SUMMARY
Enterohemorrhagic Escherichia coli (EHEC) employs a type III secretion system (T3SS) to export translocator and effector proteins required for mucosal colonization. The T3SS is encoded in a pathogenicity island called the locus of enterocyte effacement (LEE) that is organized in five major operons, LEE1 to LEE5. LEE4 encodes a regulator of secretion (SepL), translocators (EspA, D and B), two chaperones (CesD2 and L0017), a T3SS component (EscF), and an effector protein (EspF). It was originally proposed that the esp transcript is transcribed from a promoter located at the end of sepL but other authors suggested that this transcript is the result of a post-transcriptional processing event. In this study, we established that the espADB mRNA is generated by post-transcriptional processing at the end of the sepL coding sequence. RNase E is the endonuclease involved in the cleavage, but the interaction of this enzyme with other proteins through its C-terminal half is dispensable. A putative transcription termination event in the cesD2 coding region would generate the 3’ end of the transcript. Similar to what has been described for other processed transcripts, the cleavage of LEE4 seems a mechanism to differentially regulate SepL and Esp protein production.
Keywords: enterohemorrhagic Escherichia coli, LEE4, RNase E, type III secretion system, espA, sepL
INTRODUCTION
Enterohemorrhagic Escherichia coli (EHEC) O157:H7 is a causative agent of bloody diarrhea, non-bloody diarrhea and haemolytic uremic syndrome (HUS) in humans. EHEC causes an ‘attaching and effacing’ (A/E) lesion characterized by localized destruction of brush border microvilli, intimate attachment to the eukaryotic cell membrane, and formation of a pedestal-like structure in the host cell. Most EHEC strains contain a ca. 43-kb pathogenicity island called the locus of enterocyte effacement (LEE), which encodes many of the virulence proteins necessary for the A/E phenotype (McDaniel et al., 1995; Perna et al., 1998). Other pathogens including enteropathogenic E. coli (EPEC), Citrobacter rodentium, and rabbit EPEC also contain LEE and are able to cause the A/E lesion (for reviews see Nataro and Kaper, 1998; Kaper et al., 2004).
LEE is organized into five major operons, LEE1 to LEE5, encoding a type III secretion system (T3SS), secreted proteins, chaperones, and regulators. The secreted proteins consist of effectors that are translocated into the host cell by the T3SS, and translocators required for delivering the effectors. LEE4 encodes the translocators EspA, EspB and EspD, a chaperone for EspD (CesD2) (Neves at al., 2003), the effector protein EspF (Nougayrède and Donnenberg, 2004; Guttman et al., 2006), a chaperone for EspA (L0017 or Orf29) (Su et al., 2008), and a component of the T3SS (EscF) (Figure 1A). EspA is polymerized to form a large filamentous structure extending from the bacterial surface by as much as 600 nm that connects the bacterium to the host cell membrane (Knutton et al., 1998; Sekiya et al., 2001; Shaw et al., 2001). The three dimentional structure of the EspA filament reveals a hollow tube forming an extension of the T3SS needle complex that is presumably composed of EscF (Wilson et al., 2001; Daniell et al., 2003). EspD is essential for the elaboration of mature EspA filaments and is thought to form a pore in the host cell membrane through which the effector proteins are translocated (Daniell et al., 2001; Kresse et al., 1999). EspB interacts with EspD (Ide et al., 2001) helping in pore formation (Luo and Donnenberg, 2006), and is also an effector protein that binds myosins in the eukaryotic cell facilitating microvillus lesions and evasion of phagocytosis (Iizumi et al., 2007).
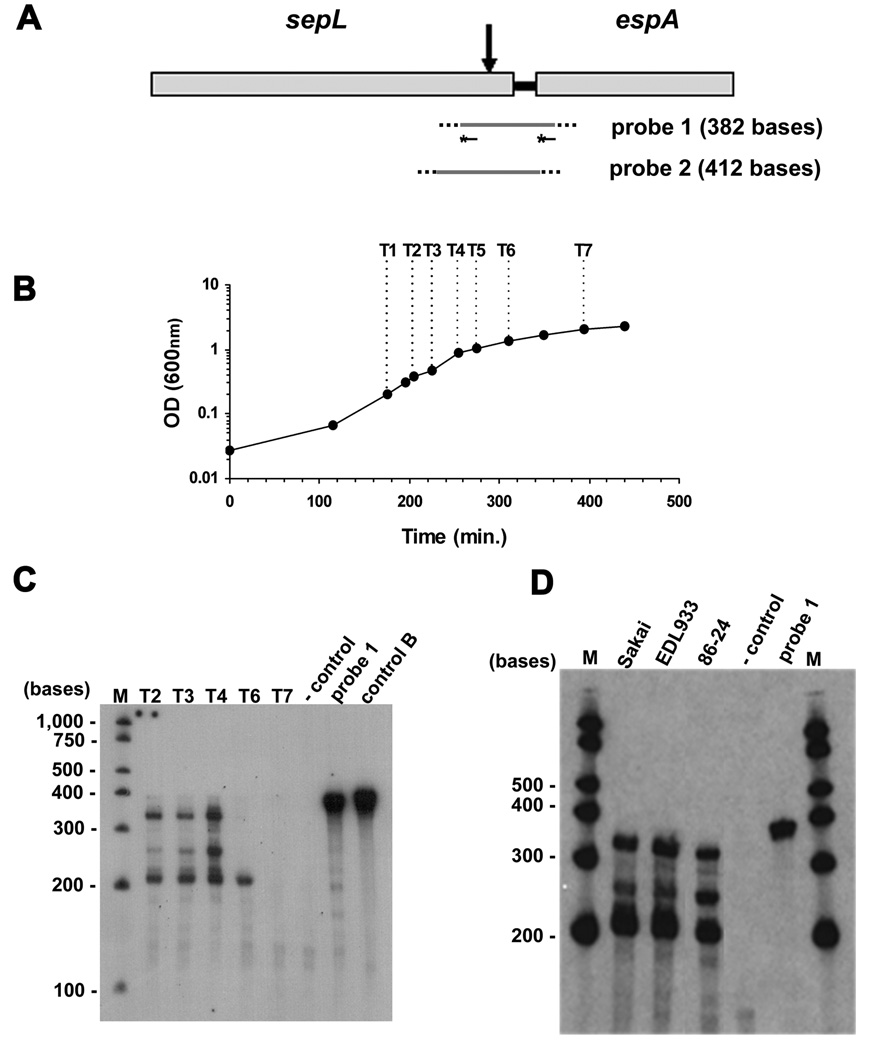
Fig. 1. Structure and transcription of the LEE4 operon.
A. Scheme depicting the structure of LEE4. ORFs are shown as grey boxes and intergenic regions as black lines. The two promoters previously proposed to drive sepL and espADB transcription in EHEC are indicated (black triangles) (Beltrametti et al, 1999). The arrows represent the primers (fLF and rLF) employed in RT-PCR in (B). The dashed line represents the probe used in Northern blot assays in (C).
B. Agarose gel electrophoresis of reverse transcription (RT) PCR products. EHEC strain Sakai was grown in LB and DMEM 1% glucose cultures. Cells were collected at mid-exponential phase (Mid-exp) and late exponential (Late-exp) for RNA extraction. Reverse transcription was performed with (+) or without (-) reverse transcriptase and PCR was performed with sepL and espF primers (fLF and rLF, respectively) (Table I). gDNA indicates that genomic DNA was used as a substrate for PCR with the same primers employed for RT-PCR. (M) λ DNA/HindIII marker.
C. Northern blot assay with a probe for espA. RNA was extracted from cultures grown in DMEM 1% glucose at different OD600 readings. T1 to T6 correspond to the time points when samples were collected and are shown in Figure 2B. The bottom panel shows rRNAs of the analyzed samples.
SepL is a non-secreted protein encoded upstream of the espA gene, and shares more than 90% identity among the different A/E pathogens. A sepL mutant shows highly reduced or undetectable secretion of translocator proteins, increased secretion of effector proteins, and is unable to form EspA filaments and A/E lesions (Kresse et al., 2000; O’Connell et al., 2004; Deng et al., 2005). SepL interacts with SepD, another component of the T3SS, probably forming a molecular switch to control the secretion of translocators and effectors by the T3SS (Deng et al., 2005). Recently, SepL was found to bind the effector protein Tir, preventing its secretion until the translocator components are exported (Wang et al., 2008). How the binding of SepL to SepD, Tir, and possibly to other proteins, regulates the kinetics of secretion of translocators and effectors is not currently understood.
In EHEC, espADB is proposed to be transcribed from a promoter (‘esp’) located at the end of the sepL coding region whereas sepL is transcribed independently (Beltrametti et al. 1999; Kresse at al., 2000; Sharma and Zuener, 2004). However, in EPEC sepL is cotranscribed with the other genes of LEE4 (Mellies et al., 1999). Experiments with chromosomal lacZ transcriptional fusions to the proposed ‘esp’ promoter failed to detect promoter activity in several different EHEC strains (Roe et al., 2003). In the same study, a transcript of 3 kb was detected with probes for espA and espD by Northern blot. These authors proposed that a promoter upstream of sepL drives the transcription of LEE4, and a post-transcriptional processing of this primary transcript generates the espADB mRNA.
In this study we define how the espADB mRNA is generated using a variety of techniques including reverse transcription (RT) -PCR, Northern Blot, Ribonuclease Protection Assay (RPA) and 5’ and 3’ Rapid Amplification of cDNA Ends (RACE). We show that sepL is part of the LEE4 operon, and that a post-transcriptional processing of the LEE4 transcript generates the espADB mRNA. The processing of LEE4 occurs in an E. coli K-12 background as well, and the endonuclease RNase E, which has a central role in RNA metabolism in participating in the degradation, processing and maturation of a variety of RNA molecules, is involved in the cleavage. This post-transcriptional processing of the LEE4 transcript has important implications in SepL and EspADB gene expression.
RESULTS
Genes of the LEE4 operon are cotranscribed
We used RT-PCR to determine whether sepL and the genes of the LEE4 operon are part of the same transcriptional unit. EHEC strain Sakai was grown in DMEM and LB media and cells were collected for RNA extraction. cDNA was synthesized, and amplified using a forward primer located at the end of sepL and a reverse primer located at the beginning of espF, the first and final genes in LEE4 (Fig 1A and Table I). The expected size of the PCR product was 4,113 bp, and a PCR product of approximately this size was obtained from all cDNA samples analyzed (Fig 1B, lanes (+)) consistent with the transcription process including the entire LEE4 operon. The expected size of the entire LEE4 transcript is approximately 5.7 kb considering the position of the sepL promoter reported previously (Kresse et al., 2000). However, Northern blot analysis using a probe for espA, revealed a major transcript of approximately 3 kb and two shorter transcripts of approximately 2.8 kb and 1.4 kb (Fig 1C). To gain further insight into the mechanism that generates the major esp transcript, we studied the sepL-espA intergenic region by RPA.
TABLE I.
DNA oligonucleotides used in this study
| Name | Directiona | Sequence 5’ to 3’ |
|---|---|---|
| fLF | F | tccttctcgggtatcgattg |
| rLF | R | tcgcattaacattcgacgag |
| F-T3LEE4b | F | aattaaccctcactaaagggaagtatgtcacgagtgcgatccttctcgggtatcgattg |
| R-T7LEE4c | R | taatacgactcactataggctgaatcgtatgaatcggaaccacctcatccttcgaca |
| F-T3LEE4b | F | aattaaccctcactaaagggaagtatgtcacgagtgcgacacttgataactatgcagatat |
| R-T7LEE4c | R | taatacgactcactataggctgaatcgtatgaatcgggtatccatctatatacctcttg |
| oligo-sepL | F | gggtatcgattgtcgaagataaacata |
| oligo-espA | F | gatctatgacttaggtaatatgtcgaag |
| roEspA | R | gcttaaatcaccactaagatcacga |
| riEspA | R | ttgtgccgtggttgacgctttagat |
| f3’RACE | F | gaccagtcttgctgaaggtacga |
| fespA | F | gctgatgttcagagtagcactg |
| respA | R | gaacgtgcactcgttaagag |
| frne-sd | F | caccggtagctatctggttctgatg |
| rrne-s | R | gtgaggttaacggtgattatgt |
| fdel-rne-s | F | ggtaaaggtgccgcaggtggt |
| rdel-rne-s | R | atcagacgcagaatagagag |
| f-rrnBe | F | agtagggatccgccaggcat |
| r-rrnBf | R | ggcagtgcccgcaaggcaatta |
| fΔsepL | F | aggaggaaattatttgatatta |
| rΔsepL | R | gagaaggactctccagcaac |
| fsep-esp | F | aatggctaatggtattgaattt |
| rsep-esp | R | taattaaattcggccactaaca |
| fsep-esp2 | F | atggctaatggtattgaatttaa |
| BamHI-LEE4-2g | R | taaggatcctaattaaattcggccactaaca |
| RACE-S20-r | R | agcttcagcaaattggcaattaagc |
F, forward; R, reverse
The sequence of the T3 promoter is underlined. A non-homologous sequence is in italics.
The sequence of the T7 promoter is underlined. A non-homologous sequence is in italics.
The first 4 nucleotides (underlined) were added for directional cloning into pENTR/SD/D-TOPO.
BamHI restriction site is underlined.
BglI restriction site is underlined.
Heterogeneous population of transcripts for esp genes
To analyze the transcripts in the sepL-espA intergenic region by RPA, a RNA probe was designed that spans the 3’ end of sepL, the intergenic region, and the 5’ end of espA (probe 1, Fig. 2A). A protected fragment of 326 bases would be generated if the probe hybridizes to a complementary transcript including the entire protected region. In addition, shorter protected fragments would be expected if transcripts initiate, terminate or are cleaved within this region. Sakai was grown in DMEM and cells were collected at different OD600 readings for RNA extraction and RPA (Fig. 2B). A protected fragment of about 320 bases and several shorter protected fragments were detected (Fig. 2C, lanes ‘T2’, ‘T3’ and ‘T4’). The highest level of expression was at the end of the exponential or beginning of stationary phase, and diminished in stationary phase to undetectable levels under these experimental conditions (Fig. 2C, lane ‘T7’) which is consistent with the Northern Blot analysis (Fig. 1C). In a previous study, quantification of espA mRNAs by real-time RT-PCR showed increasing levels from early to late exponential phase, but levels in stationary phase were not reported (Walters and Sperandio, 2006). In addition, the proportion of cells expressing EspA filaments in a bacterial population increased during the exponential phase, but then dropped off rapidly as the bacteria entered the stationary phase (Roe et al., 2003).
Fig. 2. A pattern of different ‘esp’ transcripts are detected by RPA.
A. Schematic representation of the probes used for RPAs. Both probes (grey lines) span the 3’ end of sepL, the intergenic region, and the 5’ end of espA (Table I and Experimental Procedures). They also contain non-complementary sequences at the ends (dotted lines). Lines with stars represent DNA oligomers complementary to sepL and espA sequences of probe 1 (Table I). These oligomers were labeled with [γ32P]ATP and used in the experiment shown in Fig. 3. The vertical arrow indicates the main processing site found at the end of sepL.
B. Sakai was grown in DMEM 1% glucose and samples were collected at different OD600 readings (arrows) for RNA extraction.
C. RPA was performed with probe 1 and total RNA extracted from bacteria at different OD600 readings corresponding to points labeled as T1 to T7 (B). Probe 1 was mixed with total yeast RNA and RPA was performed without RNase T1 treatment (lane ‘probe 1’). As a negative control, an RPA including RNase T1 treatment was done with yeast RNA and probe 1. An RPA including RNase T1 treatment was done with probe 1 and an in vitro transcribed complementary RNA (lane ‘control B’).The products were resolved in a 5% polyacrylamide/urea gel. (M) Century Plus Marker (Ambion).
D. RPA was performed with probe 1 and total RNA extracted from different EHEC strains as indicated. Bacteria were grown in DMEM 0.2% glucose to the end of exponential phase, and samples were collected for RNA extraction. After RPA, the products were resolved in a 5% polyacrylamide/urea gel.
An RPA using yeast RNA as substrate did not yield protected fragments indicating that the hybridization was specific to the sepL-espA region (Fig. 2C, lane ‘- control’, also see Fig. 5A for other negative controls). As an additional control, the complementary strand of probe 1, which is the sense strand, was transcribed in vitro and used as a substrate in an RPA instead of total RNA. In this case, the fragment corresponding to the entire sepL-espA protected region but none of the smaller protected fragments (Fig. 2C, lane ‘control B’) was detected, indicating the smaller fragments are due to mRNA differences and not to an RPA technical problem. RPAs performed with RNA extracted from EHEC strains EDL933 and 86-24 showed the same pattern of protected fragments (Fig. 2D).
Fig. 5. Effect of a thermosensitive (ts) mutation of RNase E in the processing of ‘esp’ transcripts.
A. Analysis by RPA of ‘esp’ transcripts in E. coli K-12. Plasmid pPL1 carries sepL-espAD genes that are transcribed from a vector (pACYC184) promoter. pPL1 was introduced into E. coli MG 1655 (lanes 2 and 3) and E. coli N3433 (lanes 5 and 6). RNA was extracted from LB cultures grown to OD600 0.58 (lane 2), 0.94 (lane 3), 0.57 (lane 5) and 0.94 (lane 6). RPA was performed with probe 1, and the products were resolved in a 5% polyacrylamide/urea gel. As negative controls, the same assays were done with RNA extracted from strains MG1655 and N3433 transformed with pACYC184 (lanes 4 and 7, respectively). (M) Century Plus Marker (Ambion)
B. Analysis by RPA of esp transcripts in an E. coli strain carrying a ts mutation in RNase E (N3431) and in its isogenic wild-type strain (N3433). Both strains were transformed with plasmid pPL1 and grown in LB at 35 °C up to the end of the exponential phase (time 0). Then, the cultures were transferred to the restrictive temperature (43 °C) and samples were collected at different times for RNA extraction and RPA analysis as indicated. Strain N3433 was also transformed with pACYC184 as a negative control for RPA. Products were resolved in a 5% polyacrylamide /urea gel. (M) Century Plus Marker (Ambion)
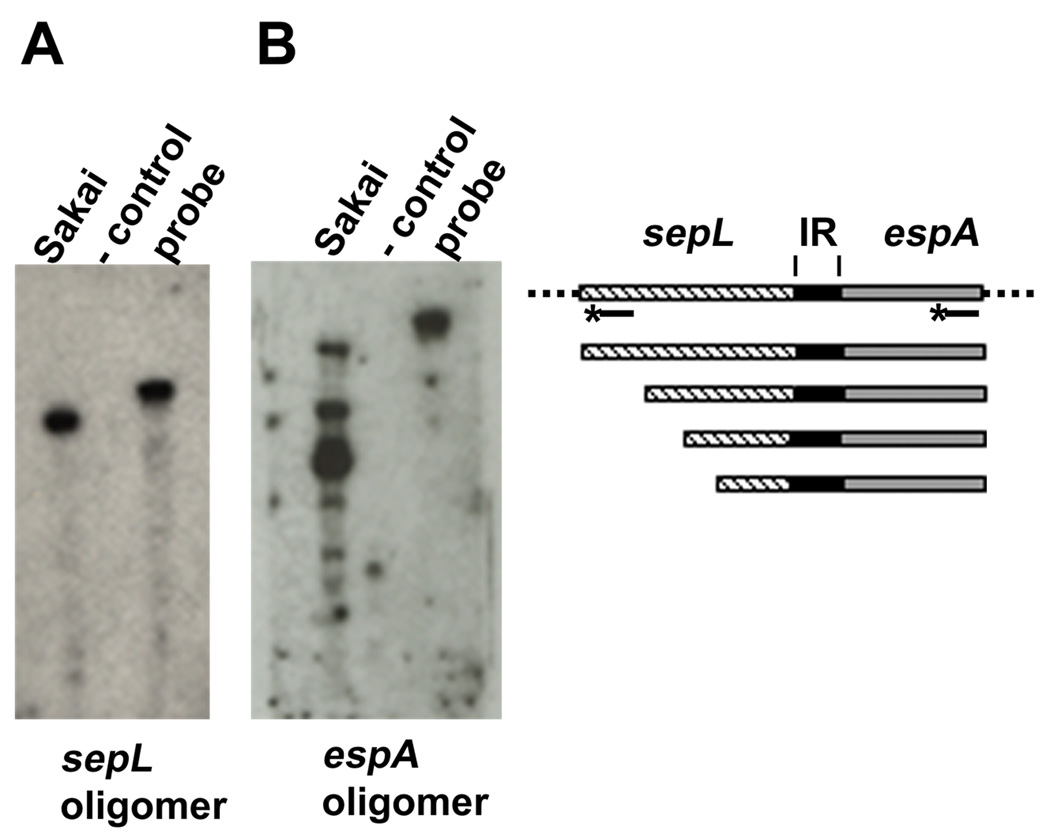
The smaller protected fragments could include the 5’ ends of ‘esp’ transcripts but also the 3’ end of sepL transcripts which could be generated by endonucleolytic cleavage or transcription termination. To discriminate between these possibilities, we designed the following assay. Duplicate RNA samples were subjected to RPA as described before but with unlabeled probe 1. Then, the protected RNA fragments were transferred and fixed to a membrane, and two separate hybridizations were done with two 5’-end labeled DNA oligomers complementary to sepL and espA, respectively (Fig. 2A). The small fragments hybridized to espA and not to sepL oligomer (Fig. 3), indicating a population of espADB transcripts with heterogeneous 5’ ends resulting from transcription initiation or endonucleolytic cleavage.
Fig. 3. The espADB mRNAs have heterogeneous 5’ ends.
Total RNA was extracted from cells collected from a DMEM culture of the Sakai strain. RPA was performed with probe 1, but in this case the probe was unlabeled. The RPA reaction products were resolved in a denaturing polyacrylamide gel, and transferred to a membrane. To detect the products, the membrane was cut into two halves. One half was hybridized with a DNA oligomer specific for sepL and the other was hybridized with a DNA oligomer specific for espA. Both oligomers were labeled with [γ-32P]ATP at their 5’ ends (represented as lines with a star). On the right side of the figure, a scheme represents the probe and protected fragments detected in this assay (IR, intergenic region).
5’ ends of esp transcripts are mono-phosphorylated
Primary transcripts can be discriminated from processed transcripts on the basis of the phosphorylation state of their 5’ ends, triphosphorylated or monophosphorylated, respectively. Briefly, total RNA was treated with specific phosphatases to modify the 5’ ends, then the monophosphorylated 5’ ends were ligated to an RNA adapter and subjected to 5’ RNA Ligase Mediated Rapid Amplification of cDNA Ends (RLM-RACE) (Bensing et al., 1996; Sawers, 2005; and see Experimental Procedures). When RLM-RACE is done with an RNA sample untreated with phophatases, only processed transcripts can yield a PCR product whereas transcripts with tri-phosphorylated 5’ ends cannot be ligated to the RNA adapter and detected. The same pattern of PCR products was obtained from an RNA sample treated to detect primary and processed transcripts, and from an RNA sample that was untreated with phosphatases (Fig. 4A, compare lanes 1 and 2 with lanes 7 and 8). This result suggests that the ‘esp’ transcripts are generated by endonucleolytic cleavage and not transcription initiation. We observed faint products from the sample treated to detect only primary transcripts (Fig. 4A, lanes 3 and 4) that might be due to inefficient monophosphate removal (see Experimental Procedures). Even though pyrophosphate can be removed from the 5’ ends of primary transcripts (Deana et al, 2008), these transcripts should have been detected by RLM-RACE because only a fraction of the population is converted to a monophosphorylated form (Celesnik et al., 2007). To determine the exact 5’ end of the ‘esp’ transcripts the PCR products were cloned and sequenced. Several 5’ ends were located in a region including ca. 90 nucleotides upstream of the sepL stop codon (Fig. 4B).
Fig. 4. RLM-RACE analysis of esp transcripts.
A. Agarose gel electrophoresis of the products obtained after RLM-RACE. esp transcripts were analyzed by 5’-RLM-RACE employing as a substrate a RNA sample extracted from a DMEM-grown culture of Sakai. The RNA sample was treated with calf intestine phosphatase (CIP) and tobacco acid pyrophosphatase (TAP) in different combinations to detect primary (Pri) and processed (Pro) transcripts (see Experimental Procedures). After the indicated treatments and reverse transcription, nested PCR was performed with 5’-RACE and espA specific primers (Table I). Lanes 1, 3, 5 and 7 correspond to the first PCR and lanes 2, 4, 6, and 8 correspond to the second nested PCR. (M) 1 kb Plus DNA Ladder (Invitrogen)
B. 5’ end locations of ‘esp’ transcripts. The products obtained after 5’-RLM-RACE were cloned and sequenced (reactions shown in lanes 2 and 8 in (A)). The 5’ ends are shown in capital letters and their distribution in 47 clones is indicated by a large arrow (18 clones), small arrow (11 clones), underlined nucleotides (5 clones each nucleotide), and double underlined nucleotides (4 clones each nucleotide). The horizontal arrows indicate the region analyzed by RPA.
RNAse E is involved in the endonucleolytic processing of LEE4
The processed region, including the intergenic sepL-espA region, is 72% A + T rich, which suggested that RNase E could be a good candidate for LEE4 processing because it cleaves preferentially in A+T rich sequences with no defined consensus sequence (McDowall et al., 1994). RNase E is a large enzyme of 1,061 amino acids that is essential for cell viability (Goldblum and Apririon, 1981; Taraseviciene et al., 1995; Kido et al., 1996). The conditionally lethal rne-3071 mutation is a C→T transition at position 742 that causes phenylalanine to be substituted for leucine and makes RNase E thermosensitive (ts) (Apirion, 1978; McDowall et al., 1993). We examined the role of RNase E in LEE4 processing using an E. coli K-12 rne-3071 strain N3431 and an isogenic rne+ strain N3433. For this purpose, sepL-espADB genes were cloned in the plasmid pACYC184 under the control of the tet promoter, replacing the tetracycline resistance gene (see Experimental Procedures). This plasmid, pPL1, was introduced into E. coli MG1655, N3431 and N3433 and analyzed for LEE4 processing with probe 1. Figure 5A shows that the pattern of bands seen in EHEC is also obtained with E. coli K-12 (pPL1) (compare lanes 1, with lanes 2, 3, 5 and 6). In contrast, when these strains were transformed with the vector only, no protected fragments were obtained (Fig. 5A, lanes 4 and 7). Next, the endonucleolytic processing was evaluated in E. coli N3431 and N3433 at the restrictive temperature. LB cultures from both strains transformed with pPL1 were grown up to the end of the exponential phase at 35°C, and then switched to 43°C. Samples were taken at different times after the temperature shift; RNA was extracted and analyzed by RPA with probe 1. At the restrictive temperature the processing was observed in the rne+ strain whereas processing was greatly reduced in the rne-3071 strain (Fig 5B) indicating that RNase E is required for the cleavage of the LEE4 transcript to generate the espADB mRNA products. An increased concentration of the transcripts was observed after the temperature shift in both rne+ and rne-3071 strains that was unrelated to the endonucleolytic processing being studied. This effect might be due to increased transcription from the vector promoter or an increased plasmid copy number at 43°C, among other possibilities.
The C-terminal domain of RNase E is not required for LEE4 cleavage
While the N terminus of RNAse E is essential for viability, the C terminal domain, which organizes a multiprotein complex called RNA degradosome, is not essential for cell viability (Taraseviciene et al., 1995; Kido et al., 1996). In addition to RNase E, the degradosome is composed of the exonuclease PNPase, the helicase RhlB, and the glycolytic enzyme enolase (for reviews see Marcaida et al., 2006; Carpousis, 2007).
An alignment of RNase E from EHEC and E. coli MG1655 shows 14 amino acid changes that are mostly conservative and located in the C-terminal half (Fig. 6A). To determine whether the interaction of RNase E with other proteins was required for LEE4 transcript cleavage, we constructed a derivative of the Sakai strain in which the 3’ end of rne gene was deleted. This mutant RNase E consists of 426 N-terminal amino acids including the domain sufficient for ribonuclease activity (Caruthers et al., 2006), plus 28 additional amino acids due to a frameshift mutation introduced with the deletion (Fig. 6B). The deletion of the scaffolding region was confirmed by PCR (Fig. 6C). RPA using probe 1 showed that there was no difference in the pattern of RNA protected fragments between the parental strain and the mutant (Fig. 6D), indicating that the degradosome assembly or the interaction of RNase E with other proteins is not required for LEE4 cleavage.
Fig. 6. The C-terminal half of RNase E is not involved in LEE4 processing.
A. Alignment of E. coli MG1655 and Sakai RNase E from amino acid 541. Both sequences are conserved up to amino acid 563. Conserved (‘:’), semi-conserved (‘.’), and non-conserved substitutions are enclosed in rectangles. Alignment was performed with ClustalW2 (Chenna et al., 2003).
B. Diagram that represents RNase E polypeptide in Sakai rne426 mutant. This mutant retains 426 N-terminal amino acids (grey box) while the C-terminal region involved in protein-protein interactions and degradosome assembly is missing (‘scaffolding region’, dashed line). The introduced deletion causes a frameshift in the reading frame (arrow and capital letters) adding 28 amino acids (stippled box) to the catalytic domain. Amino acids are shown in one letter code and some of the 28 additional amino acids are shown underlined.
C. Agarose gel electrophoresis of products obtained after colony PCR of Sakai and the mutant Sakai rne426. For PCR, specific DNA oligomers for rne were employed (frne-s and rrne-s, Table I). Lanes 2 and 3 show the amplification product of two mutant clones. (M) 1 kb Plus DNA Ladder (Invitrogen)
D. Analysis by RPA of ‘esp’ transcripts in Sakai and its mutant rne426. Samples for RNA extraction were collected from DMEM 0.2% glucose cultures of the parent (WT) and two mutant clones grown to the end of exponential phase. RPA was performed with probe 1, and then the products were resolved by denaturing 5 % polyacrylamide/urea gel. Total yeast RNA was employed in the negative control.
The 5’ product after transcript cleavage is degraded
The 5’ product after endonucleolytic cleavage, which includes the sepL gene, is not detected by RPA with probe 1 (Fig. 3). One possibility is that the 5’ product is rapidly degraded while the 3’ products – espADB mRNAs- are stable. Alternatively, the 5’ product could be undetectable because of inefficient ethanol precipitation in RPA due to its small size. To distinguish between these two possibilities we designed a second probe (probe 2) for RPA. In probe 2, a longer sequence spanned sepL whereas a shorter sequence spanned espA, in comparison with former probe 1 (Fig. 2A). Thus, if the 5’ product after cleavage was as stable as the 3’ product, RPA with probe 2 would give a protected RNA fragment of approximately 235 bases. Figure 7 (lane 4) shows that a fragment of this size was undetectable whereas a protected smaller fragment that corresponds to the 3’ portion of probe 2 was detected. When RNA was hybridized with probe 1, the usual protected fragments were obtained (Fig. 7, lane 3). Therefore, LEE4 is cleaved at the end of the sepL coding region and the 5’ product containing the sepL sequence is efficiently degraded.
Fig. 7. The 5’ product after LEE4 transcript processing is undetectable.
RNA was extracted from cells collected from a DMEM culture of the Sakai strain and analyzed by RPA using probe 1 (lane ‘Sakai (probe1)’) or probe 2 (lane ‘Sakai (probe 2)’) (see also Figure 2A). Negative controls with both probes were performed with total yeast RNA. Products were resolved in a 5% polyacrylamide/urea gel. (M) Century Plus Marker (Ambion).
The endonucleolytic cleavage diminishes SepL cellular concentration without affecting EspA level
The endonucleolytic cleavage of the LEE4 transcript at the end of the sepL coding sequence could diminish the cellular concentration of SepL without affecting EspA levels. To test this hypothesis, we compared SepL protein levels when expressed from pPL1 and a derivative plasmid, pΔSepL3, which lacks the RNase E processed sequence. Plasmid pΔSepL3 carries an in-frame deletion of 141 nucleotides and would generate a SepL polypeptide with a predicted molecular weight of 36.3 kDa whereas pPL1 would produce a SepL polypeptide of 41.9 kDa. A derivative of EHEC strain EDL933 in which the entire LEE region is deleted (EDL933ΔLEE) (Pósfai et al., 1997) was transformed with pPL1, pΔSepL3, or the vector alone. Cell extracts were prepared from DMEM cultures to analyze SepL and EspA protein levels by SDS-PAGE and immunoblot. EPEC strain UMD878 (EPECsepL) (O’Connell et al., 2004) with a kanamycin cassette insertion in sepL, and its complemented strain (EPECsepL(pSepL)) were analyzed as negative and positive controls for SepL synthesis, respectively. SepL was detected in a cell extract from EPECsepL(pSepL) as a product of about 40 kDa (Fig. 8A). As expected, SepL was not detected in the strain EPECsepL or in EDL933 ΔLEE transformed with the vector alone. The concentration of the SepL polypeptide was 2–2.5 fold higher in EDL933 ΔLEE (pΔSepL3) than in EDL933 ΔLEE (pPL1) indicating that the processing at the end of the sepL coding sequence diminishes SepL production. In contrast, EspA production in the same cell extracts was not affected by the deletion at the end of sepL (Fig. 8B). Similar experiments with the Sakai strain also show that under these growth conditions SepL was undetectable while EspA was clearly detected. Thus, our results suggest that this imbalance between SepL and EspA may be due in part to RNase E processing.
Fig. 8. Processing of the LEE4 transcript at the end of sepL diminishes SepL protein level.
Bacteria were grown in DMEM 0.2 % glucose and cellular proteins from bacterial pellets were subjected to SDS-PAGE and Western blotting with the indicated antisera raised against SepL (A) or EspA(B). The amount of protein loaded in the gel was standardized by OD600 readings for strains UMD878 (EPECsepL) and the complemented strain UMD878 (pCO24) (EPECsepL(pSepL)). For the other samples, protein concentration was measured and adjusted before loading the gel. After immunoblot, the membranes were stained with Brilliant Blue R (Sigma) to show total protein (right panels in (A) and (B)).
3’ end mapping of espADB mRNAs
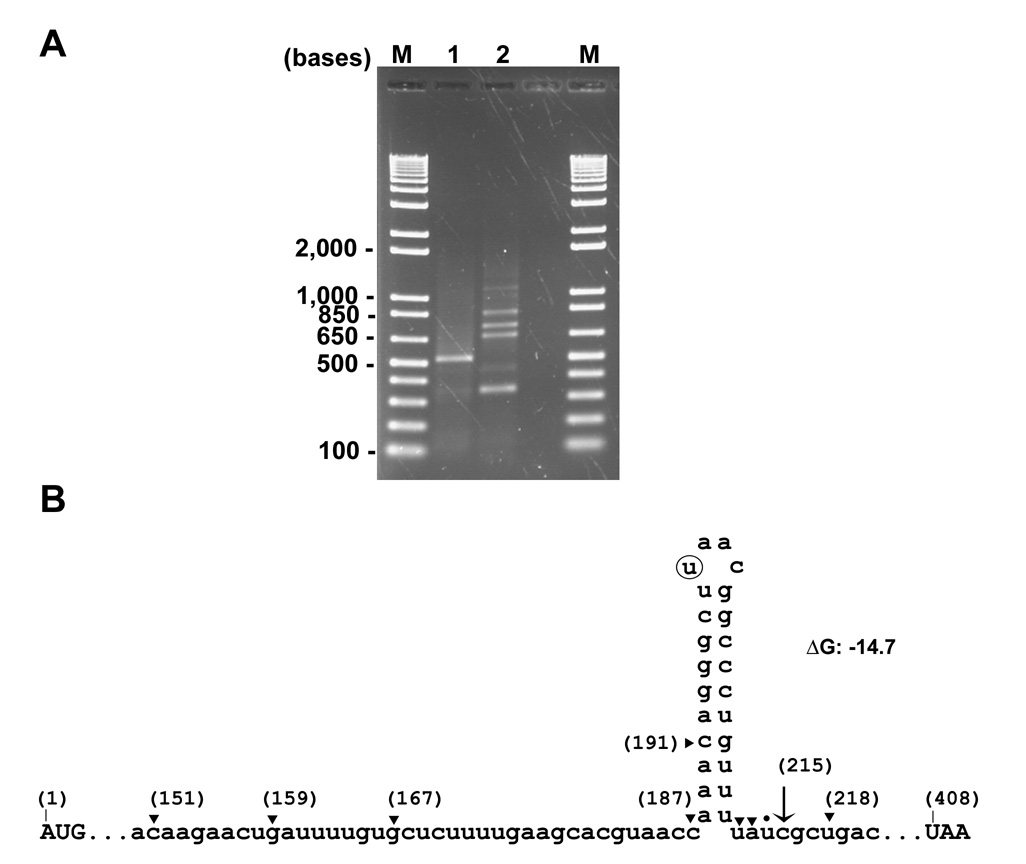
To determine the 3’ end of the espADB mRNAs, total RNA was extracted from the Sakai strain, polyA tails were added to the 3’ ends using poly(A)polymerase (PAP), and 3’-RACE was employed to amplify fragments containing the modified 3’ ends (see Experimental Procedures). A major PCR product was obtained (Fig. 9A lane 1) that was cloned and sequenced. The major terminus of the espADB transcripts was at position 215 considering the ‘A’ of cesD2 start codon as position 1 with other 3’ end sites found in a minority of clones (Fig. 9B). The size of the espADB transcript determined by Northern Blot is ~3 kb which is in good agreement with the size determined by 5’ and 3’ mapping. The sequence at the 3’ end of the espADB transcript can be folded into a putative secondary structure with features of an intrinsic transcriptional terminator (d’Aubenton Carafa et al., 1990) (Fig. 9B). The cesD2 nucleotide sequence is highly conserved (90% identity or higher) among EHEC, EPEC (strains E2348/69, 0181-6/86 and 3431-4/86), Citrobacter rodentium, and rabbit EPEC. In the loop of the hairpin, only a single nucleotide change was seen in the Citrobacter rodentium sequence.
Fig. 9. 3’ -end mapping of espADB mRNAs.
A. Agarose gel electrophoresis of the products obtained after 3’ RACE. RNA was isolated from Sakai strain grown in DMEM 0.2% glucose, treated with poly A polymerase (PAP) to add poly A tails (lane 1) or non-treated (lane 2), and then subjected to 3’ RACE and PCR.
B. Sequence of cesD2 showing the location of the 3’ ends found after PAP and 3’ RACE. The PCR product shown in (A) was gel purified and cloned. The major end is indicated by an arrow (10 out of 20 clones). Minor ends are indicated by a triangle (1 clone found for each end out of 20 clones), and a dot (two clones out of twenty). The 3’ end of the transcript might be folded in a predicted hairpin obtained with the Mfold program (Mathews et al., 1999; Zuker, 2003) that might function as an intrinsic transcriptional terminator. The sequence of this structure is conserved in EPEC, and in Citrobacter rodentium there is one base change (T→A, circled) located in the loop.
The same RNA sample was the substrate for a 3’ RACE reaction but without prior treatment with PAP. In this case, a different pattern of amplification products was obtained (Fig. 9A, lane 2). The products shown in Fig 9A lane 2 corresponded to mRNA 3’ ends located in cesD2 or espB and they might be the result of polyadenylated transcripts that are intermediates of degradation. Polyadenylation in E. coli serves to facilitate the exonucleolytic degradation of structured RNA molecules (Dreyfus and Régnier, 2002), and can occur on fragments resulting from endo or exonucleases cleavages (Haugel-Nielsen et al., 1996).
Discussion
In this study we established that transcription of the LEE4 operon in EHEC proceeds from sepL to espF, the first and the last genes of this operon, respectively. This result is consistent with previous results from our laboratory for the transcription of LEE4 in EPEC (Mellies et al., 1999). Our current work demonstrates that the LEE4 transcript is processed to generate an espADB transcript. First, E. coli K-12 (pPL1) harboring a ts mutation in rne does not produce the ‘esp’ transcripts at the restrictive temperature, indicating they are generated by RNase E-dependent processing. Second, the 5’ ends of the espADB transcripts in EHEC are monophosphorylated, which indicates formation by processing rather than transcription initiation. Third, EspA levels were unaffected by deletion of the processing region, ruling out a role for the deleted regions in transcription initiation. Even though the observed espADB transcripts are the products of processing, we cannot definitively rule out the existence of a weak promoter at the end of sepL. For example, in Salmonella typhimurium processed mRNAs were detected in the his operon and also an internal weak promoter (P2) close to a processing site (Alifano et al, 1992). Using 3’ RACE we mapped a major end of espADB transcripts inside the coding region of cesD2. A predicted stem-loop structure at the 3’ end of these transcripts resembles a Rho-independent transcription terminator in that it has a G + C -rich stem, a poly U tail at the 3’ end, a tetraloop, and the presence of As at the 5’ end of the stem-loop (d’Aubenton Carafa et al., 1990). Interestingly, a bioinformatic analysis of the E. coli K-12 genome predicted more than 40 intrinsic transcriptional terminators residing in ORFs (Lesnik et al., 2001), and a functional intragenic Rho-independent transcriptional terminator was found in the coding region of the bmpB gene in Borrelia burgdorferi (Ramamoorthy et al., 2005).
Even though the function of the potential intrinsic terminator has to be tested experimentally, its location inside a coding region raise the question of how the genes located downstream of the terminator could be transcribed. One possibility is that transcription could be imperfectly terminated at this particular terminator and thus, the genes located downstream might be transcribed as a result of readthrough by the RNA polymerase. In this respect, previous studies have stated that termination efficiency of different terminators varies greatly both in vitro and in vivo (Reynolds et al, 1992; Nojima et al., 2005) based not only on the terminator structure but also upon the context of the transcription unit in which it is placed. The context affecting termination efficiency includes both early transcribed sequences (Goliger et al., 1989; King et al., 1996) and terminator-distal sequences (Telesnitsky and Chamberlin, 1989). Transcription anti-termination could also be the result of protein factors that interact with RNA polymerase changing its properties (for reviews see Bailey et al.,1997; Nudler and Gottesman, 2002; Borukhov et al., 2005) or protein factors with RNA chaperone activity that melt RNA secondary structures (Phadtare et al., 2007). We did not map the 3’ end of the primary transcript, but by visual inspection or the use of RNA folding programs we could not find a structure that might resemble a prototypical intrinsic terminator at the end of espF. Thus, transcription of the whole operon-from sepL to espF- could terminate by a Rho-dependent transcription termination mechanism.
We found that RNase E is the endonuclease involved in LEE4 processing. Mutations of other genes encoding RNases -P, III, and G- had no effect (data not shown). RNase E cleaves its substrates in A + U rich-regions with no defined consensus sequence recognition (McDowall et al., 1994; Callaghan et al., 2005) which is consistent with our results. However, the endonucleolytic cleavage by RNase E should be tested by in vitro experiments. Our results with a deletion mutant of the C-terminal half of RNase E in EHEC establish that the degradosome assembly or the interaction of RNase E with other proteins like Hfq (Morita et al., 2005) is not necessary for LEE4 processing. In this regard, the C-terminal part of RNase E is dispensable for the maturation of 5S rRNA as well (Lopez et al., 1999; Ow et al., 2000). As far as we know, other studies have not evaluated the participation of the RNase E C-terminal half in RNA processing or maturation.
RNase E has a central role in RNA metabolism that ultimately controls gene expression. It is involved in bulk chemical decay of RNAs (Mudd et al., 1990) and in the degradation of specific RNAs (Tomcsányi and Apirion, 1985; Spickler et al., 2001; Régnier and Hajnsdorf, 1991; Aiba, 2007). RNase E not only cleaves RNAs that are then degraded but also cleaves precursor RNAs to yield functional mature products. Examples include: 5S rRNA (Apirion and Lassar, 1978), 16 S rRNA (Li et al., 1999), tRNAs (Li and Deutscher, 2002; Ow and Kushner, 2002), the RNA subunit of RNAse P (Lundberg and Altman, 1995), and ssrA RNA which adds a tag to truncated peptides for its degradation (Lin-Chao et al., 1999). Processing of precursor mRNAs by RNase E can also yield transcripts of different stability contributing to a different molar ratio of the encoded proteins as appear to be the case here. Examples of this kind of processing include: precursor mRNAs of phage f1 (Kokoska and Steege, 1998), the polycistronic ftsQAZ transcript encoding essential cell division proteins (Cam et al., 1996; Tamura et al., 2006), and mRNAs from operons such as pap responsible for the synthesis of pili for adhesion to mammalian cells (Nilsson and Uhlin, 1991; Nilsson et al., 1996).
Our results are reminiscent of what has been reported for the regulation of gene expression of pap and other operons encoding fimbriae in pathogenic E. coli (Jordi et al., 1993; Loomis and Moseley, 1998; Balsalobre, et al., 2003; Koo et al., 2004). The major structural subunit of fimbriae is needed in large excess relative to minor structural subunits or regulatory proteins encoded in the same operon. In the pap operon, the most-promoter proximal gene, papB, encodes a transcriptional activator that is cotranscribed with the gene for the major pilus subunit, papA. Processing by RNase E in the papB-papA intercistronic region of the mRNA B-A produces the stable transcript mRNA-A leading to high-level expression of PapA (Båga et al., 1988; Nilsson and Uhlin, 1991; Nilsson et al., 1996).
In A/E pathogens, EspA, D and B are secreted proteins forming the translocation apparatus whereas SepL is an intracellular protein that regulates secretion of translocators and effectors. The processing at the end of sepL would be a coordinate way to synthesize higher levels of EspA, D, and B relative to SepL, which might be needed in a lower concentration. In our experimental conditions SepL was undetectable in immunoblots whereas EspA was easily detected. This result, combined with the efficient cleavage in the sepL coding region, suggests that poor translation of sepL mRNA might expose it to RNase E binding and degradation. We noticed the absence of a prototypic Shine-Dalgarno sequence for sepL, which suggests inefficient translation initiation. In this respect, it was shown that inefficient translation of epd mRNA, which encodes the enzyme erythrose-4-phosphate dehydrogenase in E. coli, exposes an RNase E entry site that results in mRNA cleavage and contributes to epd low level expression (Bardey et al., 2005). In contrast, translation of espADB mRNA would increase its stability by hiding entry sites for RNase E as it was reported for other transcripts (Iost and Dreyfus, 1995; Baker and Mackie, 2003).
The function of SepL as a regulator has not yet been fully elucidated, and thus, why the expression of different levels of SepL and Esp proteins would be necessary for infection is unknown. In EPEC, mutant strains of sepL showed a defect in A/E lesion and EspA filament formation in vitro, and this effect was reversed with the expression of very little SepL whereas its overexpression did not interfere with its function (O’Connell, 2004). EspA forms the extracellular filament of the T3SS essential for the injection of virulence proteins to the host cell. The intracellular availability of EspA subunits limits the length of the filaments (Crepin et al., 2005), and thus, a constant supply of EspA subunits might be necessary to maintain the integrity of the filaments to allow a successful colonization of the epithelium. In a similar way, EspD is required for efficient EspA filament formation because mutations in espD affect filament formation in EHEC (Kresse et al., 1999) and EPEC (Knutton et al., 1998). EspB is targeted to the cytosol and membrane of the host cell (Knutton et al., 1998; Ide et al., 2001). It is a translocator protein necessary for the injection of proteins by the T3SS, and also is an effector that interferes with myosin and actin interaction in EPEC (Iizumi et al., 2007). The myosin binding domain of EPEC EspB shows 75% and 38 % identity with the same region of Citrobacter rodentium EspB and EHEC EspB, respectively (Iizumi et al., 2007). EspB was proposed to be delivered in highly concentrated doses to its site of action to prevent myosin binding to the highly abundant protein actin (Mattoo et al., 2008).
In Yersinia pseudotuberculosis, a strain with attenuated RNase E function showed a reduction in infectivity of culture macrophages and reduced secretion of one effector protein-YopE- by the T3SS. However, as YopE expression levels were not affected in this strain, the authors proposed that RNase E should control the levels of other transcripts required for optimal T3SS functioning (Yang et al., 2008). As far as we know, our study shows for the first time the RNase E-dependent processing of an operon necessary for the biogenesis of the T3SS in a pathogen. Finally, an additional post-transcriptional mechanism whose factors are encoded outside LEE, seems to control the heterogeneous production of EspA filaments in populations of EHEC cells (Roe et al., 2003). We did not analyze the expression of the remaining genes of LEE4, but previously published information suggests that post-transcriptional regulation would control the production of proteins encoded by those genes as well. For example EspF is detected in whole bacterial pellets and supernatants of cultures from both EHEC and EPEC (Viswanathan et al., 2004) while two other proteins – EscF and Orf29- were reported undetectable in immunoblots suggesting very low concentrations (Wilson et al., 2001; Su et al, 2008). In summary, previous studies and our results establish that there is a differential expression among the genes encoded in LEE4, and a tight regulation of protein production kinetics and secretion. However, we have just begun to unravel the post-transcriptional mechanisms controlling gene expression of this operon whose controlled expression presumably helps assure the efficient colonization of the intestine.
Experimental Procedures
Bacterial strains, culture conditions and recombinant DNA techniques
The EHEC O157:H7 strains used in this study were RIMD 0509952 (a Δstx Kanr derivative of the Sakai outbreak strain referred to in this study as simply ‘Sakai’) (Hayashi et al., 2001; Dahan et al., 2004), EDL933 (Perna et al., 2001), EDL933ΔLEE (Pósfai et al., 1997), and 86-24 (Griffin et al., 1988). The E. coli strain N3431 harbors a thermosensitive mutation in the rne gene that encodes RNase E, and N3433 is an isogenic rne+ strain (Apirion, 1978). Both strains were supplied by The Coli Genetic Stock Center at University of Yale. Other E. coli strains employed were MG1655 (Blattner et al., 1997), DH5α λ pir (Ménard et al., 1993) and E. coli SM10 λ pir (Simon et al, 1983). Bacteria were grown routinely in Luria broth (LB) (Sambrook and Russell, 2001). Antibiotics were added to the media when required in the following final concentrations: ampicillin (Ap), 100 µg ml−1; chloramphenicol (Cm), 20 µg ml−1.
Plasmid purification, DNA gel electrophoresis, genomic DNA extraction, transformation, ligation, and PCR were performed using standard methods (Sambrook and Russell, 2001). Easy-A™ Hi-Fi PCR cloning enzyme (Stratagene) or Pfu Ultra™ II Fusion HS DNA Polymerase (Stratagene) were used for PCR and cloning, and all constructs were confirmed by sequencing.
RNA extraction and reverse transcription-polymerase chain reaction (RT-PCR)
Cultures of EHEC strains were grown aerobically in Luria-Bertani broth (LB broth) overnight at 37°C and diluted (1:1,000 or 1:500) in pre-warmed Dulbecco’s modified Eagle’s medium (DMEM) (Invitrogen, catalog number 11885) supplemented with 1 or 0.2% glucose, grown aerobically at 37°C and shaken at 250 rpm. Samples were collected, and mixed with 2 volumes of RNAprotect Bacteria Reagent (Qiagen). Then, the cells were sedimented by centrifugation, and stored at −80 °C until RNA extraction. RNA was extracted with RNeasy Midi extraction kit (Qiagen), and treated twice with DNase (Qiagen and Ambion). RNA was quantified by UV absorbance, and its integrity was verified by visualization in denaturing agarose gel. cDNA was synthesized from 2 µg of total RNA using SuperScript III First-Strand (Invitrogen) following manufacturer’s instructions. PCR to detect LEE4 expression was performed with specific primers for sepL and espF genes (fLF and rLF, respectively) (Table I) and Easy-A Hi-Fi PCR cloning enzyme (Stratagene).
Northern blot
Electrophoresis of 20 µg of total RNA was performed on 1.2 % agarose formaldehyde-morpholinepropanesulfonic acid as previously described (Sambrook and Russell, 2001). After electrophoresis, the RNA was blotted onto a Hybond-N nylon membrane (Amersham Biosciences) by downward transfer (Chomcynski, 1992) and fixed by UV crosslinking (UV Stratalinker 1800, Stratagene). Prehybridization and hybridization were done at 42°C using NorthernMax Prehybridization/Hybridization Buffer (Ambion) following manufacturer’s instructions. The espA probe, a PCR fragment of 260 bp amplified with primers fEspA and rEspA (Table I), was labeled with [32P]dCTP and Ready-To-Go DNA labeling beads (GE Healthcare). After hybridization, the membrane was washed twice for 5 min at room temperature with 2X SSC, 0.1% SDS and twice for 15 min at 42°C in 0.1X SSC, 0.1% SDS.
In vitro transcription and Ribonuclease Protection Assay (RPA)
Probes for RPAs were synthesized by in vitro transcription using Maxiscript T7/ T3 kit (Ambion). Templates for in vitro transcription were PCR products generated with primers that incorporated a T7 promoter and a T3 promoter sequence at the 5’ ends. In addition, each primer included a non-homologous sequence to allow discrimination between the probe and the full-length protected fragment in RPAs. Primer pairs F-T3LEE4/ R-T7LEE4 and F-T3LEE4b /R-T7LEE4b were used to amplify the template for in vitro transcription of probe 1 and probe 2, respectively (Table I). Probes were labeled using Biotin-16-UTP (Roche) to UTP ratio of 40:60, and were gel purified after in vitro transcription. The RPA III Ribonuclease Protection Assay kit (Ambion) was employed for RPAs following the manufacturer’s recommendations. 5 µg of total RNA and 700 pg of labeled probe were used in overnight hybridization reactions at 42°C, except in the experiment shown in Figure 2C where 40 µg of total RNA and 800 pg of labeled probe were employed. As negative controls, hybridizations were performed with yeast total RNA provided with the kit, or E. coli K-12 RNA. Digestion of unhybridized RNA was performed with 1:30 dilution of RNase T1 supplied with the kit. The reactions were resolved in a 5% polyacrylamide/8 M urea denaturing gel, and transferred to a positively charged nylon membrane (Hybond-N+, Amersham Biosciences) using a semi-dry transfer system (Trans-Blot SD Semi-Dry electrophoretic Transfer Cell, Bio Rad) in 0.5X TBE. After transfer, the RNA was immediately fixed by UV crosslinking. The detection of the protected fragments was performed with CDP-Star substrate (Tropix, Applied Biosystems) following manufacturer’s recommendations with some modifications. The blot was washed twice for 5 min in 0.5 ml cm−2 Blocking Buffer (1X PBS, 0.2% I-Block reagent, 0.5% SDS), and blocked in the same buffer for 1 h (1 ml cm−2), with the exception of the experiment shown in Fig. 2C where skim milk was employed as a blocking agent. Then, the blot was incubated for 20 min with agitation in Streptavidin-AP conjugate (Roche) diluted 1:5,000 in Blocking Buffer (0.1 ml cm−2). The following washes were performed: 5 min in Blocking Buffer (0.5 ml cm−2), three 10 min washes in 1 ml cm−2 Washing Buffer (1X PBS, 0.5 % SDS), and two 2 min washes in Assay Buffer (20 mM Tris-HCl (pH 9.8), 1 mM MgCl2). After washing, the blot was incubated for 3–4 min in the substrate (3 ml 100 cm−2), and exposed to film X-Omat Blue XB-1 (Kodak).
Identification of protected fragments
For the experiment shown in Figure 3, RPA was performed with 40 µg of total RNA and 800 pg of unlabeled probe 1. The products were run in a denaturing polyacrylamide gel and transferred to a membrane as described above. To determine whether the protected fragments corresponded to sepL or espA coding regions, hybridizations were performed with specific DNA oligomers, oligo-sepL and oligo-espA (Table I) labeled with [γ-32P]ATP (GE Healthcare) using T4 Polynucleotide Kinase (Invitrogen). For hybridizations Rapid-hyb buffer (GE Healthcare) was employed. After hybridization, the membranes were washed following manufacturer’s recommendations.
RNA ligase mediated rapid amplification of cDNA ends (RLM-RACE)
The strategy described by Bensing et al. (1996) and Sawers (2005) was used to discriminate between mono or triphosphorylated 5’ ends. Total RNA was treated with phosphatases, and then 5’-RLM-RACE was performed using FirstChoice RLM-RACE kit (Ambion) following the manufacturer’s protocol. Briefly, a RNA sample was subjected to 4 separate treatments with tobacco acid pyrophosphatase (TAP) digestion, which removes pyrophosphate from the 5’ end of primary transcripts, and calf intestine phosphatase (CIP), which removes monophosphate groups from processed transcripts. In the first treatment, RNA was treated only with TAP and thus, processed and primary transcripts yield a product. In the second treatment, RNA was first treated with CIP and then with TAP, and thus only primary transcripts yield a product. In the third reaction, the RNA sample was untreated and thus only processed transcripts can be detected. Finally, the negative control is an RNA sample treated only with CIP. 900 ng of total RNA that were previously treated with CIP and TAP, CIP only, TAP only, or not treated, were ligated to the 5’ RACE adapter. After ligation and reverse transcription, the first PCR was done with the primers 5’-RACE outer primer, which hybridizes the adapter sequence, and roEspA (Table I). Then, an aliquot of the first PCR was employed as substrate for the second, nested PCR, with the primers 5’RACE inner primer and riEspA (Table I). The PCR products shown in lanes 2 and 8 of Figure 4A were cloned using the TOPO TA Cloning system (Invitrogen) and sequenced. As a positive control for the 5’ RLM-RACE technique, we analyzed the 5’ ends generated by transcription initiation of rpsT mRNAs which encode the ribosomal protein S20 (Fig. 1S). Primers 5’-RACE outer primer and RACE-S20-r (Table I) were employed in this assay.
The 3’ end of the espADB transcripts was mapped by 3’-RLM-RACE using the FirstChoice RLM-RACE kit (Ambion). Prior to 3’-RLM-RACE, 2 µg of total RNA were treated with poly (A) polymerase (PAP) (Ambion) to add a poly A tail to the 3’ ends. 3’-RLM-RACE was also done with 2 µg of total RNA without poly A to show that the amplification product obtained with PAP treatment was a specific product resulting from poly A addition to the 3’ end of espADB mRNAs. Reverse transcription was performed with the 3’ RACE Adapter, which is provided with the kit, and has a poly T sequence for annealing to poly A tails at the 3’ ends. Then, PCR was performed with the primers 3’ RACE Outer primer, which hybridizes the adapter, and f3’RACE (Table I). The PCR product was gel purified and cloned using the TOPO TA Cloning system.
PAGE and immunoblot analysis of proteins
Bacterial cells were collected from DMEM cultures and frozen for later analysis. Cell extracts were prepared by boiling cell pellets and protein concentration was measured by DC Protein Assay (Bio-Rad) to load equal amounts in gels. Cell extracts were mixed with NuPAGE LDS sample buffer 4X, and heated for 10 min at 75 °C before loading gels. Proteins were resolved on PAGE gels (Laemmli, 1970) using Next Gel (Amresco) and Next Gel Running Buffer (Amresco). After electrophoresis, proteins were transferred to Immobilon-FL membranes (Millipore). For immunoblotting and detection, the Odyssey Infrarred Imaging System (Li-Cor) was used following manufacturer’s protocols. EspA was detected with anti-EPEC EspA polyclonal antibodies raised in rabbits (Elliott et al., 2000) and diluted 1:10,000 in Odyssey Blocking Buffer, 0.1 % Tween 20. SepL was detected with polyclonal antibodies against EPEC SepL raised in rabbit (O’Connell et al., 2004) and diluted 1:5,000 in Odyssey Blocking Buffer, 0.05 % Tween 20. Anti-rabbit Alexa Fluor 680 was used as a secondary antibody diluted 1:10,000 in Odyssey Blocking Buffer, 0.1 % Tween 20.
Construction of RNase E mutant
Genomic DNA from Sakai Δstx strain was used as template to amplify the last 2,817 bp of the rne gene plus 709 bp downstream of the TAA stop codon using the primers frne-s and rrne-s (Table I). This product was cloned into pENTR/SD/D-TOPO (Invitrogen), and was the template for inverse PCR to create a deletion in the rne gene using the primers fdel-rne-s and rdel-rne-s. This mutant sequence, named rne426, would encode a polypeptide of 426 amino acids of the N-22 terminal region plus 28 additional amino acids (rne426). The mutant rne sequence was cloned in pJMM112 (a gift from Jane Michalski) that is a derivative of pCVD442 (Donnenberg and Kaper, 1991), and the resulting plasmid was named pJrne426. pJMM112 has an Ap resistance marker, the gene sacB and a module with the following determinants: attR1- cat-ccdB- attR2.
The attR sites are bacteriophage λ -derived DNA recombination sequences, cat specifies a Cm resistance marker, and ccdB allows negative selection of the plasmid. The rne426 sequence was moved to pJMM112 using the Gateway LR clonase II Enzyme Mix (Invitrogen) following the manufacturer’s directions. E. coli DH5α λ pir was transformed with the clonase reaction. Transformants were selected for Ap resistance and tested for Cm sensitivity. The presence of the rne426 deletion in transformants was confirmed by PCR. Plasmid pJrne426 was introduced into the donor E. coli SM10 λ pir and transfer to Sakai Δstx by mating overnight in LB agar. The cells were resuspended in PBS, and Sakai exconjugants were selected in MacConkey agar containing Ap (100 µg ml−1), sorbitol (10 g l−1) and potassium tellurite (2.5 µg ml−1). Several white colonies (non-fermenting sorbitol) were inoculated in LB cultures, and after overnight growth, dilutions were prepared and plated on LB agar containing no NaCl and 5% (w/v) sucrose. Sucrose-resistant colonies were tested for Ap sensitivity indicative of the loss of vector sequences. Colony PCR was done to detect the mutated rne allele. Approximately half of the sucrose resistant and Ap sensitive colonies carried the rne426 deletion, which was confirmed by sequencing.
Plasmid constructions
To construct the plasmid pPL1, the coding sequences of the first three genes of the LEE4 operon- sepL, espA and espD- were cloned into plasmid pACYC184 replacing the tetracycline resistance gene but leaving the coding sequence for the first 19 amino acids of Tet. Thus, transcription of the cloned genes is controlled by the tet promoter and SepL is synthesized as a fusion protein with 19 N-terminal amino acids of the Tet protein. In addition, the rrnB T1 and T2 transcription termination sequences were cloned downstream of espD. To accomplish this, the primers f-rrnB and r-rrnB, carrying BamHI and BglI restriction sites, respectively, were used to amplify rrnB T1 and T2 sequences from plasmid pENTR/SD/D-TOPO. This product was cloned into the BamHI and BglI sites of plasmid pACYC184 to generate pACYC184rrnB. Then the sepLespADB sequence was amplified with primers fsep-esp and rsep-esp using genomic DNA of Sakai Δstx as template, and cloned into the vector pCR 2.1-TOPO (Invitrogen). This construction was used as a template to re-amplify sepLespAD with primers f-sepesp2, and BamHI-LEE4-2 to introduce a BamHI site downstream of the espD stop codon. The PCR product sepLespAD was digested with BamHI, and ligated to BamHI and ScfI-digested plasmid pACYC184 rrnB. The ligation product was gel purified, the ScfI 5’ overhang was filled with DNA polymerase I, Large (Klenow) fragment (New England Biolabs) to create a blunt end, and then the plasmid blunt ends were ligated by T4 DNA ligase (New England Biolabs).
To construct plasmid pΔSepL3 carrying a deletion of the processing region, plasmid pPL1 was used as template in a PCR with primers fΔsepL and rΔsepL. The product was gel purified, phosphorylated by T4 kinase (New England Biolabs), cleaned using QIAquick PCR purification kit (Qiagen) and ligated with T4 DNA ligase (New England Biolabs). Restriction enzyme digestions, PCR and other enzyme treatments were done following the manufacturer’s protocols.
Acknowledgments
We thank Dr Mary Jane Lombardo and Dr Paul S. Lovett for the critical reading of the manuscript. We thank Jane Michalski for the construction of the pJMM112 plasmid, and Dr M. S. Donnenberg for providing SepL antiserum and the EPEC strains UMD878 and UMD878(pCO24). This study was supported by NIH grants DK58957 and AI21657 to J.B.K.
Footnotes
Accession Numbers for LEE sequence
Previously deposited sequences cited in this work are from atypical EPEC (AJ633129 and AJ633130), typical EPEC (AF022236), Citrobacter rodentium (AF311901), rabbit EPEC (AF461393 and AF200363), EHEC O157:H7 strain Sakai (BA000007), EHEC O157:H7 strain EDL933 (AE005174)
REFERENCES
- Aiba H. Mechanism of RNA silencing by Hfq-binding small RNAs. Curr Opin Microbiol. 2007;10:134–139. doi: 10.1016/j.mib.2007.03.010. [DOI] [PubMed] [Google Scholar]
- Alifano P, Piscitelli C, Blasi V, Rivellini F, Nappo AG, Bruni CB, Carlomagno MS. Processing of a polycistronic mRNA requires a 5’ cis element and active translation. Mol Microbiol. 1992;6:787–798. doi: 10.1111/j.1365-2958.1992.tb01529.x. [DOI] [PubMed] [Google Scholar]
- Apirion D. Isolation, genetic mapping and some characterization of a mutation in Escherichia coli that affects the processing of ribonucleic acid. Genetics. 1978;90:659–671. doi: 10.1093/genetics/90.4.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apirion D, Lassar AB. A conditional lethal mutant of Escherichia coli which affects the processing of ribosomal RNA. J Biol Chem. 1978;253:1738–1742. [PubMed] [Google Scholar]
- Båga M, Göransson M, Normark S, Uhlin BE. Processed mRNA with differential stability in the regulation of E. coli pilin gene expression. Cell. 1988;52:197–206. doi: 10.1016/0092-8674(88)90508-9. [DOI] [PubMed] [Google Scholar]
- Baker KE, Mackie GA. Ectopic RNase E sites promote bypass of 5’-end-dependent mRNA decay in Escherichia coli. Mol Microbiol. 2003;47:75–88. doi: 10.1046/j.1365-2958.2003.03292.x. [DOI] [PubMed] [Google Scholar]
- Balsalobre C, Morschhäuser J, Jass J, Hacker J, Uhlin BE. Transcriptional analysis of the sfa determinant revealing mRNA processing events in the biogenesis of S fimbriae in pathogenic Escherichia coli. J Bacteriol. 2003;185:620–629. doi: 10.1128/JB.185.2.620-629.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardey V, Vallet C, Robas N, Charpentier B, Thouvenot B, Mougin A, et al. Characterization of the molecular mechanisms involved in the differential production of erythrose-4-phosphate dehydrogenase, 3-phophoglycerate kinase and class II fructose-1,6-bisphosphate aldolase in Escherichia coli. Mol Microbiol. 2005;57:1265–1287. doi: 10.1111/j.1365-2958.2005.04762.x. [DOI] [PubMed] [Google Scholar]
- Bailey MJA, Hughes C, Koronakis V. RfaH and the ops element, components of a novel system controlling bacterial transcription elongation. Mol Microbiol. 1997;26:845–851. doi: 10.1046/j.1365-2958.1997.6432014.x. [DOI] [PubMed] [Google Scholar]
- Beltrametti F, Kresse AU, Guzmán CA. Transcriptional regulation of the esp genes of enterohemorrhagic Escherichia coli. J Bacteriol. 1999;181:3409–3418. doi: 10.1128/jb.181.11.3409-3418.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensing BA, Meyer BJ, Dunny GM. Sensitive detection of bacterial transcription initiation sites and differentiation from RNA processing sites in the pheromone-induced plasmid transfer system of Enterococcus faecalis. Proc Natl Acad Sci USA. 1996;93:7794–7799. doi: 10.1073/pnas.93.15.7794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borukhov S, Lee J, Laptenko O. Bacterial transcription elongation factors: new insights into molecular mechanism of action. Mol Microbiol. 2005;55:1315–1324. doi: 10.1111/j.1365-2958.2004.04481.x. [DOI] [PubMed] [Google Scholar]
- Blattner FR, Plunkett G, 3rd, Bloch CA, Perna NT, Burland V, Riley M, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- Callaghan AJ, Marcaida MJ, Stead JA, McDowall KJ, Scott WG, Luisi BF. Structure of Escherichia coli RNase E catalytic domain and implications for RNA turnover. Nature. 2005;437:1187–1191. doi: 10.1038/nature04084. [DOI] [PubMed] [Google Scholar]
- Cam K, Rome G, Krisch HM, Bouché J-P. RNase E processing of essential cell division genes mRNA in Escherichia coli. Nucleic Acids Res. 1996;24:3065–3070. doi: 10.1093/nar/24.15.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpousis AJ. The RNA degradosome of Escherichia coli: an mRNA-degrading machine assembled on RNase E. Annu Rev Microbiol. 2007;61:71–87. doi: 10.1146/annurev.micro.61.080706.093440. [DOI] [PubMed] [Google Scholar]
- Caruthers JM, Feng Y, McKay DB, Cohen SN. Retention of core catalytic functions by a conserved minimal ribonuclease E peptide that lacks the domain required for tetramer formation. J Biol Chem. 2006;281:27046–27051. doi: 10.1074/jbc.M602467200. [DOI] [PubMed] [Google Scholar]
- Celesnik H, Deana A, Belasco JG. Initiation of RNA decay in Escherichia coli by 5' pyrophosphate removal. Mol Cell. 2007;27:79–90. doi: 10.1016/j.molcel.2007.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P. One-hour downward alkaline capillary transfer for blotting of DNA and RNA. Anal Biochem. 1992;201:134–139. doi: 10.1016/0003-2697(92)90185-a. [DOI] [PubMed] [Google Scholar]
- Crepin VF, Shaw R, Abe CM, Knutton S, Frankel G. Polarity of enteropathogenic Escherichia coli EspA filament assembly and protein secretion. J Bacteriol. 2005;187:2881–2889. doi: 10.1128/JB.187.8.2881-2889.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Aubenton Carafa Y, Brody E, Thermes C. Prediction of rho-independent Escherichia coli transcription terminators. A statistical analysis of their RNA stem-loop structures. J Mol Biol. 1990;216:835–858. doi: 10.1016/s0022-2836(99)80005-9. [DOI] [PubMed] [Google Scholar]
- Dahan S, Knutton S, Shaw RK, Crepin VF, Dougan G, Frankel G. Transcriptome of enterohemorrhagic Escherichia coli O157 adhering to eukaryotic plasma membranes. Infect Immun. 2004;72:5452–5459. doi: 10.1128/IAI.72.9.5452-5459.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell SJ, Delahay RM, Shaw RK, Hartland EL, Pallen MJ, Booy F, et al. Coiled-coil domain of enteropathogenic Escherichia coli type III secreted protein EspD is involved in EspA filament-mediated cell attachment and hemolysis. Infect Immun. 2001;69:4055–4064. doi: 10.1128/IAI.69.6.4055-4064.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell SJ, Kocsis E, Morris E, Knutton S, Booy FP, Frankel G. 3D structure of EspA filaments from enteropathogenic Escherichia coli. Mol Microbiol. 2003;49:301–308. doi: 10.1046/j.1365-2958.2003.03555.x. [DOI] [PubMed] [Google Scholar]
- Deng W, Li Y, Hardwidge PR, Frey EA, Pfuetzner RA, Lee S, et al. Regulation of type III secretion hierarchy of translocators and effectors in attaching and effacing bacterial pathogens. Infect Immun. 2005;73:2135–2146. doi: 10.1128/IAI.73.4.2135-2146.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deana A, Celesnik H, Belasco JG. The bacterial enzyme RppH triggers messenger RNA degradation by 5' pyrophosphate removal. Nature. 2008;451:355–358. doi: 10.1038/nature06475. [DOI] [PubMed] [Google Scholar]
- Donnenberg MS, Kaper JB. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect Immun. 1991;59:4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfus M, Régnier P. The poly(A) tail of mRNAs: bodyguard in Eukaryots, scavenger in bacteria. Cell. 2002;111:611–613. doi: 10.1016/s0092-8674(02)01137-6. [DOI] [PubMed] [Google Scholar]
- Elliott SJ, Sperandio V, Girón JA, Shin S, Mellies JL, Wainwright L, et al. The locus of enterocyte effacement (LEE)-encoded regulator controls expression of both LEE- and non-LEE-encoded virulence factors in enteropathogenic and enterohemorrhagic Escherichia coli. Infect Immun. 2000;68:6115–6126. doi: 10.1128/iai.68.11.6115-6126.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldblum K, Apririon D. Inactivation of the ribonucleic acid-processing enzyme ribonuclease E blocks cell division. J Bacteriol. 1981;146:128–132. doi: 10.1128/jb.146.1.128-132.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goliger JA, Yang X, Guo HC, Roberts JW. Early transcribed sequences affect termination efficiency of Escherichia coli RNA polymerase. J Mol Biol. 1989;205:331–341. doi: 10.1016/0022-2836(89)90344-6. [DOI] [PubMed] [Google Scholar]
- Griffin PM, Ostroff SM, Tauxe RV, Greene KD, Wells JG, Lewis JH, Blake PA. Illnesses associated with Escherichia coli O157:H7 infections. A broad clinical spectrum. Ann Intern Med. 1988;109:705–712. doi: 10.7326/0003-4819-109-9-705. [DOI] [PubMed] [Google Scholar]
- Guttman JA, Li Y, Wickham ME, Deng W, Vogl WA, Finlay BB. Attaching and effacing pathogen-induced tight junction disruption in vivo. Cell Microbiol. 2006;8:634–645. doi: 10.1111/j.1462-5822.2005.00656.x. [DOI] [PubMed] [Google Scholar]
- Haugel-Nielsen J, Hajnsdorf E, Régnier P. The rpsO mRNA of Escherichia coli is polyadenylated at multiple sites resulting from endonucleolytic processing and exonucleolytic degradation. EMBO J. 1996;271:3144–3152. [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Makino K, Ohnishi M, Kurokawa K, Ishii K, Yokoyama K, et al. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 2001;8:11–22. doi: 10.1093/dnares/8.1.11. [DOI] [PubMed] [Google Scholar]
- Ide T, Laarmann S, Greune L, Schillers H, Oberleithner H, Schmidt MA. Characterization of translocation pores inserted into plasma membranes by type III-secreted Esp proteins of enteropathogenic Escherichia coli. Cell Microbiol. 2001;3:669–679. doi: 10.1046/j.1462-5822.2001.00146.x. [DOI] [PubMed] [Google Scholar]
- Iizumi Y, Sagara H, Kabe Y, Azuma M, Kume K, Ogawa M, et al. The enteropathogenic E. coli effector EspB facilitates microvillus effacing and antiphagocytosis by inhibiting myosin function. Cell Host Microbe. 2007;2:383–392. doi: 10.1016/j.chom.2007.09.012. [DOI] [PubMed] [Google Scholar]
- Iost I, Dreyfus M. The stability of Escherichia coli lacZ mRNA depends upon the simultaneity of its synthesis and translation. EMBO J. 1995;14:3252–3261. doi: 10.1002/j.1460-2075.1995.tb07328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordi BJ, op den Camp IE, de Haan LA, van der Zeijst BA, Gaastra W. Differential decay of RNA of the CFA/I fimbrial operon and control of relative gene expression. J Bacteriol. 1993;175:7976–7981. doi: 10.1128/jb.175.24.7976-7981.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- Kido M, Yamanaka K, Mitani T, Niki H, Ogura T, Hiraga S. RNase E polypeptides lacking a carboxyl-terminal half suppress a mukB mutation in Escherichia coli. J Bacteriol. 1996;178:3917–3925. doi: 10.1128/jb.178.13.3917-3925.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RA, Banik-Maiti S, Jin DJ, Weisberg RA. Transcripts that increase the processivity and elongation rate of RNA polymerase. Cell. 1996;87:893–903. doi: 10.1016/s0092-8674(00)81996-0. [DOI] [PubMed] [Google Scholar]
- Knutton S, Rosenshine I, Pallen MJ, Nisan I, Neves BC, Bain C, et al. A novel EspA-associated surface organelle of enteropathogenic Escherichia coli involved in protein translocation into epithelial cells. EMBO J. 1998;17:2166–2176. doi: 10.1093/emboj/17.8.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokoska RJ, Steege DA. Appropiate expression of filamentous phage f1 DNA replication genes II and X requires RNase E-dependent processing and separate mRNAs. J Bacteriol. 1998;180:3245–3249. doi: 10.1128/jb.180.12.3245-3249.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo JT, Choe J, Moseley SL. HrpA, a DEAH-box RNA helicase, is involved in mRNA processing of a fimbrial operon in Escherichia coli. Mol Microbiol. 2004;52:1813–1826. doi: 10.1111/j.1365-2958.2004.04099.x. [DOI] [PubMed] [Google Scholar]
- Kresse AU, Rohde M, Guzmán CA. The EspD protein of enterohemorrhagic Escherichia coli is required for the formation of bacterial surface appendages and is incorporated in the cytoplasmic membranes of target cells. Infect Immun. 1999;67:4834–4842. doi: 10.1128/iai.67.9.4834-4842.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kresse AU, Beltrametti F, Müller A, Ebel F, Guzmán CA. Characterization of SepL of enterohemorrhagic Escherichia coli. J Bacteriol. 2000;182:6490–6498. doi: 10.1128/jb.182.22.6490-6498.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lesnik EA, Sampath R, Levene HB, Henderson TJ, McNeil JA, Ecker DJ. Prediction of rho-independent transcriptional terminators in Escherichia coli. Nucleic Acids Res. 2001;29:3583–3594. doi: 10.1093/nar/29.17.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Pandit S, Deutscher MP. RNase G (CafA protein) and RNase E are both required for the 5' maturation of 16S ribosomal RNA. EMBO J. 1999;18:2878–2885. doi: 10.1093/emboj/18.10.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Deutscher MP. RNase E plays an essential role in the maturation of Escherichia coli tRNA precursors. RNA. 2002;8:97–109. doi: 10.1017/s1355838202014929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin-Chao S, Wei CL, Lin YT. RNase E is required for the maturation of ssrA RNA and normal ssrA RNA peptide-tagging activity. Proc Natl Acad Sci USA. 1999;96:12406–12411. doi: 10.1073/pnas.96.22.12406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loomis WP, Moseley SL. Translational control of mRNA processing in the F1845 fimbrial operon of Escherichia coli. Mol Microbiol. 1998;30:843–853. doi: 10.1046/j.1365-2958.1998.01117.x. [DOI] [PubMed] [Google Scholar]
- Lopez PJ, Marchand I, Joyce SA, Dreyfus M. The C-teminal half of RNase E, which organizes the Escherichia coli degradosome, participates in mRNA degradation but not rRNA processing in vivo. Mol Microbiol. 1999;33:188–199. doi: 10.1046/j.1365-2958.1999.01465.x. [DOI] [PubMed] [Google Scholar]
- Lundberg U, Altman S. Processing of the precursor to the catalytic RNA subunit of RNase P from Escherichia coli. RNA. 1995;1:327–334. [PMC free article] [PubMed] [Google Scholar]
- Luo W, Donnenberg MS. Analysis of the function of enteropathogenic Escherichia coli EspB by random mutagenesis. Infect Immun. 2006;74:810–820. doi: 10.1128/IAI.74.2.810-820.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackie GA, Parsons GD. Tandem promoters in the gene for ribosomal protein S20. J Biol Chem. 1983;258:7840–7846. [PubMed] [Google Scholar]
- Marcaida MJ, DePristo MA, Chandran V, Carpousis AJ, Luisi BF. The RNA degradosome: life in the fast lane of adaptive molecular evolution. Trends Biochem Sci. 2006;31:359–365. doi: 10.1016/j.tibs.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Mathews DH, Sabina J, Zucker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- Mattoo S, Alto NM, Dixon JE. Subversion of myosin function by E. coli. Dev Cell. 2008;14:8–10. doi: 10.1016/j.devcel.2007.12.016. [DOI] [PubMed] [Google Scholar]
- McDaniel TK, Jarvis KG, Donnenberg MS, Kaper JB. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc Natl Acad Sci USA. 1995;92:1664–1668. doi: 10.1073/pnas.92.5.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowall KJ, Hernandez RG, Lin-Chao S, Cohen SN. The ams-1 and rne-3071 temperature-sensitive mutations in the ams gene are in close proximity to each other and cause substitutions within a domain that resembles a product of the Escherichia coli mre locus. J Bacteriol. 1993;175:4245–4249. doi: 10.1128/jb.175.13.4245-4249.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowall KJ, Lin-Chao S, Cohen SN. A+U content rather than a particular nucleotide order determines the specificity of RNase E cleavage. J Biol Chem. 1994;269:10790–10796. [PubMed] [Google Scholar]
- Mellies JL, Elliott SJ, Sperandio V, Donnenberg MS, Kaper JB. The Per regulon of enteropathogenic Escherichia coli: identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler) Mol Microbiol. 1999;33:296–306. doi: 10.1046/j.1365-2958.1999.01473.x. [DOI] [PubMed] [Google Scholar]
- Ménard R, Sansonetti PJ, Parsot C. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J Bacteriol. 1993;175:5899–5906. doi: 10.1128/jb.175.18.5899-5906.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita T, Maki K, Aiba H. RNase E-based ribonucleoprotein complexes: mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes Dev. 2005;19:2176–2186. doi: 10.1101/gad.1330405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudd EA, Krisch HM, Higgins CF. RNase E, an endoribonuclease, has a general role in the chemical decay of Escherichia coli mRNA: evidence that rne and ams are the same genetic locus. Mol Microbiol. 1990;4:2127–2135. doi: 10.1111/j.1365-2958.1990.tb00574.x. [DOI] [PubMed] [Google Scholar]
- Nataro JP, Kaper JB. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11:142–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves BC, Mundy R, Petrovska L, Dougan G, Knutton S, Frankel G. CesD2 of enteropathogenic Escherichia coli is a second chaperone for the type III secretion translocator protein EspD. Infect Immun. 2003;71:2130–2141. doi: 10.1128/IAI.71.4.2130-2141.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson P, Uhlin BE. Differential decay of a polycistronic Escherichia coli transcript is initiated by RNase E-dependent endonucleolytic processing. Mol Microbiol. 1991;5:1791–1799. doi: 10.1111/j.1365-2958.1991.tb01928.x. [DOI] [PubMed] [Google Scholar]
- Nilsson P, Naureckiene S, Uhlin BE. Mutations affecting mRNA processing and fimbrial biogenesis in the Escherichia coli pap operon. J Bacteriol. 1996;178:683–690. doi: 10.1128/jb.178.3.683-690.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojima T, Lin AC, Fujii T, Endo I. Determination of the termination efficiency of the transcription terminator using different fluorescent profiles in green fluorescent protein mutants. Anal Sci. 2005;21:1479–1481. doi: 10.2116/analsci.21.1479. [DOI] [PubMed] [Google Scholar]
- Nougayrède JP, Donnenberg MS. Enteropathogenic Escherichia coli EspF is targeted to mitochondria and is required to initiate the mitochondrial death pathway. Cell Microbiol. 2004;6:1097–1111. doi: 10.1111/j.1462-5822.2004.00421.x. [DOI] [PubMed] [Google Scholar]
- Nudler E, Gottesman ME. Transcription termination and anti-termination in E. coli. Genes Cells. 2002;7:755–768. doi: 10.1046/j.1365-2443.2002.00563.x. [DOI] [PubMed] [Google Scholar]
- O’onnell CB, Creasey EA, Knutton S, Elliott S, Crowther LJ, Luo W, et al. SepL, a protein required for enteropathogenic Escherichia coli type III translocation, interacts with secretion component SepD. Mol Microbiol. 2004;52:1613–1625. doi: 10.1111/j.1365-2958.2004.04101.x. [DOI] [PubMed] [Google Scholar]
- Ow MC, Liu Q, Kushner SR. Analysis of mRNA decay and rRNA processing in Escherichia coli in the absence of RNase E-based degradosome assembly. Mol Microbiol. 2000;38:854–866. doi: 10.1046/j.1365-2958.2000.02186.x. [DOI] [PubMed] [Google Scholar]
- Perna NT, Mayhew GF, Pósfai G, Elliott S, Donnenberg MS, Kaper JB, Blattner FR. Molecular evolution of a pathogenicity island from enterohemorrhagic Escherichia coli O157:H7. Infect Immun. 1998;66:3810–3817. doi: 10.1128/iai.66.8.3810-3817.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perna NT, Plunkett G, 3rd, Burland V, Mau B, Glasner JD, Rose DJ, et al. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001;409:529–533. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- Phadtare S, Kazakov T, Bubunenko M, Court DL, Pestova T, Severinov K. Transcription antitermination by translation initiation factor IF1. J Bacteriol. 2007;189:4087–4093. doi: 10.1128/JB.00188-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pósfai G, Koob MD, Kirkpatrick HA, Blattner FR. Versatile insertion plasmids for targeted genome manipulations in bacteria: isolation, deletion, and rescue of the pathogenicity island LEE of the Escherichia coli O157:H7 genome. J Bacteriol. 1997;179:4426–4428. doi: 10.1128/jb.179.13.4426-4428.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamoorthy R, McClain NA, Gautam A, Scholl-Meeker D. Expression of the bmpB gene of Borrelia burgdorferi is modulated by two distinct transcription termination events. J Bacteriol. 2005;187:2592–2600. doi: 10.1128/JB.187.8.2592-2600.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Régnier P, Hajnsdorf E. Decay of mRNA encoding ribosomal protein S15 of Escherichia coli is initiated by an RNase E-dependent endonucleolytic cleavage that removes the 3’ stabilizing stem and loop structure. J Mol Biol. 1991;217:283–292. doi: 10.1016/0022-2836(91)90542-e. [DOI] [PubMed] [Google Scholar]
- Reynolds R, Bermúdez-Cruz RS, Chamberlin MJ. Parameters affecting transcription termination by Escherichia coli RNA polymerase. I. Analysis of 13 Rho-independent terminators. J Mol Biol. 1992;224:31–51. doi: 10.1016/0022-2836(92)90574-4. [DOI] [PubMed] [Google Scholar]
- Roe AJ, Yull H, Naylor SW, Woodward MJ, Smith DG, Gally DL. Heterogeneous surface expression of EspA translocon filaments by Escherichia coli O157:H7 is controlled at the posttranscriptional level. Infect Immun. 2003;71:5900–5909. doi: 10.1128/IAI.71.10.5900-5909.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: a laboratory manual. 3rd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 2001. [Google Scholar]
- Sawers RG. Evidence for novel processing of the anaerobically inducible dicistronic focA-pfl mRNA transcript in Escherichia coli. Mol Microbiol. 2005;58:1441–1453. doi: 10.1111/j.1365-2958.2005.04915.x. [DOI] [PubMed] [Google Scholar]
- Sekiya K, Ohishi M, Ogino T, Tamano K, Sasakawa C, Abe A. Supermolecular structure of the enteropathogenic Escherichia coli type III secretion system and its direct interaction with the EspA-sheath-like structure. Proc Natl Acad Sci USA. 2001;98:11638–11643. doi: 10.1073/pnas.191378598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma VK, Zuerner RL. Role of hha and ler in transcriptional regulation of the esp operon of enterohemorrhagic Escherichia coli O157:H7. J Bacteriol. 2004;186:7290–7301. doi: 10.1128/JB.186.21.7290-7301.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw RK, Daniell S, Ebel F, Frankel G, Knutton S. EspA filament-mediated protein translocation into red blood cells. Cell Microbiol. 2001;3:213–222. doi: 10.1046/j.1462-5822.2001.00105.x. [DOI] [PubMed] [Google Scholar]
- Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- Spickler C, Stronge V, Mackie GA. Preferential cleavage of degradative intermediates of rpsT mRNA by the Escherichia coli RNA degradosome. J Bacteriol. 2001;183:1106–1109. doi: 10.1128/JB.183.3.1106-1109.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su MS, Kao HC, Lin CN, Syu WJ. Gene l0017 encodes a second chaperone for EspA of enterohaemorrhagic Escherichia coli O157:H7. Microbiology. 2008;154:1094–1103. doi: 10.1099/mic.0.2007/013946-0. [DOI] [PubMed] [Google Scholar]
- Tamura M, Lee K, Miller CA, Moore CJ, Shirako Y, Kobayashi M, Cohen SN. RNase E maintenance of proper FtsZ/FtsA ratio required for nonfilamentous growth of Escherichia coli cells but not for colony-forming ability. J Bacteriol. 2006;188:5145–5152. doi: 10.1128/JB.00367-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taraseviciene L, Björk GR, Uhlin BE. Evidence for an RNA binding region in the Escherichia coli processing endoribonuclease RNase E. J Biol Chem. 1995;270:26391–26398. doi: 10.1074/jbc.270.44.26391. [DOI] [PubMed] [Google Scholar]
- Telesnitsky A, Chamberlin MJ. Terminator-distal sequences determine the in vitro efficiency of the early terminators of bacteriophages T3 and T7. Biochemistry. 1989;28:5210–5218. doi: 10.1021/bi00438a044. [DOI] [PubMed] [Google Scholar]
- Tomcsányi T, Apirion D. Processing enzyme ribonuclease E specifically cleaves RNA I. An inhibitor of primer formation in plasmid DNA synthesis. J Mol Biol. 1985;185:713–720. doi: 10.1016/0022-2836(85)90056-7. [DOI] [PubMed] [Google Scholar]
- Viswanathan VK, Koutsouris A, Lukic S, Pilkinton M, Simonovic I, Simonovic M, Hecht G. Comparative analysis of EspF from enteropathogenic and enterohemorrhagic Escherichia coli in alteration of epithelial barrier function. Infect Immun. 2004;72:3218–3227. doi: 10.1128/IAI.72.6.3218-3227.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters M, Sperandio V. Autoinducer 3 and epinephrine signaling in the kinetics of locus of enterocyte effacement gene expression in enterohemorrhagic Escherichia coli. Infect Immun. 2006;74:5445–5455. doi: 10.1128/IAI.00099-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Roe AJ, McAteer S, Shipston MJ, Gally DL. Hierarchal type III secretion of translocators and effectors from Escherichia coli O157:H7 requires the carboxy terminus of SepL that binds to Tir. Mol Microbiol. 2008 doi: 10.1111/j.1365-2958.2008.06377.x. [DOI] [PubMed] [Google Scholar]
- Wilson RK, Shaw RK, Daniell S, Knutton S, Frankel G. Role of EscF, a putative needle complex protein, in the type III protein translocation system of enteropathogenic Escherichia coli. Cell Microbiol. 2001;3:753–762. doi: 10.1046/j.1462-5822.2001.00159.x. [DOI] [PubMed] [Google Scholar]
- Yang J, Jain C, Schesser K. RNase E regulates the Yersinia type 3 secretion system. J Bacteriol. 2008;190:3774–3778. doi: 10.1128/JB.00147-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]