Abstract
PELP1 (proline-rich, glutamic acid–rich, and leucine-rich protein-1) is a potential proto-oncogene that functions as a coregulator of estrogen receptor (ER), and its expression is deregulated during breast cancer progression. Emerging evidence suggests growth factor signaling crosstalk with ER as one possible mechanism by which breast tumors acquire resistance to therapy. In this study, we examined mechanisms by which growth factors modulate PELP1 functions, leading to activation of ER. Using in vivo labeling assays, we have found that growth factors promote phosphorylation of PELP1. Utilizing a panel of substrate-specific phosphorylated antibodies, we discovered that growth factor stimulation promotes phosphorylation of PELP1 that is recognized by a protein kinase A (PKA) substrate–specific antibody. Accordingly, growth factor–mediated PELP1 phosphorylation was effectively blocked by PKA-specific inhibitor H89. Utilizing purified PKA enzyme and in vitro kinase assays, we obtained evidence of direct PELP1 phosphorylation by PKA. Using deletion and mutational analysis, we identified PELP1 domains that are phosphorylated by PKA. Interestingly, site-directed mutagenesis of the putative PKA site in PELP1 compromised growth factor–induced activation and subnuclear localization of PELP1 and also affected PELP1-mediated transactivation function. Utilizing MCF-7 cells expressing a PELP1 mutant that cannot be phosphorylated by PKA, we provide mechanistic insights by which growth factor signaling regulates ER transactivation in a PELP1-dependent manner. Collectively, these findings suggest that growth factor signals promote phosphorylation of ER coactivator PELP1 via PKA pathway, and such modification may have functional implications in breast tumors with deregulated growth factor signaling.
Introduction
The steroid hormone 17β-estradiol plays an important role in controlling the expression of genes involved in a wide variety of biological processes, including development, differentiation, and homeostasis in a wide variety of tissues, including bone, brain, breast, and uterus (1, 2). The biological functions of estrogen are mediated by the estrogen receptor (ER), a ligand-dependent transcription factor that modulates gene transcription via direct recruitment to the target gene chromatin (3, 4). In addition, the ER also participates in cytoplasmic and membrane-mediated signaling events (nongenomic signaling) and generally involves the stimulation of Src kinase, mitogen-activated protein kinase (MAPK), and phosphatidylinositol-3-kinase, and protein kinase A (PKA; refs. 5, 6).
In the past decade, it has become increasingly clear that the recruitment of coregulatory proteins to ERs is required for ER-mediated optimal transcriptional and biological activities and that coregulators provide an additional level of complexity in ER action (7, 8). Coregulators seem to function as multitasking molecules, and their actions include chromatin modifications, remodeling, RNA splicing, and protein degradation (8, 9). It is suspected that deregulation of ER coregulators could influence target gene expression and thus participate in the development of hormone-responsive cancers (7, 10). However, the molecular mechanisms that modulate coregulator functions remain elusive.
ER signaling has also been shown to play a role in the progression of breast cancer with ~ 70% of ER-positive breast tumors (11, 12). Women with ER-positive tumors are commonly treated using ER-targeted therapy with selective estrogen blockers, such as tamoxifen, that target ER interactions with coregulators and/or with aromatase inhibitors that inhibit peripheral estrogen synthesis (13, 14). Although these treatments are effective in the initial period, many patients eventually acquire resistance to these endocrine therapies (10). The causes of resistance to estrogen-targeted therapy remain elusive. Deregulated epidermal growth factor (EGF) factor receptor signaling (15) and constitutive activation of cytosolic pathways are suggested as one possible mechanism by which tumors acquire resistance to therapy (16, 17). The mechanism, by which growth factor signaling cross-talks with ER, is not completely understood and is an active area of investigation.
PELP1 [proline-rich, glutamic acid–rich, and leucine-rich protein-1; also termed modulator of nongenomic actions of ER (MNAR)] is a recently identified ER coregulator (18, 19). PELP1 is a unique coactivator that plays an important role in both the genomic and nongenomic actions of the ER (20). PELP1 recruits to the ER target gene promoter, interacts with histones and histone-modifying enzymes, and is suggested to play a role in chromatin remodeling activity of the ligand-bound ER (21). The ability of PELP1 to interact and couple cytosolic kinases c-Src and phosphatidylinositol-3-kinase to the ER highlights a novel role for PELP1 in nongenomic ER signaling (19, 22, 23). Recent evidence suggests that PELP1 is a potential proto-oncogene; its expression is deregulated during cancer progression (24). Although substantial information is available on the potential role of PELP1 as a ER coregulator, no studies have examined the role of PELP1 as a regulatory target of growth factor signaling.
In this study, we show that growth factor signaling promoted phosphorylation of PELP1 under physiologic conditions. We used kinase substrate–specific antibodies to show that growth factor signaling promoted phosphorylation of PELP1 via PKA. Furthermore, we identified PELP1 amino acids S350, S415, and S613 as the substrate sites of PKA and show that PKA-mediated PELP1 phosphorylation had functional consequences. Our results provide evidence that growth factors modulate PELP1 nuclear functions via phosphorylation, and such modification enhances the hormone-independent activation of the ER.
Results
PELP1 Is a Phosphorylated Protein
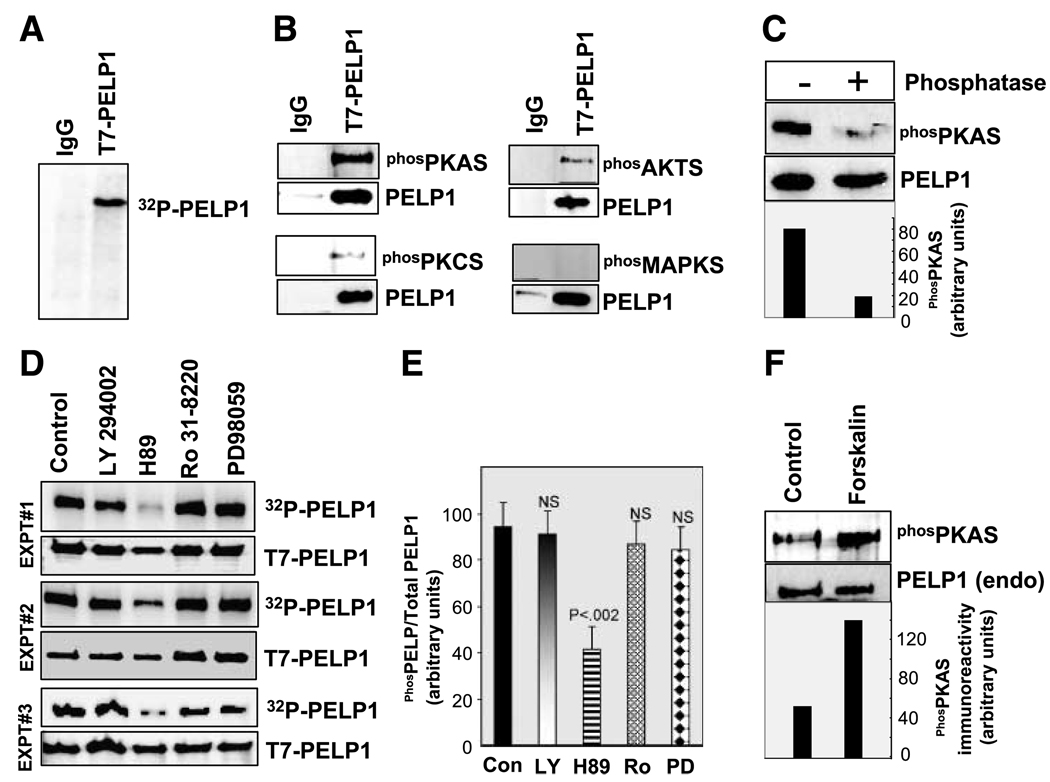
To study the possibility that PELP1 is phosphorylated in vivo, we immunoprecipitated the T7-epitope–tagged PELP1 from MCF-7 cell lysates that were metabolically labeled with [32P]orthophosphoric acid. Autoradiography of the precipitated PELP1 showed that it was phosphorylated in vivo (Fig. 1A). Earlier studies showed that PELP1 is phosphorylated on tyrosine (22, 23); however, whether PELP1 is phosphorylated on serines/threonines is not known. Mutation of the previously identified Y920 site in PELP1 did not completely abolish the in vivo phosphorylation of PELP1, raising the possibility that PELP1 may be phosphorylated on additional sites and by other kinases (data not shown). To identify other putative kinases that may be phosphorylating PELP1 in vivo, we did Western blotting of immunoprecipitated PELP1 with a panel of substrate-specific phosphorylated antibodies that uniquely recognize PKA, PKC, MAPK, or AKT phosphorylated proteins. Interestingly, Western analysis of PELP1 immunoprecipitates indicated a strong intensity band with PKA substrate–specific phosphorylated antibody and weak intensity band with AKT and PKC substrate antibodies (Fig. 1B). No detectable staining was observed with MAPK substrate–specific antibody. To confirm the phosphorylation specificity of the substrate-specific phosphorylated antibodies, we did phosphatase treatment after immunoprecipitation. Western blot analysis revealed that treatment of the immunoprecipitated PELP1 with alkaline phosphatase abolished the PKA substrate antibody recognition, suggesting that PELP1 phosphorylation is needed for the recognition of the PKA substrate–specific antibody (Fig. 1C). Treatment of cells with PKA, phosphatidylinositol-3-kinase, PKC, or MAPK pathway specific inhibitors and subsequent immunoprecipitation of the in vivo [32P]-labeled PELP1 showed that blockage of PKA signaling substantially reduced in vivo phosphorylation of PELP1 (Fig. 1D and E). To further confirm the specificity of the PKA-mediated phosphorylation of PELP1, MCF-7 cells were treated with forskolin, a specific activator of PKA pathway. Immunoprecipitation of endogenous PELP1 indeed confirmed that forskolin treatment enhanced PELP1 phosphorylation that can be recognized by the PKA substrate–specific antibody (Fig. 1F). Collectively these results suggest that PELP1 is phosphorylated in vivo on serine/threonine residues and the PKA pathway constitutes a major signaling pathway that modulates PELP1 serine/threonine phosphorylation. We have therefore limited our focus in this study to characterize the PKA regulation of PELP1.
Figure 1.
PELP1 is phosphorylated by serine/threonine kinases in vivo. A. MCF-7-PELP1 cells were labeled with [32P]PI in vivo, and PELP1 phosphorylation status was analyzed by immunoprecipitation and autoradiography. B. PELP1 was immunoprecipitated using a control or the T7-epitope – tagged antibody from total lysates prepared from MCF-7-PELP1 clones and Western blotted with phosphorylated (serine/threonine) substrate – specific antibodies that uniquely recognize PKA, PKC, AKT, or MAPK. C. T7-PELP1 was immunoprecipitated from MCF-7-PELP1 cells and treated with phosphatase, and phosphorylation status was analyzed by using the phosphorylated PKA substrate antibody. D. MCF-7-PELP1 cells were cultured in 10% serum, in vivo labeled with [32P]PI, and treated for 1 h with various inhibitors: LY294002 (phosphatidylinositol-3-kinase inhibitor), H89 (PKA inhibitor), Ro 31-8220 (PKC inhibitor), or PD98059 (MAPK pathway inhibitor). T7-PELP1 was immunoprecipitated, and phosphorylation status was measured by autoradiography. These in vivo experiments are repeated thrice and are shown as experiments 1, 2, and 3. E. Intensity of the bands in D were quantified by densitometry and are shown as bar graph. Columns, average of three experiments and the differences in the intensity were analyzed by t test. P, significance level; NS, not significant. F. MCF-7 cells were cultured in serum-free medium and stimulated with PKA activator forskolin, and the phosphorylation status of endogenous PELP1 was analyzed by immunoprecipitation and autoradiography.
Growth Factor Signaling Modulates PELP1 Phosphorylation
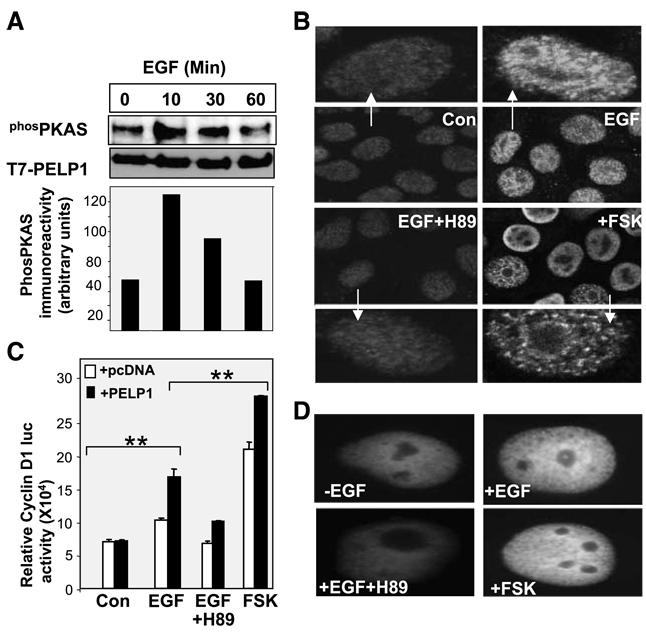
Earlier studies indicated that PELP1 participates in growth factor signaling cross-talk with the ER (20). We examined whether growth factor signaling promoted phosphorylation of PELP1 via the PKA pathway. MCF-7 cells were treated with EGF, and PELP1 phosphorylation was measured using the PKA substrate–specific antibody. The results showed that an increase in the phosphorylation of PELP1 occurred upon EGF stimulation, with maximal phosphorylation occurring at 10 min after stimulation (Fig. 2A). Because substantial amount of PELP1 resides in the nuclear compartment, we examined whether PKA-mediated phosphorylation modulates its localization. Confocal microscopy revealed that EGF stimulation did not significantly alter the nuclear localization of PELP1; however, EGF stimulated the reorganization of the subnuclear localization of PELP1. In the absence of EGF stimulation, PELP1 localization seemed diffuse whereas growth factor stimulation induced PELP1 localization into prominent foci/speckle-like structures. Interestingly, pretreatment of cells with the PKA inhibitor H89 substantially reduced the growth factor–induced foci/speckle-like distribution of PELP1 in the nuclear compartment (Fig. 2B). To examine the biological significance of EGF phosphorylation of PELP1, we used Cos1 cells to examine whether PKA phosphorylation of PELP1 is required for PELP1-mediated up-regulation of cyclin D1 expression. For these assays, we have used Cos1 cells because these cells express very low levels of endogenous PELP1 and thus represent a good model cells to study the effect of PKA inhibitor PELP1 transactivation functions in transient Growth Factor Modulation of PELP1 Functions transfection assays. In reporter gene assays using the cyclin D1 promoter luciferase, cotransfection of PELP1 substantially enhanced the growth factor–mediated activation of cyclin D1 promoter. PELP1-mediated enhancement was abolished when the cells were pretreated with H89 (Fig. 2C). Pretreatment of the cells with forskolin, a PKA-specific activator, substantially enhanced PELP1-mediated activation of cyclin D1. Similar to MCF-7 cells, blockage of PKA signaling in Cos1 cells also affected the subnuclear redistribution of PELP1 upon growth factor stimulation (Fig. 2D). Collectively, these results suggest that growth factors use PKA to phosphorylate PELP1 and that such phosphorylation may play a role in the subnuclear localization and biological functions of PELP1.
Figure 2.
Growth factors promote phosphorylation of PELP1 via PKA. A. MCF-7-PELP1 cells were serum starved for 24 h and stimulated with EGF (100 ng/mL) for the indicated periods of time, and T7-PELP1 was immunoprecipitated. The status of PKA-mediated phosphorylation on PELP1 was analyzed by using the PKA antibody. B. MCF-7 cells were treated with EGF in the presence or absence of the PKA inhibitor H89, and the cellular localization of PELP1 was determined by using confocal microscopy. C. Cos1 cells were transfected with the cyclin D1 luciferase reporter, along with pcDNA vector or pcDNA-PELP1. Cells were serum starved for 24 h, treated with EGF, EGF + H89, or forskolin for 12 h. The luciferase activity was then measured. *, P < 0.05; **, P < 0.001. Columns, mean from three independent experiments done in triplicate wells; bars, SE. D. Cos1 cells were transfected with GFP-PELP1-WT vector and treated with EGF (100 ng/mL) in the presence or absence of the PKA inhibitor H89, and the cellular localization of GFP-PELP1 was determined by using confocal microscopy.
PELP1 Is a Substrate of PKA
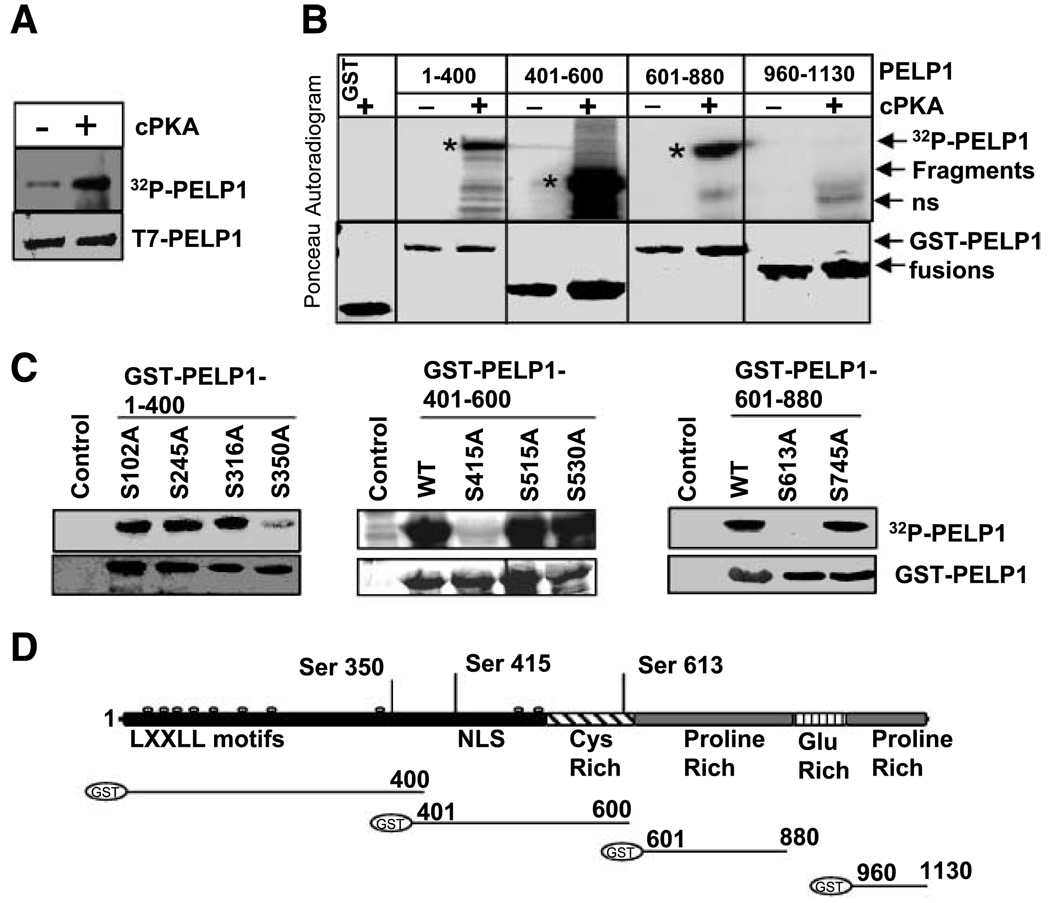
To determine whether PELP1 is a direct substrate of PKA, we did in vitro kinase assays using the purified PKA enzyme and immunoprecipitated T7-epitope–tagged PELP1 as a substrate using a published protocol (25). The results showed that PKA phosphorylates full-length T7-PELP1 in vitro (Fig. 3A). To identify the domains of PELP1 that are phosphorylated by PKA, we examined the ability of the purified PKA enzyme to phosphorylate glutathione S-transferase (GST) fusion proteins made with different domains of PELP1. PKA specifically phosphorylated the PELP1-GST fusions containing amino acids 1–400, 401–600, and 601–880; however, purified PKA enzyme failed to phosphorylate GST-PELP1 fusion containing amino acids 960–1130 (Fig. 3B). Analysis of the PELP1 1–880 amino acid sequence indicated that there are nine consensus PKA serine phosphorylation sites. To map the PKA phosphorylation sites in PELP1, we mutated serines to alanine in these nine potential PKA phosphorylation sites. Mutation of PELP1 S350, S415, and S613 to alanine completely abolished the ability of PKA to phosphorylate PELP1-1-400, PELP1-400-600, and PELP1-601-866 domains, respectively (Fig. 3C). Collectively, these deletion and mutation studies suggest that PKA phosphorylates PELP1 at three distant sites, including S350, S415, and S613.
Figure 3.
PELP1 is a substrate of PKA. A. T7-PELP1 was immunoprecipitated from serum-starved MCF-7-PELP1 cells and subjected to in vitro kinase assays using the purified catalytic subunit of PKA (cPKA). B. PELP1-GST fusions of indicated length were subjected to in vitro kinase reaction with [32P]γ-ATP in the presence or absence of catalytic subunit of PKA. *, specific PELP1 GST proteins that are phosphorylated by PKA; ns, nonspecific band. C. In vitro kinase assay of GST-PELP1 mutants that lack specific PKA phosphorylation sites. Mutations were created by using site-directed mutagenesis with GST-PELP1 1–400, 401–600, 601–880 and 960–1130 plasmids as backbones. D. Schematic representation of GST-PELP1 fusions of various PELP1 domains used in this study. Putative PKA phosphorylation sites identified in PELP1 (top).
PKA Phosphorylation Is Required for Optimal Coactivation Functions of PELP1
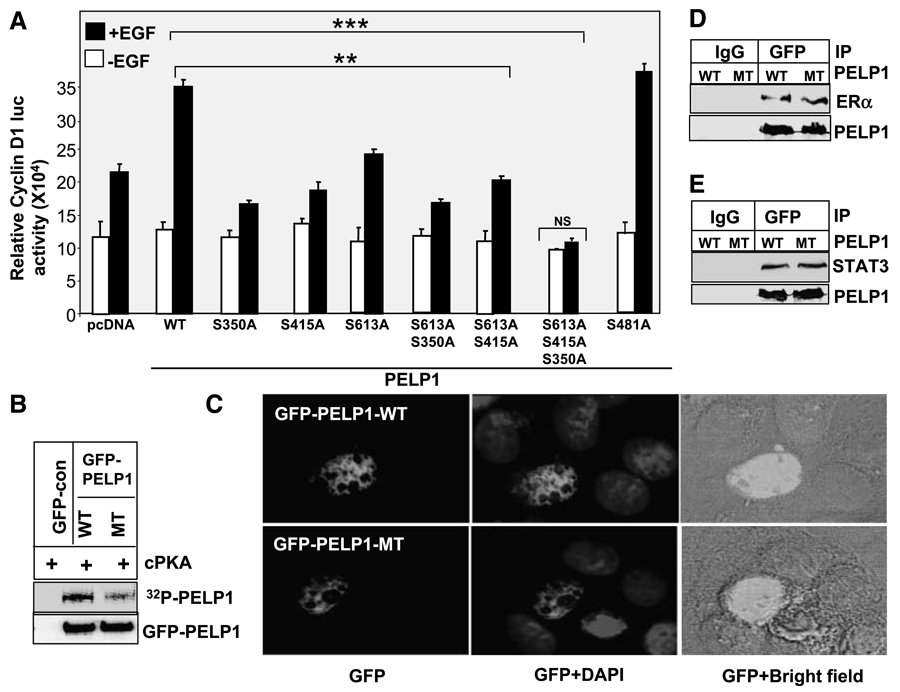
To determine the significance of PKA phosphorylation in the mechanism of PELP1 action, we examined the transcriptional activity of PELP1 mutants that lack PKA phosphorylation sites. We previously showed that PELP1 selectively activates the cyclin D1 promoter by functioning as a coactivator of transcription factors, such as the ER and signal transducers and activators of transcription 3 (STAT3; refs. 18, 26). We used the cyclin D1 reporter gene assay and growth factor stimulation to establish the significance of PKA phosphorylation sites in PELP1. Compared with PELP1-WT, individual mutation of S350, S450, or S613 to alanine reduced PELP1-mediated induction of the cyclin D1 reporter gene upon growth factor stimulation (Fig. 4A). However, these effects of individual site mutation are not identical. Mutation of S351 and S415 sites have more effect on decreasing PELP1 coactivation function than that of S613 site. Of all the three sites, S613 seems to be weak. We have also tested a couple of double mutations; however, their effect is very similar to single mutations, suggesting that the remaining third site can rescue some of the mutation effects by compensation. On the contrary, mutation of all three sites completely abolished in the growth factor induced PELP1 coactivation function. Mutation of irrelevant serine site (S481) has no effect on growth factor–mediated PELP1 coactivation functions (Fig. 4A). To confirm that the triple mutant is indeed not phosphorylated by PKA, we did in vitro kinase assay using immunoprecipitated GFP-PELP-WT or GFP-PELP1-MT. In these assays, PKA efficiently phosphorylated GFP-PELP1-WT; however, PKA failed to phosphorylate GFP-PELP1-MT (Fig. 1B). To confirm that the GFP-PELP1 triple mutant is expressed and localized similarly as GFP-PELP1-WT, we transiently transfected GFP-PELP1 and GFP-PELP1-MT and determined localization by using confocal microscopy. The results show that both PELP1-WT and PELP1-MT proteins were localized predominantly in the nuclear compartment (Fig. 4C). To evaluate whether mutation of PKA sites affected PELP1 interaction with either the ER or STAT3, we did immunoprecipitation of PELP1 after treating MCF-7 cells with EGF. Western analysis of the immunoprecipitates showed that mutation of the PKA sites in PELP1 did not significantly affect PELP1 binding to either the ER or STAT3 (Fig. 4D and E). However, PKA phosphorylation is required for optimal PELP1 activation of cyclin D1 under growth factor stimulation (Fig. 4A), suggesting that PKA phosphorylation may play a role in other nuclear functions of PELP1.
Figure 4.
Growth factor – mediated PELP1 coactivation functions require functional PKA phosphorylation sites. A. Cos1 cells were cotransfected with cyclin D1 – luciferase reporter along with the cDNAs for the pcDNA vector, pcDNA-PELP1-WT, or the PELP1 mutants that lack specific PKA phosphorylation sites. Cells were serum starved for 24 h and stimulated with 100 ng/mL of EGF for 12 h, and the luciferase activity was measured. *, P < 0.05; **, P < 0.001. Columns, mean from three independent experiments done in triplicate wells; bars, SE. B. GFP-PELP1 or GFP-PELP1-MT was transfected into Cos1 cells, and 72 h later, GFP-PELP1-WT and GFP-PELP1-MT were immunoprecipitated from the serum-starved Cos1 cells and subjected to in vitro kinase assays using catalytic subunit of PKA. C. MCF-7 cells were transiently transfected with GFP-tagged PELP1-WT or GFP-tagged PELP1-MT (S350, S415, S613A), and the cellular localization of GFP-PELP1 was determined by using confocal microscopy. D and E. Cos1 were cotransfected with PELP1-WT or PELP1-MT (S350, S415, S613A) plasmids along with ER or STAT3 expression plasmids. After 48 h, cells were stimulated with EGF (100 ng/mL) and PELP1 was immunoprecipitated. The association of the ER and STAT3 with PELP1 was analyzed by Western blot analysis.
PKA Phosphorylation of PELP1 Is Required for Nuclear Distribution
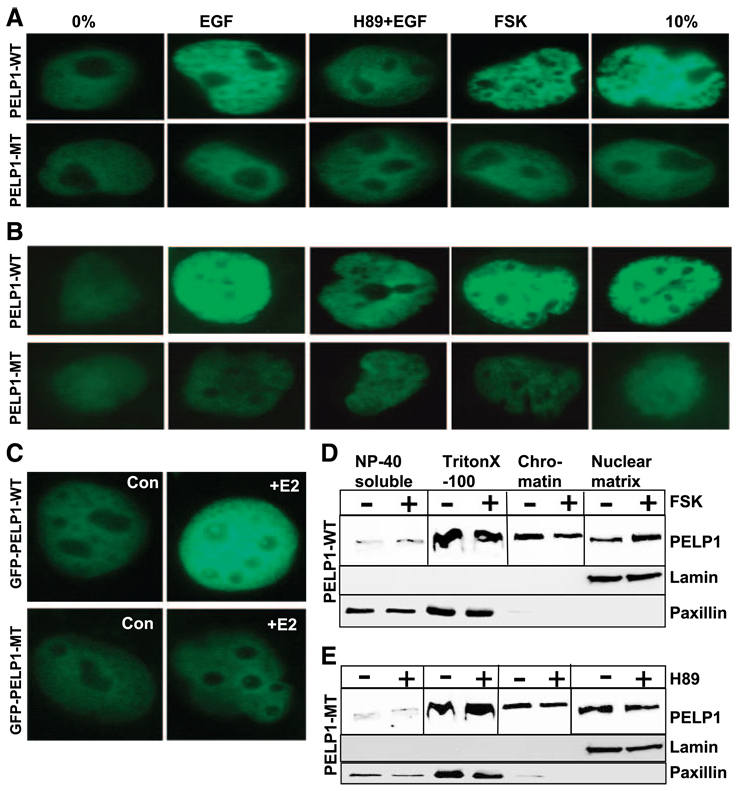
Because PELP1 exhibited punctuate distribution in growth factor–stimulated cells (Fig. 2B), we examined whether PKA phosphorylation modulates intranuclear distribution of PELP1 upon growth factor stimulation. MCF-7 cells were transfected with GFP-tagged PELP1-WT or PELP-MT (that lacks all three PKA phosphorylation sites) and stimulated with serum, EGF, or forskolin. The nuclear distribution of PELP1 was determined by using fluorescence imaging. When not stimulated, PELP1 exhibited diffuse staining throughout the nucleus. Stimulation with serum or EGF increased PELP1 localization into discrete foci/speckle-like structures in the nucleus. Interestingly, pretreatment of cells with the PKA inhibitor H89 diminished EGF-induced PELP1-related foci/speckles. Activation of the PKA pathway by addition of forskolin promoted increased formation of PELP1-related foci/speckles (Fig. 5A, top). PELP1-MT that lacks all three PKA phosphorylation sites failed to form PELP1-related foci/speckles upon growth factor, serum, or forskolin stimulation and exhibited very diffuse staining that was similar to the pattern seen in serum-starved cells (Fig. 5A, bottom). To confirm the changes in subnuclear localization of PELP1, we repeated these experiments in another commonly used breast cancer model cell, T47D. Confocal analysis in T47D cells also revealed that growth factors or serum promoted PELP1-WT localization into foci/speckle-like structures (Fig. 5B, top), whereas these stimuli have no significant effect on the nuclear distribution of PELP1-MT (Fig. 5B, bottom). Similarly, E2 treatment of MCF-7 also promoted accumulation of PELP1-WT into a foci-like structure, whereas E2 had no significant effect on PELP1-MT localization (Fig. 5C). We also tested the effects of selective activation or inhibition of PKA pathway using forskolin and H89, respectively, on the subcellular localization of endogenous PELP1 using sequential fractionation of MCF-7 cells. Forskolin or H89 treatment did not substantially affect the amount of PELP1 localized in the cytoplasmic or cytoskeleton fractions. However, forskolin treatment enhanced the PELP1 fractionation within the nuclear matrix (Fig. 5D), whereas H89 reduced amount of PELP1 in the nuclear matrix (Fig. 5E). These results suggest that PKA-mediated phosphorylation of PELP1 may play a role in the formation of the PELP1-related foci/speckles in the nuclear compartment and may also influence PELP1 association with the nuclear matrix.
Figure 5.
PKA promotes nuclear redistribution of PELP1. MCF-7 cells (A) or T47D cells (B) were transfected with GFP-PELP1-WT or GFP-PEFP1-MT (S350, S415, S613A). After 48 h, cells were serum starved for a further 24 h, stimulated with serum, EGF, EGF + H89, or forskolin. Localization of GFPPELP1-WT and GFP-PELP1-MT (S350, S415, S613A) was determined by using confocal microscopy. C. MCF-7 cells were transfected with GFP-PELP1-WT and GFP-PELP1-MT (S350, 415, 613A) and treated with or without E2. The cellular localization of GFP-PELP1 was determined by using confocal microscopy. D and E. MCF-7cells were serum starved, treated with either forskolin (D) or H89 (E), and serially fractionated into NP40-soluble, cyto/ nucleoplasmic, chromatin, and nuclear matrix fraction. Equal amounts of protein were analyzed for localization of PELP1 by using Western blot analysis.
PKA Signaling Enhances PELP1-Mediated Tamoxifen Agonist Actions in Breast Cancer Cells
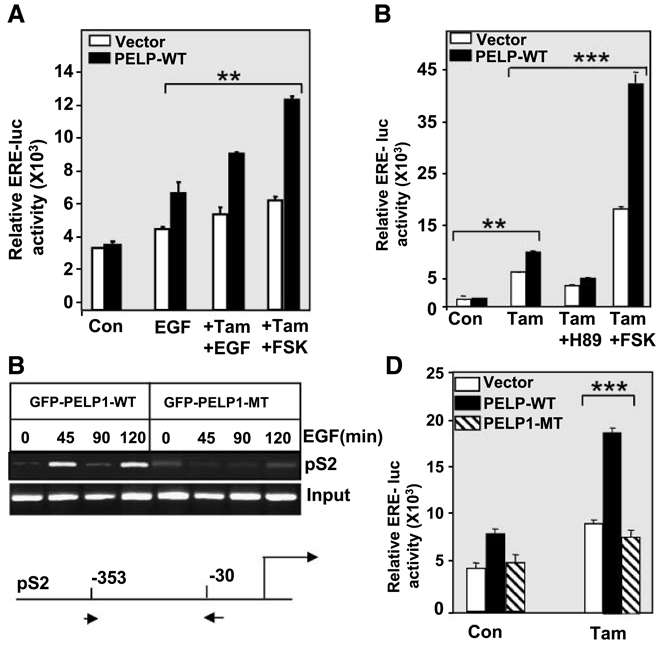
Growth factor pathways were shown to modify the ER and its coactivators by phosphorylation, and such modifications are shown to promote ligand-independent activation of ER target genes and/or differential responses to selective ER modulators. Similarly PELP1 deregulation was also shown to promote the agonistic action of tamoxifen. To determine whether PKA signaling promotes PELP1-mediated ligand-independent activation of ER target genes or PELP1 recruitment to ER target chromatin, we did estrogen-responsive element (ERE) reporter gene and chromatin immunoprecipitation assays. Growth factor stimulation promoted the PELP1-mediated activation of ERE reporter gene in the absence of estrogen (Fig. 6A). Transfection of PELP1 alone in MCF-7 cells did not significantly affected the tamoxifen-mediated activation of ERE reporter. Curiously, forskolin or EGF treatment substantially enhanced PELP1-mediated partial agonistic action of tamoxifen.
Figure 6.
PELP1-mediated growth factor signaling cross-talk with the ER requires a functional PKA pathway. A. MCF-7 cells were transfected with ERE-luciferase reporter along with pcDNA vector or pcDNA-PELP1 expression vector. After 48 h, cells were stimulated with EGF (100 ng/mL), tamoxifen (10−8 mol/L) in the presence or absence of forskolin for 12 h, and then luciferase activity was measured. B. MCF-7 cells expressing GFP-PELP1-WT or GFP-PELP1-MT (S350, S415, S613A) were treated with EGF (100 ng/mL) for indicated periods of time, chromatin was prepared and immunoprecipitated with anti-GFP monoclonal antibody to precipitate GFP-PELP1, and chromatin immunoprecipitation analysis was done using primers specific for pS2. C. Ishikawa cells were transfected with ERE-luciferase reporter with the pcDNA vector or the pcDNA-PELP1 expression vector. After 48 h, cells were stimulated with tamoxifen (10−8 mol/L) in the presence or absence of H89 or forskolin for 12 h, and then luciferase activity was measured. D. Ishikawa cells were transfected with ERE-luciferase reporter with or without the GFP vector, GFP-PELP1-WT, or GFP-PELP1-MT (S350, S415, S613A). After 48 h, cells were stimulated with tamoxifen (10−8 mol/L) for 12 h, and then luciferase activity was measured. *, P < 0.05; **, P < 0.001. Columns, means from three independent experiments done in triplicate wells; bars, SE.
We examined whether PKA phosphorylation of PELP1 played a role in PELP1 recruitment to the ER target promoter upon growth factor stimulation. MCF-7 cells were transfected with PELP1-WT or PELP1-MT expression plasmids. Transfected cells exhibited similar transfection efficiency as seen when using GFP florescence. After 48 hours, the cells were serum starved and treated with EGF. The chromatin was subjected to immunoprecipitation using the GFP antibody. Chromatin immunoprecipitation results show PELP1-WT recruitment to the promoter of pS2 upon growth factor stimulation in a cyclical manner (Fig. 6B). However, PELP1-MT that cannot be phosphorylated by PKA showed decreased ability to recruit to the pS2 promoter up on growth factor stimulation. These results suggested that PKA phosphorylation of PELP1 may have a role in growth factor–mediated activation of the ER target genes. To further confirm these findings, we have used Ishikawa cells, which are commonly used to document tamoxifen-mediated partial agonist actions that occur in pathologic conditions. As reported earlier, PELP1 induced activation of the ERE reporter in these cells and tamoxifen treatment further enhanced PELP1 actions. However, H89 pretreatment blocked PELP1-mediated activation of ERE reporter whereas forskolin addition significantly enhanced PELP1 coactivation functions in the presence of tamoxifen (Fig. 6C). Mutation of PKA phosphorylation sites in PELP1 abolished its ability to activate the ERE reporter in the presence of tamoxifen (Fig. 6D). Collectively, these results suggest that growth factor signaling has the potential to modulate ER transactivation functions via phosphorylation of PELP1, and this pathway has the potential to contribute to some extent to the partial agonistic actions of tamoxifen.
Discussion
Nuclear receptor (NR) coregulators are proteins that interact with an NR to modulate their transactivation functions (9, 27). Recent evidence suggests that NR coregulators function as major regulators and coordinators of hormone receptor physiology, which includes the determination of tissue specificity of hormone action, the integration of membrane and nuclear signaling, and the integration of growth factor and physiologic signaling to NRs (9). Within the past decade, it has become clear that recruitment of coregulatory proteins to NRs is required for NR-mediated optimal transcriptional and biological activities. PELP1, a novel NR coactivator, interacts with several NRs, including the ER. However, little is known about the molecular mechanisms that regulate the functions of PELP1. The results from this study reveal that growth factor signaling promotes phosphorylation of PELP1 via PKA. A reduction of PELP1 phosphorylation in vivo by H89 (PKA inhibitor), an increase in PELP1 phosphorylation in vivo by forskolin (PKA activator), and the ability of the purified PKA enzyme to phosphorylate PELP1 in vitro clearly show that PELP1 is a bonafide substrate of PKA. Furthermore, we identified three novel sites in PELP1 (S350, S415, and S613) that are phosphorylated by PKA and showed that mutation of these sites severely affects PELP1 coactivation functions upon growth factor stimulation.
PKA is widely expressed in mammalian cells and is implicated in the regulation of a wide variety of processes, including growth, development, metabolism, and gene expression (28). PKA holoenzymes are composed of two regulatory and two catalytic subunits (29). A number of hormones and growth factors have been shown to stimulate PKA in target cells via production of cyclic AMP (cAMP). Upon cAMP binding, PKA catalytic subunits dissociate from cAMP-saturated regulatory subunits and promote the phosphorylation of specific nuclear factors (28). PKA phosphorylates a wide variety of substrates, and such phosphorylation is reported to elicit differential responses depending on the substrate. PKA phosphorylation of cAMP-responsive element binding protein promotes cellular gene expression via recruitment of coactivator paralogues, such as cAMP-responsive element binding protein–binding protein and p300 (30). PKA modulates β-catenin signaling through direct phosphorylation and promoting its interaction with the cAMP-responsive element binding protein–binding protein (31). Phosphorylation by PKA potentiates retinoic acid receptor α activity by means of increasing interaction with and phosphorylation by cyclin H/cyclin-dependent kinase 7 (32). PKA phosphorylation regulates degradation and subcellular localization of the NR coactivator GRIP1 (33). Our results suggest that PELP1 is another novel substrate of PKA. Growth factor stimulation promotes phosphorylation of PELP1. Inhibition of PKA activity by H89 or mutation of PKA phosphorylation sites in PELP1 substantially reduced growth factor–induced coactivation functions of PELP1. However, we failed to see an effect of PKA phosphorylation either on PELP1 interactions with the ER and STAT3 (Fig. 4D and E) or its degradation/stability (data not shown). Because PELP1 functions as a scaffolding protein and interacts with several proteins (~ 22 identified interacters; ref. 20), it is possible that the PKA phosphorylation may affect PELP1 interactions with proteins other than ER and STAT3. Additional studies are needed to determine how PKA signaling modulates PELP1 functions and interactions with other cellular proteins.
Foci formation of NRs is shown to correlate with the functionally active state of the NRs. Earlier studies show that PKA signaling influences the transcriptional activation of NRs via phosphorylation of coregulators and by altering subnuclear localization. For example, activation of PKA signaling is shown to enhance steroidogenic factor transactivation functions by enhancing nuclear foci formation (34). One of the notable findings of this study is the recruitment of PELP1 to discrete subnuclear foci after stimulation of the PKA signaling pathway. In our earlier study using subnuclear fractionation, we showed that substantial amount of PELP1 associated with the chromatin and nuclear matrix fractions (21). In this study, inhibition of PKA signaling substantially reduced the amount of PELP1 in the nuclear matrix fractions. Similarly mutation of PKA phosphorylation sites in PELP1 affected its localization in the discrete foci upon forskolin stimulation. The specificity of cellular and nuclear responses to cAMP, in part, is mediated by targeting of the regulatory subunit of PKA to discrete subcellular loci through associations with A-kinase anchoring proteins (35). A-kinase anchoring protein 95 is a zinc-finger protein, which binds and anchors PKA, and is found in the nuclear matrix of a wide variety of mammalian cells. Because inhibition of PKA signaling reduced the PELP1 attachment to the nuclear matrix and because PKA is present within the nuclear matrix via A-kinase anchoring protein, PKA-mediated phosphorylation may be involved in the stabilization of PELP1 interactions with the nuclear matrix (36). We conclude that the underlying mechanism for the PKA phosphorylation–induced increase in PELP1 coactivation functions is likely due to PELP1 nuclear redistribution upon PKA phosphorylation. The localization of PKA within the foci/nuclear matrix may play a role in optimal coactivation functions of PELP1.
PKA signaling promotes ligand-independent activation of the ER and also regulates ligand-dependent ER activation (37, 38). PKA phosphorylation at S236 in the DNA-binding domain of the ER promotes dimerization of the ER, and the PKA signaling is shown to promote alteration in the agonist/antagonist balance of antiestrogens in breast cancer cells (38). Similarly, phosphorylation of S305 in the hinge region of the ERα by PKA is shown to induce resistance to tamoxifen (39). It was also shown that down-regulation of the negative regulator of PKA, PKA-RIα, was associated with tamoxifen resistance before clinical treatment, implicating PKA deregulation in tamoxifen resistance (39) Our study extends these earlier studies that PKA signaling promotes tamoxifen agonistic actions and further suggests that mechanism of PKA action also involve ER coregulator PELP1 phosphorylation as the PELP1 mutation that prevents PKA phosphorylation substantially reduced PKA-mediated tamoxifen actions.
Like peptide growth factors, estrogens cause activation of various protein kinases, such as MAPKs, and increases in the levels of second messengers, such as cAMP within minutes (37, 38). In this study, we found that PKA phosphorylates PELP1 and such modification enhances PELP1 coactivation functions. These results also raise a possibility that cross-talk between the second messengers generated via nongenomic ER pathways could probably have important roles in estrogen genomic responses via phosphorylation of coactivators. Because PELP1 plays an important role in nongenomic ER actions, the capability of PELP1 to get phosphorylated by these pathways suggest that this could be one mechanism to amplify the estrogen signaling via an autocrine signaling loop. Such cross-talk between ER genomic and nongenomic pathways may have implications for therapy.
NR coregulators are suggested to likely function as “master genes,” sensing physiologic signals and activating the appropriate set of genes using a wide variety of NRs (40, 41). Phosphorylation of coregulators seems to be the key regulatory mechanism that controls the localization, specificity, and function of coregulators and probably allow coregulators to sense the physiologic signals (42). It seems that PELP1 has the capacity to sense a wide variety of signals, and phosphorylation may allow PELP1 to cross-talk between NRs and growth factor pathways. Earlier studies have shown that growth factor signaling induces the phosphorylation of PELP1 on tyrosine. EGF promotes PELP1/MNAR association with the EGF receptor, resulting in the tyrosine phosphorylation of PELP1/MNAR (22). PELP1 tyrosine phosphorylation is implicated in the activation of AKT pathway in a nongenomic manner by the ER (23). In this study, we have identified additional kinases (PKA, PKC, AKT) that phosphorylate PELP1 upon growth factor stimulation. Using inhibitors and phosphorylated state–specific antibodies, we found that PKA contributes to the substantial amount of PELP1 phosphorylation upon growth factor stimulation.
PELP1 seems to function as a scaffolding protein, has potential binding sites for several key molecules involved in the breast cancer progression including ER, EGF receptor, Src, phosphatidylinositol-3-kinase, and therefore is a potential candidate for the signaling cross-talk in cancer cells. Earlier studies showed that PELP1 promotes growth factor–mediated activation of ER target genes. We found that mutation of PKA phosphorylation sites in PELP1 substantially reduced the ability of PELP1 to activate ER target gene upon growth factor stimulation and reduced PELP1 recruitment to the ER target gene promoter. These findings complement the earlier observation that PELP1 participates in growth factor–ER cross-talk and further suggest that growth factor–mediated phosphorylation of PELP1 via PKA plays an important role in facilitating PELP1 cross-talk with ER signaling.
Endocrine therapy using tamoxifen, a selective ER modulator, has been shown to improve relapse-free and overall survival (43); however, initial or acquired resistance to endocrine therapies frequently occurs. Although the mechanisms for hormone therapy resistance remains elusive, recent studies suggest the presence of alternative signaling pathways that activate ER may account for hormonal therapy resistance (19, 44, 45). Growth factor–mediated phosphorylation of ER and ER coregulatory proteins have been shown to have a role in tamoxifen resistance (15). High levels of the coactivator AIB1 alone or together with high levels of HER-2/neu were associated with worse disease-free survival in tamoxifen-treated patients (46). PELP1 enhances the agonist actions of tamoxifen in endometrial cells, suggesting that PELP1 plays a role in the tamoxifen-mediated partial agonist action observed in the endometrium (47). Findings from this study suggest that PKA phosphorylation of PELP1 is one mechanism by which growth factor signaling promotes tamoxifen partial agonistic actions.
In summary, our results show that growth factors promote phosphorylation of the ER coregulator PELP1 via the PKA pathway and provide the first in vivo evidence that PKA phosphorylates PELP1 in breast cancer cells. This phosphorylation plays an important role in growth factor cross-talk with the ER pathway and identifies a novel mechanism for modulation of ER signaling through direct phosphorylation of the ER coregulator PELP1 by PKA. Furthermore, these findings suggest that deregulation of PKA and PELP1 pathways has potential to contribute to tamoxifen resistance via upregulation of its agonistic actions.
Experimental Procedures
Cell Cultures and Reagents
MCF-7 human breast cancer cells were maintained in RPMI supplemented with 10% FCS (28). Cos-1 cells were obtained from the American Type Culture Collection. The human endometrial Ishikawa cell line, a model of well-differentiated endometrial adenocarcinoma, was obtained from Dr. Bruce A. Lesley (University of North Carolina at Chapel Hill; ref. 48). MCF-7 cells overexpressing PELP1 (clone 20) were described previously (49). Phosphorylated (serine/threonine) kinase substrate antibodies were obtained from Cell Signaling Technology. Charcoal-stripped serum (DCC serum), forskolin, and PKA enzyme were purchased from Sigma. The anti–T7-epitope antibody was purchased from Novagen. Antibodies against PELP1 were purchased from Bethyl Laboratories. Paxillin antibody was obtained from Neomarkers. Lamin B was obtained from Oncogene. PKA inhibitor H89 was procured from Biomol.
Plasmids and Reporter Genes
The wild-type PELP1, GST-PELP1, and ERE reporter constructs were described previously (21). The PELP1 S102A, S245A, S316A, S350A, S415A, S515A, S530A, and S613A mutations were generated by site-directed mutagenesis (Quick Change Mutagenesis kit, Stratagene) using GST-PELP1-1-400, GST-PELP1-400-600, GST-PELP1-600-880, and pcDNA-PELP1 as backbones. GFP-PELP1-WT was generated by PCR-based cloning of the open reading frame of PELP1 into GFP vector (Clontech). GFP-PELP1-MT (S315, S415A, S613A) was generated by using site-directed mutagenesis of GFP-PELP1. Cyclin D1 – luciferase reporter (1745-CD1-Luc) used in this study was described previously (49).
Cell Extracts, Immunoblotting, and Immunoprecipitation
MCF-7 or MCF-7-PELP1 cell lines were serum starved for 48 h in phenol red–free RPMI serum containing 3% DCC serum and treated with estrogen (10−9 mol/L) for the indicated periods of time. For the EGF treatment, the cells were cultured in serum-free medium for at least 24 h before treatment with 100 ng/mL EGF for the indicated periods of time. To prepare cell extracts, the cells were washed thrice with PBS and then lysed in Triton X-100 buffer [50 mmol/L Tris-HCl (final pH 7.5), 100 mmol/L NaCl, 10% glycerol, 0.5% Triton X-100, 1 × protease inhibitor mixture (Roche), and 1 mmol/L sodium vanadate] for 15 min on ice. Cell lysates containing ~ 200 µg protein were resolved on 8% SDS-polyacrylamide gels, transferred to nitrocellulose membranes, probed with the appropriate antibodies, and developed using either the enhanced chemiluminescence method or the alkaline phosphatase–based color reaction method. For PELP1 immunoprecipitation, MCF-7 cells were lysed in high salt extraction buffer [20 mmol/L HEPES (final pH 7.9), 1.5 mmol/L MgCl2, 420 mmol/L NaCl, 0.2 mmol/L EDTA, 0.5 mmol/L DTT, 25% glycerol, 1 mmol/L NaVO4, and 1 × protease inhibitor and phosphatase inhibitor] for at least 1 h on ice. The lysate was diluted to a salt concentration of 100 mmol/L before adding PELP1 antibody–coupled protein A beads. For cell fractionation, MCF-7 cells were serum starved for at least 24 h and treated with forskolin or H89 and fractionated into cytoplasmic, nucleoplasmic, chromatin, and nuclear matrix fractions as described (21). Equal amounts of protein were analyzed for localization of PELP1 by Western blotting.
In vivo and In vitro Phosphorylation Assays
For measuring in vivo phosphorylation, MCF-7 cells stably expressing T7-PELP1 were cultured overnight in phosphate-free medium. Cells were labeled with [32P]Pi for 6 h followed by treatment with various inhibitors, including H89 (48 µmol/L), LY294002 (20 µmol/L), PD098059 (20 µmol/L), and Ro 31–8220 (10 nmol/L) for 1 h. Cells were lysed in Triton X-100 lysis buffer and immunoprecipitated with anti-T7 monoclonal antibody. The phosphorylation status was measured by autoradiography. In vitro kinase assays were done in kinase buffer (20 mmol/L HEPES, 20 mmol/L MgCl2, 1 mmol/L DTT, 0.5 mmol/L EGTA, 0.2 mmol/L NaV) containing 5 units of PKA enzyme (Sigma), 10 µCi of [γ-32P]ATP and 100 µmol/L cold ATP. T7-tagged full-length PELP1, GST-PELP-1-400, GST-PELP-400-600, GST-PELP-600-880, and GST-PELP-800-1130 were used as a substrates for the in vitro PKA kinase assays. Each reaction was carried out in a final volume of 30 µL for 30 min at 30°C and stopped by the addition of 10 µL of SDS buffer. Reaction products were analyzed by SDS-PAGE and visualized by autoradiography with a PhosphorImager.
Reporter Gene Assays and Chromatin Immunoprecipitation Assays
ERE-luciferase (200 ng) or cyclin D1 – luciferase (200 ng) reporter constructs were cotransfected with or without PELP1-WT or PELP1-MT plasmids (100 ng) using FuGENE6. After 48 h, cells were serum starved or cultured in 3% DCC medium without phenol red for 48 h and then treated with EGF (100 ng/mL), H89 (10 mmol/L), forskolin (20 mmol/L), or tamoxifen (10−8 mol/L) for 12 h. Cells were lysed with passive lysis buffer, and the luciferase activity was measured using a luciferase reporter assay kit (Promega). The total amount of DNA used in the transfections was kept constant by adding a parental vector. Each transfection was carried out using six-well plates in triplicate and normalized with β-galactosidase activity or the total protein concentration. Chromatin immunoprecipitation assays and pS2 primers used were described previously (21).
Immunofluorescence and Confocal Microscopy Studies
Cellular localization of PELP1 WT or MT was determined by indirect immunofluorescence, as described previously (21). Briefly, MCF-7 cells were grown on glass coverslips and maintained in serum-free medium for at least 24 h followed by treatment with H89, forskolin, or EGF. The cells were rinsed twice with PBS and fixed in 3.7% paraformaldehyde for 15 min at room temperature. The cells were incubated with the PELP1 antibody (1:200) for 2 h at room temperature, washed thrice in PBS, and then incubated with secondary antibodies conjugated with Alexa 488 dye (green) from Molecular Probes. The DNA dye 4′,6-diamidino-2-phenylindole (Molecular Probes) was used to costain DNA (blue). Confocal scanning analysis was done using Olympus FV300 laser scanning confocal microscope in accordance with established methods, using sequential laser excitation to minimize the possibility of fluorescent emission bleed through.
Acknowledgments
Grant support: NIH/National Cancer Institute grant CA095681 (R.K. Vadlamudi).
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Deroo BJ, Korach KS. Estrogen receptors and human disease. J Clin Invest. 2006;116:561–570. doi: 10.1172/JCI27987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnes CJ, Vadlamudi RK, Kumar R. Novel estrogen receptor coregulators and signaling molecules in human diseases. Cell Mol Life Sci. 2004;61:281–291. doi: 10.1007/s00018-003-3222-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jensen EV, Jordan VC. The estrogen receptor: a model for molecular medicine. Clin Cancer Res. 2003;9:1980–1989. [PubMed] [Google Scholar]
- 4.McDonnell DP, Norris JD. Connections and regulation of the human estrogen receptor. Science. 2002;296:1642–1644. doi: 10.1126/science.1071884. [DOI] [PubMed] [Google Scholar]
- 5.Losel R, Wehling M. Nongenomic actions of steroid hormones. Nat Rev Mol Cell Biol. 2003;4:46–56. doi: 10.1038/nrm1009. [DOI] [PubMed] [Google Scholar]
- 6.Bjornstrom L, Sjoberg M. Mechanisms of estrogen receptor signaling: convergence of genomic and nongenomic actions on target genes. Mol Endocrinol. 2005;19:833–842. doi: 10.1210/me.2004-0486. [DOI] [PubMed] [Google Scholar]
- 7.O’Malley BW. Coregulators: from whence came these “master genes”. Mol Endocrinol. 2007;21:1009–1013. doi: 10.1210/me.2007-0012. [DOI] [PubMed] [Google Scholar]
- 8.Collingwood TN, Urnov FD, Wolffe AP. Nuclear receptors: coactivators, corepressors and chromatin remodeling in the control of transcription. J Mol Endocrinol. 1999;23:255–275. doi: 10.1677/jme.0.0230255. [DOI] [PubMed] [Google Scholar]
- 9.Lonard DM, O’Malley BW. The expanding cosmos of nuclear receptor coactivators. Cell. 2006;125:411–414. doi: 10.1016/j.cell.2006.04.021. [DOI] [PubMed] [Google Scholar]
- 10.Schiff R, Massarweh S, Shou J, Osborne CK. Breast cancer endocrine resistance: how growth factor signaling and estrogen receptor coregulators modulate response. Clin Cancer Res. 2003;9:447S–454S. [PubMed] [Google Scholar]
- 11.Ali S, Coombes RC. Endocrine-responsive breast cancer and strategies for combating resistance. Nat Rev Cancer. 2002;2:101–112. doi: 10.1038/nrc721. [DOI] [PubMed] [Google Scholar]
- 12.Ariazi EA, Ariazi JL, Cordera F, Jordan VC. Estrogen receptors as therapeutic targets in breast cancer. Curr Top Med Chem. 2006;6:195–216. [PubMed] [Google Scholar]
- 13.Turgeon JL, McDonnell DP, Martin KA, Wise PM. Hormone therapy: physiological complexity belies therapeutic simplicity. Science. 2004;304:1269–1273. doi: 10.1126/science.1096725. [DOI] [PubMed] [Google Scholar]
- 14.Pietras RJ. Biologic basis of sequential and combination therapies for hormone-responsive breast cancer. Oncologist. 2006;11:704–717. doi: 10.1634/theoncologist.11-7-704. [DOI] [PubMed] [Google Scholar]
- 15.Shou J, Massarweh S, Osborne CK, et al. Mechanisms of tamoxifen resistance: increased estrogen receptor-HER2/neu cross-talk in ER/HER2-positive breast cancer. J Natl Cancer Inst. 2004;96:926–935. doi: 10.1093/jnci/djh166. [DOI] [PubMed] [Google Scholar]
- 16.Moy B, Goss PE. Estrogen receptor pathway: resistance to endocrine therapy and new therapeutic approaches. Clin Cancer Res. 2006;12:4790–4793. doi: 10.1158/1078-0432.CCR-06-1535. [DOI] [PubMed] [Google Scholar]
- 17.Schiff R, Massarweh SA, Shou J, et al. Advanced concepts in estrogen receptor biology and breast cancer endocrine resistance: implicated role of growth factor signaling and estrogen receptor coregulators. Cancer Chemother Pharmacol. 2005;56 Suppl 1:10–20. doi: 10.1007/s00280-005-0108-2. [DOI] [PubMed] [Google Scholar]
- 18.Vadlamudi RK, Wang RA, Mazumdar A, et al. Molecular cloning and characterization of PELP1, a novel human coregulator of estrogen receptor α. J Biol Chem. 2001;276:38272–38279. doi: 10.1074/jbc.M103783200. [DOI] [PubMed] [Google Scholar]
- 19.Wong CW, McNally C, Nickbarg E, Komm BS, Cheskis BJ. Estrogen receptor-interacting protein that modulates its nongenomic activity-crosstalk with Src/Erk phosphorylation cascade. Proc Natl Acad Sci U S A. 2002;99:14783–14788. doi: 10.1073/pnas.192569699. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Vadlamudi RK, Kumar R. Functional and biological properties of the nuclear receptor coregulator PELP1/MNAR. Nucl Recept Signal. 2007;5:e004. doi: 10.1621/nrs.05004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nair SS, Mishra SK, Yang Z, Balasenthil S, Kumar R, Vadlamudi RK. Potential role of a novel transcriptional coactivator PELP1 in histone H1 displacement in cancer cells. Cancer Res. 2004;64:6416–6423. doi: 10.1158/0008-5472.CAN-04-1786. [DOI] [PubMed] [Google Scholar]
- 22.Vadlamudi RK, Manavathi B, Balasenthil S, et al. Functional implications of altered subcellular localization of PELP1 in breast cancer cells. Cancer Res. 2005;65:7724–7732. doi: 10.1158/0008-5472.CAN-05-0614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greger JG, Fursov N, Cooch N, et al. Phosphorylation of MNAR promotes estrogen activation of phosphatidylinositol 3-kinase. Mol Cell Biol. 2007;27:1904–1913. doi: 10.1128/MCB.01732-06. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Rajhans R, Nair S, Holden AH, Kumar R, Tekmal RR, Vadlamudi RK. Oncogenic potential of the nuclear receptor coregulator proline-, glutamic acid-, leucine-rich protein 1/modulator of the nongenomic actions of the estrogen receptor. Cancer Res. 2007;67:5505–5512. doi: 10.1158/0008-5472.CAN-06-3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cosentino C, Di DM, Porcellini A, et al. p85 regulatory subunit of PI3K mediates cAMP-PKA and estrogens biological effects on growth and survival. Oncogene. 2007;26:2095–2103. doi: 10.1038/sj.onc.1210027. [DOI] [PubMed] [Google Scholar]
- 26.Manavathi B, Nair SS, Wang RA, Kumar R, Vadlamudi RK. Proline-, glutamic acid-, and leucine-rich protein-1 is essential in growth factor regulation of signal transducers and activators of transcription 3 activation. Cancer Res. 2005;65:5571–5577. doi: 10.1158/0008-5472.CAN-04-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith CL, O’Malley BW. Coregulator function: a key to understanding tissue specificity of selective receptor modulators. Endocr Rev. 2004;25:45–71. doi: 10.1210/er.2003-0023. [DOI] [PubMed] [Google Scholar]
- 28.Montminy M. Transcriptional regulation by cyclic AMP. Annu Rev Biochem. 1997;66:807–822. doi: 10.1146/annurev.biochem.66.1.807. [DOI] [PubMed] [Google Scholar]
- 29.Kim C, Xuong NH, Taylor SS. Crystal structure of a complex between the catalytic and regulatory (RIa) subunits of PKA. Science. 2005;307:690–696. doi: 10.1126/science.1104607. [DOI] [PubMed] [Google Scholar]
- 30.Flammer JR, Popova KN, Pflum MK. Cyclic AMP response element-binding protein (CREB) and CAAT/enhancer-binding protein β (C/EBPβ) bind chimeric DNA sites with high affinity. Biochemistry. 2006;45:9615–9623. doi: 10.1021/bi052521a. [DOI] [PubMed] [Google Scholar]
- 31.Taurin S, Sandbo N, Qin Y, Browning D, Dulin NO. Phosphorylation of β-catenin by cyclic AMP-dependent protein kinase. J Biol Chem. 2006;281:9971–9976. doi: 10.1074/jbc.M508778200. [DOI] [PubMed] [Google Scholar]
- 32.Gaillard E, Bruck N, Brelivet Y, et al. Phosphorylation by PKA potentiates retinoic acid receptor α activity by means of increasing interaction with and phosphorylation by cyclin H/cdk7. Proc Natl Acad Sci U S A. 2006;103:9548–9553. doi: 10.1073/pnas.0509717103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoang T, Fenne IS, Cook C, et al. cAMP-dependent protein kinase regulates ubiquitin-proteasome-mediated degradation and subcellular localization of the nuclear receptor coactivator GRIP1. J Biol Chem. 2004;279:49120–49130. doi: 10.1074/jbc.M409746200. [DOI] [PubMed] [Google Scholar]
- 34.Fan W, Yanase T, Wu Y, et al. Protein kinase A potentiates adrenal 4 binding protein/steroidogenic factor 1 transactivation by reintegrating the subcellular dynamic interactions of the nuclear receptor with its cofactors, general control nonderepressed-5/transformation/transcription domain-associated protein, and suppressor, dosage-sensitive sex reversal-1: a laser confocal imaging study in living KGN cells. Mol Endocrinol. 2004;18:127–141. doi: 10.1210/me.2003-0110. [DOI] [PubMed] [Google Scholar]
- 35.Colledge M, Scott JD. AKAPs: from structure to function. Trends Cell Biol. 1999;9:216–221. doi: 10.1016/s0962-8924(99)01558-5. [DOI] [PubMed] [Google Scholar]
- 36.Coghlan VM, Langeberg LK, Fernandez A, Lamb NJ, Scott JD. Cloning and characterization of AKAP 95, a nuclear protein that associates with the regulatory subunit of type II cAMP-dependent protein kinase. J Biol Chem. 1994;269:7658–7665. [PubMed] [Google Scholar]
- 37.Al-Dhaheri MH, Rowan BG. Protein kinase A exhibits selective modulation of estradiol-dependent transcription in breast cancer cells that is associated with decreased ligand binding, altered estrogen receptor α promoter interaction, and changes in receptor phosphorylation. Mol Endocrinol. 2007;21:439–456. doi: 10.1210/me.2006-0059. [DOI] [PubMed] [Google Scholar]
- 38.Fujimoto N, Katzenellenbogen BS. Alteration in the agonist/antagonist balance of antiestrogens by activation of protein kinase A signaling pathways in breast cancer cells: antiestrogen selectivity and promoter dependence. Mol Endocrinol. 1994;8:296–304. doi: 10.1210/mend.8.3.7517003. [DOI] [PubMed] [Google Scholar]
- 39.Michalides R, Griekspoor A, Balkenende A, et al. Tamoxifen resistance by a conformational arrest of the estrogen receptor α after PKA activation in breast cancer. Cancer Cell. 2004;5:597–605. doi: 10.1016/j.ccr.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 40.O’Malley BW. Molecular biology. Little molecules with big goals. Science. 2006;313:1749–1750. doi: 10.1126/science.1132509. [DOI] [PubMed] [Google Scholar]
- 41.Rosenfeld MG, Lunyak VV, Glass CK. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 2006;20:1405–1428. doi: 10.1101/gad.1424806. [DOI] [PubMed] [Google Scholar]
- 42.Wu RC, Qin J, Yi P, et al. Selective phosphorylations of the SRC-3/AIB1 coactivator integrate genomic reponses to multiple cellular signaling pathways. Mol Cell. 2004;15:937–949. doi: 10.1016/j.molcel.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 43.Lewis-Wambi JS, Jordan VC. Treatment of Postmenopausal Breast Cancer with Selective Estrogen Receptor Modulators (SERMs) Breast Dis. 2005;24:93–105. doi: 10.3233/bd-2006-24108. [DOI] [PubMed] [Google Scholar]
- 44.Adjei AA, Hidalgo M. Intracellular signal transduction pathway proteins as targets for cancer therapy. J Clin Oncol. 2005;23:5386–5403. doi: 10.1200/JCO.2005.23.648. [DOI] [PubMed] [Google Scholar]
- 45.Barletta F, Wong CW, McNally C, Komm BS, Katzenellenbogen B, Cheskis BJ. Characterization of the interactions of estrogen receptor and MNAR in the activation of cSrc. Mol Endocrinol. 2004;18:1096–1108. doi: 10.1210/me.2003-0335. [DOI] [PubMed] [Google Scholar]
- 46.Osborne CK, Bardou V, Hopp TA, et al. Role of the estrogen receptor coactivator AIB1 (SRC-3) and HER-2/neu in tamoxifen resistance in breast cancer. J Natl Cancer Inst. 2003;95:353–361. doi: 10.1093/jnci/95.5.353. [DOI] [PubMed] [Google Scholar]
- 47.Vadlamudi RK, Balasenthil S, Broaddus RR, Gustafsson JA, Kumar R. Deregulation of estrogen receptor coactivator proline-, glutamic acid-, and leucine-rich protein-1/modulator of nongenomic activity of estrogen receptor in human endometrial tumors. J Clin Endocrinol Metab. 2004;89:6130–6138. doi: 10.1210/jc.2004-0909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Apparao KB, Lovely LP, Gui Y, Lininger RA, Lessey BA. Elevated endometrial androgen receptor expression in women with polycystic ovarian syndrome. Biol Reprod. 2002;66:297–304. doi: 10.1095/biolreprod66.2.297. [DOI] [PubMed] [Google Scholar]
- 49.Balasenthil S, Vadlamudi RK. Functional interactions between the estrogen receptor coactivator PELP1/MNAR and retinoblastoma protein. J Biol Chem. 2003;278:22119–22127. doi: 10.1074/jbc.M212822200. [DOI] [PMC free article] [PubMed] [Google Scholar]