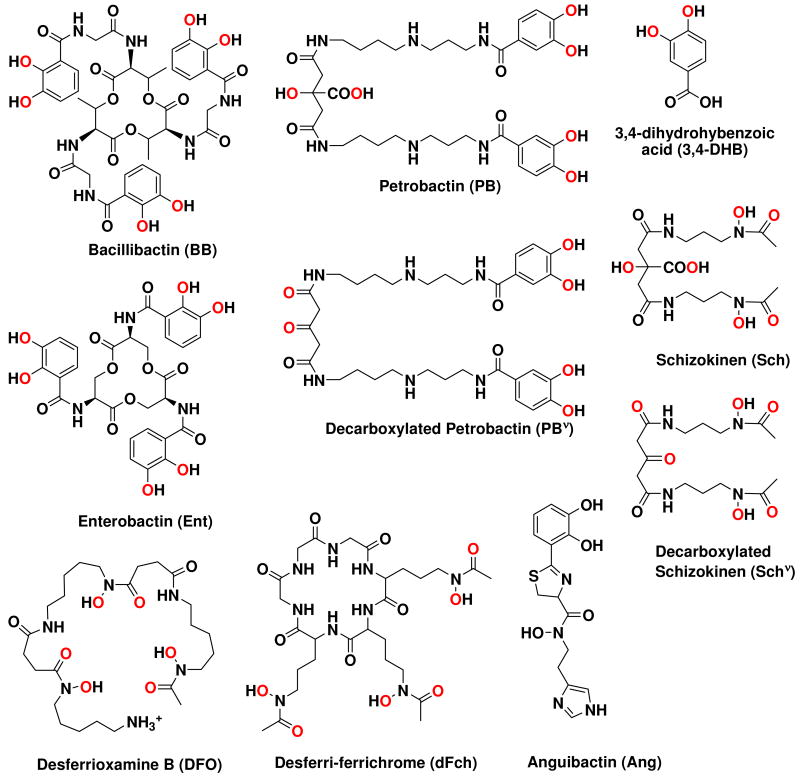
Abstract
During growth under iron limitation, Bacillus cereus and Bacillus anthracis, two human pathogens from the Bacillus cereus group of Gram-positive bacteria, secrete two siderophores, bacillibactin (BB) and petrobactin (PB), for iron acquisition via membrane-associated substrate-binding proteins (SBPs) and other ABC transporter components. Since PB is associated with virulence traits in B. anthracis, the PB-mediated iron uptake system presents a potential target for antimicrobial therapies; its characterization in B. cereus is described here. Separate transporters for BB, PB, and several xenosiderophores are suggested by 55Fe-siderophore uptake studies. The PB precursor, 3,4-dihydroxybenzoic acid (3,4-DHB), and the photoproduct of FePB (FePBν) also mediate iron delivery into iron-deprived cells. Putative SBPs were recombinantly expressed, and their ligand specificity and binding affinity assessed using fluorescence spectroscopy. The noncovalent complexes of the SBPs with their respective siderophores were characterized using ESI-MS. The differences between solution phase behavior and gas phase measurements are indicative of noncovalent interactions between the siderophores and the binding sites of their respective SBPs. These studies combined with bioinformatics sequence comparison identify SBPs from five putative transporters specific for BB and enterobactin (FeuA), 3,4-DHB and PB (FatB), PB (FpuA), schizokinen (YfiY), and desferrioxamine and ferrichrome (YxeB). The two PB receptors show different substrate ranges: FatB has the highest affinity for ferric 3,4-DHB, iron-free PB, FePB, and FePBν, whereas FpuA is specific to only apo- and ferric PB. The biochemical characterization of these SBPs provides the first identification of the transporter candidates that most likely play a role in the B. cereus group pathogenicity.
The Bacillus cereus group of Gram-positive bacteria encompasses closely genetically related organisms, ranging from soil inhabitants to pathogenic species, and has high economic, medical, and biodefense significance (1). Members of this group include: Bacillus cereus, which is very widespread in our environment and is associated with human food poisoning and systemic and local infections in immunocompromised patients; Bacillus anthracis, which is the causative agent of anthrax; and Bacillus thuringiensis, an insect pathogen. Like all aerobic microbes, they must, in order to thrive, overcome low iron availability (10-18 M) in a pH neutral, oxic environment; or within the body of a host, where iron is bound by carrier and storage proteins (2). One of the most widely utilized mechanisms of microbial iron acquisition is the production and excretion of siderophores, low molecular weight iron chelators that bind ferric iron with extremely high affinity and shuttle it into the cells (3). All members of the B. cereus group secrete bacillibactin (BB), a 2,3-dihydroxybenzoyl-Gly-Thr trilactone siderophore, which is a similar counterpart of the enterobactin (Ent) produced by Gram-negative bacteria. However, B. anthracis and B. cereus primarily produce petrobactin (PB) (Fig. 1) (4, 5). Although it was recently shown that PB is also produced by innocuous B. cereus and B. thuringiensis isolates (6), PB biosynthesis was demonstrated to be required both for growth of B. anthracis in macrophages and for virulence in mice (7). The rationale for such redundancy of iron acquisition systems has been explained by the discovery of the innate immune protein siderocalin, which binds several siderophores and their iron complexes rendering them unavailable for pathogenic bacteria (8, 9, 10). Siderophores that are not recognized by siderocalin (stealth siderophores) can then be regarded as a virulence trait. Since siderocalin sequesters BB, but not PB, both synthesis and uptake of PB presents a good target for limiting the development of the bacteria during infection. The surface-exposed PB transporter in particular can be a target for bacteria typing and a potential vaccine candidate (11). While the biosynthetic pathways of PB in B. anthracis were recently elucidated (12, 13, 14), the PB-mediated iron transport systems in B. cereus group remain unidentified.
Figure 1.
Molecular structures of siderophores produced by the B. cereus group of organisms (BB, PB, and 3,4-DHB), other siderophores used in the study, including Ent, Ang, Sch, DFO, and Fch, and the products of ferric PB and Sch photolysis, PBν and Schν.
PB, an unusual siderophore in that it contains two 3,4-catecholate moieties and a citrate-based backbone, was first isolated from the marine bacterium Marinobacter hydrocarbonoclasticus (15). Ferric PB undergoes photolysis initiated by ligand-to-metal charge transfer of the Fe(III)-citrate complex that results in iron reduction, decarboxylation and oxidation of the ligand; a 3-ketoglutarate residue is formed at the former site of the citryl moiety (Fig. 1) (15). The PB photoproduct (PBν) also coordinates FeIII with similar affinity as PB at physiological pH and removes iron from transferrin efficiently (16). The photoreactivity of ferric complexes formed with citrate-based siderophores produced by marine bacteria, including PB and aerobactin, was suggested to be a mechanism for the production of biologically available ferrous iron (17). However, the possible role of such photoreactivity in siderophores produced by soil or pathogenic bacteria such as Bacilli remains unclear. We have shown that the ferric complex of PB and its photoproduct can both support the growth of Bacillus subtilis under iron-limited conditions, indicating that the photoreactivity of PB has little effect on its function as a siderophore and implying the presence of a PB-specific transport system for this exogenous siderophore that is different from the BB receptor (16). In addition to PB, B. anthracis and B. cereus also secrete large quantities of the PB precursor, 3,4-dihydroxybenzoic acid (3,4-DHB) (18). Although the role of 3,4-DHB as a siderophore was suggested for a magnetotactic bacterium, Magnetospirillum magneticum (19), it was never previously confirmed in the B. cereus group.
In Gram-positive bacteria the iron-siderophore complexes are retrieved from the extracellular milieu by specific membrane-tethered receptors and transport proteins belonging to the ABC-type transporters (20). The prototypical Gram-positive bacterium, B. subtilis has at least five substrate-binding lipoproteins anchored to the cell membrane that recognize and bind the endogenous BB and a whole array of xenosiderophores (21, 22). These substrate-binding proteins (SBPs) scavenge iron-loaded siderophores, which upon interaction with the transmembrane permease components of the ABC-transporters are channeled into the cell. Transport is facilitated by cytoplasmic ABC subunits that provide energy for siderophore translocation. The SBPs act as receptors and resemble the periplasmic binding proteins of Gram-negative bacteria, which have a kidney-shaped bilobal structure; although the SBPs show more ligand specificity and superior affinity for ferric ligands (3). The B. subtilis FeBB-binding protein FeuA recognizes both BB and the related triscatecholate Ent (23). Ferric citrate, ferrichromes, ferrioxamines, and photoreactive citrate-based hydroxamates, such as schizokinen (Sch) and arthrobactin, are recognized by distinct SBPs (YfmC, FhuD, YxeB, and YfiY, respectively). Some B. subtilis SBPs (FhuD and YxeB) use the same ABC transporter FhuBGC, similar to the ferric hydroxamate uptake system in Staphylococcus aureus (24). The only siderophore uptake system identified within the B. cereus group is a ferric dicitrate transporter that was shown to be required for full virulence of B. cereus 569 in a lepidopteran infection model (25).
In this study, a biochemical approach has been used to characterize siderophore transporters in B. cereus ATCC 14579 with an emphasis on virulence-related siderophore receptors. The high degree of genetic relatedness within the B. cereus group makes B. cereus a good representative to study genome-encoded iron acquisition systems in the whole group (26). We determined the scope of siderophores utilized by B. cereus for iron delivery and characterized the ligand specificity of five recombinantly produced SBPs. Electrospray ionization-mass spectrometry (ESI-MS) was utilized to monitor formation of noncovalent complexes of SBPs with siderophores, a new application of this technique. The comparison of the complex stability upon transition into the gas phase provides information about noncovalent interactions between the proteins and the siderophores. This report presents the first identification and characterization of PB-binding receptor proteins.
Experimental Procedures
Bacterial strains and growth media. B. cereus
ATCC 14579 was obtained from American Type Culture Collection and maintained on nutrient agar plates. Iron-limited medium was prepared as described previously (27).
55Fe-siderophore transport assay and uptake kinetics
Iron siderophore complexes were prepared as described previously (27). Briefly, the iron complex was formed by mixing 55FeCl3 (3.7 Ci mmol-1, 5 μL, 19 nmol) and FeCl3 (2.9 μL, 72 nmol) with the corresponding ligand (100 nmol) in a ratio of 0.9:1 and ddH2O to the total volume of 200 μL. This solution was incubated at room temperature for 2 h, after which 100 μL of 1 M sodium phosphate buffer at pH 7.4 was added. Free iron was removed by centrifugation at 14,000 rpm for 1 min. One hundred-μL of this concentrated stock solution was diluted with 9.9 mL of the iron-limited medium to yield the working ferric complex solution. The concentrations of the ferric siderophore solutions were quantified by determining their specific radioactivity.
For siderophore uptake studies, 50 mL of iron-limited medium was inoculated with 1 mL of overnight iron-limited culture of B. cereus, and the cultures were grown to the late exponential phase at 37 °C with shaking at 250 rpm. Then the cells were washed three times and suspended in iron-limited medium to the optical density (600 nm) of 0.62±0.02. Transport assay was performed by the addition of 55Fe siderophore (0.9 μM, 1 mL) to 9 mL of cells incubated at 37 °C for 10 min. To test the 55Fe siderophore uptake in the presence of inhibitors, 15-fold excess of non-labeled iron siderophore complex was added at 2 min. Aliquots (1 mL) were removed at 0, 2, 5, 10, 15, and 20 min, filtered through 0.45 μm membrane filters (Millipore), and washed with 10 mL of ice-cold 0.1 M sodium citrate. Filters were dried and 6 mL of liquid scintillation cocktail (Ecolume, ICN) was added. The vials were shaken, stored for 12 h, and radioactivity was measured using the liquid scintillation analyzer Tri-Carb 2800TR (Perkin-Elmer, Waltham, MA). Data were normalized to 1 mL of bacterial cultures.
To obtain kinetic parameters of iron uptake, aliquots of 55Fe siderophore (0.1 mL) diluted in iron-limited medium were added to 0.9 mL of bacterial culture prepared the same way as for the transport assay to yield a distribution of concentrations of 0.1, 0.25, 0.5, 1, 2, and 5 μM. The samples were incubated for 2 and 5 min at 37 °C, filtered through 0.45 μm membrane filters (Millipore), and washed with 0.1 M sodium citrate (10 mL). Filters were dried and 6 mL of liquid scintillation cocktail (Ecolume, ICN) was added. The vials were shaken, stored for 12 h, and radioactivity was measured. A difference between the values recorded at 2 and 5 min indicated that uptake was occurring and the initial rate was determined. Analysis of the experimental data with SigmaPlot and Excel yielded Km and vmax values.
Cloning of SBPs
The complete genome sequence of B. cereus ATCC 14579 (GenBank AE016877) was searched for putative SBPs genes and 10 such genes were identified. The genes were analyzed using SignalP 3.0 (28) to predict the presence of signal peptides (Table 2). The genes encoding the SPBs without signal peptide sequences were amplified by PCR from the total DNA of B. cereus ATCC 14579 using the primers designed by Invitrogen's OligoPerfect Designer (Table 1). The primers were designed to enable directional cloning into a pET101/D-TOPO expression vector introducing a C-terminal His6-tag (Invitrogen, Carlsbad, CA). The TEV protease cleavage site was incorporated into the BC3738 reverse primer to allow for tag removal. The PCR products were purified using a QIAquick PCR purification kit (Qiagen, Inc., Valencia, CA) and used for cloning. The PCR products were cloned into a pET101/D-TOPO vector using a Champion pET Directional TOPO expression kit (Invitrogen), and used to transform One Shot TOP10 chemically competent Escherichia coli cells. Clones with plasmids containing the expected insert sizes, as confirmed by PCR with M13 primers and Taq polymerase (Invitrogen), were selected and the correct orientation of the inserts and the exact insert sequences were confirmed by sequencing. The plasmid DNA from the selected clones was purified using a Wizard Plus SV Minipreps DNA Purification System (Promega). To express the recombinant fusion proteins, chemically competent BL21(DE3) E. coli cells (Invitrogen) were transformed with the purified plasmid DNA.
Table 2.
Sequence analysis of the B. cereus ATCC 14579 SBP genes included in the study.
| Locus tag | ORF product description in GenBank | SP/CDS (aa)a | Similarityb | Identity/Similarity (%) | E value | Genomic context (related ORFs) |
|---|---|---|---|---|---|---|
| BC_3738 | Iron(III) dicitrate-binding protein | 21/321 | Iron compound binding proteins of four strains of the B. cereus group, B. anthracis BAS3583 |
≥98/99 97/99 |
0.0 9e-140 |
BC_3737 permease fecD, BC_3736 permease fecC, BC_3735 ATP-binding protein fecE |
| B. subtilis FeuA | 49/68 | 2e-77 | ||||
| BC_5106 | Ferric anguibactin-binding protein FatB | 24/338 | Iron compound binding proteins of thirteen strains of the B. cereus group, B. anthracis BA5330 |
≥93/96 95/97 |
0.0 0.0 |
BC_5105 permease fatD, BC_5104 permease fatC, BC_5103 ATP-binding protein |
| B. subtilis YclQ | 31/51 | 8e-27 | ||||
| BC_4528 | Ferrichrome-binding protein FhuD | 23/324 | Iron compound binding proteins of seven strains of the B. cereus group, B. anthracis BA4766 |
≥99/99 99/99 |
0.0 0.0 |
BC_4529 similar to ABC transporter permease BA4767 (E= 0.0, 97/98% identity/similarity) |
| B. subtilis YfmC | 32/53 | 8e-35 | ||||
| BC_0616 | Iron(III) dicitrate binding protein FhuD | 26/322 | Iron compound binding proteins of fourteen strains of the B. cereus group, B. anthracis BA0615 |
≥95/98 95/97 |
≥2e-154 3e-176 |
BC_0617 permease fecD, BC_0618 permease fecC, BC_0619 ATP-binding protein fecE |
| B. subtilis YfiY | 56/72 | 3e-97 | ||||
| BC_0383 | Ferrichrome-binding protein FeuA | 20/306 | Iron compound binding proteins of eleven strains of the B. cereus group, B. anthracis BA0351 |
≥93/97 95/99 |
≥1e-164 1e-171 |
BC_0382 permease fhuB, BC_0381 permease fhuG |
| B. subtilis YxeB | 37/61 | 3e-44 |
Length of SP – signal peptide and length of CDS – coding sequence of the pre-protein; the recombinant proteins were expressed without SP;
The similarities of the predicted SBPs were determined using BLASTP. Since the proteins have several highly similar orthologues within the strains of the B. cereus group, the number of the hits is reported along with examples of B. anthracis SBPs. Also the B. subtilis SBP orthologues are included.
Table 1.
Oligonucleotides used for cloning.
| Primer namea | Sequenceb (5′→3′) |
|---|---|
| BC0383 F | CACCATGAATAGCTCTACAGACAAAAAGAACGA |
| BC0383 R | TTTACCAAGAAAGCTCTTCTTGAA |
| BC0616 F | CACCATGAAATCAAATAACAAAGAAGAATCTACAA |
| BC0616 R | TTTAACAAAGCGTTTTTCGATG |
| BC3738 F | CACCATGAATAGTGCGGATAAAACTTCAGG |
| BC3738 R | TTTAGATAAATTTTTCACGGCTTCT |
| BC3738TEVR | TCCCTGAAAATACAGGTTTTCTTTAGATAAATTTTTCACGGCTTCT |
| BC4528 F | CACCATGGAGAAAAAAGAAACAAAAGCGG |
| BC4528 R | CTTCTTCGCTGTCATTACATCTG |
| BC5106 F | CACCATGGATTCAAGTAAAGAAACTTCGAAGG |
| BC5106 R | CTTAGCTAAAGTTTTTGCCATATTTC |
Oligonucleotide names refer to the gene locus tag from the GenBank database.
The underlined sequences of forward primers represent the sequence required for directional cloning and the following start codon; the TEV protease recognition sequence is italicized.
Expression of recombinant SBPs
An overnight culture of E. coli BL21(DE3) was inoculated (10% v/v) into 200 mL LB medium containing ampicillin (100 μg mL-1) and bacteria were grown at 37 °C with shaking at 250 rpm for 2 hours. Expression of the recombinant protein was induced with 0.5 mM of IPTG when the cultures reached an OD600 of 0.6-0.8. After induction, cultures were incubated an additional 4-5 h at 37 °C. To confirm enzyme expression, 1-mL aliquots of non-induced and induced cultures were removed, bacteria were harvested by centrifugation (18,000 × g for 10 min) and the pellets were resuspended in 100 μl of 1× sample buffer, vortexed, boiled for 5 min, centrifuged briefly, and 10 μl was loaded onto an SDS-PAGE gel to examine protein expression. Cells were harvested by centrifugation at 10,000 × g for 25 min and the pellets frozen at -20 °C.
Purification of recombinant SBPs
The harvested frozen cells were suspended in BugBuster Protein Extraction Reagent Master Mix (5 mL per 1 g of wet cells) (Novagen, EMD Biosciences). The protease inhibitor cocktail for purification of His-tagged proteins (Sigma-Aldrich, Saint Louis, MI) was added (50 μL to cell lysate from 1 g wet weight) and the mixture was incubated at room temperature. After 10 min of incubation, 50% glycerol was added to the final concentration of 5% together with NaCl (300 mM final concentration) and imidazole (5 mM; Sigma) and the mixture was incubated for an additional 10 min. The cell lysate was centrifuged for 20 min at 16,000 × g at 4 °C. Recombinant SBPs were purified from the clarified cell extract using HIS-Select nickel affinity agarose gel (Sigma) using native conditions and a batch purification method according to the manufacturer's instructions with some modifications. The equilibration and wash buffer consisted of 50 mM sodium phosphate, pH 8.0, 0.5 M NaCl, 10% glycerol, and 10 mM imidazole. The elution buffer contained 50 mM sodium phosphate, pH 8.0, 0.5 M NaCl, 10% glycerol, and 250 mM imidazole. Cell lysate from 1 g of wet cell pellet was mixed with 0.5 mL of equilibrated agarose gel and purified according to the batch protocol, but centrifugation was substituted with gravity flow purification in 5-mL disposable columns and the target protein was eluted without mixing the gel two times with 1 mL of elution buffer. The recombinant BC_3738 containing TEV protease recognition site enabling removal of His6-tag after protein purification was treated with AcTEV protease (Invitrogen) according to the manufacturer's instructions and purified using HIS-Select nickel affinity agarose gel. The concentration of eluted proteins was measured by bicinchoninic acid (BCA) protein assay (Pierce, Rockford, IL). SDS-PAGE analysis was performed in 4-15% polyacrylamide gels (BioRad) and the gels were stained with Coomasie Brilliant Blue R250. The purified SBPs were stored frozen at −20 °C in the elution buffer. Molecular masses of the expressed proteins and their ligand binding properties were examined using ESI-MS. Siderophore binding was evaluated using spectrofluorimetric assay.
Electrospray ionization-mass spectrometry (ESI-MS)
Cloned and purified recombinant SBPs were analyzed in positive mode using ESI-MS (Q-TOF Premier, Waters) to determine the molecular weight of the free proteins and of the noncovalent complexes between the proteins and siderophores. For siderophore binding experiments, the proteins (5 μM) in 10 mM ammonium acetate (pH 6.8) were equilibrated with two equivalents of the ligand in its iron-free and ferric forms 2-4 h prior to the analysis. Samples were infused into the ESI source using a syringe pump. The source parameters were as follows: capillary voltage: 2.98 kV, sampling cone voltage: 40 V, source temperature: 80 °C, and desolvation temperature: 150°C. MassLynx software was used for data analysis.
Fluorescence spectroscopy
Fluorescence quenching of recombinant SBPs upon binding of siderophores was measured on a Jobin Yvon fluoroLOG-3 fluorometer or Varian Cary Eclipse fluorescence spectrophotometer using the excitation wavelength λexc = 280 nm and emission wavelength individually selected for each protein (λem = 324-329 nm) and with the slit band pass (3-4.5 nm) adjusted to give optimal emission intensity (8). Measurements were performed at a protein concentration of 100 nM in Tris-buffered saline, pH 7.0, 5% DMSO, and 32 μg/mL ubiquitin (Sigma-Aldrich). The ligand solutions were freshly prepared in situ; an aliquot of a DMSO stock solution of the iron-free ligand (4 mM, 25 μL) was combined with ferric chloride solution (27 mM, 1 eq.), vortexed and diluted with TBS buffer to form the ferric iron complex at a concentration of 0.1 mM. No metal was added in the case of apo-siderophores. The solutions were equilibrated for 2 h at 20 °C and diluted to a final concentration of 6-18 μM in 5% DMSO, TBS buffer, pH 7.2. To obtain photoproducts of the ferric-PB and ferric-Sch complexes, the ferric siderophores were exposed to light for 1 h and their conversion to photoproducts was confirmed by measuring the UV-Vis absorption spectra. The protein solutions were titrated with siderophore solutions and fluorescence intensity was measured after equilibration of the mixture at 20 °C. Fluorescence values were corrected for dilution upon addition of ligands. Fluorescence data were analyzed by nonlinear regression analysis of fluorescence response versus ligand concentration using a one-site binding model as implemented in DYNAFIT (29). The Kd values are the results of at least three independent titrations.
Siderophores
BB was extracted from iron-limited cultures of B. subtilis ATCC 6051 and purified using reverse phase HPLC as previously described (30). PB was obtained from Prof. B. Byers (University of Mississippi Medical Center) as crude extract from B. anthracis str. Sterne and purified by HPLC using conditions similar to the BB purification protocol. Ent was synthesized as previously described (31). Ferrichrome (Fch) and desferrioxamine (DFO) were obtained in iron-free form from Sigma-Aldrich and CIBA Pharmaceutical Co., respectively. Iron-free Sch was obtained from EMC Microcollections GmbH Tuebingen.
Results
Specificity of iron transport systems
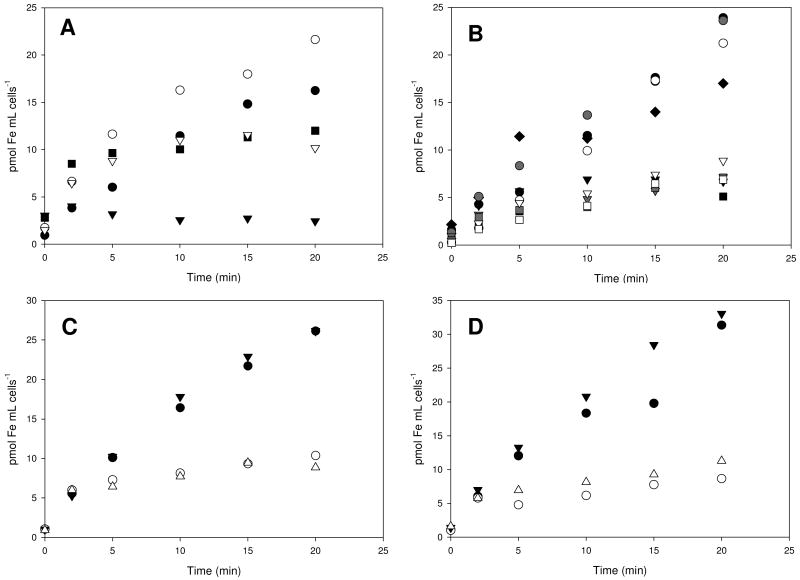
The ability of several siderophores to mediate iron uptake in B. cereus ATCC 14579 was studied using 55Fe-siderophore complexes. All siderophores endogenous to B. cereus (BB, PB, and 3,4-DHB) as well as xenosiderophores (Ent, DFO, Fch, and Sch) were found to mediate 55Fe transport in iron-starved B. cereus (Fig. 2). Additionally, the photoproducts of the ferric PB and ferric Sch complexes were also transported into bacterial cells. To investigate whether single or multiple transport systems are responsible for this uptake, the incorporation of 55Fe-siderophore complexes was monitored in the presence of an excess of nonradiolabeled ferric siderophores, used as inhibitors. Uptake of radioactive ferric BB was completely blocked by cold ferric Ent, whereas incorporation of radioactive ferric Ent was only slightly affected by excess of cold ferric BB and PB (Fig. 2A). Thus, it appears that B. cereus transports BB and Ent through the same permease but also expresses a separate receptor for Ent and PB. Uptake of radioactive ferric PB, PBν, and 3,4-DHB was inhibited by excess of cold ferric complexes of PBν and 3,4-DHB, PB and 3,4-DHB, and PB and PBν, respectively (Fig. 2B). On the other hand, uptake of 55FePB was not affected by FeBB added as an inhibitor. This suggests a common transport route for ferric PB, PBν, and 3,4-DHB, which is separate from the BB transporter. In the case of 55FeSch uptake, a nonradioactive ferric complex of its decarboxylated photoproduct was used as an inhibitor and vice versa. Since ferric complexes of Sch and its photoproduct blocked each other's transport, the two forms of the siderophore must be transported via the same receptor (Fig. 2C). The ferric tris-hydroxamates DFO and Fch were also incorporated into B. cereus (Fig. 2D). The addition of excess of nonradioactive FeFch inhibited uptake of 55FeDFO and the presence of cold FeDFO inhibited transport of 55FeFch, suggesting that both ligands enter the cells through the same transport system.
Figure 2.
Iron transport mediated by 55Fe siderophore complexes in B. cereus at 37°C in iron-limited medium. All inhibitors (I) were cold iron siderophore complexes added in 15-fold excess at 2 min: A) black circles 55FeBB, black triangles 55FeBB + I (FeEnt), black squares 55FeBB + I (FePB), white circles 55FeEnt, and white triangles 55FeEnt + I (FeBB); B) black circles 55FePB, black triangles 55FePB + I (FePBν), black squares 55FePB + I (Fe(3,4-DHB)2), black diamonds 55FePB + I (FeBB), gray circles 55FePBν, gray triangle 55FePBν + I (FePB), gray squares 55FePBν + I (Fe(3,4-DHB)2), white circles 55Fe(3,4-DHB)2, white triangle 55Fe(3,4-DHB)2 + I (FePB), and white squares 55Fe(3,4-DHB)2 + I (FePBν); C) black circles 55FeSch, white circles 55FeSch + I (FeSchν), black triangles 55FeSchν, and white triangles 55FeSchν + I (FeSch); D) black circles 55FeDFO, white circles 55FeDFO + I (FeFch), black triangles 55FeFch, and white triangles 55FeFch + I (FeDFO). Data presented are the average of two independent experiments.
Additionally, the transport kinetic parameters of the representative ferric siderophores were determined (Table 3). Ferric BB transport had a slightly higher Km value (0.3 μM) than ferric Ent (0.2 μM), but 55FeBB was transported at a higher maximum uptake rate. Surprisingly, the B. cereus uptake system specific for PB and 3,4-DHB has about three times lower affinity for ferric PB than for ferric PBν and 3,4-DHB. At the same time ferric PB is transported with a rate twice higher than ferric PBν and 3,4-DHB.
Table 3.
Summary of Michaelis-Menten kinetics of uptake of 55Fe-siderophore complexes. The standard error is given in parentheses.
| Ferric complex | Km (μM) | vmax (pmol Fe mL-1 min-1) |
|---|---|---|
| Bacillibactin | 0.3 ± 0.1 | 4.7 ± 0.2 |
| Enterobactin | 0.2 ± 0.1 | 6.4 ± 0.5 |
| Petrobactin | 0.9 ± 0.6 | 5.8 ± 0.2 |
| Petrobactin photoproduct | 0.3 ± 0.1 | 2.9 ± 0.2 |
| 3,4-DHB | 0.4 ± 0.1 | 2.3 ± 0.1 |
Identification of potential iron transport systems
Since SBPs are responsible for ligand recognition and determine transport specificity we characterized the B. cereus SBPs putatively involved in iron-siderophore binding. The B. cereus ATCC 14579 complete genome sequence revealed the presence of at least ten genes related to periplasmic iron-siderophore-binding proteins. The SBPs were recombinantly produced in E. coli as C-terminal His6-tag fusions. Their signal peptides including cystein residues predicted for lipid modifications were omitted which resulted in generating unacetylated and untethered proteins. Only seven proteins were successfully expressed using our system. After purification the proteins were screened for ligand binding using fluorescence spectroscopy and mass spectrometry that allowed for characterization of five SBPs. Ligand specificity of the remaining two proteins could not be determined among siderophores available to us at the time of this study and they were not included in this publication.
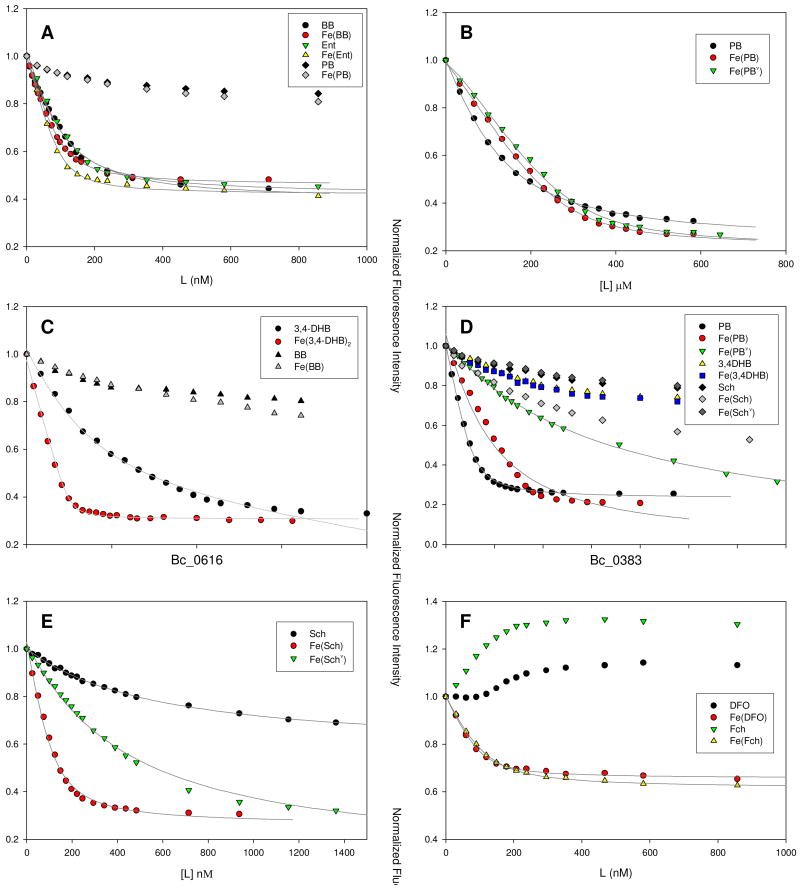
Intrinsic protein fluorescence quenching upon ligand binding was used to determine affinity of recombinant SBPs for different siderophores. The protein solutions at the near neutral pH were titrated with iron-free and ferric solutions of BB, PB, Ent, DFO, Fch, Sch, and citrate (Fig. 3). When protein fluorescence quenching was observed, the equilibrium dissociation constants (Kd) were determined by nonlinear regression analysis of the fluorescence response versus ligand concentration, using a one-site binding model (29). Mass spectrometry was used to determine the molecular weight of the recombinant receptor proteins and of the noncovalent complexes between the proteins and siderophores (Fig. 4). The mass measurements determined the stoichiometry of the noncovalent complexes as 1:1. Based on the specific interactions of the recombinant proteins with siderophores, the following SBPs were identified: FeuA as a BB and Ent-binding protein, FatB and FpuA as PB-binding proteins, YfiY as a Sch-binding protein, and YxeB as a DFO and Fch-binding protein (Table 4).
Figure 3.
Fluorescence quenching analyses of recombinant B. cereus SBPs: A) FeuA with apo- and ferric bacillibactin (BB) and enterobactin (Ent), B) FatB with apo- and ferric forms of petrobactin (PB) and the photoproduct of FePB (FePBν), C) FatB with apo- and ferric 3,4-dihydroxybenzoic acid (3,4-DHB), D) FpuA with PB, FePB, FePBν, and apo- and ferric 3,4-DHB, E) YfiY with apo- and ferric forms of schizokinen and its photoproduct (Sch and Schν), and F) YxeB with apo- and ferric forms of desferrioxamine (DFO) and ferrichrome (Fch). The lines give the calculated fits.
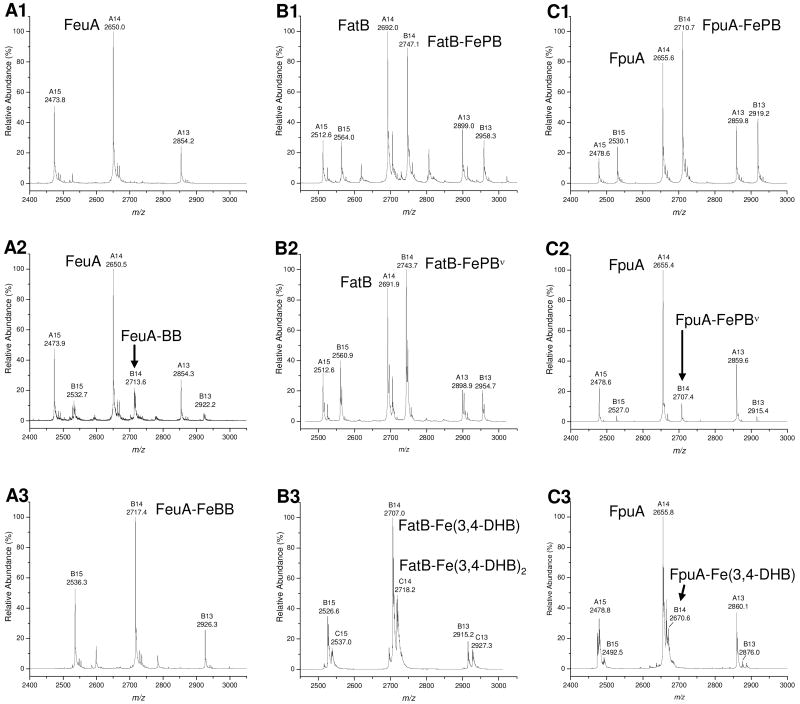
Figure 4.
Representative mass spectra of B. cereus SBPs and their noncovalent complexes with siderophores showing the most intense charge states from +15 to +13: A1) FeuA, A2) complex of FeuA with bacillibactin (BB), B series of peaks, A3) complex of FeuA with FeBB, B1) complex of FatB with ferric petrobactin (PB), B series of peaks, B2) complex of FatB with photoproduct of FePB (FePBν), B series of peaks, B3) complex of FatB with ferric 3,4-dihydroxybenzoic acid (Fe(3,4-DHB) and Fe(3,4-DHB)2, B and C series of peaks, respectively), C1) complex of FpuA with FePB, B series of peaks, C2) complex of FpuA with FePBν, B series of peaks, C3) complex of FpuA with Fe(3,4-DHB), B series of peaks.
Table 4.
Ligand binding by recombinant SBPs determined using fluorescence emission quenching of proteins upon binding of the apo- and ferric form of ligands and molecular weights of SBPs and complexes of proteins with the siderophores determined by ESI-MS.
| Protein | Liganda | Expected ligand charge at pH 7 | Kd (nM)b | Molecular weight (Da) | |
|---|---|---|---|---|---|
| Calculatedc | Measuredd | ||||
| FeuA (Bc_3738) | 37094.6 | 37093 | |||
| BB | [BB]0 | 59 (11) | 37976.6 | 37976 | |
| FeBB | [FeIII(BB)]3- | 19 (1) | 38029.6 | 38029 | |
| Ent | [Ent] 0 | 36 (3) | 37763.6 | 37763 | |
| FeEnt | [FeIII(Ent)]3- | 12 (3) | 37816.6 | 37817 | |
| FatB (Bc_5106) | 37675.4 | 37675 | |||
| PB | [PB]0 | 77 (9) | 38393.4 | NDf | |
| FePB | [FeIII(PB)]3- | 127 (1) | 38446.4 | 38445 | |
| FePBν | [FeIII(PBν)]2- | 135 (47) | 30401.4 | 38400 | |
| 3,4-DHB | [3,4-DHB]1- | 191 (69) | 37829.4 | ND | |
| Fe(3,4-DHB) | [FeIII(3,4-DHB)]0 | nde | 37884.4 | 37884 | |
| Fe(3,4-DHB)2 | [FeIII(3,4-DHB)2]3- | 1.2 (0.7) | 38037.4 | 38038 | |
| FpuA (Bc_4528) | 37166.3 | 37165 | |||
| PB | [PB]0 | 23 (8) | 37885.3 | ND | |
| FePB | [FeIII(PB)]3- | 175 (35) | 37937.3 | 37937 | |
| FePBν | [FeIII(PBν)]2- | 699 (169) | 37891.3 | 37889 | |
| 3,4-DHB | [3,4-DHB]0 | nd | 37320.3 | ND | |
| Fe(3,4-DHB) | [FeIII(3,4-DHB)]1- | nd | 37375.3 | 37374 | |
| YfiY (Bc_0616) | 37118.3 | 37116 | |||
| Sch | [Sch]0 | 495 (33) | 37537.3 | 37535 | |
| FeSch | [FeIII(Sch)]0 | 34 (3) | 37590.3 | 37589 | |
| FeSchν | [FeIII(Schν)]1+ | 298 (114) | 37545.3 | ND | |
| YxeB (Bc_0383) | 36093.8 | 36094 | |||
| DFO | [DFO]1+ | nd | 36654.1 | ND | |
| FeDFO | [FeDFO]1+ | 18 (4) | 36706.8 | 36707 | |
| Fch | [Fch]0 | nd | 36781.1 | ND | |
| FeFch | [FeFch]0 | 30 (1) | 36833.8 | 36833 | |
bacillibactin (BB), enterobactin (Ent), petrobactin (PB), petrobactin photoproduct (PBν), 3,4-dihydroxybenzoic acid (3,4-DHB), schizokinen and its photoproduct (Sch and Schν, respectively), desferrioxamine (DFO), and ferrichrome (Fch)
The dissociation constants (Kd) were determined from fluorescence data and are the mean of at least three independent measurements with SD given in parenthesis,
Calculated masses of noncovalent complexes were computed using masses of neutral ligand species,
Mass measured ± 1 Da,
nd – not determined,
ND – not detected.
Bacillibactin and enterobactin receptor FeuA
The analysis of the recombinant BC_3738, which has 49% sequence identity to the B. subtilis BB-binding protein FeuA (Table 2) revealed that the protein specifically binds apo and ferric forms of BB and Ent, but not PB, and therefore can be regarded as a B. cereus BB/Ent-binding protein, FeuA. The dissociation constants, determined using protein fluorescence quenching to monitor ligand binding, showed three-fold higher affinity for the ferric form of the ligands than for the apo siderophores (Fig. 3A, Table 4). The Kd value for FeBB was slightly higher than for FeEnt, indicating stronger affinity of FeuA towards the exogenous siderophore Ent. The formation of noncovalent complexes between FeuA and both siderophores was demonstrated using ESI-MS (Fig. 4). The charge state distribution was not shifted by adding the ligands and the most abundant charge states were +15, +14, and +13, with +14 being the most intense. When the protein was incubated with two equivalents of ferric BB or Ent, the peaks belonging to the free protein disappeared and only the signal from the complexes of FeuA with the ligands was observed (Fig. 4A1 and 3). For the complexes formed with iron-free BB and Ent, the abundance of the complex peaks was lower and the free protein peaks were the dominant species present (Fig. 4A2). The results obtained for the BC_3738 protein devoid of the His6-tag were not significantly different than for the His-tagged protein and therefore all studies were performed using His6-tag containing recombinant proteins.
Petrobactin receptors FatB and FpuA
The screening of the recombinant receptors for specific binding of ligands using fluorescence spectroscopy revealed two proteins that showed high affinity for PB: BC_5106 and BC_4528, which CDS are predicted to produce a ferric anguibactin-binding protein FatB and a citrate-binding protein YfmC, respectively. We propose to name the BC_4528 protein FpuA (ferric PB uptake) not to confuse it with the B. subtilis ferric citrate receptor. In addition to iron free and ferric complexes of PB, the affinity of the identified PB-binding receptors towards photoproduct of FePB was examined to assess the possible physiological role of FePB photoreactivity. Other ligands, including PB precursor 3,4-DHB, citrate, anguibactin, Fch, BB, and Sch were used for binding studies to further probe the aspects of ligand recognition by the receptors (Figs. 3 and 4). The two proteins FatB and FpuA were found to bind iron-free PB with low Kd values and the affinity of both receptor proteins for ferric PB was within the same nanomolar range (Fig. 3B-E, Table 3). However, they differed significantly in their affinity toward ferric PBν and 3,4-DHBA. While dissociation constants of FatB were similar for iron (III) complexes of PB and its photoproduct, FpuA had much lower affinity for FePBν than FePB (Table 4).
To test whether 3,4-catechol residues are recognized by the receptor proteins and if these receptors can also recognize and participate in uptake mediated by 3,4-DHB, which is excreted in large quantities by B. anthracis (18), binding of 3,4-DHB was examined. First, the stoichiometry of ferric complex formation by 3,4-DHB in neutral pH was determined to be 1:2 and the ratio of 1:3 to 1:4 of iron to ligand was found to result in the highest concentration of the ML2 complex. The ESI-MS analysis confirmed that the predominant ferric complex observed at pH 6.8 was [FeIII(3,4-DHB)2]-1 (m/z 360.0) accompanied by minor peaks of m/z 277.9 and 279.9 corresponding to FeIIICl2(3,4-DHB)-1 species. Therefore the 1:3 ratio of iron to ligand was used in the experiments. FatB binds both iron-free 3,4-DHB and its ferric complex (Fig. 6B). Only one equivalent of Fe(3,4-DHB)2 was required to quench the protein fluorescence and the corresponding dissociation constant of 1.2 ± 0.7 nM indicates that the receptor may be at least two orders of magnitude more specific for ferric 3,4-DHB than [FeIII(PB)]3-(Fig. 3C, Table 3). Also, the FatB affinity for iron-free 3,4-DHB was much lower. On the other hand, FpuA fluorescence was not specifically quenched by ferric or apo-3,4-DHB. Additionally, the affinity of both receptor proteins toward ferric citrate was assayed and showed no recognition. Binding of anguibactin and ferrichrome to the respective proteins FatB and FpuA was also tested. The proteins did not bind the aposiderophores whereas the addition of ferric complexes resulted in unspecific protein emission quenching below 10-20% without saturation, indicating binding incompatibility (data not shown). Similar results were obtained when FatB was titrated with apo and ferric BB and FpuA with Sch, FeSch, and FeSchν (Fig. 3C and D) indicating binding incompatibility. Only FeSch quenched FpuA fluorescence more than Fe(3,4-DHB), suggesting some possible interaction of intact citrate moiety-iron center in FeSch with the FpuA binding pocket. This binding was still much less specific than what was observed for FePB or even FePBν.
Mass spectrometry was used to study formation of noncovalent complexes of FatB and FpuA with siderophores (Fig. 4 B and C). Mass spectral analysis gave molecular weight determination of the recombinant proteins and of the noncovalent complexes formed by incubation with two molar equivalents of siderophores (Table 4). When the proteins were incubated with ferric PB, the intensity of the free protein peaks decreased and the peaks corresponding to the protein-ferric ligand complexes grew in; the ratio of free protein to complex peaks was similar for both FatB and FpuA (Fig. 4B1 and C1). The ratios of the protein and complex peaks were different in the case of other ligands. FatB formed a complex with ferric PBν with a similar intensity to that seen with FePB (Fig. 4B2). ESI-MS spectrum of FpuA after incubation with FePBν however, revealed a protein-ligand complex peak of much lower intensity indicating weaker or different interactions (Fig. 4C2). Similarly, when two equivalents of ferric 3,4-DHB were added to FatB, only the protein complex with the ligand (as Fe(3,4-DHB) and Fe(3,4-DHB)2) was observed with no free protein detectable (Fig. 4B3). On the other hand, only very low intensity signal of FpuA and ferric 3,4-DHB complex was observed, consistent with the lack of specific binding detected by fluorescence measurements (Fig. 4C3). Formation of the FatB and FpuA complexes with iron-free PB and 3,4-DHB was not observed using ESI-MS even after increasing the instrument backing pressure to promote detection of weak noncovalent complexes. Additionally, the samples of the two proteins after incubation with ferric citrate, FatB with ferric anguibactin, and FpuA with Fch were analyzed but no protein-ligand complexes were detected.
Schizokinen receptor YfiY
The BC_0616 protein was shown to specifically bind Sch and since it has high homology (55% identity) to the B. subtilis Sch/arthrobactin receptor YfiY, it can be considered a B. cereus Sch-binding receptor YfiY (Table 2). Protein fluorescence quenching measurements showed that YfiY has a threefold higher affinity to ferric Sch than to the apo form of this xenosiderophore (Fig. 3E, Table 3). Sch, similarly to PB, has a citrate backbone and its ferric complex is photoreactive (32). Photolysis of FeSch resulted in a ten-fold decrease in protein binding affinity. Both complexes of the YfiY with free and ferric Sch were identified by ESI-MS (Table 3). However, the photoproduct of ferric Sch complex (FeSchν) was not detected, consistent with the large Kd value and low protein affinity estimated from fluorescence measurements.
Ferrioxamine and ferrichrome receptor YxeB
The protein BC_0383, which is orthologous to B. subtilis ABC transporter-binding protein YxeB (BSU39610, 38% identity) and ferrichrome-binding protein FhuD (BSU33320, 37% identity) (Table 2), was shown to bind FeDFO and FeFch as well as their iron-free forms and therefore can be considered a B. cereus DFO and Fch receptor YxeB. While the binding of the respective ferric complexes caused protein fluorescence quenching, the binding of apo-siderophores caused increase in protein fluorescence (Fig. 3F). Dissociation constants were calculated for FeDFO and FeFch and indicate a two-fold higher affinity of the receptor toward FeDFO (Table 3). The noncovalent complexes of YxeB with ferric Fch were detected using ESI-MS in standard optimized conditions but the noncovalent YxeB complexes with FeDFO were only observed by increasing the backing pressure (Table 3). In the case of the iron-free ligands, their complexes with YxeB were not detected even after increasing the backing pressure and the protein peaks were broadened in the presence of the ligands.
Discussion
Pathogenic bacteria produce siderophores and multiple uptake systems as virulence determinants in their interaction with the host in an effort to adapt to iron inaccessibility (2). All members of the B. cereus group synthesize BB, but this siderophore is deactivated by the innate immune protein siderocalin (9). Like other bacterial pathogens that synthesize secondary siderophores, B. cereus and B. anthracis produce PB (3). PB, although inferior to BB in its affinity for iron (16, 30), evades the host immune system response and is necessary for outgrowth of B. anthracis from spores and the development of infection (7, 12). The delivery of the ferric siderophore complexes into the cells depends on the expression of siderophore specific receptors. The presence of multiple receptors is advantageous to pathogenic bacteria that cannot only utilize their endogenous siderophores but can also pirate exogenous chelates (20). We have shown that, like B. subtilis (22, 27), B. cereus can utilize the whole array of siderophores for iron delivery in addition to BB, PB, and its precursor 3,4-DHB, including Ent, Sch, DFO, and Fch (Fig. 2). In inhibition studies we determined that BB is transported via the same transporter as Ent (Fig. 2A), which similarly to what was found for B. subtilis, appears to have another unidentified transporter (30). However, PB has a separate receptor from BB, since the two siderophores did not display reciprocal inhibition (Fig. 2A and B). In contrast, mutual uptake inhibition of 55Fe complexes of PB, its photoproduct and 3,4-DHB established common transport routes for both forms of PB and its precursor (Fig. 2B). Photolysis of ferric PB had no significant effect on its siderophore function in B. cereus, similar to what was shown for B. subtilis (16). A similar observation was made for the marine bacterium Vibrio sp., which was able to incorporate the ferric photoproduct of citrate-based aerobactin (33). Moreover, uptake studies revealed the presence of separate receptors for Sch and other hydroxamates, such as DFO and Fch (Fig. 3 C and D). Similarly, photolysis of ferric Sch, did not impair its ability to deliver iron into the cells. The incorporation of a citrate backbone may therefore be vestigial for soil and pathogenic Bacilli. However, it is remarkable that photoreactive siderophores are produced by apparently disparate bacteria: PB by marine Marinobacter spp. and soil and pathogenic Bacillus spp., aerobactin by marine Vibrio sp. and enteric bacteria, and Sch by cyanobacteria, rhizobia, and B. megaterium (6, 15, 34, 35). We can speculate that photolysis of photoreactive metabolites may serve in some instances as a signal of an exposure to light and participate in cell cycle regulation (34). Finally, 3,4-DHB mediated 55Fe uptake and thus appears to be able to serve as a siderophore; though since it is a bidentate ligand that has affinity for iron lower than hexadentate ligands, like another Bacillus species metabolite itoic acid, its role in solubilizing iron or removing iron from host transport proteins may depend on milieu conditions.
The examination of ligand specificity of recombinant B. cereus SBPs and bioinformatics sequence comparison yielded candidates for five receptors that were further characterized by fluorescence spectroscopy and ESI-MS. FeuA, expressed from the feuABCD locus coding for a complete ABC transporter, is an orthologue of B. subtilis FeuA and demonstrated similar ligand binding properties (23). B. cereus FeuA had high affinity for both FeBB and FeEnt with slightly lower Kd for FeEnt (Table 4) suggesting that it recognizes these ferric siderophore complexes independently of their metal center chirality, as the B. subtilis FeuA does (36). The two proteins FatB and FpuA were identified as PB-binding proteins of distinct ligand specificity. FatB has homology to the B. subtilis siderophore receptor YclQ recently characterized by our laboratory (Zawadzka et al., In preparation) and FpuA to the B. subtilis citrate receptor YfmC (Table 2). The affinity of FatB was about 100-fold higher for ferric 3,4-DHB than for ferric PB or its photoproduct (Table 4, Fig. 3B and C) and Fe(3,4-DHB)2 had a Kd with FatB as low as 1.2 nM, which is in the range of affinities of outer membrane transporters (OMTs) of Gram-negative bacteria. Such high receptor affinity for ferric 3,4-DHB may compensate for the low iron affinity of a bidentate ligand that was shown to mediate iron transport. Photoinduced decarboxylation of ferric PB had no significant effect on the ligand affinity for FatB. On the other hand, FpuA was clearly most specific for ferric PB with a Kd of 175 nM, similar to that of FatB (Table 4). The affinity of FpuA for FePBν was about four-fold lower than for the parent ligand and the receptor showed no specific affinity for ferric 3,4-DHB (Fig. 3D). Also FatB had no affinity for the 2,3-catecholate BB and FeBB (Fig. 3C). These observations suggest a different type of ligand recognition in the protein binding pocket between the two PB receptors: FatB senses the 3,4-catecholate moieties and the citrate part of PB is not involved in recognition while FpuA discriminates the ligands based on the presence of citrate residue. In the latter case, citrate needs to be a part of the siderophore scaffold, because Fe citrate was not bound by FpuA. A similar trend was observed for the Sch-binding protein YfiY. Ferric Sch was bound by the protein with the highest affinity (Kd = 34 nM) and photoinduced decarboxylation lowered binding affinity by almost ten-fold (Tab. 3, Fig. 3E).
Despite differences in ligand specificity, it is likely that both PB-binding proteins utilize the same ATP-binding protein since a complete ABC-transporter is present only in the FatB transcriptional unit, whereas FpuA is accompanied only by a permease (Table 2). Such transporter configuration would be similar to B. subtilis BB/Ent and Sch/arthrobactin uptake systems (22). Observations of the common transport route for ferric PB, its photoproduct, and 3,4-DHB in our 55Fe-siderophore uptake experiments support such a hypothesis. We note that the transport rate for FePB is higher than that for FePBν and 3,4-DHB, which would suggest that FePB, which is bound by both FatB and FpuA with similar affinity, is transported through those two receptors independently. Therefore, if FpuA uses the same ATP-binding protein as FatB, it seems not to constitute a limiting step in the transport process, assuming that the higher transport rate of FePB is due to the presence of two SBPs participating in the process.
Significantly, both PB-binding proteins were found to bind apo-PB with appreciable Kd values (Table 4) that were almost two and seven-fold lower than for ferric PB. Binding of iron-free siderophores to their cognate receptors with close affinities to those of iron-loaded ligands was observed in other Gram-positive bacteria but only the iron-loaded form is transported into the cell (23). In the periplasmic binding proteins of Gram-negative bacteria related to the SBPs, the conformational changes upon siderophore binding are less pronounced than for the binding to OMTs (37). However, these changes should also allow for the discrimination of the iron-loaded and non-loaded ligands. In the case of apo-PB binding, different mechanisms of protein fluorescence quenching may also account for the discrepancy with what is usually observed for other siderophore receptors (3). It would be consistent with the trend observed for YxeB protein binding DFO and Fch, which binding caused increase of SBP fluorescence (Fig. 3F). Higher affinity of the receptor for the apo-ligand may also be implicated in the mechanism of an alternative iron uptake mechanism via siderophore shuttle (38). The biological significance of apo-siderophore binding to their receptors is unknown, it can possibly increase siderophore concentration in the cell vicinity or have some regulatory function analogous to that showed by Gaballa and Helmann in B. subtilis (39). In their study the presence of free BB enhanced activation of the BB receptor transcription by binding to the novel intracellular Btr transcription activator.
Several noncovalent complexes of B. cereus SBPs with siderophores were characterized using ESI-MS. The formation of noncovalent complexes of proteins with various ligands has been widely studied by ESI-MS (40). The mass measurements of the noncovalent SBP-siderophore complexes confirmed the 1:1 stoichiometry of these complexes and showed that the protein-bound decarboxylated ferric siderophore was easily distinguished from the protein-bound intact ferric siderophore since the mass difference between the two noncovalent complexes equals 45 Da. In some instances we observed binding of an additional siderophore molecule to the protein (Fig. 4A3 and B1). The titration experiments of FeuA with FeBB and FatB with FePB established that the formation of the SBP complexes with two ligands could be attributed to the nonspecific binding of the second ligand (41). Higher ligand concentration (the 1:8 ratio of protein to ligand) did not appear to increase the fraction of the SBP-(ligand)2 (data not shown). The charge state distribution of the free SBPs (+13 to +15) was preserved after adding the ligands and formation of the noncovalent complexes. Binding of the ligands did not seem to prevent protonation of basic residues in or near the binding site and did not affect the charge state and ionization efficiency of the ligand-bound proteins as observed earlier for other protein-ligand interactions (42). Therefore we compared the relative ratio of the peaks corresponding to the proteins and to the noncovalent complexes of ferric siderophores. In the case of FatB and FpuA, the intensity of the peaks of noncovalent complexes appeared to correlate with the binding affinities determined in solution by fluorescence measurements (Table 4, Fig. 4B and C). This phenomenon was reflected by the observation that only 2 equivalents of FeBB or FeEnt were required to fully quench fluorescence of FeuA and only the protein-ligand complex was detected by ESI-MS (Figs. 3A and 4A3), whereas only about half of the fluorescence intensity was quenched and half of the FatB and FpuA proteins formed complex at the same protein to ligand ratio (Figs. 3B and C and 4B1 and C1).
Although gas-phase measurements do not always correlate with the solution-phase behavior, the type of bonding that govern noncovalent interactions in solution could be assessed by ESI-MS (40, 42). The most striking observation was that the complexes of SBPs with ferric siderophores were easily detectable, whereas the protein complexes with iron-free ligands were detected only in the cases of FeuA (Fig. 4B) and YfiY (Table 4). It is indicative of the differences in the types and strength of noncovalent interactions in solution compared with those that occur in the gas phase (43, 44). Since electrostatic and dipolar interactions are strengthened in the absence of solvent (unlike hydrophobic interactions which are strong in the presence of water and labile in the gas phase), the easily detectable complexes of SBPs with ferric ligands must be maintained primarily by electrostatic and polar forces. For iron-free ligands, ionic and polar interactions appear to have a minor role. It is likely that negatively charged ferric catecholate sidereophores are interacting with positively charged amino acid residues in their respective receptor protein hydrophilic binding pockets, similarly to the Fe-triscatecholate binding periplasmic protein CeuE from Campylobacter jejuni (45). The residues participating in ligand binding by FeuA may be similar to the principal recognition and binding residues of the siderocalin binding pocket despite structural dissimilarity between the two proteins. Siderocalin specifically interacts with negatively charged ferric catecholate siderophores through electrostatic forces and cation-π interactions in the positively charged calyx (9, 10) and the protein complex formed with FeEnt was observed using ESI-MS (46). The interactions of SBPs with uncharged apo-catecholate siderophores may be much less electrostatic in nature and therefore not always detectable by ESI-MS. On the other hand, owing to their different chemical structure, the dominant noncovalent interactions between hydroxamate siderophores and SBPs should be different. X-ray crystallographic analysis of the E. coli periplasmic binding protein FhuD bound to hydroxamate siderophores showed that recognition occurs through the iron-hydroxamate centers of the ligand and the side chains of several amino acid residues in the hydrophobic binding pocket and is stabilized by hydrogen bonds (47). In our ESI-MS measurements we were able to detect only the complexes of neutral ligands with the proteins: Fch with YxeB and apo- and ferric Sch with YfiY (Table 4), which overall were less intense than the complexes of FeuA, FatB, and FpuA with their respective ferric catecholate ligands. The positively charged FeDFO bound to YxeB was only observed under increased backing pressure, whereas the photoproduct of FeSch (FeSchν) bound to YfiY was never observed. The presented results open the possibility of further comparative structural studies on the basis of ligand recognition by their cognate transporters and design of siderophore-mimicking inhibitors of iron transport specifically targeting relevant siderophore receptors.
The genome comparison between B. cereus ATCC 14579 and B. anthracis shows that a genomic island containing a ferric anguibactin transport system is absent in the genome of B. anthracis (48). However, the BLASTN search for BC_5106 and BC_4528 revealed the presence of highly similar orthologues in the B. anthracis Ames Ancestor strain genome: GBAA5330 is 95% identical to BC_5106 and GBAA4766 is 98% identical to BC_4528 (49). Similarly to the B. cereus PB-binding proteins, the BC_5106 orthologue GBAA5330 is followed by the permease and ATP-binding protein, whereas the BC_4528 orthologue GBAA4766 does not have all four ABC-transporter genes in its vicinity. These orthologous genes were overexpressed by B. anthracis Sterne 34F2 strain in vivo during infection in host macrophages (50). The BC_5106 orthologue GBAA5330 was one of the highly up-regulated genes during germination and early outgrowth in host macrophages, along with the putative BB receptor GBAA3867. During the later infection stages, the situation was reversed and it was the putative BB-binding receptor GBAA3867 that was the most highly up-regulated iron transport component but the BC_5106 orthologue was also overexpressed. On the other hand, the BC_4528 orthologue was not listed in the comparison of the B. anthracis gene expression patters, but the permease neighboring this gene, GBAA4767 was shown to be up-regulated between one and two hours post-infection during growth within mouse macrophages. Since PB was shown to be an important virulence factor during B. anthracis growth in macrophages, it is very likely that the newly identified receptor proteins are parts of the PB-mediated uptake system in the B. cereus group. The elucidation of the physiological functions of these receptors in vivo will be possible using transporter deletion mutants and the effort to generate B. cereus siderophore transporter knock-out strains is ongoing in our laboratory and will be presented elsewhere.
Gram-positive bacteria encompass many human pathogens, including members of the B. cereus group, which produce siderophores as a part of efficient iron acquisition systems that constitute a requisite for establishing infection within the host. The human immune system protein siderocalin interrupts this essential iron acquisition process by binding several siderophores. Of the siderophores secreted by the B. cereus group, PB is not recognized by siderocalin and it is considered a virulence factor for these bacteria. The second component of siderophore-mediated iron acquisition systems involves protein receptors, which recognize and transport the ferric siderophore complexes across the membrane. Significantly, among the five siderophore-binding proteins that were identified and characterized in this study, two (FatB and FpuA) bind PB. These newly described PB receptor candidates can be considered a part of the PB-mediated iron acquisition system contributing to virulence and are potential targets of preventive and therapeutic strategies against infections caused by organisms from the B. cereus group, including Bacillus anthracis.
Acknowledgments
Anguibactin was a generous gift from Prof. Jorge Crosa (Oregon Health and Sciences University) and petrobactin crude extract was obtained from Prof. B. Rowe Byers (University of Mississippi Medical Center). Helpful discussions on data analysis with Dr. Petr Kuzmic and Trisha Hoette are acknowledged.
Footnotes
Paper number 85 in the series “Coordination Chemistry of Microbial Iron Transport”. For the previous paper, see ref 10.
Abbreviations: BB, bacillibactin; PB, petrobactin; SBPs, substrate binding proteins; 3,4-DHB, 3,4-dihydroxybenzoic acid; Ent, enterobactin; Sch, schizokinen; Fch, ferrichrome; DFO, desferrioxamine B; ESI-MS, electrospray ionization-mass spectrometry; OMTs, outer membrane transporters.
This work was supported by the National Institutes of Health grants AI11744 to K.N.R. and 1S10RR022393-01 for the acquisition of the Q-TOF mass spectrometer.
References
- 1.Rasko DA, Altherr MR, Han CS, Ravel J. Genomics of the Bacillus cereus group of organisms. FEMS Microbiol Rev. 2005;29:303–329. doi: 10.1016/j.femsre.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 2.Andrews SC, Robinson AK, Rodriguez-Quinones F. Bacterial iron homeostasis. FEMS Microbiol Rev. 2003;27:215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- 3.Miethke M, Marahiel MA. Siderophore-based iron acquisition and pathogen control. Microbiol Mol Biol Rev. 2007;71:413–451. doi: 10.1128/MMBR.00012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koppisch AT, Browder CC, Moe AL, Shelley JT, Kinkel BA, Hersman LE, Iyes S, Ruggiero CE. Petrobactin is a primary siderophore synthesized by Bacillus anthracis str. Sterne under conditions of iron starvation. Biometals. 2005;18:577–585. doi: 10.1007/s10534-005-1782-6. [DOI] [PubMed] [Google Scholar]
- 5.Wilson MK, Abergel RJ, Raymond KN, Arceneaux JEL, Byers BR. Siderophores of Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis. Biochem Biophys Res Commun. 2006;348:320–325. doi: 10.1016/j.bbrc.2006.07.055. [DOI] [PubMed] [Google Scholar]
- 6.Koppisch AT, Dhungana S, Hill KK, Boukhalfa H, Heine HS, Colip LA, Romero RB, Shou Y, Ticknor LO, Marrone BL, Hersman SI, Ruggiero CE. Petrobactin is produced by both pathogenic and non-pathogenic isolates of the Bacillus cereus group of bacteria. Biometals. 2008;21:581–589. doi: 10.1007/s10534-008-9144-9. [DOI] [PubMed] [Google Scholar]
- 7.Cendrowski S, MacArthur W, Hanna P. Bacillus anthracis requires siderophore biosynthesis for growth in macrophages and mouse virulence. Mol Microbiol. 2004;51:407–417. doi: 10.1046/j.1365-2958.2003.03861.x. [DOI] [PubMed] [Google Scholar]
- 8.Abergel RJ, Wilson MK, Arceneaux JEL, Hoette TM, Strong RK, Byers BR, Raymond KN. The anthrax pathogen evades the mammalian immune system through stealth siderophore production. Proc Natl Acad Sci USA. 2006;103:18499–18503. doi: 10.1073/pnas.0607055103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goetz DH, Holmes MA, Borregaard N, Bluhm ME, Raymond KN, Strong RK. The neutrophil lipocalin NGAL is a bacteriostatic agent that interferes with siderophore-mediated iron acquisition. Mol Cell. 2002;10:1033–1043. doi: 10.1016/s1097-2765(02)00708-6. [DOI] [PubMed] [Google Scholar]
- 10.Hoette TM, Abergel RJ, Xu J, Strong RK, Raymond KN. The role of electrostatics in siderophore recognition by the immunoprotein siderocalin. J Am Chem Soc. 2008;130:17584–17592. doi: 10.1021/ja8074665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown JS, Ogunniyi AD, Woodrow MC, Holden DW, Paton JC. Immunization with components of two iron uptake ABC transporters protect mice against systemic Streptococcus pneumoniae infection. Infect Immun. 2001;69:6702–6706. doi: 10.1128/IAI.69.11.6702-6706.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee JY, Janes BK, Passalacqua KD, Pfleger BF, Bergman NH, Liu H, Håkansson K, Somu RV, Aldrich CC, Cendrowski S, Hanna PC, Sherman DH. Biosynthetic analysis of the petrobactin siderophore pathway from Bacillus anthracis. J Bacteriol. 2007;189:1698–1710. doi: 10.1128/JB.01526-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oves-Costales D, Kadi N, Fogg MJ, Song L, Wilson KS, Challis GL. Enzymatic logic of anthrax stealth siderophore biosynthesis: AsbA catalyzes ATP-dependent condensation of citric acid and spermidine. J Am Chem Soc. 2007;129:8416–8417. doi: 10.1021/ja072391o. [DOI] [PubMed] [Google Scholar]
- 14.Pfleger BF, Lee JY, Somu RV, Aldrich CC, Hanna PC, Sherman DH. Characterization and analysis of early enzymes for petrobactin biosynthesis in Bacillus anthracis. Biochemistry. 2007;46:4147–4157. doi: 10.1021/bi6023995. [DOI] [PubMed] [Google Scholar]
- 15.Barbeau K, Zhang G, Live DH, Butler A. Petrobactin, a photoreactive siderophore produced by the oil-degrading marine bacterium Marinobacter hydrocarbonoclasticus. J Am Chem Soc. 2002;124:378–379. doi: 10.1021/ja0119088. [DOI] [PubMed] [Google Scholar]
- 16.Abergel RJ, Zawadzka AM, Raymond KN. Petrobactin-mediated iron transport in pathogenic bacteria: coordination chemistry of an unusual 3,4-catecholate/citrate siderophore. J Am Chem Soc. 2008;130:2124–2125. doi: 10.1021/ja077202g. [DOI] [PubMed] [Google Scholar]
- 17.Barbeau K. Photochemistry of organic iron(III) complexing ligands in oceanic systems. Photochem Photobiol. 2006;82:1505–1516. doi: 10.1562/2006-06-16-IR-935. [DOI] [PubMed] [Google Scholar]
- 18.Garner BL, Arceneaux JEL, Byers BR. Temperature control of a 3,4-dihydroxybenzoate (protocatechuate)-based siderophore in Bacillus anthracis. Curr Microbiol. 2004;49:89–94. doi: 10.1007/s00284-004-4286-7. [DOI] [PubMed] [Google Scholar]
- 19.Calugay RJ, Takeyama H, Mukoyama D, Fukuda Y, Suzuki T, Kanoh K, Matsunaga T. Catechol siderophore excretion by magnetotactic bacterium Magnetospirillum magneticum AMB-1. J Biosci Bioeng. 2006;101:445–447. doi: 10.1263/jbb.101.445. [DOI] [PubMed] [Google Scholar]
- 20.Brown JS, Holden DW. Iron acquisition by Gram-positive bacterial pathogens. Microbes Infect. 2002;4:1149–1156. doi: 10.1016/s1286-4579(02)01640-4. [DOI] [PubMed] [Google Scholar]
- 21.Moore CM, Helmann JD. Metal homeostasis in Bacillus subtilis. Curr Opin Microbiol. 2005;8:188–195. doi: 10.1016/j.mib.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 22.Ollinger J, Song KB, Antelmann H, Hecker M, Helmann JD. Role of the Fur regulon in iron transport in Bacillus subtilis. J Bacteriol. 2006;188:3664–3673. doi: 10.1128/JB.188.10.3664-3673.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miethke M, Klotz O, Linne U, May JJ, Beckering CL, Marahiel MA. Ferri-bacillibactin uptake and hydrolysis in Bacillus subtilis. Mol Microbiol. 2006;61:1413–1427. doi: 10.1111/j.1365-2958.2006.05321.x. [DOI] [PubMed] [Google Scholar]
- 24.Sebulsky MT, Heinrichs DE. Identification and characterization of fhuD1 and fhuD2, two genes involved in iron-hydroxamate uptake in Staphylococcus aureus. J Bacteriol. 2001;183:4994–5000. doi: 10.1128/JB.183.17.4994-5000.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harvie DR, Ellar DJ. A ferric dicitrate uptake system is required for the full virulence of Bacillus cereus. Curr Microbiol. 2005;50:246–250. doi: 10.1007/s00284-004-4442-0. [DOI] [PubMed] [Google Scholar]
- 26.Helgason E, Økstad OA, Caugant DA, Johansen HA, Fouet A, Mock M, Hegna I, Kolstø AB. Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis - One species on the basis of genetic evidence. Appl Environ Microbiol. 2000;66:2627–2630. doi: 10.1128/aem.66.6.2627-2630.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dertz EA, Stintzi A, Raymond KN. Siderophore-mediated iron transport in Bacillus subtilis and Corynebacterium glutamicum. J Biol Inorg Chem. 2006;11:1087–1097. doi: 10.1007/s00775-006-0151-4. [DOI] [PubMed] [Google Scholar]
- 28.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 29.Kuzmic P. Program DYNAFIT for the analysis of enzyme kinetic data: application to HIV proteinase. Anal Biochem. 1996;237:260–273. doi: 10.1006/abio.1996.0238. [DOI] [PubMed] [Google Scholar]
- 30.Dertz EA, Xu J, Stintzi A, Raymond KN. Bacillibactin-mediated iron transport in Bacillus subtilis. J Am Chem Soc. 2005;128:22–23. doi: 10.1021/ja055898c. [DOI] [PubMed] [Google Scholar]
- 31.Meyer M, Telford JR, Cohen SM, White DJ, Xu J, Raymond KN. High-yield synthesis of the enterobactin trilactone and evaluation of derivative siderophore analogs. J Am Chem Soc. 1997;119:10093–10103. [Google Scholar]
- 32.Butler A. Marine siderophores and microbial iron mobilization. Biometals. 2005;18:369–374. doi: 10.1007/s10534-005-3711-0. [DOI] [PubMed] [Google Scholar]
- 33.Kupper FC, Carrano CJ, Kuhn JU, Butler A. Photoreactivity of iron(III)-aerobactin: photoproduct structure and iron(III) coordination. Inorg Chem. 2006;45:6028–6033. doi: 10.1021/ic0604967. [DOI] [PubMed] [Google Scholar]
- 34.Byers BR, Powell MV, Lankford CE. Iron-chelating hydroxamic acid (schizokinen) active in initiation of cell division in Bacillus megaterium. J Bacteriol. 1967;93:286–294. doi: 10.1128/jb.93.1.286-294.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matzanke BF. Structures, coordination chemistry and functions of microbial iron chelates. In: Winkelmann G, editor. CRC Handbook of microbial iron chelates. CRC Press; Boca Raton, FL: 1991. pp. 15–64. [Google Scholar]
- 36.Bluhm ME, Kim SS, Dertz EA, Raymond KN. Corynebactin and enterobactin: related siderophores of opposite chirality. J Am Chem Soc. 2002;124:2436–2437. doi: 10.1021/ja016651s. [DOI] [PubMed] [Google Scholar]
- 37.Krewulak KD, Shepherd CM, Vogel HJ. Molecular dynamics simulations of the periplasmic ferric-hydroxamate binding protein FhuD. Biometals. 2005;18:375–386. doi: 10.1007/s10534-005-3712-z. [DOI] [PubMed] [Google Scholar]
- 38.Stintzi A, Barnes C, Xu J, Raymond KN. Microbial iron transport via a siderophore shuttle: a membrane ion transport paradigm. PNAS. 2000;97:10691–10696. doi: 10.1073/pnas.200318797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gaballa A, Helmann JD. Substrate induction of siderophore transport in Bacillus subtilis mediated by a novel one-component regulator. Mol Microbiol. 2007;66:164–173. doi: 10.1111/j.1365-2958.2007.05905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loo JA. Electrospray ionization mass spectrometry: a technology for studying noncovalent macromolecular complexes. Int J Mass Spec. 2000;200:175–186. [Google Scholar]
- 41.Sun J, Kitova EN, Wang W, Klassen JS. Method for distinguishing specific from nonspecific protein-ligand complexes in nanoelectrospray ionization mass spectrometry. Anal Chem. 2006;78:3010–3018. doi: 10.1021/ac0522005. [DOI] [PubMed] [Google Scholar]
- 42.Jecklin MC, Touboul D, Bovet C, Wortmann A, Zenobi R. Which electrospray-based ionization method best reflects protein-ligand interactions found in solution? A comparison of ESI, nanoESI, and ESSI for the determination ofdissociation constants with mass spectrometry. J Am Soc Mass Spectrom. 2007;19:332–343. doi: 10.1016/j.jasms.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 43.Daniel JM, Friess SD, Rajagopalan S, Wendt S, Zenobi R. Quantitative determination of noncovalent binding interactions using soft ionization mass spectrometry. Int J Mass Spec. 2002;216:1–27. [Google Scholar]
- 44.Rockwood AL, Busman M, Smith RD. Coulombic effects in the dissociation of large highly charged ions. Int J Mass Spectrom Ion Processes. 1991;111:103–129. [Google Scholar]
- 45.Müller A, Wilkinson AJ, Wilson KS, Duhme-Klair AK. An [{Fe(mecam)}2]6-bridge in the crystal structure of a ferric enterobactin binding protein. Angew Chem Int. 2006;45:5132–5136. doi: 10.1002/anie.200601198. [DOI] [PubMed] [Google Scholar]
- 46.Doneanu CE, Strong RK, Howald WN. Characterization of a noncovalent lipocalin complex by liquid chromatography/electrospray ionization mass spectrometry. J Biomol Tech. 2004;15:208–212. [PMC free article] [PubMed] [Google Scholar]
- 47.Clarke TE, Braun V, Winkelmann G, Tari LW, Vogel HJ. X-ray crystallographic structures of the Escherichia coli periplasmic protein FhuD bound to hydroxamate-type siderophores and the antibiotic albomycin. J Biol Chem. 2002;277:13966–13972. doi: 10.1074/jbc.M109385200. [DOI] [PubMed] [Google Scholar]
- 48.Zhang R, Zhang CT. Identification of genomic islands in the genome of Bacillus cecreus by comparative analysis with Bacillus anthracis. Physiol Genomics. 2003;16:19–23. doi: 10.1152/physiolgenomics.00170.2003. [DOI] [PubMed] [Google Scholar]
- 49.Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- 50.Bergman NH, Anderson EC, Swenson EE, Janes BK, Fisher N, Niemeyer MM, Miyoshi AD, Hanna PC. Transcriptional Profiling of Bacillus anthracis during infection of host macrophages. Infect Immun. 2007;75:3434–3444. doi: 10.1128/IAI.01345-06. [DOI] [PMC free article] [PubMed] [Google Scholar]