Abstract
Dynamic nuclear polarization can create hyperpolarized compounds with magnetic resonance (MR) signal-to-noise ratio enhancements on the order of 10,000-fold. Both exogenous and normally occurring endogenous compounds can be polarized, and their initial concentration and downstream metabolic products can be assessed using MR spectroscopy. Given the transient nature of the hyperpolarized signal enhancement, fast imaging techniques are a critical requirement for real-time metabolic imaging. We report on the development of an ultrafast, multi-slice, spiral chemical shift imaging sequence, with subsecond acquisition time, achieved on a clinical MR scanner. The technique was used for dynamic metabolic imaging in rats, with measurement of time-resolved spatial distributions of hyperpolarized 13C1-pyruvate and metabolic products 13C1-lactate and 13C1-alanine, with a temporal resolution of as fast as 1 second. Metabolic imaging revealed different signal time courses in liver from kidney. These results demonstrate the feasibility of real-time, hyperpolarized metabolic imaging and highlight its potential in assessing organ-specific kinetic parameters.
Keywords: dynamic nuclear polarization, pyruvate, dynamic metabolic imaging
Introduction
The recent development of hyperpolarized magnetic resonance imaging (MRI) agents, i.e., MRI-visible compounds whose magnetization is much higher than that normally achieved at in vivo temperatures, presents both unprecedented opportunities and new technical challenges. Signal-to-noise ratio (SNR) enhancements on the order of the 10,000-fold can be achieved using either dynamic nuclear polarization (DNP) (1,2) or para-hydrogen induced polarization based on the para-hydrogen and synthesis allow dramatically enhanced nuclear alignment (PASADENA) effect (3,4). Polarization of metabolically active substrates permits in vivo chemical shift imaging (CSI), or metabolic imaging, of both the injected agent and downstream metabolic products, which can be differentiated from the substrate based on their chemical shifts (5). This feature of hyperpolarized CSI provides a unique method for noninvasive monitoring of critical dynamic metabolic processes under both normal and pathologic conditions. As a noninvasive in vivo tool it is ideally suited for longitudinal studies in animals, and, ultimately, in clinical applications.
Numerous compounds have been identified as potential candidates for in vivo hyperpolarized studies, with long T1 nuclear magnetic resonance relaxation times and low toxicity as the primary constraints. Among the viable substrates, pyruvate is so far the most researched candidate. Recent studies have demonstrated that both anaerobic and aerobic metabolism can be studied in vivo following the bolus injection of 13C1-pyruvate hyperpolarized with DNP (5,6). Potential applications include tumor diagnosis (7,8) and treatment monitoring (9), cardiovascular pathologies (10–12), neurovascular and neurological diseases (13), and metabolic disorders (14).
Because the enhanced magnetization is non-recoverable and decaying toward its thermal equilibrium value, fast imaging techniques are critical to follow dynamic in vivo processes. Moreover, in metabolic imaging of hyperpolarized compounds, a reduction in scan time does not necessarily lead to a loss in SNR because the longitudinal magnetization is not replenished between excitations. When the data acquisition is properly timed, in the case of hyperpolarisation the SNR for fast sequences can actually be higher than that for the gold standard phase-encoded CSI (14). Recently, several fast CSI techniques have been developed for the application to metabolic imaging of hyperpolarized substrates (15–18). In this work, we focus on spiral imaging because of its very high spatial encoding efficiency, which is achieved by applying gradients simultaneously in two dimensions. In combination with the SNR increase afforded by hyperpolarization, this technique provides the spatial and temporal resolution required for real-time metabolic imaging.
Material and Methods
Spiral CSI (19) is a fast spectroscopic imaging technique that simultaneously encodes spectral and 2D spatial information with each excitation. Therefore, it potentially allows the complete acquisition of a given slice after a single excitation. However, for most applications, multiple excitations are necessary to achieve the desired spectral bandwidth (SW) due to limitations in gradient hardware. In the absence of anaplerosis, the metabolic products of 13C1-labeled pyruvate (Pyr) comprise 13C1-lactate (Lac), 13C1-alanine (Ala), and 13C-bicarbonate (Bic). Additionally, 13C1-pyruvate hydrate (Pyh) is present in dynamic equilibrium with pyruvate (< 8%) and itself is not metabolically active (5). The corresponding sparse spectrum can be exploited to reduce the number of interleaves by undersampling the data in the spectral domain (15). If the spectral bandwidth is chosen appropriately, spectral overlap with aliased signals can be reduced or completely avoided. The evolution of the chemical shift leads to a phase accrual during the data acquisition in the presence of the spiral readout gradients. Therefore, a linear phase correction has to be performed to avoid image-blurring artifacts. As this cannot simultaneously be done for spectral components that have been spectrally aliased a different number of times, multiple reconstructions are necessary in which only components with resonance frequencies within a selected bandwidth are reconstructed “in-focus” while components outside that band are severely blurred (“spectral tomosynthesis”). Here we have implemented a multi-slice variant of undersampled spiral CSI with a total acquisition time per slice of only 375 ms on a clinical MR scanner and applied the method to dynamic metabolic imaging of the rat after injection of hyperpolarized 13C1-pyruvate. As there is no recovery of the longitudinal magnetization for hyperpolarized compounds, there is a trade-off between the number of temporal samples and the SNR for each sample. Therefore, we performed two experiments with different temporal resolutions and sampling rates.
Substrate and Polarization Procedure
The polarized substrate consists of a mixture of 14-M 13C-labeled pyruvic acid enriched to 99% in the C1 position mixed with 15-mM Oxo63 trityl radical. After adding 3 µL of a 50:1 dilution of ProHance (Bracco Diagnostics Inc., Princeton, NJ) to 40 mg of the pyruvate mixture to increase the degree of polarization by reducing the T1 relaxation time of the unpaired electrons, the sample was polarized using a HyperSense system (Oxford Instruments Molecular Biotools, Oxford, UK) operating at 3.35 T. The polarization and dissolution process was similar to that described by Ardenkjaer-Larsen et al. (1). Dynamic nuclear polarization was achieved over approximately 90 min by microwave irradiation at the electron resonance frequency of 94.1 GHz of the sample cooled to 1.4 K. The polarized sample was then dissolved with 4.56 g of a 100-mM NaOH solution, mixed with 40-mM TRIS buffer and 0.1-g/L EDTA-Na2, heated to 185°C at 10 bar, leading to a 100-mM solution of hyperpolarized pyruvate with a pH of 7.5 ± 0.3 (mean ± s.d., n = 6). The final liquid-state polarization was between 20 and 25% as estimated from the solid-state polarization buildup curve (scale in arbitrary units) for each sample and independent calibration experiments in which the liquid-state polarization of a sample was measured directly after dissolution in a separate bench top spectrometer. This estimation assumed that the loss of polarization that occurred during the dissolution process did not change appreciably since performing the calibration measurements. The time delay between dissolution to injection into the animal ranged from 16 to 20 s. All the chemicals required for the polarization and dissolution process remained in the pyruvate solution, and, hence, were injected into the animal.
Animal Handling
Each of the healthy male Wistar rats (410–455 g body weight) was anesthetized with 1–3% isoflurane in oxygen (~1.5 L/min). A microbore catheter (0.5 mL volume) was inserted into a tail vein for administration of the hyperpolarized substrate. The animals were held in an MR-invisible plastic structure with a nose cone for delivery of isoflurane anesthesia and were placed in the radio frequency (RF) coil in a prone position. Respiration, rectal temperature, heart rate, and oxygen saturation from the hind limb were monitored throughout the experiment. Rectal temperature was kept at 37°C by heating the animal with a temperature-controlled warm water blanket. Before each injection of hyperpolarized pyruvate, 0.5 mL of saline with 1% heparin was injected to prevent catheter clotting. For each 13C-CSI experiment, 1.6 mL of the final pyruvate solution were injected manually through the tail vein catheter at a rate of approximately 0.25 mL/s. Due to the dead volume of the catheter, this resulted in 1.1 mL being injected into each animal. Immediately following the CSI measurements, the residual pyruvate in the catheter was cleared by injecting another 0.5 mL of saline/heparin. At the end of the experiment, the animals were euthanized by a combination of isoflurane overdose and a lethal injection of pentobarbital. All procedures were approved by the Institutional Animal Care and Use Committees at Stanford University and SRI International.
MR Hardware and Acquisition Parameters
All experiments were performed on a clinical 3T Signa MR scanner (GE Healthcare, Waukesha, WI) equipped with self-shielded gradients (40 mT/m, 150 mT/m/ms). A custom-built dual-tuned (1H/13C) quadrature rat coil (inner diameter: 80 mm, length: 90 mm), operating at 127.7 MHz and 32.1 MHz, respectively, was used for both RF excitation and signal reception. The coil was based on a previously published design (20) with a second half-Helmholtz unit added to provide quadrature operation in the proton mode.
Single-shot fast spin-echo proton MR images were acquired in axial, sagittal, and coronal orientations as anatomical references for prescribing the 13C-CSI experiments. In each direction, up to 45 2-mm slices were acquired with 0-mm separation and a nominal in-plane resolution of 0.47 mm (256×192 matrix, echo time (TE)/repetition time (TR) = 38.6/1492 ms).
The implemented spiral CSI sequence (15) consisted of a slice(z)-selective excitation and a spiral readout gradient for combined spatial(xy)-spectral(f) encoding. The spiral waveforms were designed for a field-of-view (FOV) of 80×80 mm2 with a nominal 5×5-mm2 in-plane resolution. After each excitation, 32 gradient echoes were acquired with an interval of 3.6 ms corresponding to a SW of 276.2 Hz. With three spatial interleaves and a TR of 125 ms, the total acquisition time per slice was 375 ms. In each CSI experiment, the excitation flip angle was progressively increased as (21) where ntot denotes the total number of excitations per slice and 1 ≤ i ≤ ntot. For each excitation, this scheme leads to the same amount of transverse magnetization as created by the previous excitation if there is no additional change in longitudinal magnetization between the excitations. This is necessary to avoid or reduce artifacts that can occur when multiple spatial interleaves are necessary in spiral imaging to acquire an entire slice. Considering the TR of only 125 ms, any changes in longitudinal magnetization, e.g., loss of polarization due to longitudinal relaxation, change of concentration due to metabolic conversion and slice in- or out-flow, should be very small. Thus, any changes that occur between consecutive measurements of any given slice will correctly be reflected in the time courses of the measured signal intensities for the various metabolites. As there is no recovery of longitudinal magnetization for hyperpolarized compounds, the applied variable flip angle scheme leads to a trade-off between SNR and the number of temporal samples N. For a given observation duration, the SNR for each sample is inversely proportional to √N when neglecting any change in magnetization other than the depletion due to RF excitation. Therefore, we performed two experiments with different sampling rates and number of samples; one experiment with high SNR at a temporal resolution of 6 s with only 6 temporal samples (ntot = 18) and a second experiment with a higher temporal resolution of 1 s, i.e., close to the maximum sampling rate of the sequence when measuring two slices, and 20 time points (ntot = 60). The acquisition slice order was inferior-to-superior, i.e., bowel–kidney–liver in experiment 1 (cf. Fig. 1a) and kidney-liver in experiment 2 (cf. Fig. 6a). The transmit RF power was calibrated using a reference phantom containing a 1.77-M solution of 13C1-lactate that was placed on top of the animal. The 13C spectrometer frequency was adjusted to 200 Hz upfield of the lactate reference signal.
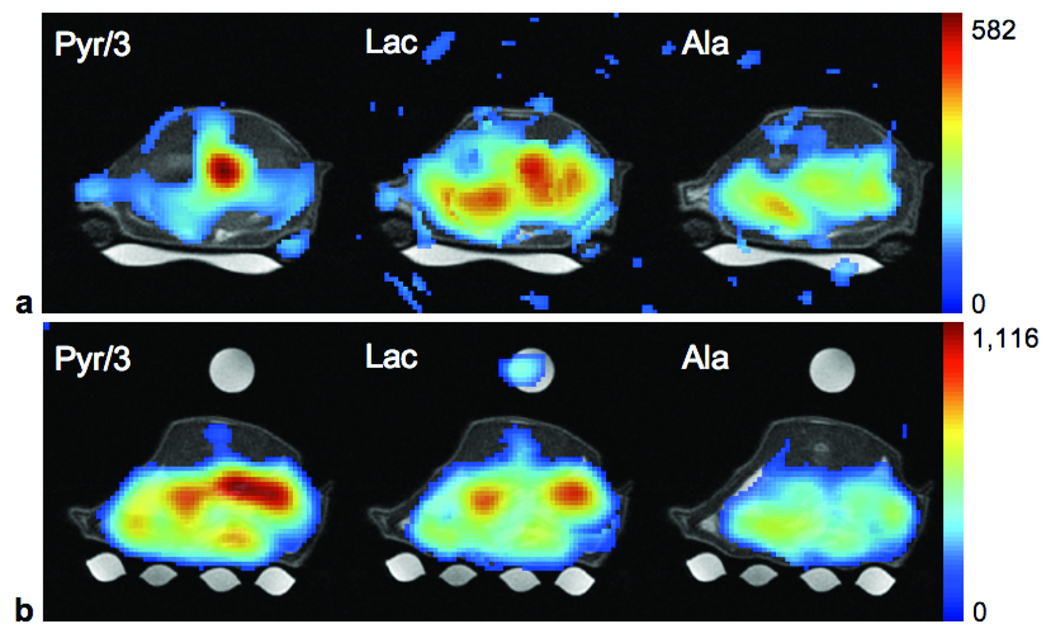
Figure 1.
High-resolution proton MRI used for anatomical reference. The coronal image (a) shows the location of the slices acquired in the 13C-CSI data sets with a temporal resolution of 6 s. The yellow squares (interpolated nominal size in-plane: 1.25×1.25 mm2) in the axial images from slices through the liver (b) and kidney (c) indicate the origin of the spectra shown in Fig. 2. Also shown in green are the ROIs evaluated for measuring the time courses of the individual metabolites (Fig. 4). The axial images have been cropped to the size of the metabolic images. The middle slice (c) shows the Lac phantom on top of the animal, below the animal is the water-filled blanket (for heating of the animal).
Figure 6.
(a) Coronal proton MRI showing the location of the slices acquired in the 13C-CSI data set with 1-s temporal resolution. Axial images from slices through the liver (b) and kidney (c) indicating the ROIs used to calculate the time courses for Pyr (black □), Lac (blue ×), and Ala (red ○) from ROIs in the liver (d) and left kidney (e). Also plotted are the time courses of Pyr from vascular ROIs in the two slices (dashed magenta ◊). The time scale is relative to start of injection. Note that the Pyr signal is scaled down by a factor of 3. (f) Time-resolved Lac/Pyr (blue) and Ala/Pyr (red) ratios for the liver (dashed) and kidney (solid) ROI. (g) Time-resolved Lac/Pyr (blue) and Ala/Pyr (red) ratios from ROIs encompassing the entire liver (dashed) and kidney (solid) slices.
Data Processing and Analysis
All data processing was performed with custom software using Matlab (MathWorks Inc., Natick, MA). First, for each time point, the data for the individual interleaves were multiplied by a correction factor calculated as the average signal from eleven data points of the first spiral readout (points 15 to 25) from all three interleaves. This was done to correct for any rapid signal changes occurring during the 125 ms between the excitations for the individual interleaves. The data were then apodized in the spectral dimension with a 7.5-Hz Gaussian line broadening and zero-padding up to 128 points. After fast Fourier transform (FFT) along kf, a frequency-dependent linear phase correction was applied along the readout direction to remove the chemical-shift artifact. When imaging 13C1-pyruvate and its metabolic products, three spectral tomosynthesis reconstructions were performed, one for Pyr at 171.5 ppm and Ala at 177 ppm, one for Lac at 183.5 ppm, and one for Bic at 161.5 ppm. After gridding, the data were apodized with a generalized Hamming window (αapo = 0.66) and zero-padded up to 64×64 pixels. Finally, a 2D-FFT was performed and the data were corrected for nonuniform spatial frequency sampling density. Spectra from each time point were corrected for B0 inhomogeneity using a frequency offset map calculated from the Pyr data averaged over all time points. Metabolic maps for Pyr, Lac, and Ala were calculated by integrating the signal within a 48-Hz interval around each peak. Each resonance was phase-corrected separately by multiplying the spectrum with a constant phase factor so that the main peak appears in absorption mode. The baseline was estimated by a straight line through the spectral points at the beginning and end of the integration interval. For the display of individual spectra, a manual first-order phase correction was performed and the baseline was subtracted by fitting a spline to signal-free regions of the smoothed spectrum. Organ-specific metabolite time courses were calculated as the total signal in the respective metabolic maps in regions of interest (ROIs) as defined on the corresponding proton reference image. Additionally, time courses were calculated for ROIs encompassing the vena cava and aorta from each slice, and for large ROIs circumscribing the whole animal in the respective slice. The time courses were normalized to the total 13C signal from the liver and kidney slices.
Results and Discussion
In CSI experiment 1, three 10-mm slices with equidistant separation were selected with the most superior slice predominantly through the liver and the middle slice through the right kidney (Fig. 1). The CSI data from the most inferior slice (mostly bowel) were not evaluated. Time-resolved CSI was performed with six time frames and a 6-s sampling interval beginning 5 s after the start of the 6-s pyruvate injection.
Representative spectra from the third time frame, from voxels in the liver and the right kidney, are shown in Fig. 2. As the slice order was inferior-to-superior with an acquisition time per slice of 375 ms, the acquisition of the kidney spectrum started at t = 17.4 s and of the liver spectrum at t = 17.8 s from the start of injection. The positions of the voxels are indicated on the axial proton images in Fig. 1b and 1c, respectively. Due to the spectral undersampling, multiple reconstructions are necessary for the different spectral components. Relative to Lac, both Pyr and Ala are aliased once, whereas Bic is aliased three times. With the chosen SW of 276.2 Hz, the signal from Pyh at 180 ppm, which is not aliased relative to Lac, partially overlaps with the aliased signal from Pyr. As the ratio of Pyh and Pyr is less than 10%, this leads only to minor artifacts in the metabolic maps for Pyr, which are visible in CSI measurements early after injection when the Pyr signal is concentrated in the vena cava and/or aorta. These artifacts can potentially be reduced by using prior knowledge of resonance frequencies, scalar coupling constants, and transverse relaxation times, and estimating the amplitudes with a minimum least-squares solution (22). Whereas Pyr, Lac, and Ala are detected in both voxels, a small Bic peak was visible only in the kidney, possibly attributable to a higher rate of oxidative decarboxylation in the kidney than liver.
Figure 2.
Single spectra from voxels (interpolated nominal size: 1.25×1.25×10 mm3) in the liver and kidney (cf. Fig. 1b and 1c). The spectrum from the liver (a) was acquired at 17.8 s from the start of injection and the spectrum from the right kidney (b) was acquired at 17.4 s with a total measurement time per spectrum of 375 ms. For each voxel, the data were reconstructed using a 7.5-Hz Gaussian line broadening and a frequency offset relative to Lac of 0 Hz (blue), 1*SW (red), and 3*SW (black) with SW = 276.2 Hz. The vertical dashed lines indicate the frequencies of Bic (aliased) and Pyh. For aliased signals the true frequency difference to the Lac resonance is given by the frequency in the spectrum minus nalias*SW with nalias the number of how often the signal has been aliased, e.g., for Ala: 65 Hz – 1*276 Hz = −211 Hz.
Metabolic maps of Pyr, Lac, and Ala computed from the third temporal frame for the liver and kidney slices are shown in Fig. 3. The maps are superimposed onto the conventional proton images for anatomical reference, and the intensity scale is the same for all three metabolites with the Pyr intensity scaled down by factor of 3. At this time point, the highest Pyr signal in the liver slice is still within the vasculature. With a nominal in-plane resolution of 5 mm for the CSI data, it was not possible to differentiate the signals from the vena cava and the aorta. Signals for Lac and Ala show heterogeneous spatial distributions, likely due to multiple lobes of the liver. In some regions, the signal at 17 s was comparable to or even higher than that in the vena cava/aorta. Given a slice thickness of 10 mm, partial volume effects add to the heterogeneity in the metabolic maps. In the slice through the kidneys, high levels of both Pyr and Lac are found in the kidneys, and Pyr is also high in the vena cava/aorta. The highest Ala signal is found anterior to the left kidney, which could be a lobe of the liver or the spleen.
Figure 3.
Metabolic maps of Pyr, Lac, and Ala from slices predominantly through the liver (a) and kidneys (b) from acquisitions at 17.8 s and 17.4 s from the start of injection, respectively. The metabolic maps are superimposed onto the proton images for anatomical reference (threshold at 20% in liver slice and 15% in kidney slice) with Pyr scaled down by a factor of 3. The small misregistration of the Lac signal from the reference phantom is probably due to movement of the phantom caused by respiratory motion of the animal.
This experiment was performed three times in the same scan session but with the animal taken out and put back into the scanner between injections 1 and 2. The time courses of the mean metabolite signals from ROIs in the liver and right kidney averaged over all three experiments are shown Fig 4. The position and shape of the ROIs are indicated on the corresponding proton images in Fig. 1. A prominent difference is the much faster rise of Pyr levels between 5 s and 11 s post-bolus injection in the kidney than liver. To a lesser degree, the faster rise can also be seen in the signals for Lac and Ala. The delayed peak/plateau time for these two metabolites in the rat liver compared to the rat kidney has also been reported in a study using dynamic slice-selective 13C-MRS acquisitions in rat liver and kidney after injection of hyperpolarized 13C1-pyruvate (23). Also shown in the two plots are the time courses of the Pyr signal from ROIs encompassing the vena cava and aorta in the respective slices. The large interexperimental standard deviation of the intravascular Pyr at the first time point in both liver and kidney is due to the rapidly changing Pyr concentration at the start of the bolus. Small variations in timing of the injection and start of data acquisition can lead to large signal variations with a coarse temporal sampling interval of 6 s. These data demonstrate that the major contribution to the early Pyr signal intensity in dynamic slice-selective 13C-MRS data is from the intravascular component, rather from Pyr within the kidneys or liver. While the time-resolved ratio of metabolic product to substrate is a parameter that is influenced by several different factors, e.g., different longitudinal relaxation times and different rates of in-flow and clearance of the ROI, it also depends on the metabolic turnover and has the advantage of being independent of the polarization level of the injected compound. For example, Day et al. (9) report that the decreased flux between pyruvate and lactate in drug-treated versus non-treated tumors could be seen in the time-resolved lactate/pyruvate ratios. A comparison of the time courses of Lac/Pyr and Ala/Pyr from liver and right kidney ROIs (Fig. 4c) suggests faster production of both lactate and alanine in the liver than kidney. However, small contributions of the renal vein and artery in the kidney ROI could lead to a slight overestimation of the kidney Pyr signal. To illustrate the limitations of estimating organ-specific kinetics and reaction rates based on only slice-selective dynamic data, Fig. 4d shows the product-to-substrate ratios as calculated from ROIs that encompass the entire respective slices. Although the data also indicate higher conversion rates for both Pyr-to-Lac and Pyr-to-Ala in the liver compared with the kidney, the rates are underestimated due to the large vascular component of Pyr. This finding illustrates the importance of acquiring spatially resolved dynamic CSI as compared to unresolved slices.
Figure 4.
The time courses averaged over three acquisitions in the same animal for Pyr (black □), Lac (blue ×), and Ala (red ○) from ROIs in the liver (a) and right kidney (b). Also plotted are the time courses of Pyr from vascular ROIs in the two slices (dashed magenta ◊). The error bars correspond to the standard deviation. The time scale is relative to start of injection. Note that Pyr from both the liver and kidney ROI is scaled down by a factor of 3, whereas vascular Pyr is scaled down by a factor of 6. (c) Time-resolved Lac/Pyr (blue) and Ala/Pyr (red) ratios for the liver (dashed) and kidney (solid) ROI. (d) Time-resolved Lac/Pyr (blue) and Ala/Pyr (red) ratios from ROIs encompassing the entire liver (dashed) and kidney (solid) slices.
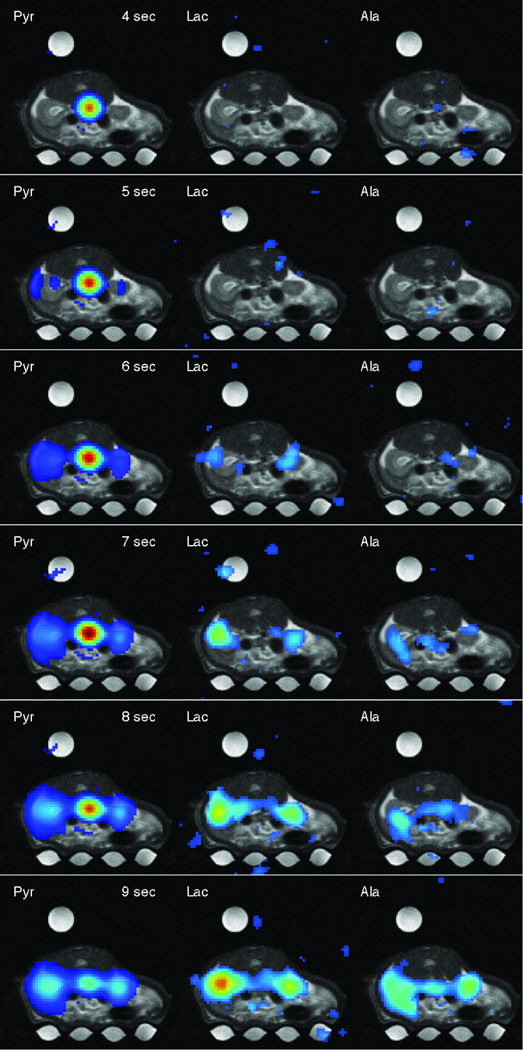
CSI experiment 2 (rat 2) had a higher temporal resolution with the data acquired using a 1-s sampling interval from two 10-mm slices predominantly through the left kidney and liver, respectively. Twenty CSI acquisitions were performed to follow the metabolic time courses over the first 19 s following the start of pyruvate injection. Figure 5 shows six frames of a movie reconstructed from the 13C-CSI data set in which the pyruvate bolus arrives in the aorta, perfuses into the kidneys, and then is converted to lactate and alanine (the complete movie is available online at http://www.stanford.edu/~dirkm/MRMmov/rat_kidney.mov). The time courses of the metabolites from liver and left kidney ROIs and Pyr from the ROIs encompassing vena cava and aorta at each slice are shown in Fig. 6. Because of the saline in the dead volume of the catheter, Pyr is first detected in the vena cava at the 3 s mark both in the kidney and liver slice. Note the two peaks in the vascular Pyr time course in the liver slice at 5 and 7 s, respectively. This is consistent with the hepatic dual blood supply through the hepatic artery and the portal vein. The second peak is due to a delayed bolus from the alimentary tract (24), which contributes to the signal from the vascular ROI. These two phases of the vascular component of Pyr were also repeatedly measured using slice-selective 13C-MRS (data not shown) and were also in Hu et al. (23) using the same technique. Detection of Pyr in the kidney ROI is delayed by approximately 2 to 3 s with respect to vascular Pyr. The earlier rise detected in the liver ROI is due to artifacts caused by partial overlap of the Pyr signal with the aliased signal of Pyh leading to blurring artifacts. The higher variability in liver data is due to lower SNR and respiratory artifacts, with the liver ROI located close to the diaphragm. Breathing artifacts likely can be reduced by synchronizing the data acquisition to the respiratory cycle. Figure 6f shows the time-resolved Lac/Pyr and Ala/Pyr ratios for the liver and kidney ROIs. Similar to the results from the experiment with lower temporal resolution, the data indicate higher conversion rates for both Pyr-to-Lac and Pyr-to-Ala in the liver than kidney. The product-to-substrate ratios calculated from large ROIs encompassing each of the respective slices underestimate the conversion rates because of non-target regions (Fig. 6g).
Figure 5.
Six frames of a time series of metabolic maps of Pyr, Lac, and Ala (superimposed onto the corresponding proton MRI) acquired from 10-mm slice through the kidneys. Each metabolic time series is individually scaled (threshold at 8% for Pyr and 20% for Lac and Ala). The time stamps are relative to time of injection. The complete movie is online at http://www.stanford.edu/~dirkm/MRMmov/rat_kidney.mov.
Conclusion
The presented data demonstrate the feasibility of dynamic multi-slice metabolic imaging with a temporal resolution as high as 1 s using undersampled spiral CSI. The short acquisition time per slice of only 375 ms allows greater flexibility in both choosing the sampling rate appropriate to monitor a specific dynamic process and trading off between temporal resolution and spatial coverage. When using the applied variable flip angle scheme in hyperpolarized MRI, there is a trade-off between the total number of temporal samples and the SNR for each sample that has to be taken into account when designing an experiment. The comparison of the time-resolved product-to-substrate ratios calculated from organ-specific ROIs with the ratios calculated from whole-slice ROIs illustrates the major advantage of the proposed imaging method. Due to the large vascular component of the substrate, whole-slice data underestimate conversion rates. The implementation of the technique on a clinical scanner will greatly facilitate the translation to larger animal models and, ultimately, humans. The use of localized phased-array receiver coils and parallel imaging techniques will allow acquisition times on the same order as presented here for rat imaging. Furthermore, reducing readout times under 400 ms is particularly beneficial for human cardiac imaging, given typical diastolic time intervals of approximately 400 ms. Combining this fast acquisition technique with metabolic modeling tools to estimate organ-specific kinetic parameters will permit the quantitative investigation of metabolism under normal and pathologic conditions, the longitudinal monitoring of disease processes, and the early evaluation of therapeutic interventions. The noninvasive nature of the technology facilitates the translation of this new metabolic imaging capability from the laboratory to the clinic.
Acknowledgments
We thank Oliver Hsu, B.A., and Evan Nunez, B.A., for assistance in animal handling and monitoring, and Dr. Edith V. Sullivan for critical review of the manuscript. This study was supported by the Lucas Foundation and National Institutes of Health Grants RR09784, AA05965, AA13521-INIA, and CA114747.
Funded by:
Lucas Foundation
NIH, Grant Number: RR09784, AA05965, AA13521-INIA, and CA114747
References
- 1.Ardenkjaer-Larsen JH, Fridlund B, Gram A, Hansson G, Hansson L, Lerche MH, Servin R, Thaning M, Golman K. Increase in signal-to-noise ratio of > 10,000 times in liquid-state NMR. Proc Natl Acad Sci U S A. 2003;100(18):10158–10163. doi: 10.1073/pnas.1733835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Golman K, Ardenkjaer-Larsen JH, Petersson JS, Mansson S, Leunbach I. Molecular imaging with endogenous substances. Proc Natl Acad Sci U S A. 2003;100(18):10435–10439. doi: 10.1073/pnas.1733836100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowers CR, Weitekamp DP. Transformation of Symmetrization Order to Nuclear-Spin Magnetization by Chemical Reaction and Nuclear Magnetic Resonance. Physical Review Letters. 1986;57(21):2645. doi: 10.1103/PhysRevLett.57.2645. [DOI] [PubMed] [Google Scholar]
- 4.Bowers CR, Weitekamp DP. Parahydrogen and synthesis allow dramatically enhanced nuclear alignment. J Am Chem Soc. 1987;109(18):5541–5542. [Google Scholar]
- 5.Golman K, in 't Zandt R, Thaning M. Real-time metabolic imaging. Proc Natl Acad Sci U S A. 2006;103(30):11270–11275. doi: 10.1073/pnas.0601319103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kohler SJ, Yen Y, Wolber J, Chen AP, Albers MJ, Bok R, Zhang V, Tropp J, Nelson S, Vigneron DB, Kurhanewicz J, Hurd RE. In vivo 13 carbon metabolic imaging at 3T with hyperpolarized 13C-1-pyruvate. Magn Reson Med. 2007;58(1):65–69. doi: 10.1002/mrm.21253. [DOI] [PubMed] [Google Scholar]
- 7.Golman K, Zandt RI, Lerche M, Pehrson R, Ardenkjaer-Larsen JH. Metabolic imaging by hyperpolarized 13C magnetic resonance imaging for in vivo tumor diagnosis. Cancer Res. 2006;66(22):10855–10860. doi: 10.1158/0008-5472.CAN-06-2564. [DOI] [PubMed] [Google Scholar]
- 8.Chen AP, Albers MJ, Cunningham CH, Kohler SJ, Yen YF, Hurd RE, Tropp J, Bok R, Pauly JM, Nelson SJ, Kurhanewicz J, Vigneron DB. Hyperpolarized C-13 spectroscopic imaging of the TRAMP mouse at 3T-initial experience. Magn Reson Med. 2007;58(6):1099–1106. doi: 10.1002/mrm.21256. [DOI] [PubMed] [Google Scholar]
- 9.Day SE, Kettunen MI, Gallagher FA, Hu DE, Lerche M, Wolber J, Golman K, Ardenkjaer-Larsen JH, Brindle KM. Detecting tumor response to treatment using hyperpolarized 13C magnetic resonance imaging and spectroscopy. Nat Med. 2007;13(11):1382–1387. doi: 10.1038/nm1650. [DOI] [PubMed] [Google Scholar]
- 10.Olsson LE, Chai CM, Axelsson O, Karlsson M, Golman K, Petersson JS. MR coronary angiography in pigs with intraarterial injections of a hyperpolarized 13C substance. Magn Reson Med. 2006;55(4):731–737. doi: 10.1002/mrm.20847. [DOI] [PubMed] [Google Scholar]
- 11.Merritt ME, Harrison C, Storey C, Jeffrey FM, Sherry AD, Malloy CR. Hyperpolarized 13C allows a direct measure of flux through a single enzyme-catalyzed step by NMR. Proc Natl Acad Sci U S A. 2007;104(50):19773–19777. doi: 10.1073/pnas.0706235104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Golman K, Petersson JS, Magnusson P, Johansson E, Akeson P, Chai CM, Hansson G, Mansson S. Cardiac metabolism measured noninvasively by hyperpolarized 13C MRI. Magn Reson Med. 2008;59(5):1005–1013. doi: 10.1002/mrm.21460. [DOI] [PubMed] [Google Scholar]
- 13.Månsson S, Johansson E, Magnusson P, Chai C-M, Hansson G, Petersson J, Ståhlberg F, Golman K. 13C imaging—a new diagnostic platform. European Radiology. 2006;16(1):57–67. doi: 10.1007/s00330-005-2806-x. [DOI] [PubMed] [Google Scholar]
- 14.Golman K, Petersson JS. Metabolic imaging and other applications of hyperpolarized 13C1. Acad Radiol. 2006;13(8):932–942. doi: 10.1016/j.acra.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 15.Mayer D, Levin YS, Hurd RE, Glover GH, Spielman DM. Fast metabolic imaging of systems with sparse spectra: application for hyperpolarized 13C imaging. Magn Reson Med. 2006;56(4):932–937. doi: 10.1002/mrm.21025. [DOI] [PubMed] [Google Scholar]
- 16.Cunningham CH, Chen AP, Albers MJ, Kurhanewicz J, Hurd RE, Yen YF, Pauly JM, Nelson SJ, Vigneron DB. Double spin-echo sequence for rapid spectroscopic imaging of hyperpolarized 13C. J Magn Reson. 2007;187(2):357–362. doi: 10.1016/j.jmr.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 17.Reeder SB, Brittain JH, Grist TM, Yen YF. Least-squares chemical shift separation for 13C metabolic imaging. Journal of Magnetic Resonance Imaging. 2007;26(4):1145–1152. doi: 10.1002/jmri.21089. [DOI] [PubMed] [Google Scholar]
- 18.Hu S, Lustig M, Chen AP, Crane J, Kerr A, Kelley DA, Hurd R, Kurhanewicz J, Nelson SJ, Pauly JM, Vigneron DB. Compressed sensing for resolution enhancement of hyperpolarized (13)C flyback 3D-MRSI. J Magn Reson. 2008;192(2):258–264. doi: 10.1016/j.jmr.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adalsteinsson E, Irarrazabal P, Topp S, Meyer C, Macovski A, Spielman DM. Volumetric spectroscopic imaging with spiral-based k-space trajectories. Magn Reson Med. 1998;39(6):889–898. doi: 10.1002/mrm.1910390606. [DOI] [PubMed] [Google Scholar]
- 20.Derby K, Tropp J, Hawryszko C. Design and evaluation of a novel dual-tuned resonator for spectroscopic imaging. J Magn Reson. 1990;86(3):645–651. [Google Scholar]
- 21.Zhao L, Mulkern R, Tseng CH, Williamson D, Patz S, Kraft R, Walsworth RL, Jolesz FA, Albert MS. Gradient-echo imaging considerations for hyperpolarized 129Xe MR. J Magn Reson B. 1996;113:179–183. [PubMed] [Google Scholar]
- 22.Levin YS, Mayer D, Yen YF, Hurd RE, Spielman DM. Optimization of fast spiral chemical shift imaging using least squares reconstruction: application for hyperpolarized (13)C metabolic imaging. Magn Reson Med. 2007;58(2):245–252. doi: 10.1002/mrm.21327. [DOI] [PubMed] [Google Scholar]
- 23.Hu S, Chen AP, Zierhut ML, Bok R, Van Criekinge M, Yen YF, Schroeder MA, Hurd RE, Nelson SJ, Kurhanewicz J, Vigneron DB. In Vivo Carbon-13 Dynamic MRS and MRSI of Rat Liver with Hyperpolarized 13C-1-Pyruvate. Toronto, Canada: 2008. p. 1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Munk OL, Bass L, Feng H, Keiding S. Determination of regional flow by use of intravascular PET tracers: microvascular theory and experimental validation for pig livers. J Nucl Med. 2003;44(11):1862–1870. [PubMed] [Google Scholar]