Abstract
The complete genome consensus sequence was determined for avian paramyxovirus (APMV) serotype 9 prototype strain PMV-9/domestic Duck/New York/22/78. The genome is 15,438 nucleotides (nt) long and encodes six non-overlapping genes in the order of 3′-N-P/V/W-M-F-HN-L-5′ with intergenic regions of 0–30 nt. The genome length follows the “rule of six” and contains a 55-nt leader sequence at the 3′ end and a 47-nt trailer sequence at the 5′ end. The cleavage site of the F protein is I-R-E-G-R-I↓F, which does not conform to the conventional cleavage site of the ubiquitous cellular protease furin. The virus required exogenous protease for in vitro replication and grew only in a few established cell lines, indicating a restricted host range. Alignment and phylogenetic analysis of the predicted amino acid sequences of APMV-9 proteins with the cognate proteins of viruses of all five genera of family Paramyxoviridae showed that APMV-9 is more closely related to APMV-1 than to other APMVs. The mean death time in embryonated chicken eggs was found to be more than 120 h, indicating APMV-9 to be avirulent for chickens.
Keywords: APMV, Avian paramyxovirus, Paramyxoviridae, Paramyxovirinae, Genome sequence
1. Introduction
The family Paramyxoviridae includes a large group of viruses isolated from a wide variety of animal species. Many members of this family cause important human and animal diseases, while the disease potential of many other members is not known. The family Paramyxoviridae is divided into two subfamilies: Paramyxovirinae and Pneumovirinae. The subfamily Paramyxovirinae comprises five genera: Rubulavirus (including mumps viruses [MuV], human parainfluenza virus types [HPIV 2 and 4], simian virus type 5 [SV5]), Respirovirus (including Sendai viruses [SeV], human parainfluenza virus types [HPIV 1 and 3]), Morbillivirus (including measles virus [MeV] and canine distemper virus [CDV]), Henipavirus (including Hendra [HeV] and Nipah [NiV] viruses), and Avulavirus (including nine serotypes of avian paramyxoviruses [APMV-1–9]). Subfamily Pneumovirinae is divided into two genera Pneumovirus (human respiratory syncytial virus [HRSV] and bovine respiratory syncytial virus [BRSV]), and Metapneumovirus (human metapneumovirus [HMPV] and avian metapneumovirus [AMPV]) (Lamb et al., 2005).
Members of the family Paramyxoviridae are pleomorphic, enveloped viruses containing a single segment of negative-sense RNA genome. The genome is 13–19 kb in length and contains 6–10 tandemly linked genes that encode up to 12 different proteins (Lamb and Parks, 2007). The viral genes are flanked by highly conserved gene-start (GS) and gene-end (GE) sequences that serve as transcription start and stop signals, respectively. At the 3′ and 5′ ends of the genome are short extragenic leader and trailer regions, respectively. The viral RNA polymerase begins transcription from the 3′ leader sequence and proceeds downstream in a sequential manner producing individual mRNAs by a start–stop mechanism guided by the GS and GE signals. During RNA replication, the GS and GE signals are ignored and a complimentary copy of the genome (antigenome) is synthesized, which serves as the template for synthesis of the progeny genome.
All members of family Paramyxoviridae encode a nucleoprotein (N), a phosphoprotein (P), a matrix protein (M), a fusion protein (F), an attachment protein called hemagglutinin (H) or haemagglutinin-neuraminidase (HN) or glycoprotein (G), and a large polymerase protein (L) (Lamb and Parks, 2007). The N protein protects the viral genomic and antigenomic RNAs by binding to their entire lengths, forming highly stable nucleocapsids that function in transcription, replication and virus assembly. The P protein acts as a polymerase co-factor and is required for RNA synthesis (Curran et al., 1995). The M protein is the most abundant protein in the virion and lines the inner face of the viral envelope (Lamb and Choppin, 1977) and plays a major role in viral morphogenesis (Peeples, 1991). The HN glycoprotein binds to sialic acid-containing cell surface receptors and facilitates virus penetration. The F glyco-protein mediates virus penetration by inducing fusion between the viral envelope and host cell plasma membrane. The large protein (L) is the viral RNA-dependent RNA polymerase. Most members of subfamily Paramyxovirinae encode two additional proteins, V and W (or I, in the case of Rubulavirus and Newcastle disease virus [NDV] of Avulavirus), from the P gene by a mechanism called RNA editing. The V protein, which contains a cysteine-rich, C-terminal, zinc finger-like domain, has roles in regulating RNA synthesis and in blocking the host type I interferon response (Goodbourn et al., 2000; Lin et al., 2005).
Paramyxoviruses have been isolated from many different avian species throughout the world. Currently, all avian members of family Paramyxoviridae, except avian metapneumovirus, have been placed in the genus Avulavirus and designated as APMVs. APMVs have been classified into nine different serotypes (APMV-1–9) based on hemagglutination inhibition (HI) and neuraminidase inhibition (NI) assays (Alexander, 2003). NDV comprises APMV-1. Since NDV causes high mortality in chickens and the greatest economic loss to the poultry industry, it is the well-characterized virus in the genus Avulavirus. However, APMV-2 and APMV-3 have been reported to cause mild disease in poultry (Alexander, 1982; Beck et al., 2003). The pathogenicity of APMV-4–9 is generally unknown (Alexander, 1982). Due to the absence of distinct clinical signs and symptoms in chickens, it is difficult to detect APMV-2–9 infections in field conditions. The lack of proper diagnostic tests against APMV-2–9 hinders investigation of their epidemiology and economic impact on the poultry industry. APMV-9 strain PMV-9/domestic Duck/New York/22/78 was initially isolated from a domestic duck during a routine surveillance program in New York (Sandhu and Hinshaw, 1981). This virus was found to be serologically and antigenically distinct from the other previously known APMV serotypes and was designated as APMV-9. Since then, many APMV-9 strains from different parts of the world have been isolated, but strain PMV-9/domestic Duck/New York/22/78 serves as the prototype strain for this serotype.
An understanding of the molecular characteristics and pathogenicity of APMV-2–9 is not only of general interest, but is also important for developing vaccines and diagnostic tests against these viruses. To date, the complete genome sequences for APMV-1 (Krishnamurthy and Samal, 1998), APMV-2 (Subbiah et al., 2008), APMV-3 (Kumar et al., 2008), APMV-4 (Nayak et al., 2008) and APMV-6 (Chang et al., 2001) have been published. As an initial step towards understanding the molecular biology and pathogenicity of APMV-9, we have determined the complete genome sequence of the APMV-9 prototype strain PMV-9/domestic Duck/New York/22/78 (GeneBank accession no. EU910942). The sequence information of strain PMV-9/domestic Duck/New York/22/78 was compared with those of other APMV serotypes and other paramyxoviruses in order to determine its phylogenetic relationship. Our results provide valuable insight into the molecular characteristics and pathogenicity of APMV-9.
2. Materials and methods
2.1. Virus and cells
APMV-9 strain PMV-9/domestic duck/NewYork/22/78 was obtained from the National Veterinary Services Laboratory, Ames, Iowa. The virus was propagated in the allantoic cavity of 9-day-old specific pathogen free (SPF) embryonated chicken eggs (ECE). Allantoic fluid from the infected eggs was harvested 96 h post-infection and the virus titer was calculated by a hemagglutination assay (HA) using 0.5% v/v suspension of chicken erythrocytes at room temperature (Alexander, 2008). The chicken embryo fibroblast (DF-1), Madin-Darby Canine Kidney (MDCK), human epidermoid carcinoma (HEp-2), Baby Hamster Kidney (BHK 21), Bovine Turbinate (BTu), Pig Kidney (PK15), Quail fibrosarcoma (QT35), Rabbit Kidney cells (RK13), African green monkey kidney (Vero), Madin-Darby Bovine Kidney (MDBK), and duck embryo (CCL-141) cell lines were obtained from the American Type Culture Collection (ATCC, Manassas, VA). Primary cultures of chicken embryo fibroblast (CEF) and chicken embryo kidney (CEK) cells were prepared in our laboratory using 7- and 20-day-old ECE, respectively, using standard procedures. The cells were grown in Dulbecco’s minimum essential medium containing 10% fetal calf serum.
2.2. Requirement of exogenous protease
Virus propagation in the cell lines and primary cell cultures listed above was carried out with cell culture medium (without fetal calf serum) containing or lacking 10% v/v allantoic fluid, 1–5 μg/ml of acetyl trypsin/ml (Gibco), or 1–5 μg/ml of α-chymotrypsin (Sigma). The cell lines supporting viral growth were observed by corroborating cytopathic effects (CPE) in cells and HA titer in cell culture supernatant both before and after disrupting the cells by 3 cycles of freeze-thawing. The ability of the virus to produce plaques was tested in the different cell culture systems using 1% methyl-cellulose overlay medium (lacking fetal calf serum) in either the presence or absence of exogenous protease.
2.3. Virus RNA isolation and sequence analysis
Viral RNA was isolated from the allantoic fluid of infected eggs using RNeasy kit according to the manufacturers instructions (QIA-GEN, USA). As a first step, degenerate consensus PCR primers were designed using published Avulavirus, Rubulavirus, Respirovirus and Morbillivirus genome sequences (accession numbers given below) from the National Center for Biotechnology Information (NCBI). The N, P, M, F, HN and L nucleotide sequences were aligned, and highly conserved sequence segments were identified and used to design minimally degenerate primers of 10–12 nucleotides (nt) in length. Reverse transcription (RT) was performed using Superscript RT (Invitrogen, USA) primer CF1, 5′-RTANAGGRGWA-3′ (which would prime at APMV-9 nt 5939–5949, within the F gene) and the resulting first strand cDNA was PCR-amplified (Recombinant DNA Taq polymerase, Invitrogen, USA) using the same degenerate forward primer along with a mixture of three degenerate consensus reverse primers: CR1, 5′-RGCNCCAASCTG-3′ (which would prime at APMV-9 nt 6751–6762, within the HN gene), CR2, 5′-NRAAGCSYNAGC-3′ (which would prime at APMV-9 nt 6767–6778, within the HN gene), and CR3, 5′-SYNKYCAAAGCG-3′ (which would prime at APMV-9 nt 6784–6795, within the HN gene). PCR was carried out using a cycling sequence of 94 °C for 4 min followed by 30 cycles of 94 °C for 1 min, 40 °C for 1 min and 72 °C for 1.5 min, which was then followed by final extension of 72 °C for 10 min. This produced a PCR product of 800 bp that was cloned and sequenced as described below. The resulting sequence indicated that this fragment spanned the end of the F gene and the beginning of the HN gene.
From this sequence, the forward GS 5′-ATTATACGGGTAGAA-3′ and reverse GE 5′-TTTTTTCTAAAATTT-3′ primers were designed based on the APMV-9 HN GS and F GE sequence motifs. Their subsequent use as RT and PCR primers amplified the remainder of the genome except for the L gene. The L gene was reverse transcribed using the GS forward primer, and PCR was performed with the same forward primer and a mixture of four different degenerate consensus reverse primers designed to bind at increasing intervals within the L gene: CR4, 5′-NKCWAGGARTTC-3′ (which would bind to APMV-9 nt 10,214–10,225), CR5, 5′-KNACTCNYTGAG-3′ (which would bind to APMV-9 nt 11,673–11,684), CR6, 5′-NCTKGTATCCGC-3′ (which would bind to APMV-9 nt 13,143–13,154), and CR7, 5′-RKYRYSTCTTGC-3′ (which would bind to APMV-9 nt 14,608–146,198). This yielded a complex mixture of PCR products reflecting amplification involving different reverse primers. The products were gel purified, cloned and sequenced as described below. This showed that they covered almost the entire length of the L gene. The remaining regions of the L gene were completed using the gene-specific primers designed from the above known sequences. The sequences of 3′ and 5′ termini of the genome were determined by rapid amplification of cDNA ends (3′ and 5′ RACE, respectively) from the viral RNA using manufacturer’s protocol (Invitrogen, USA) with modification from Subbiah et al. (2008). All the PCR products were cloned into TOPO TA cloning vector (Invitrogen, USA) and positive clones were sequenced using M13 forward and M13 reverse primers. DNA sequencing was performed in a 3130xl genetic analyzer (ABbiosystem, USA) according to the manufacturer’s instruction. The entire genome sequence was determined twice: once by analysis of cloned cDNAs and once by direct sequencing of uncloned RT-PCR products, providing a consensus sequence.
2.4. Sequence and phylogenetic tree analysis
Primer design, sequence analysis, BLAST searches, and prediction of ORFs were carried out using the SEQMAN and EDITSEQ programs, PCR primers were also designed using the PRIMER SELECT program in DNASTAR Lasergene 7 (software suite for sequence analysis, version 7.2.1 410). Construction of a phylogenetic tree and divergence analysis was performed using the Clustal W multiple alignment algorithm of MEGALIGN program.
2.5. Database accession numbers
The complete genome sequence of APMV-9 strain PMV-9/domestic Duck/New York/22/78 was submitted in GenBank (accession number EU910942). The accession numbers of other viral sequences used for sequence comparison and primer designing are as follows: EU338414 for APMV-2, EU403085 for APMV-3, FJ177514 for APMV-4, NC_003043 for APMV-6, NC_002617 for APMV-1, NC_003443 for HPIV2, NC_006430 for SV5, NC_004074 for Tioman virus, NC_001552 for SeV, NC_001796 for HPIV3, NC_003461 for HPIV1, AY988601 for NiV, NC_001906 for HeV, NC_001498 for MeV, NC_001921 for CDV and NC_007454 for J virus.
3. Results
3.1. Growth characteristics of APMV-9
APMV-9 yielded a titer of 26–28 HA units per ml of allantoic fluid in 9-day-old SPF-ECE at 3 days post-inoculation (DPI). Eleven different cell lines and two primary cell cultures were evaluated to determine a suitable system that can support the growth of APMV-9. It was necessary to include 10% allantoic fluid for replication of the virus in cell culture, indicating a requirement of external proteases for efficient cleavage of the F protein. However, the requirement for allantoic fluid could not be met by chymotrypsin or by trypsin. The virus grew efficiently in primary CEK cells but not in CEF cells. Three cell lines (DF-1, Vero and BHK21) were also found to support the growth of APMV-9. The typical CPE observed was rounding and detachment of cells. Syncytia formation in the virus-infected cells was also observed. The peak HA titers (per ml) of the virus in different cell lines tested were: CEK; 24, BHK21; 24, Vero; 22 and DF-1; 22 at 72 h post-infection. The virus failed to produce visible plaques under methylcellulose overlay medium in the presence or absence of exogenous protease. Examination of negatively stained virions by electron microscopy showed that the particles were enveloped, pleomorphic, and ranged in diameter from 150 to 250 nm, thus displaying morphological characteristics typical of the family Paramyxoviridae (data not shown). The pathogenicity of APMV-9 virus to ECE was determined using a mean death time (MDT) assay in 9-day-old ECE (Alexander, 2008). The MDT of APMV-9 in ECE was more than 120 h, indicating its avirulent nature for chickens.
3.2. Determination of the complete genome sequence of APMV-9
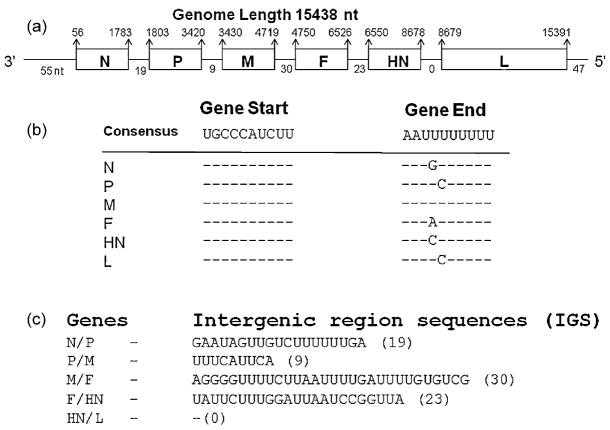
The genome of APMV-9 consists of 15,438 nt (GenBank accession no. EU910942), a length that is average for the members of Respirovirus, Morbillivirus, Rubulavirus, and Avulavirus sequenced to date (Wang et al., 2007). The genome length is a multiple of six, conforming to the ‘rule of six’ common to members of subfamily Paramyxovirinae (Calain and Roux, 1993; Samal and Collins, 1996). The viral genes are arranged in the order of 3′-N-P-M-F-HN-L-5′, similar to APMV-1–4. The length, position and characteristics of the six genes are listed in Table 1. The percentage of the genome that encodes protein is 90%, which is similar to the average coding percentage (92%) of other members of subfamily Paramyxovirinae (Miller et al., 2003). The genes are flanked by extragenic leader sequences at the 3′ end and the trailer sequences at the 5′ end.
Table 1.
Genomic features and protein characteristics of APMV-9.
| Genes | Hexamer phasing Position at gene start | mRNA characteristics (nt) |
Intergenic Sequence (nt) | Deduced protein characteristics |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Total length | 5′ UTR | ORF | 3′ UTR | Size (aa) | MW (kDa)a | p/ | |||
| N | 2 | 1728 | 66 | 1470 | 192 | 19 | 489 | 53.8 | 5.0 |
| P/V (P) | 3 | 1618 | 113 | 1260 | 245 | 9 | 419 | 45.5 | 6.4 |
| P/V (V) | 3 | 1619 | 113 | 792 | 714 | – | 263 | 28.5 | 6.6 |
| P/V (W) | 3 | 1620 | 113 | 585 | 922 | – | 194 | 21.2 | 7.4 |
| M | 4 | 1290 | 34 | 1095 | 161 | 30 | 364 | 40.0 | 9.5 |
| F | 4 | 1777 | 55 | 1656 | 66 | 23 | 551 | 58.5 | 6.5 |
| HN | 4 | 2129 | 97 | 1740 | 292 | 0 | 579 | 63.7 | 6.7 |
| L | 3 | 6713 | 11 | 6633 | 69 | – | 2210 | 250.2 | 7.4 |
Molecular weights indicated do not include any posttranslational modifications to the proteins.
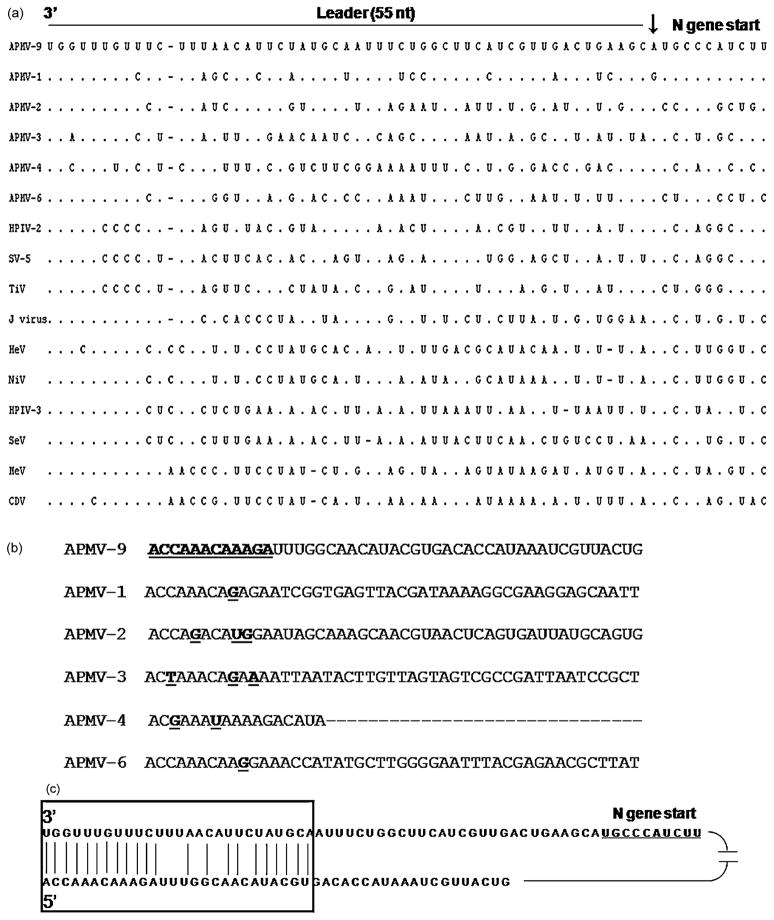
The 3′ leader sequence of APMV-9 is 55 nt, a sequence length that is conserved among most members of subfamily Paramyxovirinae (Fig. 1a). The first 8 nt of the leader sequence (3′ UGGUUUGU-5′) are exactly conserved among Avulavirus (APMV -1, -2 and -6), Respirovirus (HPIV3 and SeV), and the unclassified J virus (Fig. 1a). The first 4 nt of the leader sequence (3′-UGGU-5′) were the same for all of the members of Avulavirus sequenced to date except for a change in the third position, where the residue G was replaced with residue A in APMV-3 and residue C in APMV-4. The length of the APMV-9 5′ trailer sequence is 47 nt; the length of the trailer region has considerable variability among the members of subfamily Paramyxovirinae (Fig. 1b). The last 12 nt of the 5′ trailer region were highly conserved with other members of genus Avulavirus except for a 1 nt difference compared to APMV-6 and -1, 2 nt differences compared to APMV-4, and 3 nt changes compared to APMV-2 and -3 (Fig. 2b). In APMV-9, the first 12 nt sequences of the 3′ leader and the last 12 nt of 5′ trailer sequences showed 100% complementarity, suggesting the presence of conserved elements in the 3′ promoter regions of the genome and antigenome (Fig. 1c).
Fig. 1.
Sequences of the APMV-9 3′ leader and 5′ trailer regions. (a) Alignment of the 3′ leader and N gene start sequences of APMV-9 with those of selected paramyxoviruses. Dots indicate identity with the assignment in APMV-9. (b) Comparison of the 5′ trailer region of APMV-9 with those of other APMVs. Bold underlined letters indicate dissimilarity of nt assignments among APMVs. (c) Complementarity between the 3′ leader and 5′ trailer regions of APMV-9. All sequences are in negative-sense. Abbreviations; SV5, Simian virus 5; APMV, Avian paramyxovirus; HPIV, Human parainfluenza virus; TiV, Tioman virus; SeV, Sendai virus; NiV, Nipah virus; HeV, Hendra virus; MeV, Measles virus; CDV, Canine distemper virus.
Fig. 2.
APMV-9 gene order, putative transcription signals, and IGS. (a) Schematic representation of the APMV-9 genome with the nt coordinates of the genes shown above the line and the lengths of the 3′ leader, 5′ trailer, and IGS shown below the line. (b) APMV-9 gene-start and gene-end sequence motifs. (c) APMV-9 IGS. All sequences are in negative-sense.
The GS sequence of APMV-9 is 3 ′-UGCCCAUCUU-5 ′, which is exactly conserved among all the six genes of APMV-9 (Fig. 2b). The GE sequence is 3 ′-AA(U)6, which is less conserved among different viral genes, with 1 nt variation in all genes except the M gene (Fig. 2b). IGS of APMV-9 varied in length between 0 and 30 nt (Table 1, Fig. 2a and c). The IGS between N/P, P/M, M/F, F/HN, HN/L are 19, 9, 30, 23 and 0 nt, respectively. There was no evidence of a conserved IGS sequence motif. An IGS was absent between HN/L, which is different than other avulaviruses where the HN/L IGS vary from 3 to 63 nt. The hexamer phasing pattern was 2 for the N gene, 3 for the P and L genes, and 4 for the M, F and HN genes (Table 1).
3.2.1. The nucleoprotein (N) gene
The N gene of APMV-9 is 1728 nt long and encodes a N protein of 489 amino acids with a predicted (molecular weight) MW at 53.8 kDa (Table 1). The N protein has a highly-conserved sequence motif (conserved amino acids are underlined) 322-FAPAEYAQLYSYAMG-336, which corresponds to the F-X4-Y-X3-α-S-Y- α-AMG motif, where X is any amino acid and α is any aromatic amino acid. This motif has been identified in the central region of N protein of the other members of Paramyxovirinae and plays a role in self assembly of N with RNA or with other N monomers (Lamb and Parks, 2007). The N protein has an amino acid sequence identity of 33–66% with Avulavirus, 30% with Rubulavirus, 22% with Morbillivirus, 25% with Henipavirus, 19% with Respirovirus, and 24% with J virus (Table 2).
Table 2.
Amino acid sequence identity between APMV-9 and other members of Paramyxoviridae.
| N | P | M | F | HN | L | |
|---|---|---|---|---|---|---|
| Avulavirus | ||||||
| APMV-1 | 66.7 | 42.1 | 57.4 | 55.2 | 61.5 | 60.5 |
| APMV-2 | 38.0 | 21.2 | 27.7 | 36.5 | 32.5 | 38.9 |
| APMV-3 | 34.6 | 21.7 | 25.3 | 28.3 | 35.6 | 33.2 |
| APMV-4 | 32.9 | 21.9 | 24.7 | 28.9 | 34.0 | 31.0 |
| APMV-6 | 37.4 | 19.0 | 29.7 | 38.1 | 30.6 | 39.8 |
| Rubulavirus | ||||||
| HPIV2 | 29.7 | 16.9 | 25.3 | 26.5 | 31.1 | 36.0 |
| SV5 | 29.9 | 19.0 | 27.2 | 29.0 | 32.8 | 35.7 |
| TiV | 31.7 | 18.6 | 28.6 | 25.0 | 15.9 | 35.7 |
| Unclassified virus | ||||||
| J virus | 23.9 | 12.9 | 16.5 | 23.0 | 21.8 | 25.7 |
| Henipavirus | ||||||
| HeV | 24.9 | 11.7 | 16.8 | 24.1 | 12.6 | 25.1 |
| NiV | 25.2 | 12.6 | 16.2 | 24.3 | 14.0 | 25.9 |
| Respirovirus | ||||||
| HPIV3 | 17.4 | 10.7 | 16.2 | 22.9 | 20.4 | 26.9 |
| SeV | 20.4 | 10.5 | 15.1 | 23.8 | 20.4 | 26.2 |
| Morbillivirus | ||||||
| CDV | 21.9 | 11.7 | 15.4 | 25.0 | 11.6 | 26.2 |
| MeV | 22.5 | 11.9 | 17.0 | 23.2 | 10.9 | 25.6 |
3.2.2. The phosphoprotein (P) gene
The P gene of APMV-9 is 1618 nt long and encodes a P protein of 419 amino acids with a predicted MW at 45.5 kDa (Table 1). The predicted P protein contains 20 serine and 9 threonine residues identified as possible phosphorylation sites using NetPhos 2.0 server (http://www.cbs.dtu.dk/). The APMV-9 P protein has an amino acid sequence identity of 19–42% with Avulavirus, 17–19% with Rubulavirus, and 10.5–13% with Morbillivirus, Henipavirus, Respirovirus, and J virus (Table 2). The P gene contains a putative RNA editing site 5′ AAAAAGGG-3′ (mRNA sense) at positions 406–414 nt of P gene (positions 2322–2329 nt in the complete genome sequence), which showed a high degree of sequence identity with editing sites of other members of subfamily Paramyxovirinae (Fig. 3). The insertion of a single G residue in the editing site would form V mRNA, which would produce a V protein of 263 aa with a predicted MW of 28.5 kDa (Table 1). The V protein contains seven conserved, invariantly spaced cysteine residues located between 218 and 249 aa, as reported for other members of subfamily Paramyxovirinae (Fig. 4). Alternatively, insertion of two G residues at the RNA editing site would produce a W mRNA that encodes a W protein of 194 aa, with a predicted MW at 21.2 kDa (Table 1).
Fig. 3.
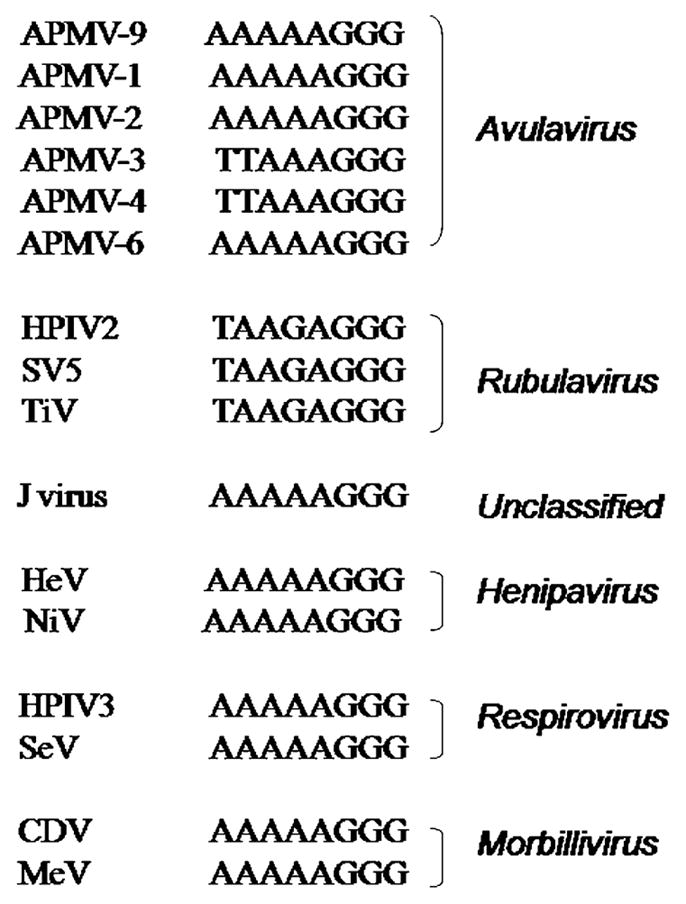
Comparison of the RNA editing site in the P gene of APMV-9 with those of selected members of subfamily Paramyxovirinae. Sequences are in positive (mRNA) sense.
Fig. 4.

Alignment of the amino acid sequence of the C-terminal end of the V protein of APMV-9 with those of different members of subfamily Paramyxovirinae. Conserved cysteine and tryptophan residues are indicated by stars. Numbers on either side of the sequence indicate the amino acid position.
3.2.3. The matrix (M) gene
The M gene of APMV-9 is 1290 nt long and encodes a M protein of 364 aa with a predicted MW at 40.0 kDa (Table 1). The M protein has an amino acid sequence identity of 24–57% with Avulavirus, 25–28% with Rubulavirus, 16.5% with J virus, 16% with Henipavirus, 15–16% with Respirovirus and 15–17% with Morbillivirus (Table 2).
3.2.4. The fusion (F) gene
The F gene of APMV-9 is 1777 nt long and encodes a F protein of 551 amino acids with a predicted MW at 58.5 kDa (Table 1). The F protein of APMV-9, like the F proteins of other paramyxoviruses, is predicted to be a type I integral membrane protein. Three potential transmembrane domains (TM) exist in the F protein, as predicted by TMHMM server v 2.0 (http://www.cbs.dtu.dk/). The first TM domain is the signal peptide at 1–20 aa, followed by the second at 113–135 aa, corresponding to the fusion peptide that becomes the N-terminus of the F1 subunit, and the third at 496–518 aa, corresponding to the predicted transmembrane anchor. The F protein (F0) becomes biologically active by being cleaved to F1 and F2 subunits by host cell proteases. The F protein has been predicted to have four potential N-linked glycosylation sites using NetNGlyc 1.0 Server (http://www.cbs.dtu.dk/). The F2 subunit has one potential glycosylation site at position 78 (NKT), and the F1 subunit has three potential N-linked glycosylation sites at positions 184 (NKT), 440 (NVS) and 464 (NSS). The putative F protein cleavage site of APMV-9 is I-R-E-G-R-I↓F (Fig. 5), which does not conform to the favored sequence motif for cleavage by the intracellular protease furin, R-X-K/R-R↓F (Hosaka et al., 1991). The APMV-9 fusion peptide contains the motif ALGVAT (amino acids 118–123) that is conserved among the family Paramyxoviridae. The F protein has an amino acid sequence identity of 28–55% with Avulavirus, 25–29% with Rubulavirus, and 23–25% with Morbillivirus, Henipavirus, Respirovirus, and J virus (Table 2).
Fig. 5.
Comparison of the F protein cleavage site of APMV-9 with those of different members of genus Avulavirus. Basic amino acid residues (R-Arginine, K-lysine) are indicated in bold.
3.2.5. The hemagglutination-neuraminidase (HN) gene
The HN gene of APMV-9 is 2129 nt long and encodes a HN protein of 579 amino acids with a predicted MW at 63.7 kDa (Table 1). Like other members of the subfamily Paramyxovirinae, the HN protein is predicted to be a type II integral membrane protein. The predicted HN protein posseses a TM domain located between 27 and 49 aa. There are five potential N-linked glycosylation sites at positions 119, 147, 341, 348 and 433, compared to APMV-2 and -4 (5 sites), APMV-1 (6 sites) and APMV-3 and -6 (7 sites). The potential sialic acid binding motif NRKSCS is located at 234–239 aa in the APMV-9 HN protein. The HN protein has an amino acid identity of 31–61% with Avulavirus, 16–33% with Rubulavirus, 11% with Morbillivirus, 13% with Henipavirus, 20% with Respirovirus, and 22% with J virus (Table 2).
3.2.6. The large polymerase (L) gene
The L gene of APMV-9 is 6713 nt long and encodes an L protein of 2210 amino acids with a predicted MW at 250.2 kDa (Table 1). The six highly conserved linear domains (I-VI) identified within the L proteins of nonsegmented negative-stranded RNA viruses (NSV) were also identified within the L protein of APMV-9 (Poch et al., 1990). The highly conserved domain GDNQ, which plays an important role during transcription of NSV (Schnell and Conzelmann, 1995), is located between 750 and 754 aa in domain III of APMV-9 L protein. Domain IV of NSV L proteins contains an ATP-binding motif (K-X18-21-G-X-G-X-G) and a similar conserved motif, K-X21-A-E-G-S-G, is located between 1759 and 1785 aa of APMV-9 L protein. The L protein has an amino acid identity of 31–60% with Avulavirus, 36% with Rubulavirus, and 26% with Morbillivirus, Henipavirus, Respirovirus and J virus, respectively (Table 2).
3.3. Phylogenetic analysis
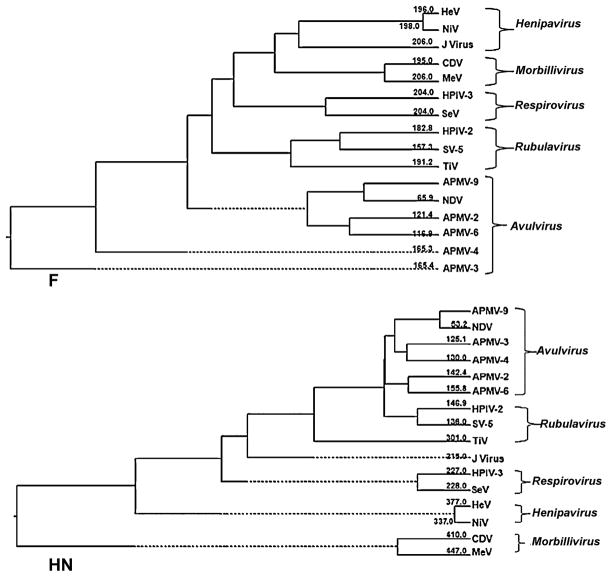
Phylogenetic trees were generated from amino acid sequence alignments of the N, P, M, F, HN and L proteins of APMV-9 with the cognate proteins of a selection of members of family Paramyxoviridae. In all of the phylogenetic trees, viruses representing the genera Avulavirus, Rubulavirus, Morbillivirus, Henipavirus, Respirovirus and J virus separated by genus into different clusters. In each tree, APMV-9 clustered with the other APMV serotypes, indicating a close genetic relationship among them. The representative phylogenetic trees generated from F and HN proteins are shown in Fig. 6. This showed that NDV was the virus most closely related to APMV-9.
Fig. 6.
Phylogenic analysis of the amino acid sequences of the F and HN proteins of APMV-9 and selected members of subfamily Paramyxovirinae. The numbers in branches represent the divergence of APMV-9 with other members of family Paramyxoviridae.
4. Discussion
The APMVs are commonly isolated from a number of avian species throughout the world. Until recently, very little was known about the pathogenicity and molecular characteristics of these viruses, apart from NDV (APMV-1). However, recent sequence analysis of prototypes of APMV-2–4 and -6 has provided molecular details of these viruses (Chang et al., 2001; Kumar et al., 2008; Nayak et al., 2008; Subbiah et al., 2008). APMV-9 is known to cause infections in ducks (Alexander et al., 1983). As a first step towards understanding the molecular characteristics of APMV-9, we have determined the complete genome sequence and growth characteristics of APMV-9. The MDT of APMV-9 in ECE was more than 120 h, indicating that the virus is most likely avirulent to chickens. APMV-9 grew only in selected cell lines, suggesting a narrow host range for this virus.
The genome of APMV-9 is 15,438 nt in length, which represents the average genome size of the members of Respirovirus, Morbillivirus, Rubulavirus and Avulavirus sequenced to date This sequence length is longer than those of APMV-2 (14,905 nt) (Subbiah et al., 2008), APMV-4 (15,054 nt) (Nayak et al., 2008) and APMV-1 (15,192 nt) (Krishnamurthy and Samal, 1998), but is shorter than those of APMV-3 (16,272 nt) (Kumar et al., 2008) and APMV-6 (16,232 nt) (Chang et al., 2001). The genome length of APMV-9 obeys the ‘rule of six’, which is a characteristic of subfamily Paramyxovirinae (Calain and Roux, 1993) but not of subfamily Pneumovirinae (Samal and Collins, 1996). The genome is composed of six genes (N, P, M, F, HN and L) that are present in all members of subfamily Paramyxovirinae, but APMV-9 lacks the SH gene that is present between the HN and F genes of APMV-6 (Chang et al., 2001). The leader sequence of APMV-9 is 55 nt long, a length that is generally conserved among members of subfamily Paramyxovirinae (Lamb and Parks, 2007). Furthermore, the first 12–13 nt in leader sequences of the members of Paramyxovirinae are usually more closely related within, rather than between, genera, and are used in the classification of new isolates. Consistent with this, the first 8 nt of the APMV-9 leader sequence were found to be conserved among APMV-1, -2, -6 and -9, consistent with a close evolutionary relationship among these viruses. APMV-3 and -4 have one nt change at position 3 and two nt changes at positions 3 and 7, respectively, indicating a diversity among the members of APMVs. The trailer sequence of APMV-9 is 47 nt long and is within the typical range (40–60 nt) for most members of family Paramyxoviridae (Shioda et al., 1986), but the recent identification of a 707 nt long trailer sequence in APMV-3 (Kumar et al., 2008) and a 17 nt short trailer sequence in APMV-4 (Nayak et al., 2008) has broadened the average length of trailer sequences in paramyxoviruses. However, the functional significance of these differences in the length of trailer sequences is still unknown. The first 12 nt of the leader sequence of APMV-9 are complementary to the last 12 nt sequence of the trailer region. This high degree of complementarity suggests conserved elements in the 3′ promoter regions of genome and antigenome.
The GS sequence of APMV-9 is identical to the GS sequence of APMV-1 except for 1 nt change in position 9, where residue U is replaced by C. The IGS of APMV-9 varies from 0 to 30 nt. The IGS length varies between 1 and 47 nt for APMV-1, 3–23 nt for APMV-2, 31–63 nt for APMV-3, 9–42 nt for APMV-4 and 2–63 nt for APMV-6. Thus, the lengths of the various IGS are not conserved among these different avulaviruses. APMV-9 is notable for the absence of an IGS between the HN and L genes. To the best of our knowledge, the only other instances of a lack of an IGS between two genes of a paramyxovirus involve the gene overlaps of HRSV and BRSV. The functional significance of the absence of IGS between the HN and L genes needs to be explored. The hexamer phasing positions of the GS of APMV-9 are, in order, 2,3,4,4,4,3, which are different from APMV-1 (2,4,4,4,3,6), APMV-2 (2,4,4,4,3,6), APMV-3 (2,5,5,2,2,1), APMV-4 (2,2,2,6,2,2) and APMV-6 (2,2,2,2,2,4,4) (Chang et al., 2001; Krishnamurthy and Samal, 1998; Kumar et al., 2008; Nayak et al., 2008; Subbiah et al., 2008). While the importance of the hexamer phasing positioning is well-known for cis-acting signals involved in RNA replication (Lamb and Parks, 2007), the importance of the phasing of transcription signals remains unclear.
The N proteins of paramyxoviruses have a conserved hydrophobic motif, F-X4-Y-X4-SYAMG, required during viral assembly and conserved among paramyxoviruses. This motif is conserved in the N protein of APMV-9. The conserved Y residue in the sequence motif is replaced by F in APMV-2 and -4 (Nayak et al., 2008; Subbiah et al., 2008). The P gene of most members of subfamily Paramyxovirinae is subject to an RNA editing mechanism that causes a frame shift that accesses additional internal ORFs; HPIV1 of genus Respirovirus is an exception that does not execute RNA editing. There are two patterns of editing within the subfamily. All viruses, except for members of genus Rubulavirus, express the P protein from the unedited mRNA and express the V protein from an mRNA that was edited to insert a single non-templated G residue. The insertion of two G residues results in an mRNA encoding the W, I or D protein, depending on the virus. Editing involves the sequence 3′-AAAAAGGG-5′, although this sequence varies somewhat in J virus and certain other unclassified viruses (Jack et al., 2005; Lamb and Parks, 2007). Members of the Rubulavirus genus follow a second pattern in which the unedited mRNA encodes the V protein, and mRNA edited by the insertion of two G residues encodes the P protein (Lamb and Parks, 2007). The insertion of a single G residue results in expression of the I protein. The editing site and organization of ORFs in the APMV-9 P gene conformed to the first pattern, consistent with the other members of Avulavirus analyzed to date (APMV-1, -2, -6 and -9). The insertion of a single residue at the predicted APMV-9 editing site would yield a V protein with conserved tryptophan and seven cysteine residues that are common across Paramyxovirinae. The predicted MW of the V protein of APMV-9 is 28.5 kDa, which is similar to that of APMV-6 (28.7 kDa), but differs from that of APMV-4 (23.9 kDa), APMV-2 (25.1 kDa) and APMV-3 (26.6 kDa). The insertion of two non-templated G residues at the editing site of APMV-9 would produce a W protein of 21.2 kDa.
The F protein of Paramyxoviridae mediates fusion between the viral envelope and host plasma membrane at neutral pH. The F protein is synthesized as an inactive protein (F0) that is cleaved by host cell protease into the active form that consists of disulfide-linked F1–F2 subunits (Lamb and Parks, 2007). The sequence of the F protein cleavage site is a major determinant of NDV pathogenicity in chickens (Panda et al., 2004; Lamb and Parks, 2007). All known virulent strains of NDV have multibasic residues (bold and underlined) that conform to the preferred cleavage site of the intracellular cellular protease furin (R-X-K/R-R↓F) present in most cell types. In contrast, avirulent NDV strains characteristically have a single basic residue immediately preceding the F protein cleavage site and are not cleaved by furin. The ability of the F protein to be cleaved by furin provides the possibility for replication in a wide variety of tissues and is essential for systemic spread of the virus. The presence of a monobasic cleavage site in an avirulent NDV strain restricts viral replication to the respiratory and enteric tracts, where secretory proteases necessary for cleavage are produced. The F cleavage site of APMV-9 (I-R-E-G-R-I↓F) does not conform to the consensus furin protease cleavage motif (R-X-K/R-R↓F). Instead, it more closely resembles the cleavage site of avirulent NDV (G-R-Q-G-R↓L) with respect to the content of basic residues. Consistent with this, we found that APMV-9 required allantoic fluid as a source of added protease for replication in cell culture, although, surprisingly, trypsin alone could not substitute for allanotic fluid. It is also possible that the APMV-9 F protein cleavage could occur between R and I residues (I-R-E-G-R↓I-F), instead of I or F residues (I-R-EG-R-I↓F), which would explain why trypsin could not substitute for allantoic fluid. Among the other APMV prototypes sequenced to date, APMV-3 (R-P-R-G-R↓L) also required exogenous protease for in vitro replication, consistent with the presence of a single basic residue immediately preceding the cleavage site; whereas, APMV-2 (K-P-A-S-R↓F) and -4 (D-I-Q-P-R↓F) did not require added protease even though there was only a single basic residue immediately preceding the cleavage site in each case. APMV-6 has also been sequenced and has the cleavage site A-P-E-P-R↓L; however, APMV-6 is generally considered to be avirulent in vivo, whether it requires exogenous protease for replication in vitro remains to be determined. Thus, although for NDV the presence of a single basic residue immediately preceding the cleavage site seems invariably to correlate with a dependence on added protease for replication in vitro, APMV-2 and -4 are exceptions to this rule.
Another distinction between avirulent and virulent strains of NDV is the presence of, respectively, a leucine or phenylalanine as the first residue of the F1 subunit. Analysis of APMV-9 identified leucine as the first residue of the F1 subunit. Consistent with this, the results of the MDT assay in ECE suggest that APMV-9 is avirulent for chickens. However, there is again inconsistency among the APMVs. Four types of APMV examined to date have phenylalanine as the first residue of F1 (virulent NDV and APMV-2, -4, and -9), but, of these, only virulent NDV and APMV-2 appear to be virulent. Conversely, three types of APMV examined to date have leucine as the first F1 residue (avirulent NDV and APMV-3 and -6), and, of these, only avirulent NDV and APMV-6 appear to be avirulent. To further investigate these structure–pathogenicity relationships, it will be important to examine additional serotypes of APMV and additional members within each serotype, to pursue more detailed pathogenesis studies, and to develop reverse genetics systems for the generation of defined mutants.
The sequence of the APMV-9 HN protein provides evidence of the expected globular head containing a sialic acid binding motif (NRKSCS) along with active neuraminidase and conserved cysteine residues that have been shown to be conserved among type II integral membrane proteins of Paramyxovirinae (Langedijk et al., 1997). Domain III of APMV-9 polymerase L protein contained the conserved catalytic motif GDNQ, which is present in all APMVs except APMV-4, where residue D is replaced with residue E. Domain IV of the APMV-9 L protein contained the ATP-binding motif K-X21-A-E-G-S-G that is identical to that of APMV-1, but differs from those of APMV-3 (R-X21-G-E-G-S-G), APMV-4 (R-X21-G-E-G-Y-G) and APMV-6 (K-X21-A-E-S-S-G).
The complete genome sequence of APMV-9 and its comparison with APMV-1, -2, -3, -4 and -6 provided additional information on the extent of sequence variation among the different APMV serotypes. The average amino acid sequence identity in all the six proteins of APMV-9 with the members of family Paramyxoviridae was 36% with Avulavirus, 27% with Rubulavirus, 20% with J virus, 19% with Henipavirus and Respirovirus and 18% with Morbillivirus, confirming a close relationship with genus Avulavirus. The overall combined average amino acid identity for all six proteins among APMV-1, -2, -3, -4, and -6 is 57, 33, 30, 28 and 32%, respectively. Based on the phylogenetic tree analysis and average amino acid identity (57%), it can be concluded that APMV-9 is more closely related to APMV-1 than other APMVs. Further sequence analysis of the remaining APMV serotypes will provide insight into the intriguing complexities and diversities among APMV serotypes. Future studies on the pathogenicity and epidemiology of APMV-9 are needed to understand the role of this virus in poultry health.
Acknowledgments
We thank Daniel Rockemann and all our laboratory members for their excellent technical assistance and help. We also thank Hamp Edwards for performing electron microscopy and Ireen Dryburgh-Barry for proofreading the manuscript. “This research was supported by NIAID contract no.N01A060009 (85% support) and NIAID, NIH Intramural Research Program (15% support). The views expressed herein do not necessarily reflect the official policies of the Department of Health and Human Services; nor does mention of trade names, commercial practices, or organizations imply endorsement by the U.S. Government.”
References
- Alexander DJ. Avian paramyxoviruses other than Newcastle disease virus. World’s Poul Sci J. 1982;38:97–104. [Google Scholar]
- Alexander DJ. Avian Paramyxoviruses. Iowa State University Press; Ames: 2003. pp. 2–9. [Google Scholar]
- Alexander DJ. OIE Manual. 5. 1 and 2. OIE; 2008. Manual of Diagnostic Tests and Vaccines for Terrestrial Animals; pp. 576–589. [Google Scholar]
- Alexander DJ, Hinshaw VS, Collins MS, Yamane N. Characterization of viruses which represent further distinct serotypes (PMV-8 and PMV-9) of avian paramyxoviruses. Arch Virol. 1983;78:29–36. doi: 10.1007/BF01310856. [DOI] [PubMed] [Google Scholar]
- Beck I, Gerlach H, Burkhardt E, Kaleta EF. Investigation of several selected adjuvants regarding their efficacy and side effects for the production of a vaccine for parakeets to prevent a disease caused by a paramyxovirus type 3. Vaccine. 2003;21:1006–1022. doi: 10.1016/s0264-410x(02)00552-2. [DOI] [PubMed] [Google Scholar]
- Calain P, Roux L. The rule of six, a basic feature for efficient replication of Sendai virus defective interfering RNA. J Virol. 1993;67:4822–4830. doi: 10.1128/jvi.67.8.4822-4830.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang PC, Hsieh ML, Shien JH, Graham DA, Lee MS, Shieh HK. Complete nucleotide sequence of avian paramyxovirus type 6 isolated from ducks. J Gen Virol. 2001;82:2157–2168. doi: 10.1099/0022-1317-82-9-2157. [DOI] [PubMed] [Google Scholar]
- Curran J, Marq JB, Kolakofsky D. An N-terminal domain of the Sendai paramyxovirus P protein acts as a chaperone for the NP protein during the nascent chain assembly step of genome replication. J Virol. 1995;69:849–855. doi: 10.1128/jvi.69.2.849-855.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodbourn S, Didcock L, Randall RE. Interferons: cell signalling, immune modulation, antiviral response and virus countermeasures. J Gen Virol. 2000;81:2341–2364. doi: 10.1099/0022-1317-81-10-2341. [DOI] [PubMed] [Google Scholar]
- Hosaka M, Nagahama M, Kim WS, Watanabe T, Hatsuzawa K, Ikemizu J, Murakami K, Nakayama K. Arg-X-Lys/Arg-Arg motif as a signal for precursor cleavage catalyzed by furin within the constitutive secretory pathway. J Biol Chem. 1991;266:12127–12130. [PubMed] [Google Scholar]
- Jack PJ, Boyle DB, Eaton BT, Wang LF. The complete genome sequence of J virus reveals a unique genome structure in the family Paramyxoviridae. J Virol. 2005;79:10690–10700. doi: 10.1128/JVI.79.16.10690-10700.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy S, Samal SK. Nucleotide sequences of the trailer, nucleo-capsid protein gene and intergenic regions of Newcastle disease virus strain Beaudette C and completion of the entire genome sequence. J Gen Virol. 1998;79 (Pt 10):2419–2424. doi: 10.1099/0022-1317-79-10-2419. [DOI] [PubMed] [Google Scholar]
- Kumar S, Nayak B, Collins PL, Samal SK. Complete genome sequence of avian paramyxovirus type 3 reveals an unusually long trailer region. Virus Res. 2008;137:189–197. doi: 10.1016/j.virusres.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb R, Parks G. In: Paramyxoviridae: The Viruses and Their Replication. Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE, editors. Lippincott Williams & Wilkins; Philadelphia: 2007. [Google Scholar]
- Lamb RA, Choppin PW. The synthesis of Sendai virus polypeptides in infected cells. III. Phosphorylation of polypeptides. Virology. 1977;81:382–397. doi: 10.1016/0042-6822(77)90154-4. [DOI] [PubMed] [Google Scholar]
- Lamb RA, Collins PL, Kolakofsky D, Melero JA, Nagai Y, Oldstone MBA, Pringle CR, Rima BK. Family Paramyxoviridae. In: Fauquet CM, editor. Virus Taxonomy: The Classification and Nomenclature of Viruses. The Eighth Report of the International Committee in Taxonomy of Viruses. 2005. [Google Scholar]
- Langedijk JP, Daus FJ, van Oirschot JT. Sequence and structure alignment of Paramyxoviridae attachment proteins and discovery of enzymatic activity for a morbillivirus hemagglutinin. J Virol. 1997;71:6155–6167. doi: 10.1128/jvi.71.8.6155-6167.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Horvath F, Aligo JA, Wilson R, He B. The role of simian virus 5 V protein on viral RNA synthesis. Virology. 2005;338:270–280. doi: 10.1016/j.virol.2005.05.014. [DOI] [PubMed] [Google Scholar]
- Miller PJ, Boyle DB, Eaton BT, Wang LF. Full-length genome sequence of Mossman virus, a novel paramyxovirus isolated from rodents in Australia. Virology. 2003;317:330–344. doi: 10.1016/j.virol.2003.08.013. [DOI] [PubMed] [Google Scholar]
- Nayak B, Kumar S, Peter L, Collins, Samal SK. Molecular characterization and complete genome sequence of avian paramyxovirus type 4 prototype strain duck/Hong Kong/D3/75. Virol J. 2008;5:124. doi: 10.1186/1743-422X-5-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda A, Huang Z, Elankumaran S, Rockemann DD, Samal SK. Role of fusion protein cleavage site in the virulence of Newcastle disease virus. Microb Pathog. 2004;36:1–10. doi: 10.1016/j.micpath.2003.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeples ME. Paramyxovirus M Proteins: Pulling it all Together and Taking it on the Road. Plenum Press; New York: 1991. [Google Scholar]
- Poch O, Blumberg BM, Bougueleret L, Tordo N. Sequence comparison of five polymerases (L proteins) of unsegmented negative-strand RNA viruses: theoretical assignment of functional domains. J Gen Virol. 1990;71 (Pt 5):1153–1162. doi: 10.1099/0022-1317-71-5-1153. [DOI] [PubMed] [Google Scholar]
- Samal SK, Collins PL. RNA replication by a respiratory syncytial virus RNA analog does not obey the rule of six and retains a nonviral trinucleotide extension at the leader end. J Virol. 1996;70:5075–5082. doi: 10.1128/jvi.70.8.5075-5082.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu T, Hinshaw V. Influenza A virus infection of domestic ducks. First Int Symp Avian Influenza. 1981:93–99. [Google Scholar]
- Schnell MJ, Conzelmann KK. Polymerase activity of in vitro mutated rabies virus L protein. Virology. 1995;214:522–530. doi: 10.1006/viro.1995.0063. [DOI] [PubMed] [Google Scholar]
- Shioda T, Iwasaki K, Shibuta H. Determination of the complete nucleotide sequence of the Sendai virus genome RNA and the predicted amino acid sequences of the F, HN and L proteins. Nucleic Acids Res. 1986;14:1545–1563. doi: 10.1093/nar/14.4.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbiah M, Xiao S, Collins PL, Samal SK. Complete sequence of the genome of avian paramyxovirus type 2 (strain Yucaipa) and comparison with other paramyxoviruses. Virus Res. 2008;137:40–48. doi: 10.1016/j.virusres.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LF, Hansson E, Yu M, Chua KB, Mathe N, Crameri G, Rima BK, Moreno-Lopez J, Eaton BT. Full-length genome sequence and genetic relationship of two paramyxoviruses isolated from bat and pigs in the Americas. Arch Virol. 2007;152:1259–1271. doi: 10.1007/s00705-007-0959-4. [DOI] [PMC free article] [PubMed] [Google Scholar]