Abstract
Voltage-activated ion channels open and close in response to changes in voltage, a property that is essential for generating nerve impulses. Studies on voltage-activated potassium (Kv) channels show that voltage-sensor activation is sensitive to the composition of lipids in the surrounding membrane. Here we explore the interaction of lipids with S1–S4 voltage-sensing domains, and find that the conversion of the membrane lipid sphingomyelin to ceramide-1-phosphate alters voltage-sensor activation in an S1–S4 voltage-sensing protein lacking an associated pore domain, and that the S3b–S4 paddle motif determines the effects of lipid modification on Kv channels. Using tarantula toxins that bind to paddle motifs within the membrane, we identify mutations in the paddle motif that weaken toxin binding by disrupting lipid-paddle interactions. Our results suggest that lipids bind to voltage-sensing domains and demonstrate that the pharmacological sensitivities of voltage-activated ion channels are influenced by the surrounding lipid membrane.
Kv channels contain a central pore domain 1,2 that opens and closes in response to conformational changes in the four surrounding voltage-sensing domains 3. Each subunit in these tetrameric channels has six membrane spanning segments (S1 through S6), with S1–S4 comprising each voltage-sensing domain and the four S5–S6 regions together forming the central pore domain. X-ray structures of these channels predict that the S1–S4 voltage-sensing domains would be extensively exposed to surrounding lipids when embedded in a membrane 1,4, and functional studies demonstrate that changing the composition of the lipid membrane can have striking effects on how these channels open and close in response to changes in voltage 5–10. In studies on the KvAP channel from Aeropyrum pernix reconstituted in defined lipid membranes, lipids with phosphate containing head groups are required for channel activation 5, leading to the proposal that Arg residues in the voltage sensor are stabilized in the membrane by interactions with phosphate lipid head groups. Experiments in native lipid membranes 6,7 indicate that the phosphate head group of sphingomyelin plays a similar role in the activation of the well-studied Shaker Kv channel from Drosophila 11 and the mammalian Kv2.1 channel from rat brain 12. Modification of sphingomyelin does not influence the gating of several other related Kv channels 6, suggesting that there is specificity in the interaction between lipids and Kv channels. In the case of Kv2.1, the effects of modification of sphingomyelin can be diminished by toxins that bind to voltage sensors 7, raising the possibility that sphingomyelin interacts directly with voltage sensors in the channel, perhaps similar to the non-annular lipids observed for other membrane proteins 13,14. In the present study we set out to determine whether sphingomyelin interacts with the voltage-sensing domains in Kv2.1 channels and to identify specific regions where the lipid might bind. Our results suggest that sphingomyelin interacts with a particular structural motif within voltages sensors and that these interactions can influence the pharmacological sensitivities of the channel.
Results
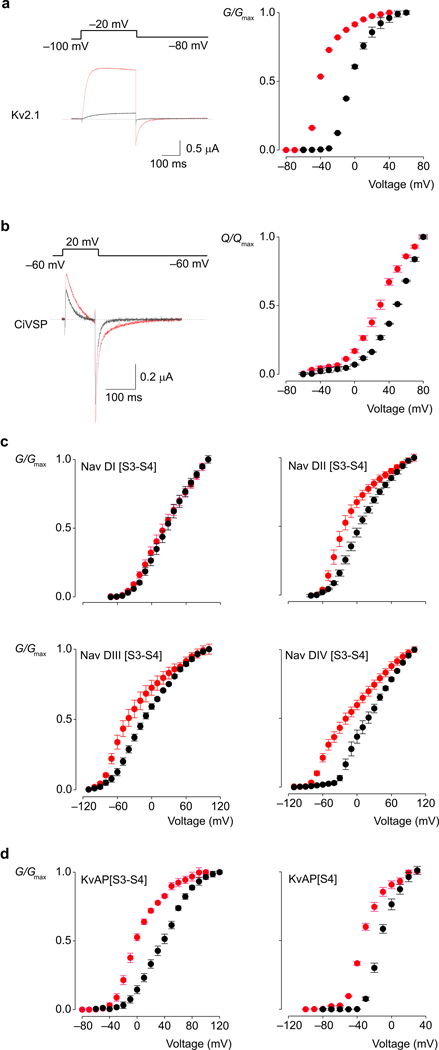
The approach we took was to investigate how sphingomyelinase D (SMaseD), an enzyme that converts sphingomyelin to ceramide-1-phosphate, influences the gating and pharmacological sensitivities of the Kv2.1 channel from rat brain 12. Lu and colleagues reported that this Kv channel is particularly sensitive to the lipase 6, with removal of the choline moiety of sphingomyelin causing the channel to open at voltages where it would otherwise be closed 6. In our experiments, application of SMaseD to the extracellular side of cells expressing Kv2.1 causes a −30 mV shift of the conductance-voltage (G–V) relation (Fig 1a) that develops over 10–15 min and is not reversible over the time frame of our recordings (>1 hr), consistent with previous results 6. Deactivation of the channel following membrane repolarization is greatly slowed after SMaseD treatment (Fig 1a), indicating that the lipid modification causes a pronounced stabilization of the open state of the channel.
Figure 1.
Membrane modification alters gating of voltage-activated ion channels and a voltage-activated phosphatase. G-V and Q-V relations obtained for native membranes are shown in black, while those after treatment with SMaseD are shown in red. (a) Ionic current traces and G-V relations for Kv2.1. (b) Gating current traces and Q-V relations for Ci-VSP. (c) G-V curves for Kv2.1 chimeras that contain individual paddle motifs from each of the four domains of rNav1.4. The holding voltage was −100mV for Nav DI and Nav DII, and −120mV for Nav DIII and NavDIV chimeras; test pulse duration was 300ms (500ms for Nav DIV); tail voltage was −60mV for Nav DI, −80mV for Nav DII, −110mV for Nav DIII and −100mV for Nav DIV. (d) G-V curves for chimeras where complete (S3–S4) or partial (S4) paddle motifs of Kv2.1 were replaced with homologous regions from KvAP. The holding voltage was −100mV, test pulse duration was 300ms, and tail voltage was −80mV. In all cases conductance was determined from normalized tail currents. For ionic currents, leak, background and capacitive currents were isolated and subtracted after blocking the Kv channels with agitoxin-2, while for gating currents, they were subtracted using a P/−4 protocol. n=3, error bars are s.e.m.
Lipid modification alters voltage-sensor activation
To explore whether the effects of SMaseD result from interactions between lipids and the S1–S4 voltage-sensing domain of the channel, we asked whether the lipase has similar effects on another protein containing an S1–S4 voltage-sensing domain but lacking a pore domain. For these experiments we used the Ciona Intestinalis voltage sensitive phosphatase (Ci-VSP), which contains a cytoplasmic phosphatase domain rather than a transmembrane pore 15, and monitored activation of its S1–S4 voltage-sensing domain by measuring charge movement in response to membrane depolarization 15–17. Extracellular application of SMaseD to cells expressing Ci-VSP produces an approximately −20 mV shift of the charge-voltage (Q-V) relationship to more negative voltages (Fig 1b). As in the case of Kv2.1, the effects of the enzyme develop over 10–15 min and are effectively irreversible. These results demonstrate that modification of sphingomyelin can influence the activity of S1–S4 voltage-sensing domains in the absence of a pore domain.
S3b–S4 paddles determine the effects of lipid modification
One interesting candidate for a region within S1–S4 voltage-sensing domains that interacts with lipids is the S3b–S4 paddle motif, a helix-turn-helix motif that moves in contact with the lipid membrane to sense changes in voltage 3,4,8,18–22. To explore the possibility that sphingomyelin interacts with the paddle motif, we measured the effects of SMaseD on Kv2.1 chimeras containing paddle motifs from a variety of different voltage-activated ion channels (Fig 1c,d). Of the constructs we studied, those containing paddle motifs from the four different voltage-sensing domains of the rat skeletal muscle Nav1.4 sodium channel23 were the most informative. These Nav channel paddle motifs are capable of gating the Kv2.1 channel and rendering the Kv channel sensitive to Nav channel toxins 21, however, SMaseD alters the G-V relations of the chimeras to different extents (Fig 1d). In particular, Kv channels containing the paddle motif from domain I of Nav1.4 are effectively insensitive to SMaseD, suggesting that the paddle motifs determine the sensitivity of the channel to lipid modification.
Lipid modification alters voltage sensor pharmacology
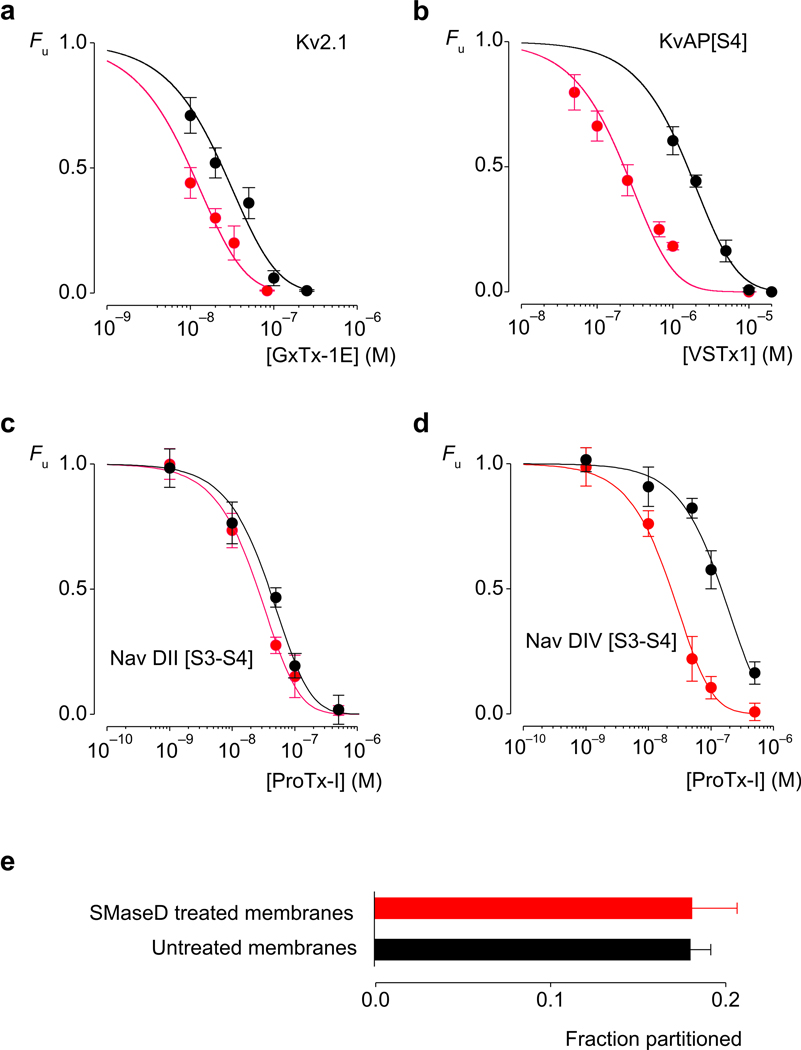
If the membrane lipids studied here actually bind to paddle motifs in Kv channels, it is possible that the binding of other ligands at the protein-lipid interface would be influenced by lipid modification. Because tarantula toxins partition into membranes 9,24–26 (Supplementary Fig 1) and bind to regions of the paddle motif facing the surrounding membrane 1,8,10,21,27–29, we investigated whether their interaction with the paddle motif is altered by lipid modification. We studied the effects of SMaseD on the interaction of GxTx-1E 30 with the Wt Kv2.1 channel, VSTx1 25,31 with a chimera produced by transplanting the S4 helix from KvAP in Kv2.1 (KvAP[S4]) 8, and ProTx-I 32 with the two Nav paddle chimeras that are sensitive to the toxin 21 (Fig 2). The lipid modification produces a 3.9-fold increase in the apparent affinity of GxTx-1E for Kv2.1 (Fig 2a), and a 6.2-fold increase in that of VSTx1 for the KvAP chimera (Fig 2b). The effects of ProTx-I on the two Nav paddle chimeras are particularly interesting, as the apparent affinity of the toxin is increased by only 50% for the domain II chimera (Nav DII [S3–S4)), but by over 6-fold for the domain IV chimera (Nav DIV [S3–S4]) (Fig 2c,d). These effects of SMaseD on the interactions of tarantula toxins with paddle motifs demonstrate that the toxins can sense the lipid modification.
Figure 2.
Membrane modification alters the apparent affinity of tarantula toxins for Kv channels without altering membrane partitioning. Concentration-dependence for tarantula toxin occupancy of wild type and chimaeric Kv channels, before (black) and after (red) SMaseD treatment. Fu represents the fraction of unbound channels and was estimated from fractional inhibition at negative voltages (see Methods; Supplementary Fig 5). Smooth curves represent fits of Fu = ([Toxin]/([Toxin] + Kd))4 to the data. The apparent Kd values for the toxin-channel pairs before and after SMaseD treatment are (a) 203±38 nM and 52±8 nM for GxTx-1E and Kv2.1. (b) 10±2.5 µM and 1.6±0.3 µM for VSTx1 and KvAP[S4]. c) 210±9 nM and 137±9 nM for ProTx-I and Nav DII [S3–S4]. (d) 780±77 nM and 122±7 nM for ProTx-I and Nav DII [S3–S4]. n = 3–5 for each toxin concentration and error bars represent s.e.m. At a concentration of 100 nM, ProTx-I had no effect on Nav DI [S3–S4] and Nav DIII [S3–S4] chimeras. (e) Partitioning of 125I-GxTx-1E into oocyte membranes before (black) and after (red) treatment with SMaseD. n=5 and error bars represent s.e.m.
The distinct effects of the lipase on ProTx-I interaction with the two Nav paddle chimeras suggest that the modification does not simply alter the way these toxins interact with lipids, for example by altering the toxin concentration in the membrane, but that the toxins can actually detect difference in the way lipids interact with distinct paddle motifs. To confirm that partitioning of tarantula toxins is not altered by SMaseD, we compared partitioning of 125I-GxTx-1E into intact oocyte membranes 9 before and after lipid modification. At a concentration of GxTx-1E (200 nM) that completely inhibits opening of Kv2.1 (Fig 2a), partitioning of the toxin is unaffected by SMaseD treatment (Fig 2e).
Tarantula toxins detect lipid-paddle interactions
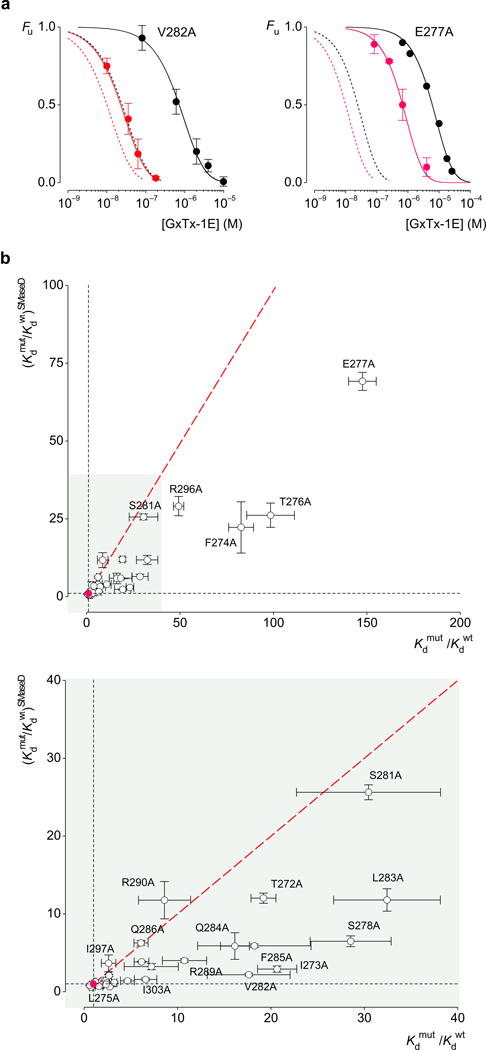
Previous studies have shown that tarantula toxins interact with the paddle motif in a specific fashion, with mutations in the paddle having pronounced effects on the apparent affinity of these toxins 8,10,21,26,27,29,33. The idea that both lipids and toxins interact with the paddle motif raises the intriguing possibility that mutations in the paddle might alter toxin affinity by disrupting lipid-paddle interactions. In search of such mutations, we substituted each residue within the paddle motif of Kv2.1 with Ala and measured the apparent affinity of GxTx-1E for these channels in control membranes and after treatment with SMaseD. Mutations such as V282A and E277A decrease the apparent affinity of the toxin when studied in untreated membranes, but to a lesser extent after lipid modification (Fig 3a). In the case of V282A, the mutant weakens toxin affinity by over 18-fold under control conditions, but by only about 2-fold after treatment with the lipase. In this instance, the extent to which the mutation weakens toxin affinity is strikingly dependent on the lipids around the channel, suggesting that the mutation actually influences how lipids interact with the paddle motif. Another way of looking at this result is that for the Wt channel, lipid modification increases toxin affinity by only about 4-fold, but after the paddle mutation has weakened toxin binding, lipid modification now increases toxin affinity by over 30-fold (Fig 3a; Supplementary Fig 2). It is as if the lipid modification can correct the disruptive effects of the mutation (see Discussion). The E277A mutant is an example where the effects of the mutation on toxin affinity is moderately different when comparing control and SMaseD-treated membranes, even though the mutant alters the affinity of the toxin by almost 150-fold when studied in control membranes (Fig 3a). In this instance, the mutant seems to weaken toxin affinity largely by altering a protein-protein interaction and the lipid modification can only modestly correct the effects of the mutation. The extent to which the lipid modification and paddle mutations are coupled can be evaluated for all mutations by plotting the mutation-induced perturbation in toxin affinity for control membranes (Kdmut/Kdwt) against that for SMaseD-treated membranes (Fig 3b,c). F285A, I273A, S278A and L283A are examples where the lipid modification largely corrects the weakening of toxins affinity, as described above for V282. In other cases, the effects of mutants are unaltered by lipid modification, regardless of whether the effects of the mutation on toxin affinity are small (e.g. I297A, Q286A, R290A) or large (e.g. S281A, R296A), indicating that there is specificity in the coupling between the lipid modification and the paddle mutations.
Figure 3.
Comparison of the effects of Kv2.1 paddle mutations on GxTx-1E affinity before and after membrane modification with SMaseD. (a) Concentration-dependence for GxTx-1E occupancy of V282A and E277A mutants of Kv2.1 before (black) and after (red) SMaseD treatment. The apparent Kd values are 3.8±0.4 µM and 120±9 nM for V282A, and 30±1.5 µM and 3.5±0.15 µM for E277A. The dotted lines represent the fits for the wild type channel data from Figure 2a. (b,c) Effects of mutations on toxin affinity (Kdmut/Kdwt) for control membranes plotted against that following treatment with SMaseD. The value for the wild type channel is indicated by the red circle. The red dotted line corresponds to identical perturbations for control and SMaseD-treated membranes (no coupling between the mutation and the lipid modification). Data from Supplementary Table 1. Each mutant was examined initially using a concentration of toxin near the Kd for the wild type; mutants with different Kd were further examined using a wider range of concentrations. n=3, error bars are s.e.m.
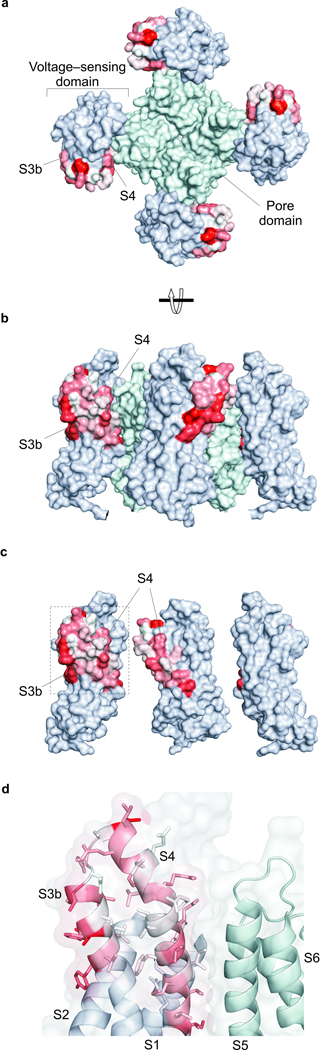
To understand where on the paddle motif these lipid interactions occur, we calculated the free energy associated with the coupling between channel mutations and the lipid modification (ΔGcoupling; Supplementary Fig 2) 34,35 and mapped them onto the X-ray structure of the Kv2.1/Kv1.2 paddle chimera 1, a structure where the voltage sensor is activated and the pore is open. Using a continuous color gradient between white and red to represent increasing |ΔGcoupling|, we can see that mutations that are coupled with the lipid modification tend to concentrate in two regions of the paddle motif (Fig 4). The first is on the surface of the S3b helix that projects out towards the surrounding bilayer (Fig 4a,b), and the second is on the surface of the S4 helix facing the S5 helix from the adjacent subunit (Fig 4c). Residues at the interface between S3b and S4 helices are notable in that their mutation does not couple to the lipid modification (Fig 4d). Further experiments using VST×1 as a detector to measure coupling between S4 mutations in the KvAP[S4] chimera and lipid modification identify a similar face of the S4 helix where coupling occurs (Supplementary Figs. 3 and 4 and Supplementary Tables 3 and 4), demonstrating that lipids interact with similar regions of S4 in distantly related Kv channels.
Figure 4.
Coupling energies mapped onto the X-ray structure of a Kv channel and a model for how lipid modification alters toxin affinity. (a) Surface representation of the Kv2.1/Kv1.2 paddle chimera with coupling energies mapped onto the paddle region, viewed from the extracellular side of the membrane. A color gradient between white and red was used to represent increasing |ΔG| values from 0 to 1.3 kcal mol−1. Residues in the pore domain are colored light cyan and those in the S1–S4 domains outside the paddle motif are colored light blue. (b) Surface representation of the Kv2.1/Kv1.2 paddle chimera viewed from the side. (c) Surface representation of the Kv2.1/Kv1.2 paddle chimera viewed from the side with front S1–S4 voltage-sensing domain and central pore domain removed. (d) Close up view of the S3b–S4 paddle motif with transparent surfaces and side chains colored as in a. Residues in the S1–S2 loop were removed for clarity. PDB accession code is 2R9R and all structures were drawn using PyMol (DeLano Scientific Inc.).
Discussion
The goal of the present study was to explore the interaction of sphingomyelin with S1–S4 voltage-sensing domains and the role of the lipid in the function and pharmacology of Kv channels. Our experiments with Ci-VSP establish that modification of sphingomyelin can influence S1–S4 voltage-sensing domains in the absence of a pore domain (Fig 1b), and the results with paddle chimeras point to interactions between the lipid and the paddle motif (Fig 2). In testing the idea that sphingomyelin may act like a non-annular lipid and bind to the paddle motif, we investigated whether tarantula toxins can sense modification of the lipid. SMaseD treatment typically increases the apparent affinity of these toxins, but the extent varies greatly for different toxin-paddle combinations (Fig 2), and in particular, for mutations on the paddle motif of Kv2.1 (Fig 3). The lipid modification does not influence the interaction of the toxin with the bulk membrane (Fig 2), suggesting that the toxin is detecting a local alteration near where it binds to the paddle motif. These results demonstrate that the pharmacological sensitivity of an ion channel can be profoundly influenced by the identity of lipids around the channel and their interactions with specific structural motifs.
How does the modification of sphingomyelin alter the sensitivity of Kv2.1 to tarantula toxins? It seems likely that the effects of SMaseD are not simply the consequence of removing sphingomyelin because conversion of the lipid to ceramide-1-phosphate with SMaseD or to ceramide with SMaseC has very different effects on the gating of Kv2.1. The former facilitates opening 6 (Fig 1), whereas the latter creates a barrier to opening of the channel 7. It is unlikely that the effects of the lipase on toxin affinity are secondary to the dissociation of sphingomyelin-rich lipid microdomains (rafts) because mutations in the paddle motif couple with the lipid modification (Fig 3). In addition, dissociation of these microdomains with cholesterol depletion does not influence activation of Kv2.1 channels 6,36 or sensitivity of the channel to tarantula toxins (not shown). One way to explain the effect of lipid modification on toxin affinity would be to postulate that tarantula toxins bind to the paddle motif with higher affinity when sphingomyelin is bound (Fig 5; Kd′<Kd)). In this way, mutations could weaken toxin binding by disrupting lipid-paddle interactions rather than by altering a protein-protein interaction between toxin and channel (Fig 3a). In the context of this model, lipid modification could rescue high affinity toxin binding after mutations weaken lipid-paddle interactions either because the interaction of the modified lipid is stronger (lower KL) or because the toxin binds more strongly to paddles interacting with ceramide-1-phosphate compared to sphingomyelin.
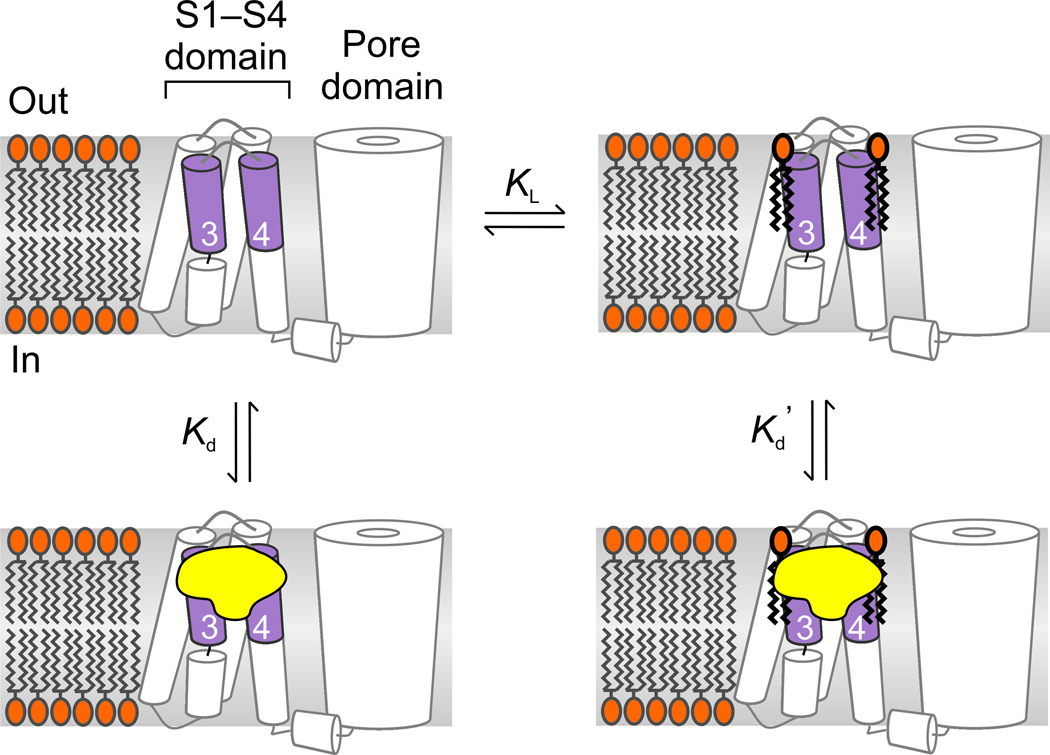
Figure 5.
Model illustrating toxin binding to lipid-associated paddle motifs. Lipids are depicted as binding and unbinding from the paddle motif and toxin affinity is higher for paddle motifs with lipids bound (Kd’<Kd). Conversion of sphingomyelin to ceramide-1-phosphate could enhance toxin affinity either because the modified lipid binds more strongly (lower KL) or because the toxin binds tighter to paddles interacting with ceramide-1-phosphate compared to sphingomyelin.
Where on the paddle motif do the lipids bind? Our experiments using tarantula toxins as detectors uncovered surfaces of both the S3b and S4 helices within the paddle motif where mutations couple with the lipid modification (Fig 4). Interactions between lipids and the S3b helix are interesting because these might help to anchor the helix near the membrane surface, which would be consistent with studies showing that residues in S3b do not exhibit substantial changes in accessibility to water soluble agents during gating 37–39. Interaction of lipids with the surface of S4 that faces S5 is an ideal location to maximize electrostatic interactions between phosphate lipid head groups and the outer S4 Arg residues to stabilize the voltage sensor in an activated state 5–7. These protein-lipid interactions at the interface between voltage-sensing and pore domains could explain why S5 mutants in this region exhibit hindered voltage sensor activation 40 even though the S4 and S5 helices do not pack together tightly in X-ray structures 1,41. The presence of bound lipids between S4 and S5 is consistent with the general picture whereby non-annular lipids bind to membrane proteins at interfaces between transmembrane helices 13,14. Although structural information on sphingolipid-binding proteins is limited, helix-turn-helix motifs reminiscent of the paddle motifs found in voltage sensors have been proposed to bind these lipids 42,43.
Our observations concerning the sensitivity of Kv channel pharmacology to lipids fit nicely with the discovery that Kv channel pharmacology can also be modulated by the mechanical properties of the membrane 10. The important implication is that the pharmacology of the ion channel is not determined by the protein alone, but by the lipids in the surrounding membrane and how they interact with the channel protein. It will be fascinating to explore whether variations in the composition and properties of membranes in different subcellular compartments, cell types, or pathophysiological conditions, influence the gating properties and pharmacological sensitivities of voltage-activated ion channels.
Methods
Methods and any associated references are available in the online version of the paper at http://www.nature.com/nsmb/.
Supplementary Material
Acknowledgments
We thank M. Holmgren, J. Kumar, M. Mayer, J. Mindell, S. Silberberg and members of the Swartz lab for helpful discussions, and the NINDS DNA sequencing facility for DNA sequencing. We thank Y. Okamura (Okazaki Center for Integrative Biosciences, Okazaki, Japan) for generously providing Ci-VSP complementary DNA, Y. Xu and Z. Lu (Univ. Penn, Philadelphia, PA) for generously supplying recombinant SMaseD and W. Schmalhofer and M. Garcia (Merck Research Labs, Rahway, NJ) for generously supplying 125I-GxTx-1E. This work was supported by the Intramural Research Program of the NINDS, NIH (K.J.S.) and by an NIH-FWO postdoctoral fellowship (F.B.).
Footnotes
Accession codes. Protein Data Bank: Coordinates for XXX have been deposited with accession code XXXX. Not relevant.
Note: Supplementary information is available on the Nature Structural and Molecular Biology webste.
References
- 1.Long SB, Tao X, Campbell EB, MacKinnon R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007;450:376–382. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- 2.Doyle DA, et al. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 3.Swartz KJ. Sensing voltage across lipid membranes. Nature. 2008;456:891–897. doi: 10.1038/nature07620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang Y, et al. X-ray structure of a voltage-dependent K+ channel. Nature. 2003;423:33–41. doi: 10.1038/nature01580. [DOI] [PubMed] [Google Scholar]
- 5.Schmidt D, Jiang QX, MacKinnon R. Phospholipids and the origin of cationic gating charges in voltage sensors. Nature. 2006;444:775–779. doi: 10.1038/nature05416. [DOI] [PubMed] [Google Scholar]
- 6.Ramu Y, Xu Y, Lu Z. Enzymatic activation of voltage-gated potassium channels. Nature. 2006;442:696–699. doi: 10.1038/nature04880. [DOI] [PubMed] [Google Scholar]
- 7.Xu Y, Ramu Y, Lu Z. Removal of phospho-head groups of membrane lipids immobilizes voltage sensors of K+ channels. Nature. 2008;451:826–829. doi: 10.1038/nature06618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alabi AA, Bahamonde MI, Jung HJ, Kim JI, Swartz KJ. Portability of paddle motif function and pharmacology in voltage sensors. Nature. 2007;450:370–375. doi: 10.1038/nature06266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Milescu M, et al. Tarantula toxins interact with voltage sensors within lipid membranes. J Gen Physiol. 2007;130:497–511. doi: 10.1085/jgp.200709869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmidt D, Mackinnon R. Voltage-dependent K+ channel gating and voltage sensor toxin sensitivity depend on the mechanical state of the lipid membrane. Proc Natl Acad Sci U S A. 2008;105:19276–19281. doi: 10.1073/pnas.0810187105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kamb A, Tseng-Crank J, Tanouye MA. Multiple products of the Drosophila Shaker gene may contribute to potassium channel diversity. Neuron. 1988;1:421–430. doi: 10.1016/0896-6273(88)90192-4. [DOI] [PubMed] [Google Scholar]
- 12.Frech GC, VanDongen AM, Schuster G, Brown AM, Joho RH. A novel potassium channel with delayed rectifier properties isolated from rat brain by expression cloning. Nature. 1989;340:642–645. doi: 10.1038/340642a0. [DOI] [PubMed] [Google Scholar]
- 13.Palsdottir H, Hunte C. Lipids in membrane protein structures. Biochim Biophys Acta. 2004;1666:2–18. doi: 10.1016/j.bbamem.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 14.Lee AG. Lipid-protein interactions in biological membranes: a structural perspective. Biochim Biophys Acta. 2003;1612:1–40. doi: 10.1016/s0005-2736(03)00056-7. [DOI] [PubMed] [Google Scholar]
- 15.Murata Y, Iwasaki H, Sasaki M, Inaba K, Okamura Y. Phosphoinositide phosphatase activity coupled to an intrinsic voltage sensor. Nature. 2005;435:1239–1243. doi: 10.1038/nature03650. [DOI] [PubMed] [Google Scholar]
- 16.Villalba-Galea CA, Sandtner W, Starace DM, Bezanilla F. S4-based voltage sensors have three major conformations. Proc Natl Acad Sci U S A. 2008;105:17600–17607. doi: 10.1073/pnas.0807387105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Villalba-Galea CA, Miceli F, Taglialatela M, Bezanilla F. Coupling between the voltage-sensing and phosphatase domains of Ci-VSP. J Gen Physiol. 2009;134:5–14. doi: 10.1085/jgp.200910215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cuello LG, Cortes DM, Perozo E. Molecular architecture of the KvAP voltage-dependent K+ channel in a lipid bilayer. Science. 2004;306:491–495. doi: 10.1126/science.1101373. [DOI] [PubMed] [Google Scholar]
- 19.Ruta V, Chen J, MacKinnon R. Calibrated measurement of gating-charge arginine displacement in the KvAP voltage-dependent K+ channel. Cell. 2005;123:463–475. doi: 10.1016/j.cell.2005.08.041. [DOI] [PubMed] [Google Scholar]
- 20.Banerjee A, MacKinnon R. Inferred motions of the S3a helix during voltage-dependent K+ channel gating. J Mol Biol. 2008;381:569–580. doi: 10.1016/j.jmb.2008.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bosmans F, Martin-Eauclaire MF, Swartz KJ. Deconstructing voltage sensor function and pharmacology in sodium channels. Nature. 2008;456:202–208. doi: 10.1038/nature07473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chakrapani S, Cuello LG, Cortes DM, Perozo E. Structural dynamics of an isolated voltage-sensor domain in a lipid bilayer. Structure. 2008;16:398–409. doi: 10.1016/j.str.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trimmer JS, et al. Primary structure and functional expression of a mammalian skeletal muscle sodium channel. Neuron. 1989;3:33–49. doi: 10.1016/0896-6273(89)90113-x. [DOI] [PubMed] [Google Scholar]
- 24.Lee SY, MacKinnon R. A membrane-access mechanism of ion channel inhibition by voltage sensor toxins from spider venom. Nature. 2004;430:232–235. doi: 10.1038/nature02632. [DOI] [PubMed] [Google Scholar]
- 25.Jung HJ, et al. Solution Structure and Lipid Membrane Partitioning of VSTx1, an Inhibitor of the KvAP Potassium Channel. Biochemistry. 2005;44:6015–6023. doi: 10.1021/bi0477034. [DOI] [PubMed] [Google Scholar]
- 26.Phillips LR, et al. Voltage-sensor activation with a tarantula toxin as cargo. Nature. 2005;436:857–860. doi: 10.1038/nature03873. [DOI] [PubMed] [Google Scholar]
- 27.Swartz KJ, MacKinnon R. Mapping the receptor site for hanatoxin, a gating modifier of voltage-dependent K+ channels. Neuron. 1997b;18:675–682. doi: 10.1016/s0896-6273(00)80307-4. [DOI] [PubMed] [Google Scholar]
- 28.Li-Smerin Y, Hackos DH, Swartz KJ. Alpha-helical structural elements within the voltage-sensing domains of a K+ channel. J Gen Physiol. 2000a;115:33–49. doi: 10.1085/jgp.115.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li-Smerin Y, Swartz KJ. Helical structure of the COOH terminus of S3 and its contribution to the gating modifier toxin receptor in voltage-gated ion channels. J Gen Physiol. 2001;117:205–218. doi: 10.1085/jgp.117.3.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herrington J, et al. Blockers of the delayed-rectifier potassium current in pancreatic beta-cells enhance glucose-dependent insulin secretion. Diabetes. 2006;55:1034–1042. doi: 10.2337/diabetes.55.04.06.db05-0788. [DOI] [PubMed] [Google Scholar]
- 31.Ruta V, Jiang Y, Lee A, Chen J, MacKinnon R. Functional analysis of an archaebacterial voltage-dependent K+ channel. Nature. 2003;422:180–185. doi: 10.1038/nature01473. [DOI] [PubMed] [Google Scholar]
- 32.Middleton RE, et al. Two tarantula peptides inhibit activation of multiple sodium channels. Biochemistry. 2002;41:14734–14747. doi: 10.1021/bi026546a. [DOI] [PubMed] [Google Scholar]
- 33.Li-Smerin Y, Swartz KJ. Localization and molecular determinants of the hanatoxin receptors on the voltage-sensing domain of a K+ channel. J Gen Physiol. 2000;115:673–684. doi: 10.1085/jgp.115.6.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hidalgo P, MacKinnon R. Revealing the architecture of a K+ channel pore through mutant cycles with a peptide inhibitor. Science. 1995;268:307–310. doi: 10.1126/science.7716527. [DOI] [PubMed] [Google Scholar]
- 35.Schreiber G, Fersht AR. Energetics of protein-protein interactions: analysis of the barnase- barstar interface by single mutations and double mutant cycles. J Mol Biol. 1995;248:478–486. doi: 10.1016/s0022-2836(95)80064-6. [DOI] [PubMed] [Google Scholar]
- 36.Martens JR, et al. Differential targeting of Shaker-like potassium channels to lipid rafts. J Biol Chem. 2000;275:7443–7446. doi: 10.1074/jbc.275.11.7443. [DOI] [PubMed] [Google Scholar]
- 37.Nguyen TP, Horn R. Movement and crevices around a sodium channel S3 segment. J Gen Physiol. 2002;120:419–436. doi: 10.1085/jgp.20028636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gandhi CS, Clark E, Loots E, Pralle A, Isacoff EY. The orientation and molecular movement of a k(+) channel voltage-sensing domain. Neuron. 2003;40:515–525. doi: 10.1016/s0896-6273(03)00646-9. [DOI] [PubMed] [Google Scholar]
- 39.Darman RB, Ivy AA, Ketty V, Blaustein RO. Constraints on Voltage Sensor Movement in the Shaker K+ Channel. J Gen Physiol. 2006 doi: 10.1085/jgp.200609624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soler-Llavina GJ, Chang TH, Swartz KJ. Functional interactions at the interface between voltage-sensing and pore domains in the Shaker K(v) channel. Neuron. 2006;52:623–634. doi: 10.1016/j.neuron.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 41.Long SB, Campbell EB, Mackinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 42.Mahfoud R, et al. Identification of a common sphingolipid-binding domain in Alzheimer, prion, and HIV-1 proteins. J Biol Chem. 2002;277:11292–11296. doi: 10.1074/jbc.M111679200. [DOI] [PubMed] [Google Scholar]
- 43.Snook CF, Jones JA, Hannun YA. Sphingolipid-binding proteins. Biochim Biophys Acta. 2006;1761:927–946. doi: 10.1016/j.bbalip.2006.06.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.