead and Ets binding sites (Hollenhorst et al., 2007; Carlsson and Mahlapuu, 2002) are denoted, as is the mutant FOX:ETS sequence used in these studies.
(B) Radiolabeled oligonucleotide probes encompassing the F10-44 ETS-A site were used in EMSA with recombinant Ets proteins. The Ets1 DNA binding domain (DBD) and Etv2 efficiently bound to the site (lanes 2, 11) and were competed by excess unlabeled self probe (wt, lanes 3, 12) but not by mutant self probe (mu, lanes 4, 13). Erg and Elf-1 displayed little or no detectable binding to ETS-A in this assay.
(C) A radiolabeled oligonucleotide probe encompassing the Mef2c F10-44 FOX-NC site was used in EMSA with recombinant Forkhead proteins. FoxA2, FoxF1, and FoxH1 showed weak or no binding to FOX-NC. FoxO1 (lanes 8, 9) showed weak binding to FOX-NC. FoxC1 (lanes 10, 11) and FoxC2 (lanes 12, 13; also lanes 14-17) exhibited robust binding. Addition of excess, unlabeled self-probe, indicated by a + sign, inhibited binding of FoxO1, FoxC1, and FoxC2 to the FOX-NC site (lanes 9, 11, 13). Additionally, inclusion of a mutant version of FOX-NC (lane 17, mu) did not inhibit binding of FoxC2 to FOX-NC at the same concentration that the wild-type self-probe completely abolished binding (lane 16, wt).
(D) Chromatin immunoprecipitation from mouse embryo fibroblasts transfected with pCDNA3.1-FoxC2-Flag (C2) or parental pCNA3.1 expression vector (ctrl). Sheared, cross-linked chromatin fragments were immunoprecipitated with anti-FLAG antibody and the region of the endogenous Mef2c locus, surrounding F10-44, was amplified by PCR. The Mef2c F10-44 region was specifically amplified in pDNA3.1-FoxC2-FLAG transfected cells (lane 5), similar to the amplification in control samples that were directly amplified without prior immunoprecipitation (input, lanes 1, 3). No amplification was detected in control transfected (lane 2) or non-specific IgG immunoprecipitated samples (lane 4).
(E) FoxC2 and Etv2 synergistically trans-activate the Mef2c F10E enhancer. FoxC2 and Etv2 each weakly activated the reporter (lanes 2, 3) compared to parental expression plasmid control transfections (lane 1). Cotransfection of the reporter with expression plasmids for FoxC2 and Etv2 together resulted in potent synergistic activation (lane 4). Mutation of the FOX:ETS motif (mutFEM) ablated activation by FoxC2 and Etv2 (lanes 5-8). Data are presented as the mean plus SEM for four independent sets of transfections and analyses.
(F) FoxC2 and Etv2 simultaneously bind the FOX:ETS motif. A radiolabeled oligonucleotide probe (Mef2c-F10 FOX:ETS) encompassing only the F10E FOX:ETS motif was used in EMSA with recombinant FoxC2 and Etv2. The labeled probe included the FOX:ETS motif plus short adjacent sequences and did not include additional potential ETS binding sites. Increasing amounts of FoxC2 in the absence of Etv2 resulted in the formation of an increasing amount of FoxC2-DNA complex (lanes 2-4). Addition of Etv2 alone resulted in the formation of an Etv2-DNA complex (lane 5). Addition of increasing amounts of FoxC2 in the presence of a constant amount of Etv2 resulted in formation of each individual protein-DNA complex as well as a slower mobility band, suggesting a FoxC2-Etv2-DNA ternary complex (lanes 6-8). Relative levels of FoxC2 and Etv2 protein and binding activity are indicated at the top of the panel. In all samples, the total amount of total protein was held constant by the addition of the appropriate amount of unprogrammed reticulocyte lysate.
(G) A 3-bp mutation (CATAACAGGAA to CATAtCtaGAA) of the FOX:ETS motif (mutFEM) or mutation of the ETS-B site in the context of Mef2c F10, which contains both neural crest and endothelial enhancers, results in loss of transgene expression in the endothelium but not the neural crest. The resultant transgenic embryos show expression patterns similar to those in which the entire 44-bp element was deleted from F10 (Mef2c F10Δ44). Representative transgenic embryos from each construct are shown.
The Mef2c F10E enhancer is bound and synergistically activated by Fox and Ets transcription factors through a novel cis-acting motif
To locate transcription factor binding sites within F10-44, we used DNaseI footprinting to identify a region at the 3′ end of F10-44 bound by an activity present in endothelial cell extracts but not in extracts from myoblasts (Supplemental Material, Fig. S2). Within this region, we identified a consensus ETS site, containing the core GGA(A/T) motif, referred to as ETS-A (Fig. 2A). While multiple Ets factors bound to ETS-A in EMSA (Fig. 2B and data not shown), Etv2 displayed the strongest binding (Fig. 2B). We also identified a second ETS site within Mef2c F10-44 (ETS-B), which was also bound by several distinct Ets proteins in EMSA, including Etv2 (data not shown). A third potential core ETS binding site (TTCC) in F10-44, located between ETS-A and ETS-B, was not bound in EMSA by Etv2 or the Ets-1 DNA binding domain (DBD) under conditions in which the control ETS site, ETS-A, and ETS-B were each robustly bound (data not shown).
In addition to the ETS sites, the footprinting studies showed an additional endothelial-specific activity immediately adjacent to the ETS-A site (Supplemental Material, Fig. S2). This adjacent sequence had weak similarity to the core Forkhead binding site RYMAAYA (Carlsson and Mahlapuu, 2002) so we performed EMSA to determine whether the footprinted region was bound by different subfamilies of Forkhead transcription factors (Fig. 2C). Indeed, FoxC1 and FoxC2 bound robustly to this non-canonical Forkhead site (FOX-NC; Fig. 2C, lanes 10-13), and this binding was disrupted by a 3-bp mutation within FOX-NC (Fig. 2C, lanes 14-17), suggesting that these Forkhead proteins bind to a broader consensus site than previously thought. FoxO1 also bound to the FOX-NC site, albeit less robustly than FoxC1 or FoxC2 (Fig. 2C, lanes 8-9). FoxA2, FoxF1, and FoxH1 did not display any detectable binding to FOX-NC in EMSA (Fig. 2C, lanes 2-7), although each protein was efficiently synthesized in vitro and each bound to its own canonical site in the same assay (Supplemental Material, Fig. S3). In addition, we performed chromatin immunoprecipitation (ChIP) analyses on primary mouse embryo fibroblasts transfected with an epitope-tagged FoxC2 construct, which demonstrated that FoxC2 could bind to the endogenous F10-44 enhancer in vivo (Fig. 2D). No binding of FoxC2 protein was detected in ChIP analyses of the skeletal muscle-specific myogenin promoter, which served as a non-specific control (Fig. 2D).
Next, we tested the ability of FoxC and Ets transcription factors to activate the F10E enhancer (Fig. 2E). Alone, FoxC2 and Etv2 activated the Mef2c F10E enhancer 3-fold and 7-fold, respectively (Fig. 2E, lanes 2, 3). Strikingly, the combination of the two factors resulted in more than 40-fold, synergistic activation (Fig. 2E, lane 4). A 3-bp mutation that disrupted the binding of Etv2 and FoxC2 to the FOX:ETS motif in EMSA resulted in nearly complete disruption of transactivation (Fig. 2E, lane 8), demonstrating the specificity of this activation.
The strong synergistic activation of the Mef2c F10E enhancer by FoxC2 and Etv2, combined with the immediate juxtaposition of the two binding sites, suggested that the two factors might be simultaneously binding to the FOX:ETS motif to cooperatively activate transcription. To determine if FoxC2 and Etv2 bound to the FOX:ETS motif simultaneously, EMSA were performed in which increasing amounts of FoxC2 were added to a constant amount of Etv2 and the Mef2c F10-44 FOX:ETS motif (Fig. 2F). As expected, Etv2 bound the FOX:ETS motif in the absence of FoxC2 (Fig. 2F, lane 5). Addition of FoxC2 to the binding reactions resulted in the presence of probe bound solely by FoxC2 and in the formation of a complex of Etv2, FoxC2, and the FOX:ETS motif (Fig. 2F, lanes 6-8). The slower mobility band suggests that FoxC2 and Etv2 form a ternary complex that requires both proteins and DNA. The ternary complex increased in abundance with increasing quantities of FoxC2 relative to Etv2 even in the presence of large amounts of excess free probe (Fig. 2F, lanes 6-8). These results support the notion that FoxC2 and Etv2 co-occupy the FOX:ETS motif simultaneously and suggests that the two proteins may function together as part of a ternary complex to synergistically activate transcription.
To define the role of the FOX:ETS motif in vivo, we introduced a 3-bp mutation into this element in the context of the full-length 5.6-kb F10 fragment and used this mutant construct to generate transgenic mouse embryos (Fig. 2G). Mef2c F10 contains both the F10E endothelial and F10N neural crest enhancers (Fig. 1A). Disruption of the FOX:ETS motif resulted in a complete loss of endothelial activity, while neural crest activity was unperturbed (Fig. 2G, F10 mutFEM). In addition, mutation of the second ETS site within F10-44 also resulted in a disruption of endothelial activity of the full-length F10 enhancer, while leaving neural crest activity undisturbed (Fig. 2G, F10 mutETS-B).
Expression of FoxC2 and Etv2 in Xenopus embryos induces ectopic vascular gene expression
To test whether FoxC2 and Etv2 were sufficient to induce endothelial-specific gene activation more generally, we co-injected mRNAs for FoxC2 and Etv2 into a single cell at the vegetal pole of Xenopus embryos at the 4-cell stage (Fig. 3). Remarkably, the two factors potently induced endothelial-specific gene expression in a normally avascular region of the endoderm in the tail region of the frog tadpole. In embryos injected with FoxC2 and Etv2, multiple regions of flk1 mRNA expression were readily observed in nearly all embryos examined (Fig. 3B). By contrast, injection of control mRNAs resulted in no induction of flk1 expression (Fig. 3A). Quantification of flk1 expression in the abdominal region of injected frog embryos by real-time PCR showed that FoxC2 and Etv2 were each able to weakly induce the expression of flk1, while the combined expression of the two factors resulted in a synergistic level of activation of flk1 expression more than 25-fold higher than in control injected embryos (Fig. 3C). Co-expression of FoxC2 and Etv2 in Xenopus embryos also resulted in strong synergistic and ectopic activation of Pecam expression (Fig. 3D). These results demonstrate that FoxC2 and Etv2 are sufficient to induce the expression of endogenous endothelial genes in vivo.
Figure 3. Misexpression of FoxC2 and Etv2 in Xenopus embryos induces ectopic endothelial gene expression.
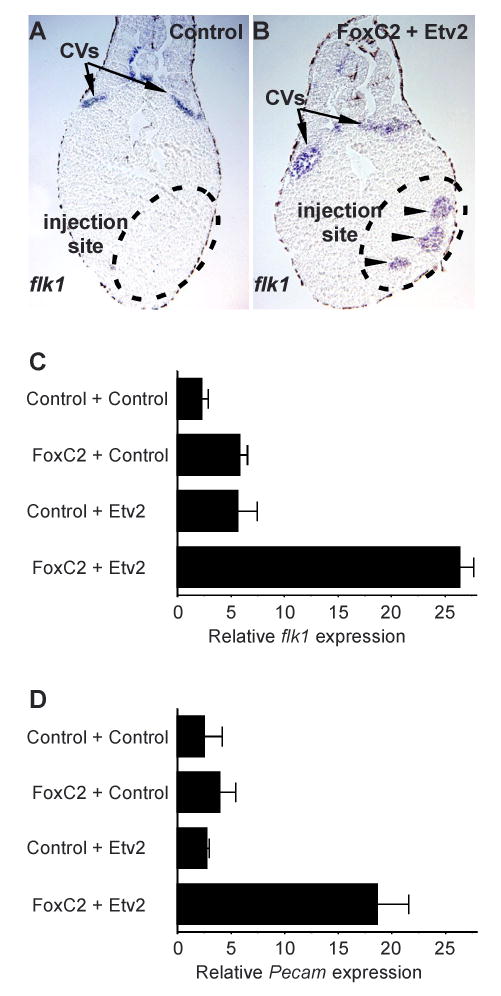
Xenopus embryos were injected with mRNAs encoding FoxC2 and Etv2 or EGFP control mRNA at the 4-cell stage and then collected at stage 36. After collection, embryos were either assayed by in situ hybridization using flk1 probe, followed by sectioning (A, B) or RNA was extracted for qPCR analysis of flk1 (C) or Pecam (D) transcripts. (A, B) flk1 expression was observed in the cardinal veins (CVs) in control (A) and FoxC2 + Etv2-injected (B) embryos. In addition, ectopic expression of flk1 was readily observed in the endoderm of the caudal region of FoxC2 + Etv2-injected embryos (B) but not in EGFP control injected embryos (A).
(C, D) Quantitative, real-time PCR shows that neither FoxC2 nor Etv2 significantly activated flk1 or Pecam expression on their own, but the combination of the two factors strongly induced expression of both endothelial-specific markers. Data are shown as the mean relative expression of flk1 or Pecam transcripts plus the SEM for three independent sets of injections and analyses.
The combined function of FoxC and Ets proteins are required for vascular development in zebrafish
Consistent with the deep conservation of F10-44, the mouse Mef2c F10E enhancer directed the expression of a GFP transgene in a vascular-specific manner in zebrafish at 48 hours post-fertilization (hpf) (Fig. 4A, B). Indeed, the expression directed by Mef2c F10E was nearly identical to the GFP expression observed in the Tg(flk1:GFP)s843 line, which is specific to endothelial cells at 48 hpf (Jin et al., 2005) (Fig. 4C, D). These observations indicate that the transcriptional pathways governing endothelial cell gene expression in zebrafish utilize the same cis-elements as in the mouse, supporting the notion that the same transcriptional code is involved in endothelial enhancer regulation in the two organisms.
Figure 4. Cooperative regulation of vascular development in zebrafish by FoxC and Ets proteins.
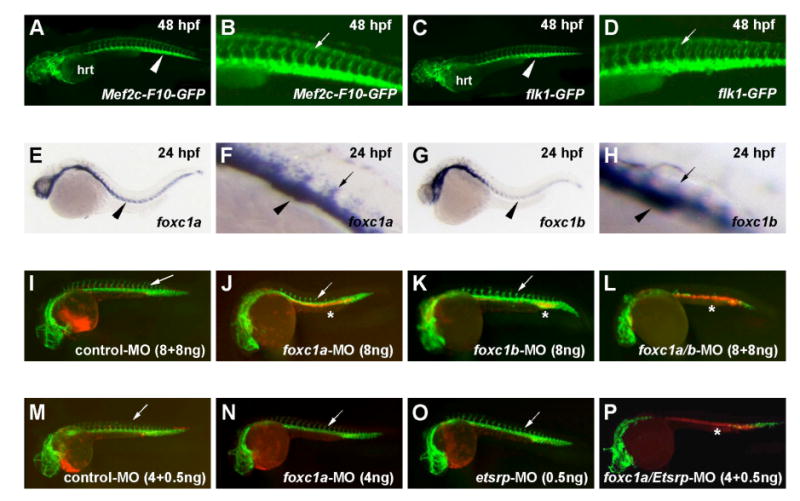
(A-D) The mouse Mef2c F10E enhancer directs expression of the GFP reporter gene in the vascular endothelium of transgenic zebrafish (A, B) in a nearly identical pattern to the endothelial-specific Tg(flk1:GFP)s843 reporter (C, D).
(E-H) In situ hybridization shows that the zebrafish foxc genes foxc1a (E, F) and foxc1b (G, H) are expressed in the developing vasculature at 24 hpf.
(I-L) Knockdown of foxc1a and foxc1b by morpholino injection alone (J, K) and in combination (L) resulted in loss of vascular structure, as detected by reduced expression of Tg(flk1:GFP)s843 (green) and the pooling of blood, as indicated by Tg(gata1:DsRed)sd2 expression (red). The combined foxc1a/foxc1b knockdown (L) resulted in a more severe perturbation of vascular development than either single knockdown. Note the normal expression of Tg(flk1:GFP)s843 and Tg(gata1:DsRed)sd2 in the control morpholino injected embryo (I).
(M-P) Injection of sub-phenotypic doses of foxc1a (N) and etsrp (O) morpholinos resulted in normal vascular development and normal expression of Tg(flk1:GFP)s843 and Tg(gata1:DsRed)sd2 in patterns identical to control injected embryos (M). Co-injection of the lower doses of foxc1a and etsrp morpholinos resulted in a nearly complete loss of vascular development (P), indicating cooperative regulation of vascular development by the two transcription factors. Asterisks mark the pooling of blood. Arrowheads point to the developing axial vessels, and arrows indicate the developing intersomitic vessels.
The orthologs of mammalian Ets factors are essential for vascular development in zebrafish (Pham et al., 2007; Sumanas and Lin, 2006), but the involvement of FoxC proteins in this process has not been described in zebrafish. We first examined the expression pattern of the zebrafish FoxC orthologs foxc1a and foxc1b (Topczewska et al., 2001). Both genes were expressed in the vasculature during the early stages of vascular development, including in the coalescing endothelial cells of the axial vessels at 24 hpf (Fig. 4E-H). We next tested the requirement of foxc genes in zebrafish by morpholino knock down (Fig. 4I-L). Control morpholino injected embryos displayed normal vascular development, which could be observed by the expression of Tg(flk1:GFP)s843 and normal accumulation of blood in the heart, as evidenced by the expression of Tg(gata1:DsRed)sd2 (Fig. 4I). Knockdown of foxc1a resulted in a decrease in intersomitic vessel sprouting, although the trunk vasculature still formed (Fig. 4J). Knockdown of foxc1b using a similar concentration of morpholino had a less profound effect on vascular development at 24 hpf (Fig. 4K). Importantly, combinatorial knockdown of both proteins resulted in a more severe vascular phenotype. No intersomitic vessel sprouts were detected at 24 hpf, and the formation of the axial vessels was severely diminished (Fig. 4L). These data suggest that FoxC proteins are required for vascular development in zebrafish.
In previous studies, it was noted that high concentration morpholino knockdown of single ets genes had some effect on vascular development in the fish, most notably with etsrp (ets1b), the zebrafish ortholog of Etv2 (Pham et al., 2007; Sumanas et al., 2008; Sumanas and Lin, 2006). However, injection of a low dose of etsrp morpholino (0.5 ng) resulted in no discernable phenotype at 24 hpf (Fig. 4O). Similarly, injection of a lower dose of foxc1a morpholino (4 ng) than that used in the experiments shown in Fig. 4J resulted in little or no vascular phenotype at 24 hpf (Fig. 4N). Remarkably, co-injection of the same sub-phenotypic doses of etsrp and foxc1a morpholinos resulted in a nearly complete ablation of vascular development, as indicated by dramatically reduced Tg(flk1:GFP)s843 expression (Fig. 4P). Since the flk1-gfp construct used to visualize the vasculature may itself be a direct target of FoxC1a and Etsrp, we also used the Tg(gata1:DsRed)sd2 line to visualize blood cells. Consistent with failed circulation due to severely disrupted vascular development, we observed massive pooling of blood in the tail in the double morpholino-injected embryos (Fig. 4P). These results provide additional strong support for an essential, cooperative role for FoxC and Ets factors in vascular development.
The FOX:ETS motif is present in many endothelial-specific enhancers
The combined requirement for FoxC and Ets factors in vascular development suggested that other endothelial-specific enhancers besides Mef2c F10E might also contain similar FOX:ETS motifs and be direct transcriptional targets of FoxC2 and Etv2. An examination of previously identified endothelial-specific enhancer elements revealed the presence of FOX:ETS motifs, including similar non-canonical Forkhead sites, in the Flk1, Tie2, Tal1, NOTCH4, and VE-CADHERIN (CDH5) enhancers (Fig. 5A) (Dube et al., 1999; Gottgens et al., 2002; Kappel et al., 1999; Prandini et al., 2005; Wu et al., 2005). Similar to the Mef2c FOX:ETS motif, each of the elements was bound robustly and specifically by FoxC2 and Ets in EMSA, and mutations within each of the FOX:ETS motifs abolished binding (Fig. 5C). ChIP analyses demonstrated that the enhancer regions of each of these additional genes were bound by FoxC2 in vivo (Fig. 5B).
Figure 5. The FOX:ETS motif is present in multiple endothelial enhancers.
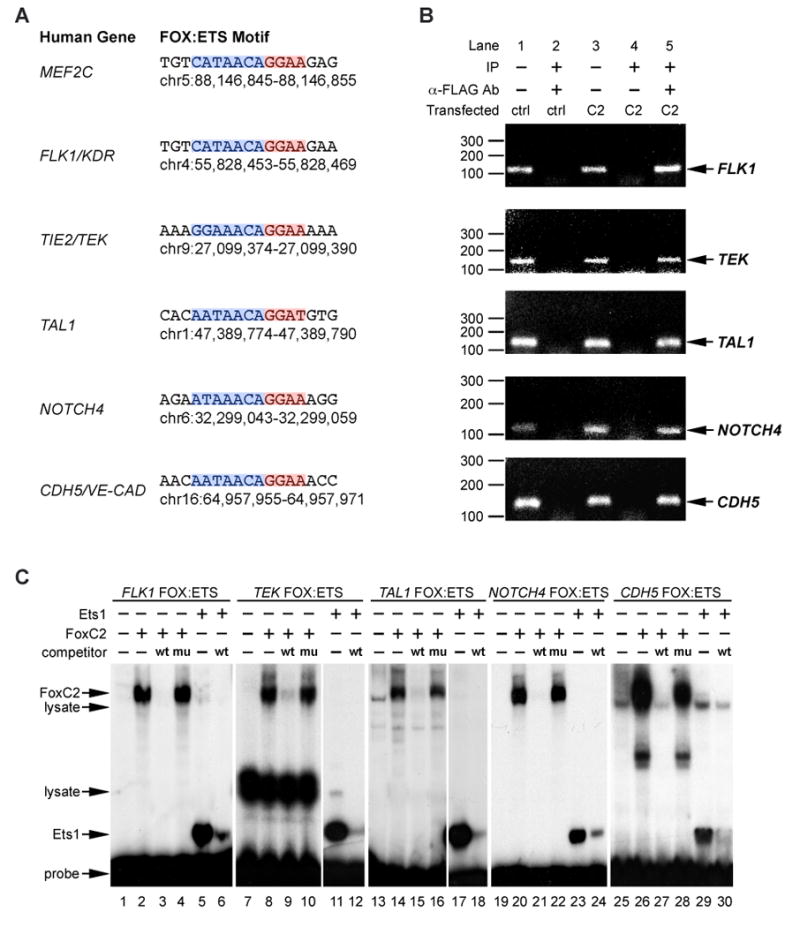
(A) Sequence and genomic location of FOX:ETS motifs in MEF2C and five other known endothelial-specific regulatory elements. The ETS sites are highlighted in red, and the FOX-NC sites are highlighted in blue. Chromosome locations refer to the May 2004 assembly of the human genome.
(B) ChIP from primary mouse embryo fibroblasts transfected with pCDNA3.1-FoxC2-FLAG (C2) or parental pCDNA3.1 expression vector (ctrl) shows that FoxC2 binds to the FOX:ETS motif in each of the previously described endothelial enhancers. In each case, the enhancer regions were specifically amplified in pDNA3.1-FoxC2-FLAG transfected cells (lane 5), similar to the amplification in control samples that were directly amplified without prior immunoprecipitation (input, lanes 1, 3). No amplification was detected in control transfected (lane 2) and non-specific IgG immunoprecipitated samples (lane 4). Note that these reactions were performed in conjunction with the ChIP for the myogenin promoter region, shown in Fig. 2D, which also serves as a non-specific control for these endogenous genes.
(C) EMSA demonstrates that FoxC2 (lanes 2, 8, 14, 20, 26) and Ets1 DBD (lanes 5, 11, 17, 23, 29) bind directly to the FOX:ETS motifs present in FLK1, TEK (Tie2), TAL1, NOTCH4, and CDH5 (VE-CADHERIN). In each case, an excess of unlabeled FOX:ETS motif self-probe (wt) efficiently competed for binding of FoxC2 and Ets1 DBD. Small mutations within the FOX-NC site (mu) disrupted competition by unlabeled probes even when added in 50× excess (lanes 4, 10, 16, 22, 28).
Next, we tested whether the Flk1, Tie2, Tal1, NOTCH4, and VE-CADHERIN enhancers were synergistically activated by the combinatorial action of FoxC2 and Etv2 in transfection analyses (Fig. 6). In all cases, FoxC2 and Etv2 by themselves only weakly activated the enhancers. However, in combination, the two transcription factors caused synergistic activation of the enhancers (Fig. 6A-E, lanes 1-4). Furthermore, mutation of the FOX:ETS motif in the VE-CADHERIN enhancer disrupted activation by FoxC2 and Etv2 in trans-activation assays (Fig. 6E, lanes 5-8), and completely abolished endothelial-specific expression of lacZ in transgenic embryos (Fig. 6F, G).
Figure 6. FoxC2 and Etv2 synergistically activate multiple endothelial enhancers.
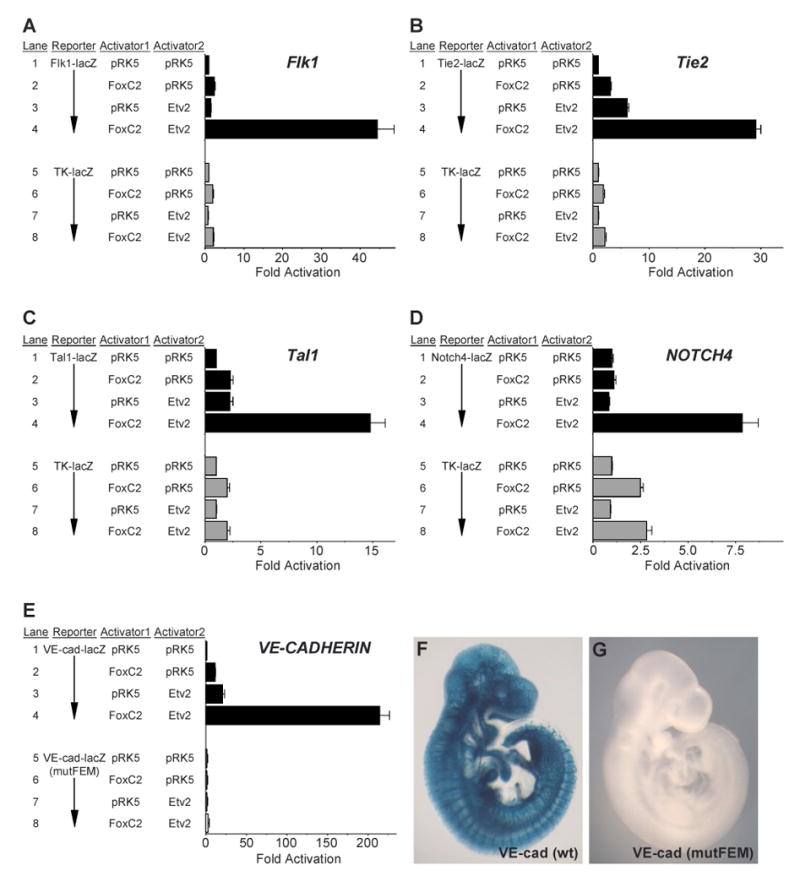
(A-E) FoxC2 and Etv2 synergistically trans-activate the Flk1 (A), Tie2 (B), Tal1 (C), NOTCH4 (D) and VE-CADHERIN/CDH5 (E) enhancers. Data are presented as the mean plus SEM for three to six independent sets of transfections and analyses. Note that in (E), a 4-bp mutation in the FOX:ETS motif (mutFEM) completely abolished activation of the VE-CADHERIN promoter/enhancer by FoxC2 and Etv2.
(F, G) Mutation of the FOX:ETS motif within the 3.5-kb VE-CADHERIN promoter/enhancer completely disrupts VE-CADHERIN-lacZ transgene expression at E9.5 (G) when compared to the strong, vascular-specific expression of the wild-type transgene (F). All five embryos transgenic for the wild-type 3.5-kb VE-CADHERIN enhancer expressed lacZ robustly in the endothelium, while none of the three embryos transgenic for the mutated enhancer showed detectable expression.
The presence of a FOX:ETS motif is sufficient to identify novel endothelial-specific enhancers
The observation that multiple, well-established endothelial-specific enhancers contained functional FOX:ETS motifs suggested that this element might be present in many enhancers of genes expressed in the endothelium. To determine if the FOX:ETS motif was overrepresented within endothelial cell expressed gene loci, we performed a computational screen to search for the presence of a FOX:ETS motif and a second ETS site within 60 bp (Fig. 7). We included the requirement for a second core ETS binding site (GGAA/T) as part of our computational screen since a second ETS site was found within 60 bp of the FOX:ETS motif in all of the enhancers listed in Fig. 5A. Fig. 7A shows a sequence logo representation of the position weight matrix used as the FOX:ETS consensus motif. Our computational screen identified the FOX:ETS motif within all six regulatory elements shown in Fig. 5A, which served as an important validation of the computational parameters of the screen. We identified 445 deeply conserved FOX:ETS motifs with a neighboring equally conserved second ETS site in the human genome. When the search was conducted such that the second ETS site only had to be conserved between mouse and human, 1500 FOX:ETS motifs, associated with 1200 genes, were identified.
Figure 7. Prediction of endothelial-specific enhancers based on the presence of a FOX:ETS motif.
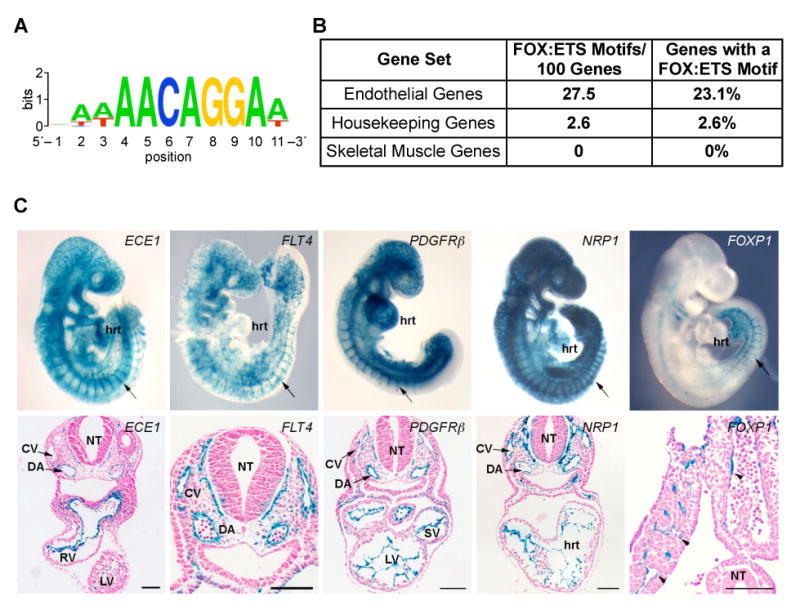
(A) Sequence logo representing the position weight matrix of the consensus FOX:ETS motif used in a genome-wide scan.
(B) The FOX:ETS motif is overrepresented in endothelial genes when compared to housekeeping and skeletal-muscle expressed genes.
(C) Identification of five novel endothelial-specific enhancers from the whole-genome screen based on the presence of a FOX:ETS motif. The upper row of photos shows representative whole-mount X-gal stained transient transgenic embryos at E9.5 from the ECE1, FLT4, PDGFRβ, NRP1, and FOXP1 genes. Each directed strong lacZ expression specifically to the endothelium, which can be clearly seen in transverse sections taken from each of the transient transgenic analyses at E9.5 (lower row of photos). CV, cardinal vein; DA, dorsal aorta; hrt, heart; LV, left ventricle; NT, neural tube; RV, right ventricle; SV, sinus venosus.
We compared the distribution of positive hits from this screen in three pre-determined sets of genes: 69 known endothelial cell-expressed genes, 305 housekeeping genes, and 75 skeletal muscle-expressed genes (Supplemental Material, Table S1). We observed a highly significant enrichment of the FOX:ETS motif and second ETS site in the endothelial gene set compared to the housekeeping and skeletal muscle gene sets (Fig. 7B; p <10-8). There was also a slight enrichment for hematopoietic genes, which were identified at 23% the frequency of endothelial genes (150/445 endothelial vs. 35/445 hematopoietic). Some association of the FOX:ETS motif with hematopoietic genes was expected given the numerous genes that are co-expressed in blood and endothelial cells and the likely existence of a common progenitor cell for the two lineages (Baron, 2003). Taken together, our observations support the idea that the FOX:ETS motif is a common feature of many endothelial-specific genes and that its presence might be used to identify endothelial specific enhancers and genes computationally.
As an initial test to determine if the presence of the FOX:ETS motif was sufficient to identify vascular enhancers, we investigated the FOX:ETS regions identified in our computational screen within 13 genes expressed in endothelial cells (Supplemental Material, Table S2). EMSA analyses demonstrated that 10 of the 13 FOX:ETS motifs were bound by FoxC2 and Etv2 proteins in vitro (data not shown). Among the 10 regions validated by EMSA, we tested eight for enhancer function in transgenic mouse embryos. In each case, a region of approximately 1 kb encompassing the FOX:ETS motif was tested in transient transgenic reporter assays for enhancer activity in mice at E9.5. Among the eight fragments tested, five functioned as endothelial-specific enhancers in transgenic embryos (Fig. 7C). The five enhancers identified using this approach, from the human FLT4 (VEGFR3), PDGFRβ, ECE1, NRP1, and FOXP1 genes, were all novel and none demonstrated extensive sequence conservation beyond the FOX:ETS motif and second ETS site, such that depth of sequence conservation alone would not have predicted these bona fide endothelial enhancers. Thus, these studies indicate the importance of the FOX:ETS motif as a predictive tool for the unbiased identification of endothelial enhancers based on its presence.
Discussion
Induction of endothelial gene expression by the combinatorial action of Forkhead and Ets factors
It is well established that members of the Ets transcription factor family are involved in the development of the endothelium, but the mechanism by which they contribute to the specificity of endothelial gene expression has been a key conundrum in vascular biology since no Ets factor is unique to the vasculature (Hollenhorst et al., 2004; Maroulakou and Bowe, 2000). In this paper, we discovered that Ets factors function cooperatively with FoxC proteins, which are also not restricted to the vasculature (Carlsson and Mahlapuu, 2002; Dejana et al., 2007). It is likely that several members of the Forkhead and Ets transcription factor families may be involved in vascular regulation via the FOX:ETS motif. In addition to Etv2, other Ets factors activated enhancers containing the FOX:ETS motif and induced ectopic vascular gene expression in Xenopus in concert with FoxC (data not shown). Foxc1/Foxc2 compound null mice have severe vascular defects (Seo et al., 2006), and we show here that combined knockdown of the two foxc genes in zebrafish also severely disrupts vascular development. However, some endothelial specification clearly still occurs in both fish and mice lacking FoxC function, supporting a possible role for other Forkhead proteins. Consistent with this notion, the Mef2c FOX:ETS motif was also bound by FoxO1 in the studies presented here, and Foxo1 null mice die by E10.5 with incomplete vascular development (Furuyama et al., 2004; Hosaka et al., 2004).
Tissue-specific enhancer prediction based on the presence of a signature cis-acting element
The haploid human genome contains nearly 3 billion base pairs, but only about 1.5% of this sequence is protein-encoding. Much of the non-protein encoding sequence has been conserved for hundreds of millions of years and performs many functions, including regulation of gene expression. However, the ability to predict functional regulatory elements within vertebrate genomes based solely on sequence information is poor. Enhancer elements have been accurately predicted in mammalian genomes only by screening for large numbers of different binding motifs or by using extreme levels of conservation (Hallikas et al., 2006; Pennacchio et al., 2006). Our recent use of deep phylogenetic conservation to identify enhancers successfully defined numerous regulatory elements, but interestingly, these studies did not identify a single enhancer directing expression to the vasculature, suggesting that using sequence conservation between species alone was not an effective approach to identify endothelial enhancers (Pennacchio et al., 2006).
The identification of a 44-bp enhancer from Mef2c that alone is sufficient to direct endothelial-specific expression is unprecedented with regard to its small size and allowed us to identify the FOX:ETS motif, a composite cis-acting element essential for enhancer function in vivo. Interestingly, although the most highly conserved 44-bp of Mef2c F10E (F10-44) was sufficient to direct endothelial specific expression throughout early embryogenesis, the activity of this small enhancer was extinguished after E10.5. By contrast, the larger 900-bp F10E construct remained active exclusively in endothelial cells of both the blood and lymphatic vasculature throughout embryogenesis and in adulthood (data not shown). It is important to note that the 900-bp F10E construct still required an intact FOX:ETS motif for activity at later stages in development. These data suggest that additional cis-acting elements are involved in endothelial-specific maintenance of the larger enhancer fragment and support a model in which the FOX:ETS motif serves as an ancient, endothelial-specific initiation element to which additional complexity has been added throughout evolution. Consistent with this notion, there is extensive homology within Mef2c F10E beyond the FOX:ETS motif, although the cross-species homology is not as deep as the FOX:ETS motif (Supplemental Material, Fig. S4). Furthermore, additional complexity may have been added to the FOX:ETS motif in enhancers with activity that restricts to distinct endothelial compartments, such as arteries, veins, and lymphatics. Testing additional putative enhancers in transgenic mice should allow for the identification of other cis-motifs that are over-represented in endothelial enhancers and are associated with the FOX:ETS motif. In addition, these studies may establish a model for enhancer prediction that may be applicable to any lineage or sub-lineage once there is adequate understanding of required cis-elements.
The Fox-Ets interaction as a target for modulation of vascular growth and remodeling
Aberrant vessel growth is an important contributor to several prevalent disease states (Carmeliet and Jain, 2000). Improper overgrowth of blood vessels is an important cause of age-related macular degeneration, and neovascularization of the retina is the hallmark of proliferative diabetic retinopathy (Andreoli and Miller, 2007; Simo et al., 2006). Likewise, growth and metastasis of solid tumors requires an adequate blood supply, and angiogenic induction of new blood vessel growth into tumors is an important component of cancer pathology (Stacker et al., 2002). Current strategies to inhibit angiogenesis are primarily based on blocking vascular endothelial growth factor (VEGF) signaling (anti-VEGF therapy), and the use of a monoclonal antibody against VEGF has been shown to be clinically effective when used in combination with other chemotherapeutic agents (Goh et al., 2007). However, drug resistance of metastatic tumors is a concern, and the identification of additional targets for blocking vessel growth remains an important goal for cancer therapy (Goh et al., 2007). The observation that the FOX:ETS motif is strongly associated with numerous endothelial genes in the human genome suggests that blocking activation via the FOX:ETS motif might sufficiently inhibit the endothelial transcriptional program to serve as a novel target for therapeutic intervention in cancer and other diseases involving aberrant vessel growth.
Experimental Procedures
Plasmids, Cloning and Mutagenesis
The 5550-bp F10 fragment from Mef2c was generated by PCR and cloned into the transgenic reporter plasmid HSP68-lacZ (De Val et al., 2004). F10-44 was created by cloning complementary oligonucleotides, corresponding to the 44-bp deeply conserved F10 sequence, into HSP68-lacZ. The mouse flk1, mouse Tie2, and human NOTCH4 enhancers were generated by PCR from genomic DNA and cloned into p-TK-β-gal. The SCL +19 enhancer, which has been described (Gottgens et al., 2002), was subcloned into p-TK-β-gal. The 3564-bp and 377-bp VE-CADHERIN promoter/enhancer fragments were generated by PCR and cloned into the promoterless lacZ reporter plasmid p-AUG-β-gal for the generation of transgenic mice and for use in transfection assays. The FLT4, FOXP1, NRP1, ECE1, PDGFRβ, NR4A3, EFNB1, and FGFR2 enhancers were amplified from human genomic DNA by PCR and cloned into HSP68-lacZ.
For zebrafish transgenesis, the 885-bp Mef2c F10E enhancer fragment was cloned upstream of an HSP70-GFP cassette. For generating foxc1a and foxc1b in situ probes, 800 bp at the 3′ end of these genes including the 3′ UTR were PCR amplified. The Xenopus flk1 in situ probe has been described previously (Cleaver et al., 1997). Expression plasmids were generated by cloning cDNAs into plasmid pRK5.
Oligonucleotides and morpholino oligonucleotides
The sequences of oligonucleotide primers for cloning, mutagenesis, morpholino knockdowns, qPCR detection of Xenopus laevis Pecam and flk1, and ChIP detection are provided in Supplemental Material, Table S3. Oligonucleotide primer sequences used in EMSA are provided in Supplemental Material, Table S4. The morpholino oligonucleotides for etsrp, foxc1a, and foxc1b have been described previously (Pham et al., 2007; Topczewska et al., 2001).
Mice, frogs, and zebrafish
Transgenic mice were generated by oocyte microinjection, and genotype analysis and X-gal staining were performed as described previously (De Val et al., 2004). Zebrafish whole-mount in situ hybridization was performed as previously described (Jin et al., 2005). To generate Mef2c-GFP transgenic zebrafish, embryos were injected with 20-50 ng of construct at the one-cell stage and analyzed at 24–48 hpf. Morpholino analyses were performed in Tg(flk1:GFP)s843; Tg(gata1:DsRed)sd2 embryos (Jin et al., 2005; Traver et al., 2003). Embryos of the frog Xenopus laevis were microinjected and incubated as described previously (Cleaver et al., 1997). RNA encoding EGFP was included in all injections as a lineage tracer. Transcript levels for flk1 and Pecam were assayed by qRT-PCR using normalized samples and SYBR-Green (Invitrogen). All experiments using animals complied with federal and institutional guidelines.
Cell culture, transfections, and Chromatin immunoprecipitation (ChIP)
For transient transfection assays, Cos1 cells were grown in DMEM supplemented with 10% FBS and seeded at 6×104 cells/2.5 cm plate. After 24 h, 750 ng each of reporter and expression plasmids were transfected using FuGENE6 (Roche) as recommended by the manufacturer. All transfections lacking an expression plasmid contained an equal amount of the parental expression vector. Following transfection, cells were cultured for 48 h, then harvested and assayed using the Luminescent β-galactosidase Detection kit II (Clontech), as previously described (Rojas et al., 2008).
For ChIP, primary mouse embryo fibroblasts (MEFs) were transfected using Lipofectamine LTX and 8 μg of either pcDNA3.1-FLAG-FoxC2 or empty pCDNA3.1 vector in 10 cm dishes. 36 h after transfection, cells were harvested as described previously (Rojas et al., 2008). ChIP was then performed using the ChIP assay kit (Upstate/Millipore) according to the manufacturer's instructions, using anti-FLAG antibody (clone M2, Sigma) and protein A-agarose. Immunoprecipitated fragments and unprecipitated lysates (input samples) were subjected to PCR using primers listed in Supplemental Material, Table S3.
Electrophoretic Mobility Shift Assay (EMSA)
EMSAs were performed as described previously (De Val et al., 2004). All recombinant proteins were generated using the TNT Quick Coupled Transcription/Translation System (Promega) according to the manufacturer's instructions. Full-length FoxC1, FoxC2, FoxA2, FoxF1, FoxH1, Erg1, Elf-1, and Etv2 were expressed from either pCS2 or pcDNA3 expression plasmids using SP6 polymerase. FoxF1, FoxO1, and the Ets1 DBD (De Val et al., 2004) were expressed from the pCITE2A in vitro expression vectors, using T7 polymerase (Novagen).
Identification of conserved sequence motifs
The sequences within and 10 kb around all human genes in the RefSeq database (Pruitt et al., 2005) were scanned utilizing rVista (Loots et al., 2002). The position weight matrix for the FOX:ETS motif, which was derived from the six FOX:ETS sequences shown in Fig. 5A plus 11 additional FOX:ETS motifs identified experimentally as bound by FoxC2 and Etv2 in EMSA, and a second consensus ETS site were used to scan mouse and human gene sequences independently. Hits in which the depth of conservation of the FOX:ETS motif was less than the surrounding 20 bp of sequence were discounted, as were those in which the depth of conservation of the second ETS site was not equal to that of the FOX:ETS motif.
Supplementary Material
Acknowledgments
We thank Peter Oettgen, Bertie Gottgens, Ralph Janknecht, Tom Kume, Robert Costa, and Deepak Srivastava for providing plasmids for these studies, Brant Weinstein for providing Ets morpholinos, Saleh Adi for advice and plasmids, Shaun Coughlin for comments on the manuscript, and Dan Palmer for assistance with MEF culture. SDV was supported in part by a postdoctoral fellowship from the American Heart Association, Western States Affiliate. NCC was supported by a K08 award from the NIH and a Fellow-to-Faculty award from the American Heart Association. This work was supported by NIH grants HL64658 to BLB and HL54737 to DYRS, by a grant from the Sandler Family Supporting Foundation to BLB, and under Department of Energy Contract DE-AC02-05CH11231 to LAP.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andreoli CM, Miller JW. Anti-vascular endothelial growth factor therapy for ocular neovascular disease. Curr Opin Ophthalmol. 2007;18:502–508. doi: 10.1097/ICU.0b013e3282f0ca54. [DOI] [PubMed] [Google Scholar]
- Baron MH. Embryonic origins of mammalian hematopoiesis. Exp Hematol. 2003;31:1160–1169. doi: 10.1016/j.exphem.2003.08.019. [DOI] [PubMed] [Google Scholar]
- Carlsson P, Mahlapuu M. Forkhead transcription factors: key players in development and metabolism. Dev Biol. 2002;250:1–23. doi: 10.1006/dbio.2002.0780. [DOI] [PubMed] [Google Scholar]
- Carmeliet P, Jain RK. Angiogenesis in cancer and other diseases. Nature. 2000;407:249–257. doi: 10.1038/35025220. [DOI] [PubMed] [Google Scholar]
- Cleaver O, Tonissen KF, Saha MS, Krieg PA. Neovascularization of the Xenopus embryo. Dev Dyn. 1997;210:66–77. doi: 10.1002/(SICI)1097-0177(199709)210:1<66::AID-AJA7>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- De Val S, Anderson JP, Heidt AB, Khiem D, Xu SM, Black BL. Mef2c is activated directly by Ets transcription factors through an evolutionarily conserved endothelial cell-specific enhancer. Dev Biol. 2004;275:424–434. doi: 10.1016/j.ydbio.2004.08.016. [DOI] [PubMed] [Google Scholar]
- Dejana E, Taddei A, Randi AM. Foxs and Ets in the transcriptional regulation of endothelial cell differentiation and angiogenesis. Biochim Biophys Acta. 2007;1775:298–312. doi: 10.1016/j.bbcan.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Dube A, Akbarali Y, Sato TN, Libermann TA, Oettgen P. Role of the Ets transcription factors in the regulation of the vascular-specific Tie2 gene. Circ Res. 1999;84:1177–1185. doi: 10.1161/01.res.84.10.1177. [DOI] [PubMed] [Google Scholar]
- Flamme I, Frolich T, Risau W. Molecular mechanisms of vasculogenesis and embryonic angiogenesis. J Cell Physiol. 1997;173:206–210. doi: 10.1002/(SICI)1097-4652(199711)173:2<206::AID-JCP22>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Furuyama T, Kitayama K, Shimoda Y, Ogawa M, Sone K, Yoshida-Araki K, Hisatsune H, Nishikawa S, Nakayama K, Nakayama K, et al. Abnormal angiogenesis in Foxo1 (Fkhr)-deficient mice. J Biol Chem. 2004;279:34741–34749. doi: 10.1074/jbc.M314214200. [DOI] [PubMed] [Google Scholar]
- Goh PP, Sze DM, Roufogalis BD. Molecular and cellular regulators of cancer angiogenesis. Curr Cancer Drug Targets. 2007;7:743–758. doi: 10.2174/156800907783220462. [DOI] [PubMed] [Google Scholar]
- Gottgens B, Nastos A, Kinston S, Piltz S, Delabesse EC, Stanley M, Sanchez MJ, Ciau-Uitz A, Patient R, Green AR. Establishing the transcriptional programme for blood: the SCL stem cell enhancer is regulated by a multiprotein complex containing Ets and GATA factors. Embo J. 2002;21:3039–3050. doi: 10.1093/emboj/cdf286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallikas O, Palin K, Sinjushina N, Rautiainen R, Partanen J, Ukkonen E, Taipale J. Genome-wide prediction of mammalian enhancers based on analysis of transcription-factor binding affinity. Cell. 2006;124:47–59. doi: 10.1016/j.cell.2005.10.042. [DOI] [PubMed] [Google Scholar]
- Hollenhorst PC, Jones DA, Graves BJ. Expression profiles frame the promoter specificity dilemma of the ETS family of transcription factors. Nucleic Acids Res. 2004;32:5693–5702. doi: 10.1093/nar/gkh906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenhorst PC, Shah AA, Hopkins C, Graves BJ. Genome-wide analyses reveal properties of redundant and specific promoter occupancy within the ETS gene family. Genes Dev. 2007;21:1882–1894. doi: 10.1101/gad.1561707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosaka T, Biggs WH, 3rd, Tieu D, Boyer AD, Varki NM, Cavenee WK, Arden KC. Disruption of forkhead transcription factor (FOXO) family members in mice reveals their functional diversification. Proc Natl Acad Sci U S A. 2004;101:2975–2980. doi: 10.1073/pnas.0400093101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SW, Beis D, Mitchell T, Chen JN, Stainier DY. Cellular and molecular analyses of vascular tube and lumen formation in zebrafish. Development. 2005;132:5199–5209. doi: 10.1242/dev.02087. [DOI] [PubMed] [Google Scholar]
- Kappel A, Ronicke V, Damert A, Flamme I, Risau W, Breier G. Identification of vascular endothelial growth factor (VEGF) receptor-2 (Flk-1) promoter/enhancer sequences sufficient for angioblast and endothelial cell-specific transcription in transgenic mice. Blood. 1999;93:4284–4292. [PubMed] [Google Scholar]
- Lin Q, Lu J, Yanagisawa H, Webb R, Lyons GE, Richardson JA, Olson EN. Requirement of the MADS-box transcription factor MEF2C for vascular development. Development. 1998;125:4565–4574. doi: 10.1242/dev.125.22.4565. [DOI] [PubMed] [Google Scholar]
- Loots GG, Ovcharenko I, Pachter L, Dubchak I, Rubin EM. rVista for comparative sequence-based discovery of functional transcription factor binding sites. Genome Res. 2002;12:832–839. doi: 10.1101/gr.225502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markstein M, Markstein P, Markstein V, Levine MS. Genome-wide analysis of clustered Dorsal binding sites identifies putative target genes in the Drosophila embryo. Proc Natl Acad Sci U S A. 2002;99:763–768. doi: 10.1073/pnas.012591199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroulakou IG, Bowe DB. Expression and function of Ets transcription factors in mammalian development: a regulatory network. Oncogene. 2000;19:6432–6442. doi: 10.1038/sj.onc.1204039. [DOI] [PubMed] [Google Scholar]
- Patan S. Vasculogenesis and angiogenesis. Cancer Treat Res. 2004;117:3–32. doi: 10.1007/978-1-4419-8871-3_1. [DOI] [PubMed] [Google Scholar]
- Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, Minovitsky S, Dubchak I, Holt A, Lewis KD, et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- Pham VN, Lawson ND, Mugford JW, Dye L, Castranova D, Lo B, Weinstein BM. Combinatorial function of ETS transcription factors in the developing vasculature. Dev Biol. 2007;303:772–783. doi: 10.1016/j.ydbio.2006.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prandini MH, Dreher I, Bouillot S, Benkerri S, Moll T, Huber P. The human VE-cadherin promoter is subjected to organ-specific regulation and is activated in tumour angiogenesis. Oncogene. 2005;24:2992–3001. doi: 10.1038/sj.onc.1208483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas A, Kong SW, Agarwal P, Gilliss B, Pu WT, Black BL. GATA4 is a direct transcriptional activator of Cyclin D2 and Cdk4 and is required for cardiomyocyte proliferation in anterior heart field-derived myocardium. Mol Cell Biol. 2008;28:5420–31. doi: 10.1128/MCB.00717-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y. Role of ETS family transcription factors in vascular development and angiogenesis. Cell Struct Funct. 2001;26:19–24. doi: 10.1247/csf.26.19. [DOI] [PubMed] [Google Scholar]
- Seo S, Fujita H, Nakano A, Kang M, Duarte A, Kume T. The forkhead transcription factors, Foxc1 and Foxc2, are required for arterial specification and lymphatic sprouting during vascular development. Dev Biol. 2006;294:458–470. doi: 10.1016/j.ydbio.2006.03.035. [DOI] [PubMed] [Google Scholar]
- Simo R, Carrasco E, Garcia-Ramirez M, Hernandez C. Angiogenic and antiangiogenic factors in proliferative diabetic retinopathy. Curr Diabetes Rev. 2006;2:71–98. doi: 10.2174/157339906775473671. [DOI] [PubMed] [Google Scholar]
- Stacker SA, Baldwin ME, Achen MG. The role of tumor lymphangiogenesis in metastatic spread. Faseb J. 2002;16:922–934. doi: 10.1096/fj.01-0945rev. [DOI] [PubMed] [Google Scholar]
- Sumanas S, Gomez G, Zhao Y, Park C, Choi K, Lin S. Interplay between Etsrp/ER71, scl and alk8 signaling controls endothelial and myeloid cell formation. Blood. 2008 doi: 10.1182/blood-2007-09-110569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumanas S, Lin S. Ets1-related protein is a key regulator of vasculogenesis in zebrafish. PLoS Biol. 2006;4:e10. doi: 10.1371/journal.pbio.0040010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topczewska JM, Topczewski J, Solnica-Krezel L, Hogan BL. Sequence and expression of zebrafish foxc1a and foxc1b, encoding conserved forkhead/winged helix transcription factors. Mech Dev. 2001;100:343–347. doi: 10.1016/s0925-4773(00)00534-7. [DOI] [PubMed] [Google Scholar]
- Traver D, Paw BH, Poss KD, Penberthy WT, Lin S, Zon LI. Transplantation and in vivo imaging of multilineage engraftment in zebrafish bloodless mutants. Nat Immunol. 2003;4:1238–1246. doi: 10.1038/ni1007. [DOI] [PubMed] [Google Scholar]
- Wu J, Iwata F, Grass JA, Osborne CS, Elnitski L, Fraser P, Ohneda O, Yamamoto M, Bresnick EH. Molecular determinants of NOTCH4 transcription in vascular endothelium. Mol Cell Biol. 2005;25:1458–1474. doi: 10.1128/MCB.25.4.1458-1474.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


