Abstract
In the 1920s, Otto Warburg observed that tumor cells consume a large amount of glucose, much more than normal cells, and convert most of it to lactic acid. This phenomenon, now known as the ‘Warburg effect,’ is the foundation of one of the earliest general concepts of cancer: that a fundamental disturbance of cellular metabolic activity is at the root of tumor formation and growth. In the ensuing decades, as it became apparent that abnormalities in chromosomes and eventually individual genes caused cancer, the ‘metabolic’ model of cancer lost a good deal of its appeal, even as emerging technologies were exploiting the Warburg effect clinically to detect tumors in vivo. We now know that tumor suppressors and proto-oncogenes influence metabolism, and that mutations in these genes can promote a metabolic phenotype supporting cell growth and proliferation. Thus, these advances have unified aspects of the metabolic and genetic models of cancer, and have stimulated a renewed interest in the role of cellular metabolism in tumorigenesis. This review reappraises the notion that dysregulated cellular metabolism is a key feature of cancer, and discusses some metabolic issues that have escaped scrutiny over the years and now deserve closer attention.
Keywords: cancer, metabolism, Warburg effect, glutamine, glucose
INTRODUCTION
The Warburg effect, also known as aerobic glycolysis, is defined as a high rate of glucose utilization and lactate production despite the presence of sufficient oxygen to oxidize glucose carbon in the mitochondria. Recognition of this unusual metabolic phenomenon stems from experiments performed by the German physiologist Otto Warburg, starting in the 1920s1, 2. In those experiments, Warburg compared the metabolism of rapidly proliferating mouse ascites tumor cells to that of differentiated, quiescent cells from organs of the adult animal. He proposed that a fundamental impairment of cellular respiratory capacity was the root cause of all cancer, a bold and controversial claim that was ultimately rejected despite his continued writing and lecturing on the subject for some 40 years. Nevertheless, appreciation of the Warburg effect as a feature of tumor cell metabolism has survived its namesake by a long stretch. Today, the glycolytic activity of tumors is not only accepted, but exploited clinically by 18F-deoxyglucose positron emission tomography (FDG-PET), which detects tumors precisely by virtue of their enhanced ability to take up and metabolize glucose compared to normal tissue. The Warburg effect remains the most frequently cited evidence that tumors display dysfunctional metabolism.
Recently, interest in tumor metabolism has enjoyed a renaissance as an ever-growing number of reports uncovers the molecular connections between transformation and cell metabolism, and as technological improvements increase the feasibility of studying tumor metabolism in vivo. The field seems poised to offer significant insights into tumor biology over the next decade. As such, it is worth re-examining the evidence for a bona fide connection between altered cellular metabolic state and tumorigenesis: does the notion of such a connection stand up to our current understanding of tumor biology and cancer genetics? If there is such a link, then the following should be true:
Tumor cells should have metabolic activities that differ from non-transformed, quiescent cells, and these activities should be required for tumor growth;
The mutations in tumor suppressors and proto-oncogenes that promote cancer should regulate the metabolic activities observed in tumors; and
Mutations in metabolic enzymes should, in at least some cases, promote tumorigenesis.
This review addresses how well these criteria are met and discusses some other issues relevant to tumor metabolism that may feature prominently in future research.
DO TUMOR CELLS HAVE METABOLIC ACTIVITIES THAT ARE DIFFERENT FROM QUIESCENT CELLS AND ARE REQUIRED FOR TUMOR GROWTH?
Most studies on tumor metabolism have been motivated by one of two general concepts about the way cell metabolism is regulated. The first is that tumor metabolism is primarily a response to stresses imposed upon cells during tumor growth. There is abundant evidence that some stresses, particularly hypoxia, exist in the tumor microenvironment and exert effects on metabolism3–5. But the high glycolytic flux in tumors can appear even when oxygen is abundant, and the metabolic consequences of hypoxia include specific impairments of protein and lipid synthesis that are counterproductive to cell growth and proliferation6–8. These observations suggest that the cellular responses to tumor hypoxia, including enhanced glycolysis, serve to facilitate tumor cell survival, not growth. This is also true in non-transformed cells, which rely on glycolysis to survive periods of hypoxia9. When oxygen delivery improves and rapid cell growth resumes, the persistence of glycolysis is likely due to other factors.
Alternatively, one can presume that tumor cell metabolic activities function primarily to support the unusually high rates of cell growth and proliferation found in tumors. Since each round of replicative cell division requires a doubling of protein, lipids and nucleic acids, it stands to reason that tumor cell metabolism must provide the energy and biosynthesis needed to meet this challenge (Fig. 1). Rapid tumor growth requires the ability to capture nutrients and process them in the appropriate metabolic pathways to convert their carbon and nitrogen into macromolecules. All of the activities discussed in this section (the Warburg effect, fatty acid synthesis and mitochondrial glutamine metabolism) occur during cell growth, and evidence suggests that the three pathways cooperate in such a way as to maximize the production of macromolecules in proliferating cells (Fig. 2)10.
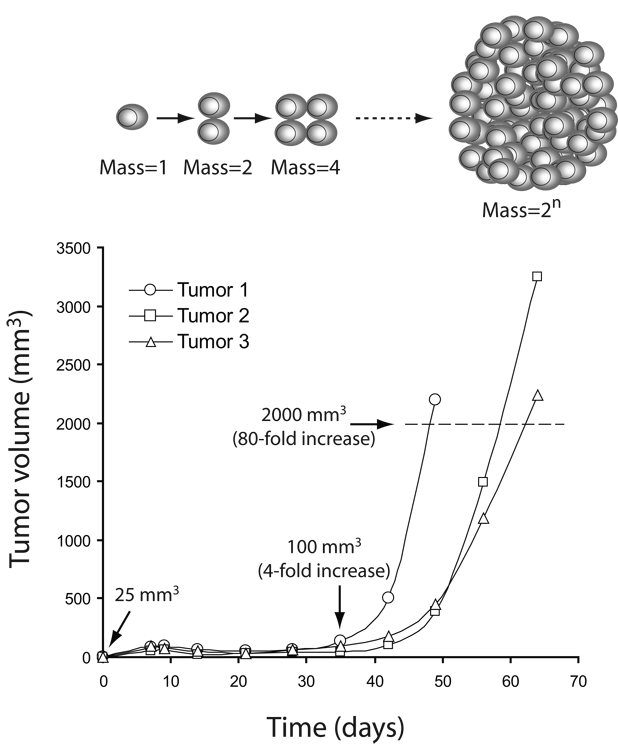
Fig. 1. A large increase in tumor cell biomass accompanies tumor growth.
Replicative cell division (top) requires that cells double their biomass (proteins, lipids, nucleic acids) each time two daughters are produced. If cell proliferation is exponential, then the production of macromolecules for the entire population is also exponential. Rapid growth of a tumor thus implies that the tumor cells have mechanisms in place to synthesize macromolecules rapidly. For example, in a typical experiment studying the growth of tumors derived from human glioblastoma cells (bottom), 10 million cells (approximately 25 mm3 total cell volume) were injected subcutaneously into nude mice, and growth of the tumor was measured each week. By the end of the fifth week, the average tumor size was 100 mm3, a four-fold increase from time zero. All tumors eventually exceeded 2000 mm3 in size (dashed line), by which time the tumor biomass had increased 80-fold from time zero. Growth of these tumors, particularly the rapid growth over the final few weeks, requires a metabolic platform supporting anabolism.
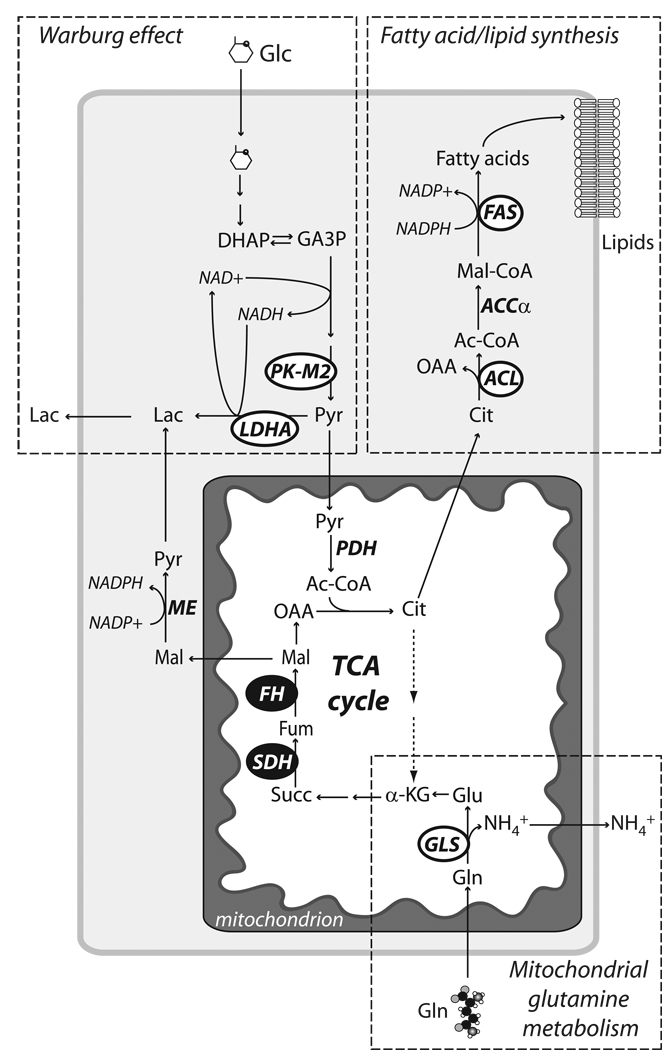
Fig. 2. Some metabolic activities are required for tumor growth.
Among the various metabolic activities that have been observed in tumors or tumor cell lines, the three with the most compelling evidence for a required role in tumor growth are aerobic glycolysis (the Warburg effect), fatty acid/lipid synthesis and mitochondrial glutamine metabolism. Current evidence suggests that these three pathways cooperate in a metabolic platform that supports cell growth and ultimately proliferation. The high rate of glycolysis, in addition to producing ATP, generates glycerol and citrate to be used to synthesize membrane lipids. Meanwhile, mitochondrial metabolism of glutamine supplies the TCA cycle with intermediates to replace those exported for lipid synthesis and other anabolic processes. Cells using this form of metabolism secrete lactate produced from both glucose and glutamine. Ammonia is also produced and secreted in abundance. Recent studies have shown that several of the enzymes participating in these pathways are required for growth of tumors in mice (white ovals), while SDH and FH function genetically as tumor suppressors in humans. Abbreviations: Glc, glucose; DHAP, dihydroxyacetone phosphate; GA3P, glyceraldehyde 3-phosphate; Pyr, pyruvate; Lac, lactate; Ac-CoA, acetyl-CoA; Cit, citrate; α-KG, α-ketoglutarate; Succ, succinate; Fum, fumarate; Mal, malate; OAA, oxaloacetate; Mal-CoA, malonyl-CoA; Gln, glutamine; Glu, glutamate, NH4+, ammonia; PK-M2, pyruvate kinase isoform M2; LDHA, lactate dehydrogenase-A; ACL, ATP citrate lyase, ACC1, acetyl-CoA carboxylase-α; FAS, fatty acid synthase; GLS, glutaminase; SDH, succinate dehydrogenase; FH, fumarate hydratase; PDH, pyruvate dehydrogenase; ME, malic enzyme
The Warburg effect is the most widely documented metabolic activity in tumors and tumor cell lines11, 12. Gene expression analysis has demonstrated the nearly ubiquitous over-expression of numerous glycolytic genes across a wide spectrum of human tumor types13, and imaging techniques like 18FDG-PET and 1H NMR spectroscopy have confirmed that the Warburg effect occurs in tumors in vivo. It is surprising, then, that there is still no clear consensus as to the function of the Warburg effect. It has been particularly difficult to explain why tumor cells with access to oxygen would deprive themselves of the majority of the ATP that can be produced from glucose metabolism by instead converting pyruvate into lactate. However, three points should be emphasized regarding the role of the Warburg effect in tumor cell metabolism. First, given that the Warburg effect is also observed during rapid proliferation of primary cells, it is more accurately viewed as a general feature of cell proliferation than as a symptom of transformation, and therefore can be assumed to contribute to anabolic metabolism14, 15. Second, rapid glucose metabolism also supplies intermediates for biosynthetic pathways that arise from glycolysis proximal to pyruvate, including ribose-5-phosphate and glycine for nucleotide biosynthesis, and glycerol for lipid synthesis. It has been suggested that one of the functions of the Warburg effect is to maintain adequate sizes of these precursor pools to maximize cell growth16. Third, cells engaged in aerobic glycolysis do not convert 100% of their pyruvate into lactate. Rather, a measurable fraction of the pyruvate is metabolized in the tricarboxylic acid (TCA) cycle, providing energy and precursors for biosynthetic pathways that consume TCA cycle intermediates15, 17, 18. Therefore, the Warburg effect serves both bioenergetic and biosynthetic roles in proliferating cells.
It may be that the Warburg effect is essentially a consequence of an imbalance between maximum rates of glycolysis and pyruvate oxidation. The rate of pyruvate oxidation is controlled by the highly regulated pyruvate dehydrogenase (PDH) complex, which converts pyruvate to acetyl-CoA (Fig. 2). During rapid cell proliferation, the glycolytic rate may exceed the Vmax of PDH by more than 10-fold19. If the PDH flux cannot match the glycolytic flux, cells must use other high-capacity systems to avoid pyruvate accumulation. Chief among these is lactate dehydrogenase-A (LDHA), which by converting pyruvate to lactate also oxidizes NADH to NAD+. Since lactate can be secreted and NAD+ is required for glycolysis, expression of LDHA allows proliferating cells to continue to reap the benefits of a high glycolytic rate even in the face of a ‘maxed-out’ PDH flux. This model is supported by studies performed in interleukin-3 (IL-3)-dependent hematopoietic cells20. In those cells, the rates of cell proliferation, glucose consumption and lactate production are directly proportional to the concentration of IL-3 in the medium. Strikingly, the ratio of lactate produced to glucose consumed was positively regulated by IL-3, but the ratio of oxygen consumed to glycolysis declined with increasing IL-3. Thus, with progressive stimulation, cells continue to metabolize glucose beyond the point of maximal pyruvate oxidation, eventually reaching a state at which a higher and higher fraction of glucose carbon is converted to lactate and the fraction of cellular ATP generated by glycolysis approaches or even exceeds that from oxidative phosphorylation, as in Warburg’s experiments21.
These data imply that factors influencing the Vmax of PDH and the balance between lactate production and pyruvate oxidation can impact the Warburg effect. Recent studies have demonstrated that this balance is regulated by hypoxia inducible factor-1α (HIF-1α), a transcription factor with multiple targets involved in glucose metabolism. HIF-1α regulates expression of pyruvate dehydrogenase kinase-1 (PDK-1), a kinase which limits PDH activity22, 23. Therefore, under hypoxic conditions, stabilization and transcriptional activity of HIF-1α results in the enhancement of glycolysis and the suppression of pyruvate oxidation by PDH. This mechanism can also contribute to the Warburg effect in well-perfused tumor cells, which may have aberrant/normoxic stabilization of HIF-1α due to a variety of genetic mechanisms24. In these cells, the resulting glycolysis is truly ‘aerobic.’
Even under conditions of HIF-1α stabilization, however, cell growth requires at least a small amount of PDH flux in order to maintain TCA cycle activity, which supplies precursors for the synthesis of fatty acids and other anabolic pathways (Fig. 2). Tumor cells often display rapid synthesis of fatty acids, cholesterol and isoprenoids, presumably because a large fraction of their membrane lipids are synthesized de novo rather than scavenged from extracellular sources25. In a metabolic flux study on human glioblastoma cells, fatty acid synthesis accounted for some 15% of cellular glucose consumption after excluding the fraction of glucose metabolized through the Warburg effect18. Thus a significant fraction of the glucose carbon that enters biosynthetic pathways is used to produce fatty acids. Consistent with that observation, the three enzymes required for fatty acid synthesis, ATP citrate lyase (ACL), acetyl-CoA carboxylase-α (ACC-α), and fatty acid synthase (FAS), are highly expressed in many human cancers and tumor cell lines26.
Fatty acid synthesis is a paradigm of tumor biosynthetic pathways because it requires the use of a TCA cycle intermediate (citrate) that might otherwise be oxidized in the mitochondria (Fig. 2). The shunting of metabolites from the TCA cycle into other pathways (cataplerosis) is part of the fundamental biochemistry of cell growth, and it emphasizes the versatility of the TCA cycle: rather than serving a purely oxidative function as a source of reducing equivalents for the electron transport chain (ETC), it can also be used as a continuous source of precursor molecules for biosynthetic pathways. Early characterization of cataplerosis in highly lipogenic hepatoma cells led to the concept of a ‘broken’ or ‘truncated’ TCA cycle because of an apparent impairment in citrate oxidation and the demonstration that the rate of citrate export was directly proportional to the rate of cell proliferation27, 28. These observations underscore the importance of cataplerosis in tumor cell growth. In addition to lipid synthesis, cataplerotic fluxes feed the synthesis of nucleotides and nonessential amino acids, and are therefore used in the synthesis of all classes of macromolecules.
Glutamine metabolism is second only to the Warburg effect in terms of historical significance to the study of tumor cell metabolism. Glutamine is the most abundant amino acid in human plasma and participates in many metabolic pathways required for normal cell function. In addition to its role in protein synthesis, it provides nitrogen for the synthesis of nonessential amino acids, purines, pyrimidines and hexosamines, and is the major source of glutamate used for glutathione synthesis. Tumor cells have long been known to consume glutamine at high rates in vivo and to require high concentrations of glutamine to survive and proliferate in vitro29, 30. Classical studies on tumor cell metabolism in culture demonstrated that glutamine is an important carbon source since most of the glutamine consumed is used as a respiratory substrate in the mitochondria rather than for protein synthesis31. More recent experiments have demonstrated that suppressing mitochondrial glutamine metabolism can alter gene expression, accelerate apoptosis and stimulate cellular differentiation32, 33. Therefore, glutamine metabolism has the potential to integrate a large number of cellular activities that support tumorigenesis.
A closer look at glutamine metabolism has revealed other vital roles in proliferating cells (Fig. 2). First, partial oxidation of glutamine to lactate (glutaminolysis) uses the cytosolic malic enzyme and therefore provides cells with NADPH for the reductive reactions of fatty acid and nucleotide biosynthesis. In some cases, the rate of glutaminolysis can match or exceed the rate of NADPH production by glucose-6-phosphate dehydrogenase (G6PD) in the pentose phosphate pathway17, 18. Second, glutamine’s conversion to α-ketoglutarate and entry into the TCA cycle can generate oxaolacetate (OAA), effectively replacing the metabolites that are removed from the cycle in cataplerotic reactions (Fig. 2). This process, termed anaplerosis, is a critical component of growth metabolism because it allows cells to maintain TCA cycle function while withdrawing intermediates for biosynthetic reactions. In some cells, glutamine metabolism is by far the most important source of anaplerosis, and depriving cells of glutamine rapidly depletes cellular pools of TCA cycle intermediates18, 34, 35. In cells simultaneously consuming both glucose and glutamine, citrate production involves the condensation of two glucose-derived carbons (as acetyl-CoA) and four glutamine-derived carbons (as OAA). Although it is the glucose carbons that are ultimately transferred to fatty acids, the process could not occur without the contribution of glutamine-based anaplerosis10.
But how strong is the evidence that any of the enzymes participating in these three pathways are required for tumor growth? A number of studies using chemical inhibitors and more recently RNA interference have addressed this question in animal models of cancer (Fig. 2). The glycolysis inhibitor 2-deoxyglucose, when given at doses that did not affect body weight or growth of the animal, significantly decreased carcinogen-induced mammary tumorigenesis in rats36. RNA interference against glycolytic enzymes or enzymes in de novo fatty acid synthesis also curtailed tumor growth in animal models37–39. Suppression of glutaminase, the first enzyme in mitochondrial glutamine metabolism, using the chemical inhibitor 6-diazo-5-oxo-L-norleucine (DON) or expression of an antisense mRNA limited tumor growth in mice40, 41. These studies are the best evidence that the metabolic activities observed in tumor cells in vitro are not simply artifacts of culture conditions, but are fundamental properties that contribute to tumor growth.
It is interesting that in addition to the enhanced metabolic rates described above, tumor cells can also display chronic suppression of the pathways that normally allow cells to utilize alternative fuels in order to survive periods of starvation. As a result, the cells have an increased reliance on specific fuels and a limited ability to compensate for fluctuations in nutrient availability. For example, constitutive activity of the oncogenic kinase Akt impairs the activation of fatty acid oxidation, resulting in abrupt glioma cell death in low-glucose conditions42. Loss of the tumor suppressor p53 diminishes the ability of colon cancer cells to engage catabolic, energy-generating pathways like autophagy and fatty acid oxidation43. Glutamine depletion selectively kills fibroblasts with enhanced c-Myc activity35. Step-wise transformation of fibroblasts with multiple oncogenes progressively increases the toxicity of a glycolysis inhibitor44. These observations add credibility to the hope that inhibiting specific metabolic pathways will be selectively toxic to tumor cells in vivo. Indeed, exploiting phenotypes of impaired metabolic flexibility may prove to be more useful in cancer therapy than inhibiting growth-promoting activities like fatty acid synthesis, since these tend to be shared with normal proliferating cells.
DO MUTATIONS IN PROTO-ONCOGENES AND TUMOR SUPPRESSORS IMPACT CELL METABOLISM?
Mutations that promote tumorigenesis often reduce or eliminate cellular dependence on extrinsic signals to maintain survival, growth and proliferation. These processes are usually under control of growth factors and other signals that originate outside of the cell and are transmitted inwards through signal transduction pathways. In cancer, this ‘outside-in’ regulation is diminished or lost, allowing cells to achieve self-sufficiency in growth and proliferation45. Cellular metabolism is also subject to external control, because lineage-specific growth factors and the signaling pathways they stimulate are required for cells to activate anabolic pathways and suppress catabolic ones46–48. Since cell proliferation cannot occur without these metabolic activities, it is not surprising that tumor cells have increased autonomy in maintaining an anabolic phenotype. A large amount of evidence now supports the idea that tumor suppressors and proto-oncogenes exert regulatory effects on metabolism in normal cells, and that tumorigenic mutations in these genes contribute to the metabolic autonomy observed in tumor cells49. Most of the regulatory mechanisms that have been described to date are focused on glucose metabolism and involve mutations in p53, phophatidylinositol 3-kinase (PI3K) signaling, Ras and Myc, the most prevalent classes of tumorigenic mutations in humans (Fig. 3).
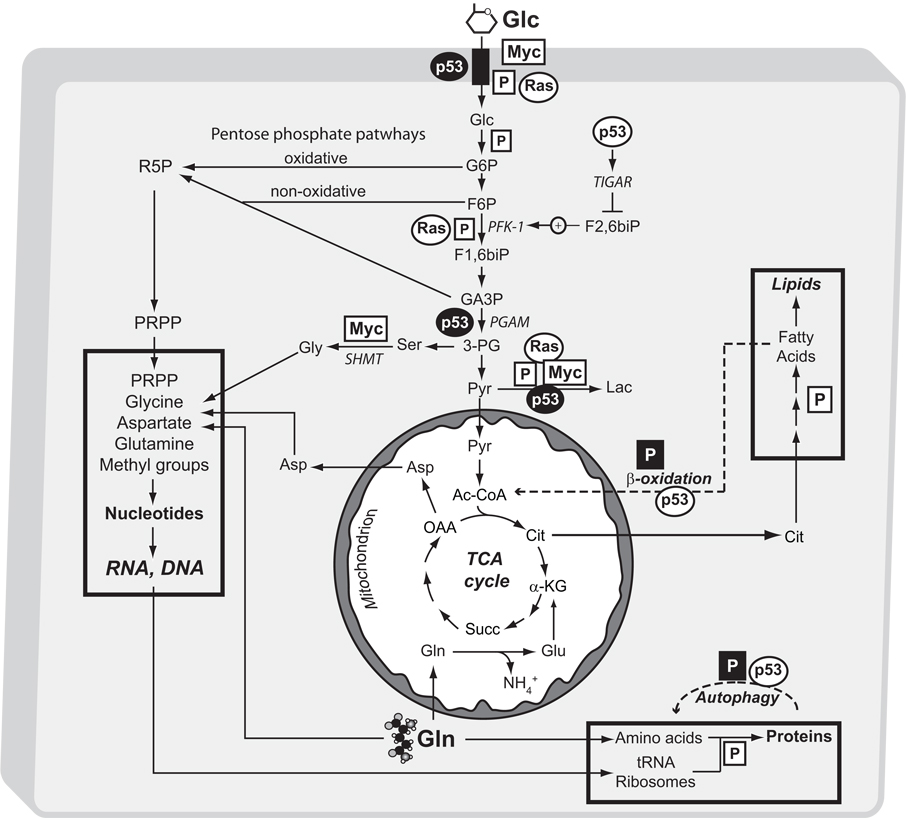
Fig. 3. Tumor suppressors and proto-oncogenes regulate the metabolic pathways involved in tumor growth.
The major biosynthetic activities used by proliferating tumor cells (synthesis of proteins, nucleic acids and lipids) are outlined in boxes. Supporting pathways, including glycolysis, the oxidative and non-oxidative arms of the pentose phosphate pathway, mitochondrial glutamine metabolism and the TCA cycle are also shown. Alternative metabolic pathways normally used during nutrient deprivation and suppressed during cell proliferation (β-oxidation of fatty acids, autophagy) are indicated by dashed lines. Selected effects of p53, Myc, Ras and the PI3K/Akt/mTOR signaling system (boxed P) are indicated; black symbols indicate supression and white symbols indicate activation. In the case of p53, loss of function mutations in tumor cells have the opposite of the effect shown here (e.g. glucose uptake and glycolysis are no longer suppressed by p53 activity). Abbreviations: Glc, glucose; G6P, glucose-6-phosphate; F6P, fructose-6-phosphate; F1,6biP, fructose-1,6-bisphosphate; F2,6biP, fructose-2,6-bisphosphate; GA3P, glyceraldehyde 3-phosphate; Pyr, pyruvate; Lac, lactate; R5P, ribose-5-phosphate; PRPP, 5-phosphoribosyl pyrophosphate; Ser, serine; Gly, glycine; Ac-CoA, acetyl-CoA; Cit, citrate; α-KG, α-ketoglutarate; Succ, succinate; OAA, oxaloacetate; Mal-CoA, malonyl-CoA; Gln, glutamine; Glu, glutamate, NH4+, ammonia; TIGAR, TP53-induced glycolysis and apoptosis regulator; PFK1, phosphofructokinase-1; PGAM, phosphoglycerate mutase; SHMT, serine hydroxymethyltransferase.
Mutations in the tumor suppressor gene TP53 occur in approximately 50% of all human cancers. Recent studies have uncovered multiple roles for p53 in glucose metabolism, revealing an inverse correlation between p53 activity and the Warburg effect. First, loss of p53 in primary fibroblasts enhances glucose transport and metabolism through IKK and NF-κB50. Second, the p53 transcriptional target TIGAR (TP53-induced glycolysis and apoptosis regulator) decreases the abundance of the glycolytic activator fructose-2,6-bisphosphate (F2,6biP), enabling cells with p53 activation after DNA damage to divert glucose-6-phosphate into the oxidative pentose phosphate pathway, bolstering production of NADPH and ribose-5-phosphate for DNA repair51. p53 also suppresses expression of the glycolytic enzyme phosphoglycerate mutase and enhances expression of the ETC assembly factor SCO2, effects that curtail glycolysis and maximize pyruvate oxidation52, 53. Therefore, loss of p53 function has multiple positive effects on aerobic glycolysis.
The phosphatidylinositol 3-kinase (PI3K) signaling pathway is a widely-expressed component of growth factor signaling in diverse organisms and cell types54. Mutations that enhance PI3K signaling are common in cancer, and include mutations in the p110α catalytic subunit of the PI3K complex, encoded by PIK3CA; loss of function of the negative regulator PTEN; gain of function mutations in growth factor receptors such as c-Kit and Her2/neu; and amplification of the downstream effector Akt55–59. PI3K signaling, largely through its effects on Akt and mammalian target of rapamycin (mTOR), regulates many of the normal metabolic consequences of growth factor stimulation49. These include glucose uptake and phosphorylation, activation of phosphofructokinase-1 (PFK-1), stimulation of the Warburg effect and enhanced synthesis of lipids and proteins60–64, all of which support cell growth and proliferation (Fig. 3). Thus, mutations that enhance PI3K signaling alleviate the dependence of the cell on extrinsic signals to activate anabolic metabolism.
The RAS proto-oncogenes (NRAS, HRAS and KRAS) encode small GTP-binding proteins that normally function in cell signaling, proliferation, differentiation and motility. Activating mutations in RAS genes, usually missense mutations that cause constitutive GTP binding, are found in approximately 30% of human tumors65 and Ras activation has a number of positive effects on glucose metabolism. Transfecting fibroblasts with oncogenic RAS alleles results in enhanced glucose transport, while Ras inhibition in glioblastoma cells suppresses the expression of LDHA and other glycolytic genes66, 67. Overexpression of mutant KRAS alleles associated with resistance to apoptosis can induce the Warburg effect in fibroblasts68. The effects of Ras on glycolysis require expression of the gene for 6-phosphofructo-2-kinase/fructose 2,6-biphosphatase (PFKFB3), which regulates abundance of the PFK-1 activator F2,6-biP, suggesting that the effects are mediated at the level of PFK-1 activity (Fig. 3)69, 70. Interestingly, Ras may also enhance mitochondrial metabolism, as transformation of bronchial epithelial cells with an oncogenic HRAS allele increases the entry of glucose carbon into the TCA cycle and various cataplerotic pathways71.
The myc family of proto-oncogenes, especially c-myc, encode regulators of gene expression and are commonly amplified in human tumor cells. As with the PI3K signaling pathway, c-Myc is required for the proliferation of normal, non-transformed cells, but enhancement of its activity in tumor cells can drive transcriptional effects independently of external stimulation. Among c-Myc’s veritable host of transcriptional targets are genes involved in glucose metabolism and the Warburg effect, including LDHA and the glucose transporter GLUT1. c-Myc also induces the expression of enzymes involved in nucleotide metabolism, including serine hydroxymethyltransferase72, which allows 3-carbon units from glycolysis to be used in purine and pyrimidine synthesis (Fig. 3). This distinguishes c-Myc from most of the other drivers of the Warburg effect, which so far seem to lack direct influence over nucleotide biosynthesis. Given c-Myc’s physiological role in facilitating the G1/S transition, it is likely that these transcriptional targets allow cells to produce the metabolites needed to complete S phase successfully.
CAN MUTATIONS IN METABOLIC ENZYMES INFLUENCE CANCER RISK?
Warburg hypothesized that the metabolism of all tumor cells was primarily due to irreversible defects in cellular respiration2. This has not turned out to be the case, as many tumor cell lines that have been carefully studied retain the capacity for normal mitochondrial metabolism11. However, there are now some very interesting exceptions in which genes for enzymes of the TCA cycle behave genetically as tumor suppressors and are severely impaired in certain human tumors (Fig. 2). First, mutations in three of the four subunits of the succinate dehydrogenase (SDH) complex have been found in pheochromocytomas and related tumors73–75. In affected families, tumor risk is inherited as a dominant trait due to loss-of-function mutations in SDH subunits, with the tumor tissue displaying loss of heterozygosity and complete absence of SDH enzyme activity76. Similarly, mutations in fumarate hydratase (FH) cause a group of dominantly-inherited cancer syndromes involving cutaneous and uterine lyomyomas77. Precisely how dysfunction of these enzymes causes tumor growth is an area of active study. One contributing factor is that these mutations lead to the accumulation of succinate and fumarate, which interfere with degradation of HIF-1α, leading to its normoxic stabilization78. This would be predicted to induce the Warburg effect, although the mechanisms of enhanced tumor growth are not clear.
The diverse roles of the mitochondria and the ETC in cell metabolism, growth, survival, apoptosis and production of reactive oxygen species have led to the idea that mutations in mitochondrial DNA (mtDNA) might also influence cancer risk. A great deal of work has attempted to correlate mutations in the mitochondrial DNA with cancer. Germline sequence variants in the mtDNA have been associated with invasive breast cancer, endometrial cancer and prostate cancer in certain populations79–81. Many other studies have reported associations of somatically acquired, tumor-specific mtDNA sequence variants in ovarian, colon, bladder, head and neck and other cancers (reviewed in ref. 82). However, many of these variants had been previously detected in large population studies and were postulated to impart adaptive functions during migration of ancestral human populations, making their contribution to tumorigenesis less obvious. To determine the effect of a specific mtDNA mutation on tumor growth, Petros et al. introduced mitochondria harboring a known pathogenic mutation into human PC3 prostate cancer cells depleted of their own mitochondrial DNA, and tested the resulting ‘cybrids’ for tumor growth in nude mice. The cybrids containing the mutant mtDNA formed tumors much more rapidly than cybrids containing wild-type mtDNA81. It should be noted that the mutant mtDNA was derived from a child with Leigh syndrome, not from a tumor, and that the recipient PC3 cells were already transformed. Nevertheless, this study proves that a maximal capacity for oxidative phosphorylation is dispensable for tumor growth, and is the strongest evidence to date that dysfunctional mtDNA can impart a growth advantage to tumor cells in vivo.
WHERE DO WE GO FROM HERE?
At present, the data on tumor cell metabolism make a compelling argument that during the process of transformation, cells acquire a fairly stereotyped set of metabolic characteristics that enable them to grow and proliferate with reduced dependence on extracellular signals, and that the mutations in tumor suppressors and proto-oncogenes that cause cancer contribute to this ‘metabolic transformation.’ There are a number of important questions that still need to be addressed before we can appreciate fully the role of metabolism in the development, progression, and treatment of cancer. Below are six areas that are in particular need of attention.
First, there is still a large gap between our understanding of tumor cell metabolism in vitro and tumor metabolism in vivo. The inhibitor and RNA interference experiments cited above demonstrate the requirement of certain enzymes for tumorigenesis, but the ability to observe the metabolism of live tumors would provide a much richer understanding of the biochemical aspects of tumor growth and would support more rigorous examination of the effect of tumorigenic mutations on metabolic flux. The use of 18FDG-PET and conventional NMR spectroscopy have provided some validation of the Warburg effect and other pathways in live tumors, but are currently best suited to give snapshots of metabolism rather than robust measurements of metabolic flux. The application of more sensitive methods such as the use of probes labeled with hyperpolarized 13C may ultimately support flux analysis in vivo83.
Second, the regulation of anaplerosis and of glutamine metabolism in general has not been carefully studied with respect to cell signaling. While glutamine consumption is a general metabolic feature of cell proliferation, it is unknown whether all tumor cells must use glutamine as the major source of carbon for anaplerosis. In a few cases, cellular biosynthesis and growth have been correlated with the induction of alternative anaplerotic pathways that do not involve glutamine metabolism19, 84, and thus the universality of glutamine-based anaplerosis is debatable. Furthermore, while stimulation with mitogens can enhance glutamine utilization in various cell types, neither the signaling pathways responsible for this activity nor the specific pathways of glutamine metabolism (e.g. glutaminolysis vs. glutamine-based anaplerosis) have been characterized15, 17. Resolving these issues will go a long way towards integrating our understanding of cell signaling and the metabolism of cell growth. Considering that glucose and glutamine play complementary roles in proliferating tumor cells, it will be interesting to test whether they are co-regulated by the same signaling pathways and affected by the same tumorigenic mutations.
Third, many studies have sought to unravel the complex relationships between oxidative stress, cellular transformation and cancer. At the level of cell metabolism, there are many open questions about how NADPH-producing systems are regulated and whether these mechanisms are altered in tumor cells. Rapidly proliferating tumor cells require NADPH both to maintain pools of reduced glutathione and to support the reductive biosynthetic reactions needed for anabolic metabolism. The two NADPH-producing systems discussed above, G6PD and cytosolic malic enzyme, are expressed in tumors of various histological types, and in some cases their activities are almost equal in proliferating tumor cells, implying equivalent contributions to the NADPH pool17, 18. It is interesting that G6PD deficiency, a common X-linked condition, does not alter the risk of death from cancer85. This suggests that malic enzyme or other systems can provide sufficient NADPH to support tumorigenesis in G6PD-deficient cells, and that selective inhibition of malic enzyme in individuals with G6PD deficiency might suppress tumor growth. Conversely, maintaining an NADPH pool and a robust response against oxidative stress is critical in preventing tumorigenesis, as emphasized by recent studies on the transcription factor NF-E2-related factor-2 (Nrf2)86. Nrf2 is sequestered in the cytosol through association with its repressor Keap1 (Kelch ECH associating protein 1).
Exposure to oxidative and other stresses allows Nrf2 to translocate to the nucleus, where it heterodimerizes with small Maf-family proteins and binds promoters containing antioxidant response elements, thus inducing expression of genes that serve antioxidant functions. This protects cells from apoptosis during exposure to oxidizing agents, UV irradiation and other stresses87–89. Knockout mice lacking Nrf2 have enhanced susceptibility to cancer when exposed to chemical mutagens90, 91. Future studies will be aimed at exploiting the Nrf2 pathway as a strategy to prevent cancer, and at understanding other mechanisms by which normal cells and tumor cells respond to oxidative stress.
Fourth, almost all previous studies on tumor cell metabolism have been performed in unsynchronized populations of cells distributed throughout the cell cycle, so that the resulting data represent average cell metabolic activity throughout the cycle. The temptation is to assign the greatest biological importance to the pathways that appear to be the most active, but such an interpretation could miss the significance of pathways that are transiently induced at specific, critical points of the cell cycle. One can envision two ways to identify these activities: by performing flux analysis in synchronized populations of cells or by using inhibitors and genetic models to determine the effect of disrupting candidate metabolic activities on progression through the cell cycle. There is already strong evidence that interfering with metabolism profoundly affects the cell cycle. In fibroblasts, glucose deprivation imposes an AMPK- and p53-dependent G1/S phase arrest92, and in Drosophila, elimination of ETC components stimulates several distinct signaling pathways that culminate in arrest at the G1/S transition93, 94. These efforts are relevant to metabolically-directed cancer therapy, especially if such therapies are used as adjuvants to existing strategies that already exert cell cycle-specific effects. For example, if ionizing radiation is most effective on cells at the G2/M transition, then a metabolic therapy targeting a G2-specific activity might increase the fraction of cells at that stage, thereby increasing the overall efficacy of treatment.
Fifth, it is possible that normal but genetically-defined variation in the expression of metabolic enzymes could influence cancer risk and progression. Microarray analysis of gene expression in cells from healthy individuals has shown that some metabolic enzymes and transporters are highly variable in their mRNA abundance, and that the variance is defined genetically95. Currently, the association between disease states and variance of expression in these genes is unknown. But the abundance of certain metabolic enzymes in tumors is correlated with poor clinical outcome in various forms of cancer96, 97. If an individual’s cells have an unusually high expression of a key enzyme(s) at baseline, then the acquisition of a transforming mutation in those cells might translate into enhanced growth and clinically aggressive features of the resulting tumor. Since the variance in expression of the enzyme would not independently cause cancer, the genes responsible for the variance would probably not be identified in linkage studies to find cancer-causing genes.
Finally, it will be interesting and important to fit tumor metabolism back into the context of the entire cancer patient. Regardless of the increased metabolic autonomy of tumor cells, tumor growth still relies on the metabolism of the host both to provide nutrients and to remove secreted waste products. Yet the issue of how tumors influence whole-body metabolism is still very much an open question, and one that relates directly to the health of cancer patients. For example, the phenomenon of cancer cachexia has been appreciated for centuries, but its causes are only partially understood and include processes that raise whole-body energy expenditure and stimulate catabolism in both the fat and muscle98, 99. Does the catabolic response function in part to provide nutrients to the tumor? In rats, tumor growth was associated with progressive increases in the synthesis and release of glutamine from muscle, ultimately resulting in decreased glutamine stores in the face of relentless tumor growth, consistent with glutamine ‘steal’ by the tumor100. Other studies have demonstrated markedly enhanced Cori cycle metabolism to convert lactate back to glucose in the liver of cancer patients who were suffering from progressive weight loss101. These studies demonstrate the interconnected nature of whole-body metabolism in patients with cancer. They also underscore the fact that any hope for rational, metabolically-directed cancer therapies depend on the development of a comprehensive understanding of metabolism in the host as well as in the tumor.
ACKNOWELDGMENTS
The author thanks his mentors, John J. Tudor, Haig H. Kazazian and Craig B. Thompson for guidance and support, and the William K. Bowes, Jr. Award Committee for their generosity. This work was supported by National Institutes of Health Grant K08 DK072565.
The author has received a K08 Award from the National Institutes of Health (Grant K08 DK072565)
REFERENCES
- 1.Warburg O. Uber den stoffwechsel der carcinomzelle. Klin. Wochenschr. 1925;4:534–536. [Google Scholar]
- 2.Warburg O. On respiratory impairment in cancer cells. Science. 1956 Aug 10;124(3215):269–270. [PubMed] [Google Scholar]
- 3.Krohn KA, Link JM, Mason RP. Molecular imaging of hypoxia. J Nucl Med. 2008 Jun;49 Suppl 2:129S–148S. doi: 10.2967/jnumed.107.045914. [DOI] [PubMed] [Google Scholar]
- 4.Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003 Oct;3(10):721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 5.Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004 Nov;4(11):891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 6.Matsumoto S, Hyodo F, Subramanian S, et al. Low-field paramagnetic resonance imaging of tumor oxygenation and glycolytic activity in mice. J Clin Invest. 2008 May 118;5:1965–1973. doi: 10.1172/JCI34928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arsham AM, Howell JJ, Simon MC. A novel hypoxia-inducible factor-independent hypoxic response regulating mammalian target of rapamycin and its targets. J Biol Chem. 2003 Aug 8;278(32):29655–29660. doi: 10.1074/jbc.M212770200. [DOI] [PubMed] [Google Scholar]
- 8.Lum JJ, Bui T, Gruber M, et al. The transcription factor HIF-1alpha plays a critical role in the growth factor-dependent regulation of both aerobic and anaerobic glycolysis. Genes Dev. 2007 May 1;21(9):1037–1049. doi: 10.1101/gad.1529107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malhotra R, Brosius FC., 3rd Glucose uptake and glycolysis reduce hypoxia-induced apoptosis in cultured neonatal rat cardiac myocytes. J Biol Chem. 1999 Apr 30;274(18):12567–12575. doi: 10.1074/jbc.274.18.12567. [DOI] [PubMed] [Google Scholar]
- 10.Deberardinis RJ, Sayed N, Ditsworth D, Thompson CB. Brick by brick: metabolism and tumor cell growth. Curr Opin Genet Dev. 2008 Feb;18(1):54–61. doi: 10.1016/j.gde.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moreno-Sanchez R, Rodriguez-Enriquez S, Marin-Hernandez A, Saavedra E. Energy metabolism in tumor cells. FEBS J. 2007 Mar;274(6):1393–1418. doi: 10.1111/j.1742-4658.2007.05686.x. [DOI] [PubMed] [Google Scholar]
- 12.Kim JW, Dang CV. Cancer's molecular sweet tooth and the Warburg effect. Cancer Res. 2006 Sep 15;66(18):8927–8930. doi: 10.1158/0008-5472.CAN-06-1501. [DOI] [PubMed] [Google Scholar]
- 13.Altenberg B, Greulich KO. Genes of glycolysis are ubiquitously overexpressed in 24 cancer classes. Genomics. 2004 Dec;84(6):1014–1020. doi: 10.1016/j.ygeno.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 14.Wang T, Marquardt C, Foker J. Aerobic glycolysis during lymphocyte proliferation. Nature. 1976 Jun 24;261(5562):702–705. doi: 10.1038/261702a0. [DOI] [PubMed] [Google Scholar]
- 15.Brand K. Glutamine and glucose metabolism during thymocyte proliferation. Pathways of glutamine and glutamate metabolism. Biochem J. 1985 Jun 1;228(2):353–361. doi: 10.1042/bj2280353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Newsholme EA, Crabtree B, Ardawi MS. The role of high rates of glycolysis and glutamine utilization in rapidly dividing cells. Biosci Rep. 1985 May;5(5):393–400. doi: 10.1007/BF01116556. [DOI] [PubMed] [Google Scholar]
- 17.Forbes NS, Meadows AL, Clark DS, Blanch HW. Estradiol stimulates the biosynthetic pathways of breast cancer cells: detection by metabolic flux analysis. Metab Eng. 2006 Nov;8(6):639–652. doi: 10.1016/j.ymben.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 18.DeBerardinis RJ, Mancuso A, Daikhin E, et al. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci U S A. 2007 Dec 4;104(49):19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Curi R, Newsholme P, Newsholme EA. Metabolism of pyruvate by isolated rat mesenteric lymphocytes, lymphocyte mitochondria and isolated mouse macrophages. Biochem J. 1988 Mar 1;250(2):383–388. doi: 10.1042/bj2500383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bauer DE, Harris MH, Plas DR, et al. Cytokine stimulation of aerobic glycolysis in hematopoietic cells exceeds proliferative demand. FASEB J. 2004 Aug;18(11):1303–1305. doi: 10.1096/fj.03-1001fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Warburg O. On the origin of cancer cells. Science. 1956 Feb 24;123(3191):309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 22.Kim JW, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006 Mar;3(3):177–185. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 23.Papandreou I, Cairns RA, Fontana L, Lim AL. Denko NC. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 2006 Mar;3(3):187–197. doi: 10.1016/j.cmet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 24.Gordan JD, Simon MC. Hypoxia-inducible factors: central regulators of the tumor phenotype. Curr Opin Genet Dev. 2007 Feb;17(1):71–77. doi: 10.1016/j.gde.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ookhtens M, Kannan R, Lyon I, Baker N. Liver and adipose tissue contributions to newly formed fatty acids in an ascites tumor. Am J Physiol. 1984 Jul;247(1 Pt 2):R146–R153. doi: 10.1152/ajpregu.1984.247.1.R146. [DOI] [PubMed] [Google Scholar]
- 26.Kuhajda FP. Fatty-acid synthase and human cancer: new perspectives on its role in tumor biology. Nutrition. 2000 Mar;16(3):202–208. doi: 10.1016/s0899-9007(99)00266-x. [DOI] [PubMed] [Google Scholar]
- 27.Parlo RA, Coleman PS. Enhanced rate of citrate export from cholesterol-rich hepatoma mitochondria. The truncated Krebs cycle and other metabolic ramifications of mitochondrial membrane cholesterol. J Biol Chem. 1984 Aug 25;259(16):9997–10003. [PubMed] [Google Scholar]
- 28.Parlo RA, Coleman PS. Continuous pyruvate carbon flux to newly synthesized cholesterol and the suppressed evolution of pyruvate-generated CO2 in tumors: further evidence for a persistent truncated Krebs cycle in hepatomas. Biochim Biophys Acta. 1986 Apr 29;886(2):169–176. doi: 10.1016/0167-4889(86)90134-5. [DOI] [PubMed] [Google Scholar]
- 29.Eagle H. Nutrition needs of mammalian cells in tissue culture. Science. 1955 Sep 16;122(3168):501–514. doi: 10.1126/science.122.3168.501. [DOI] [PubMed] [Google Scholar]
- 30.Kovacevic Z, McGivan JD. Mitochondrial metabolism of glutamine and glutamate and its physiological significance. Physiol Rev. 1983 Apr;63(2):547–605. doi: 10.1152/physrev.1983.63.2.547. [DOI] [PubMed] [Google Scholar]
- 31.Reitzer LJ, Wice BM, Kennell D. Evidence that glutamine, not sugar, is the major energy source for cultured HeLa cells. J Biol Chem. 1979 Apr 25;254(8):2669–2676. [PubMed] [Google Scholar]
- 32.Mates JM, Segura JA, Alonso FJ, Marquez J. Pathways from glutamine to apoptosis. Front Biosci. 2006;11:3164–3180. doi: 10.2741/2040. [DOI] [PubMed] [Google Scholar]
- 33.Donadio AC, Lobo C, Tosina M, et al. Antisense glutaminase inhibition modifies the O-GlcNAc pattern and flux through the hexosamine pathway in breast cancer cells. J Cell Biochem. 2008 Feb 15;103(3):800–811. doi: 10.1002/jcb.21449. [DOI] [PubMed] [Google Scholar]
- 34.Portais JC, Voisin P, Merle M, Canioni P. Glucose and glutamine metabolism in C6 glioma cells studied by carbon 13 NMR. Biochimie. 1996;78(3):155–164. doi: 10.1016/0300-9084(96)89500-9. [DOI] [PubMed] [Google Scholar]
- 35.Yuneva M, Zamboni N, Oefner P, Sachidanandam R, Lazebnik Y. Deficiency in glutamine but not glucose induces MYC-dependent apoptosis in human cells. J Cell Biol. 2007 Jul 2;178(1):93–105. doi: 10.1083/jcb.200703099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu Z, Jiang W, McGinley JN, Thompson HJ. 2-Deoxyglucose as an energy restriction mimetic agent: effects on mammary carcinogenesis and on mammary tumor cell growth in vitro. Cancer Res. 2005 Aug 1;65(15):7023–7030. doi: 10.1158/0008-5472.CAN-05-0453. [DOI] [PubMed] [Google Scholar]
- 37.Christofk HR, Vander Heiden MG, Harris MH, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008 Mar 13;452(7184):230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 38.Fantin VR, St-Pierre J, Leder P. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell. 2006 Jun;9(6):425–434. doi: 10.1016/j.ccr.2006.04.023. [DOI] [PubMed] [Google Scholar]
- 39.Hatzivassiliou G, Zhao F, Bauer DE, et al. ATP citrate lyase inhibition can suppress tumor cell growth. Cancer Cell. 2005 Oct;8(4):311–321. doi: 10.1016/j.ccr.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 40.Lobo C, Ruiz-Bellido MA, Aledo JC, Marquez J, Nunez De Castro I, Alonso FJ. Inhibition of glutaminase expression by antisense mRNA decreases growth and tumourigenicity of tumour cells. Biochem J. 2000 Jun 1;348(Pt 2):257–261. [PMC free article] [PubMed] [Google Scholar]
- 41.Ovejera AA, Houchens DP, Catane R, Sheridan MA, Muggia FM. Efficacy of 6-diazo-5-oxo-L-norleucine and N-[N-gamma-glutamyl-6-diazo-5-oxo-norleucinyl]-6-diazo-5-oxo-norleucine against experimental tumors in conventional and nude mice. Cancer Res. 1979 Aug;39(8):3220–3224. [PubMed] [Google Scholar]
- 42.Buzzai M, Bauer DE, Jones RG, et al. The glucose dependence of Akt-transformed cells can be reversed by pharmacologic activation of fatty acid beta-oxidation. Oncogene. 2005 Jun 16;24(26):4165–4173. doi: 10.1038/sj.onc.1208622. [DOI] [PubMed] [Google Scholar]
- 43.Buzzai M, Jones RG, Amaravadi RK, et al. Systemic treatment with the antidiabetic drug metformin selectively impairs p53-deficient tumor cell growth. Cancer Res. 2007 Jul 15;67(14):6745–6752. doi: 10.1158/0008-5472.CAN-06-4447. [DOI] [PubMed] [Google Scholar]
- 44.Ramanathan A, Wang C, Schreiber SL. Perturbational profiling of a cell-line model of tumorigenesis by using metabolic measurements. Proc Natl Acad Sci U S A. 2005 Apr 26;102(17):5992–5997. doi: 10.1073/pnas.0502267102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000 Jan 7;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 46.Vander Heiden MG, Plas DR, Rathmell JC, Fox CJ, Harris MH, Thompson CB. Growth factors can influence cell growth and survival through effects on glucose metabolism. Mol Cell Biol. 2001 Sep;21(17):5899–5912. doi: 10.1128/MCB.21.17.5899-5912.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lum JJ, Bauer DE, Kong M, et al. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005 Jan 28;120(2):237–248. doi: 10.1016/j.cell.2004.11.046. [DOI] [PubMed] [Google Scholar]
- 48.Deberardinis RJ, Lum JJ, Thompson CB. Phosphatidylinositol 3-kinase-dependent modulation of carnitine palmitoyltransferase 1A expression regulates lipid metabolism during hematopoietic cell growth. J Biol Chem. 2006 Dec 8;281(49):37372–37380. doi: 10.1074/jbc.M608372200. [DOI] [PubMed] [Google Scholar]
- 49.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008 Jan;7(1):11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 50.Kawauchi K, Araki K, Tobiume K, Tanaka N. p53 regulates glucose metabolism through an IKK-NF-kappaB pathway and inhibits cell transformation. Nat Cell Biol. 2008 May;10(5):611–618. doi: 10.1038/ncb1724. [DOI] [PubMed] [Google Scholar]
- 51.Bensaad K, Tsuruta A, Selak MA, et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006 Jul 14;126(1):107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 52.Kondoh H, Lleonart ME, Gil J, et al. Glycolytic enzymes can modulate cellular life span. Cancer Res. 2005 Jan 1;65(1):177–185. [PubMed] [Google Scholar]
- 53.Matoba S, Kang JG, Patino WD, et al. p53 regulates mitochondrial respiration. Science. 2006 Jun 16;312(5780):1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 54.Franke TF, Hornik CP, Segev L, Shostak GA, Sugimoto C. PI3K/Akt and apoptosis: size matters. Oncogene. 2003 Dec 8;22(56):8983–8998. doi: 10.1038/sj.onc.1207115. [DOI] [PubMed] [Google Scholar]
- 55.Samuels Y, Wang Z, Bardelli A, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004 Apr 23;304(5670):554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 56.Chian R, Young S, Danilkovitch-Miagkova A, et al. Phosphatidylinositol 3 kinase contributes to the transformation of hematopoietic cells by the D816V c-Kit mutant. Blood. 2001 Sep 1;98(5):1365–1373. doi: 10.1182/blood.v98.5.1365. [DOI] [PubMed] [Google Scholar]
- 57.Slamon DJ, Godolphin W, Jones LA, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989 May 12;244(4905):707–712. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- 58.Knobbe CB, Merlo A, Reifenberger G. Pten signaling in gliomas. Neuro Oncol. 2002 Jul;4(3):196–211. [PMC free article] [PubMed] [Google Scholar]
- 59.Pedrero JM, Carracedo DG, Pinto CM, et al. Frequent genetic and biochemical alterations of the PI 3-K/AKT/PTEN pathway in head and neck squamous cell carcinoma. Int J Cancer. 2005 Mar 20;114(2):242–248. doi: 10.1002/ijc.20711. [DOI] [PubMed] [Google Scholar]
- 60.Barata JT, Silva A, Brandao JG, Nadler LM, Cardoso AA, Boussiotis VA. Activation of PI3K is indispensable for interleukin 7-mediated viability, proliferation, glucose use, and growth of T cell acute lymphoblastic leukemia cells. J Exp Med. 2004 Sep 6;200(5):659–669. doi: 10.1084/jem.20040789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rathmell JC, Fox CJ, Plas DR, Hammerman PS, Cinalli RM, Thompson CB. Akt-directed glucose metabolism can prevent Bax conformation change and promote growth factor-independent survival. Mol Cell Biol. 2003 Oct;23(20):7315–7328. doi: 10.1128/MCB.23.20.7315-7328.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Elstrom RL, Bauer DE, Buzzai M, et al. Akt stimulates aerobic glycolysis in cancer cells. Cancer Res. 2004 Jun 1;64(11):3892–3899. doi: 10.1158/0008-5472.CAN-03-2904. [DOI] [PubMed] [Google Scholar]
- 63.Bauer DE, Hatzivassiliou G, Zhao F, Andreadis C, Thompson CB. ATP citrate lyase is an important component of cell growth and transformation. Oncogene. 2005 Sep 15;24(41):6314–6322. doi: 10.1038/sj.onc.1208773. [DOI] [PubMed] [Google Scholar]
- 64.Gingras AC, Raught B, Sonenberg N. Regulation of translation initiation by FRAP/mTOR. Genes Dev. 2001 Apr 1;15(17):807–826. doi: 10.1101/gad.887201. [DOI] [PubMed] [Google Scholar]
- 65.Roberts PJ, Der CJ. Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene. 2007 May 14;26(22):3291–3310. doi: 10.1038/sj.onc.1210422. [DOI] [PubMed] [Google Scholar]
- 66.Flier JS, Mueckler MM, Usher P, Lodish HF. Elevated levels of glucose transport and transporter messenger RNA are induced by ras or src oncogenes. Science. 1987 Mar 20;235(4795):1492–1495. doi: 10.1126/science.3103217. [DOI] [PubMed] [Google Scholar]
- 67.Blum R, Jacob-Hirsch J, Amariglio N, Rechavi G, Kloog Y. Ras inhibition in glioblastoma down-regulates hypoxia-inducible factor-1alpha, causing glycolysis shutdown and cell death. Cancer Res. 2005 Feb 1;65(3):999–1006. [PubMed] [Google Scholar]
- 68.Vizan P, Boros LG, Figueras A, et al. K-ras codon-specific mutations produce distinctive metabolic phenotypes in NIH3T3 mice [corrected] fibroblasts. Cancer Res. 2005 Jul 1;65(13):5512–5515. doi: 10.1158/0008-5472.CAN-05-0074. [DOI] [PubMed] [Google Scholar]
- 69.Kole HK, Resnick RJ, Van Doren M, Racker E. Regulation of 6-phosphofructo-1-kinase activity in ras-transformed rat-1 fibroblasts. Arch Biochem Biophys. 1991 May 1;286(2):586–590. doi: 10.1016/0003-9861(91)90084-v. [DOI] [PubMed] [Google Scholar]
- 70.Telang S, Yalcin A, Clem AL, et al. Ras transformation requires metabolic control by 6-phosphofructo-2-kinase. Oncogene. 2006 Nov 23;25(55):7225–7234. doi: 10.1038/sj.onc.1209709. [DOI] [PubMed] [Google Scholar]
- 71.Telang S, Lane AN, Nelson KK, Arumugam S, Chesney J. The oncoprotein H-RasV12 increases mitochondrial metabolism. Mol Cancer. 2007;6:77. doi: 10.1186/1476-4598-6-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nikiforov MA, Chandriani S, O'Connell B, et al. A functional screen for Myc-responsive genes reveals serine hydroxymethyltransferase, a major source of the one-carbon unit for cell metabolism. Mol Cell Biol. 2002 Aug;22(16):5793–5800. doi: 10.1128/MCB.22.16.5793-5800.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Astuti D, Latif F, Dallol A, et al. Gene mutations in the succinate dehydrogenase subunit SDHB cause susceptibility to familial pheochromocytoma and to familial paraganglioma. Am J Hum Genet. 2001 Jul;69(1):49–54. doi: 10.1086/321282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Baysal BE, Ferrell RE, Willett-Brozick JE, et al. Mutations in SDHD, a mitochondrial complex II gene, in hereditary paraganglioma. Science. 2000 Feb 4;287(5454):848–851. doi: 10.1126/science.287.5454.848. [DOI] [PubMed] [Google Scholar]
- 75.Niemann S, Muller U. Mutations in SDHC cause autosomal dominant paraganglioma, type 3. Nat Genet. 2000 Nov;26(3):268–270. doi: 10.1038/81551. [DOI] [PubMed] [Google Scholar]
- 76.Gimenez-Roqueplo AP, Favier J, Rustin P, et al. The R22X mutation of the SDHD gene in hereditary paraganglioma abolishes the enzymatic activity of complex II in the mitochondrial respiratory chain and activates the hypoxia pathway. Am J Hum Genet. 2001 Dec;69(6):1186–1197. doi: 10.1086/324413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tomlinson IP, Alam NA, Rowan AJ, et al. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat Genet. 2002 Apr;30(4):406–410. doi: 10.1038/ng849. [DOI] [PubMed] [Google Scholar]
- 78.Selak MA, Armour SM, MacKenzie ED, et al. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase. Cancer Cell. 2005 Jan;7(1):77–85. doi: 10.1016/j.ccr.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 79.Canter JA, Kallianpur AR, Parl FF, Millikan RC. Mitochondrial DNA G10398A polymorphism and invasive breast cancer in African-American women. Cancer Res. 2005 Sep 1;65(17):8028–8033. doi: 10.1158/0008-5472.CAN-05-1428. [DOI] [PubMed] [Google Scholar]
- 80.Liu VW, Wang Y, Yang HJ, et al. Mitochondrial DNA variant 16189T>C is associated with susceptibility to endometrial cancer. Hum Mutat. 2003 Aug;22(2):173–174. doi: 10.1002/humu.10244. [DOI] [PubMed] [Google Scholar]
- 81.Petros JA, Baumann AK, Ruiz-Pesini E, et al. mtDNA mutations increase tumorigenicity in prostate cancer. Proc Natl Acad Sci U S A. 2005 Jan 18;102(3):719–724. doi: 10.1073/pnas.0408894102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Brandon M, Baldi P, Wallace DC. Mitochondrial mutations in cancer. Oncogene. 2006 Aug 7;25(34):4647–4662. doi: 10.1038/sj.onc.1209607. [DOI] [PubMed] [Google Scholar]
- 83.Day SE, Kettunen MI, Gallagher FA, et al. Detecting tumor response to treatment using hyperpolarized 13C magnetic resonance imaging and spectroscopy. Nat Med. 2007 Nov;13(11):1382–1387. doi: 10.1038/nm1650. [DOI] [PubMed] [Google Scholar]
- 84.Kim BJ, Forbes NS. Flux analysis shows that hypoxia-inducible-factor-1-alpha minimally affects intracellular metabolism in tumor spheroids. Biotechnol Bioeng. 2007 Apr 15;96(6):1167–1182. doi: 10.1002/bit.21205. [DOI] [PubMed] [Google Scholar]
- 85.Cocco P, Todde P, Fornera S, Manca MB, Manca P, Sias AR. Mortality in a cohort of men expressing the glucose-6-phosphate dehydrogenase deficiency. Blood. 1998 Jan 15;91(2):706–709. [PubMed] [Google Scholar]
- 86.Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 87.Lee HR, Cho JM, Shin DH, et al. Adaptive response to GSH depletion and resistance to L:-buthionine-(S,R)-sulfoximine: involvement of Nrf2 activation. Mol Cell Biochem. 2008 Jun 28; doi: 10.1007/s11010-008-9853-y. [DOI] [PubMed] [Google Scholar]
- 88.Kawachi Y, Xu X, Taguchi S, et al. Attenuation of UVB-induced sunburn reaction and oxidative DNA damage with no alterations in UVB-induced skin carcinogenesis in Nrf2 gene-deficient mice. J Invest Dermatol. 2008 Jul;128(17):1773–1779. doi: 10.1038/sj.jid.5701245. [DOI] [PubMed] [Google Scholar]
- 89.Cho JM, Manandhar S, Lee HR, Park HM, Kwak MK. Role of the Nrf2-antioxidant system in cytotoxicity mediated by anticancer cisplatin: implication to cancer cell resistance. Cancer Lett. 2008 Feb 18;260(1–2):96–108. doi: 10.1016/j.canlet.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 90.Iida K, Itoh K, Kumagai Y, et al. Nrf2 is essential for the chemopreventive efficacy of oltipraz against urinary bladder carcinogenesis. Cancer Res. 2004 Sep 15;64(18):6424–6431. doi: 10.1158/0008-5472.CAN-04-1906. [DOI] [PubMed] [Google Scholar]
- 91.Ramos-Gomez M, Kwak MK, Dolan PM, et al. Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in nrf2 transcription factor-deficient mice. Proc Natl Acad Sci U S A. 2001 Mar 13;98(6):3410–3415. doi: 10.1073/pnas.051618798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jones RG, Plas DR, Kubek S, et al. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol Cell. 2005 Apr 29;18(3):283–293. doi: 10.1016/j.molcel.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 93.Mandal S, Guptan P, Owusu-Ansah E, Banerjee U. Mitochondrial regulation of cell cycle progression during development as revealed by the tenured mutation in Drosophila. Dev Cell. 2005 Dec;9(6):843–854. doi: 10.1016/j.devcel.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 94.Owusu-Ansah E, Yavari A, Mandal S, Banerjee U. Distinct mitochondrial retrograde signals control the G1-S cell cycle checkpoint. Nat Genet. 2008 Mar;40(3):356–361. doi: 10.1038/ng.2007.50. [DOI] [PubMed] [Google Scholar]
- 95.Cheung VG, Conlin LK, Weber TM, et al. Natural variation in human gene expression assessed in lymphoblastoid cells. Nat Genet. 2003 Mar;33(3):422–425. doi: 10.1038/ng1094. [DOI] [PubMed] [Google Scholar]
- 96.Langbein S, Zerilli M, Zur Hausen A, et al. Expression of transketolase TKTL1 predicts colon and urothelial cancer patient survival: Warburg effect reinterpreted. Br J Cancer. 2006 Feb 27;94(4):578–585. doi: 10.1038/sj.bjc.6602962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Krockenberger M, Honig A, Rieger L, et al. Transketolase-like 1 expression correlates with subtypes of ovarian cancer and the presence of distant metastases. Int J Gynecol Cancer. 2007 Jan-Feb;17(1):101–106. doi: 10.1111/j.1525-1438.2007.00799.x. [DOI] [PubMed] [Google Scholar]
- 98.Bosaeus I, Daneryd P, Svanberg E, Lundholm K. Dietary intake and resting energy expenditure in relation to weight loss in unselected cancer patients. Int J Cancer. 2001 Aug 1;93(3):380–383. doi: 10.1002/ijc.1332. [DOI] [PubMed] [Google Scholar]
- 99.Tisdale MJ. Molecular pathways leading to cancer cachexia. Physiology (Bethesda) 2005 Oct;20:340–348. doi: 10.1152/physiol.00019.2005. [DOI] [PubMed] [Google Scholar]
- 100.Chen MK, Espat NJ, Bland KI, Copeland EM, 3rd, Souba WW. Influence of progressive tumor growth on glutamine metabolism in skeletal muscle and kidney. Ann Surg. 1993 Jun;217(6):655–666. doi: 10.1097/00000658-199306000-00007. discussion 666-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Holroyde CPGT, Putnam RC, Paul P, Reichard GA. Altered glucose metabolism in metastatic carcinoma. Cancer Research. 1975;35:3710–3714. [PubMed] [Google Scholar]