Abstract
Background
Missense heterozygous mutations in the coding region of angiogenin (ANG) gene, encoding a 14 kDa angiogenic RNase, were recently found in patients of amyotropic lateral sclerosis (ALS). Functional analyses have shown that these are loss-of-function mutations, implying that angiogenin deficiency is associated with ALS pathogenesis and that increasing ANG expression or angiogenin activity could be a novel approach for ALS therapy.
Objective
Review the evidence showing the involvement of angiogenin in motor neuron physiology and function, and provide a rationale for targeting angiogenin in ALS therapy.
Methods
Review the current understanding of the mechanism of angiogenin action in connection with ALS genetics, pathogenesis and therapy.
Conclusion
ANG is the first gene whose loss-of-function mutations are associated with ALS pathogenesis. Therapeutic modulation of angiogenin level and activity in the spinal cord, either by systemic delivery of angiogenin protein or through retrograde transport of ANG-encoding viral particles, may be beneficial for ALS patients.
Keywords: amyotropic lateral sclerosis, angiogenesis, angiogenin, endothelial cells, loss-of-function gene mutation, motor neurons, ribonuclease, rRNA transcription
1. Introduction
Amyotropic lateral sclerosis (ALS) is a progressive neurodegenerative disease with specific loss of motor neurons in the brain, brain stem and spinal cord [1]. The average age of onset is 55 years with upper and lower motor neuron signs, including distal muscle weakness and wasting, increased muscle tone with hyperreflexia and at times diaphragmatic and/or bulbar weakness. A significant percentage of ALS patients (up to 50%) have evidence of cognitive impairment, and 5 – 10% of them are demented [2–7]. Death occurs from respiratory failure at average 4 years after disease onset. It is a devastating disease without cure. At present, the only recognized treatment for ALS is riluzole, which extends survival by about 3 months with no improvement in motor muscular functions [8].
The incidence of ALS is estimated at 0.6 – 2.4/100,000 population [9]. Approximately 90% of ALS cases are sporadic with no known family history, whereas the remaining 10% are familial cases inherited in either an autosomal dominant or recessive manner [1,10]. Mutations in the Cu/Zn superoxide dismutase gene 1 (ALS1; SOD1), have been identified in ~20% of familial [11–13] and in ~3% of sporadic [14–16] ALS patients.
Angiogenin (ANG) gene, encoding a 14 kDa angiogenic ribonuclease [17], seems to be the first loss-of-function gene identified in ALS. Since the original discovery of ANG as an ALS candidate gene [18], a total of 14 missense mutations in the coding region of ANG have been identified in 35 of the 3170 ALS patients of the Irish, Scottish, Swedish, North American and Italian populations [18–22]. Among the 14 mutations identified so far, 10 have been characterized in detail and shown to be loss-of-function mutations [22–24].
Mouse angiogenin is strongly expressed in the CNS during development [25]. Human angiogenin is strongly expressed in both endothelial cells and motor neurons of normal human fetal and adult spinal cords [22]. Wild type (WT) angiogenin has been shown to stimulate neurite outgrowth and pathfinding of motor neurons in culture and to protect hypoxia-induced motor neuron death, whereas the mutant angiogenin proteins not only lack these activities but also induce motor neuron degeneration [24]. Therefore, a role of angiogenin in motor neuron physiology and a therapeutic activity of angiogenin toward ALS can be foreseen.
2. ALS
2.1 ALS genetics
At present, SOD1 is the only known autosomal dominant gene in which mutations have been functionally associated with ALS, although three other loci (ALS3, ALS6 and ALS7) have been identified for typical autosomal dominant ALS by linkage analysis [1,26]. Other dominantly inherited genetic loci, associated with an atypical ALS phenotype, have also been identified (ALS with dementia/parkinsonism, MAPT; progressive lower motor neuron disease, DCTN1; and ALS8, VAPB). In autosomal dominant ALS with frontotemporal dementia (FTD), genetic linkage has been reported to 9q21 – q22 [27]. Mutations in the SETX gene have been identified in juvenile onset autosomal dominant ALS. Genetic loci identified for juvenile onset autosomal recessive disease include Alsin (ALS2) and ALS5 [1,26]. It is notable that besides SOD1, the other genes and loci described above have only been found in very few ALS patients, and often in atypical ALS with slow progression.
Genetic association studies have also identified several risk factors in ALS, including deletions or insertions in the neurofilament heavy chain gene [28–30], polymorphisms in VEGF [31], hemochromatosis gene HFE [32] and paraoxonase-1 (PON1) [33]. A 5 bp deletion in mitocondrial cytochrome c oxidase subunit I (COX1) [34] and a T4272C mutation in isoleucine tRNA synthesase (IARS2) [35] have been linked to ALS, although only in a single case. A common mitochondrial DNA deletion mutation (mt DNA4977) is increased in the brain of ALS patients [36]. The apolipoprotein E epsilon4 allele has been associated with decreased survival of ALS patients [37]. Copy number variation in survival motor neuron (SMN) has also been shown to be a susceptibility factor [38]. More recently, whole genome association studies have identified genetic variations in dipeptidyl-peptidase 6 (DPP6), [39] and inositol 1,4,5-triphosphate receptor 2 (ITPR2) [40] genes in ALS patients. Whole genome association has also identified a minor association of a single nucleotide polymorphism near the FLJ10986 gene to ALS [41]. Most recently, missense mutations in the coding region of TARDBP encoding the Tar DNA binding protein TDP-43 were found in both familial and sporadic ALS patients [42–46] following the discovery that TDP-43 is a major constituent of the neuronal cytoplasmic inclusions [3,47,48]. Five studies have independently reported TARDBP mutations in ALS patients [42–46], whereas two studies failed to identify any mutations [49,50]. A total of 15 mutations have been identified in TARDBP among 1637 ALS patients [42–46,49,50], which places TARDBP as the third most frequently mutated gene so far identified in ALS (after SOD1 and ANG).
Since 2004, ANG has emerged as an important gene in ALS [18,21]. A total of 14 different missense mutations in the coding region of ANG have been identified in 35 of 3170 patients from Irish, Scottish, Swedish, North American and Italian ALS populations [18–22]. Thus, ANG seems to be the second most frequently mutated gene in ALS (after SOD1). Importantly, although WT angiogenin induces angiogenesis, stimulates neurite outgrowth of motor neurons and protects them from hypoxia-induced death, mutant angiogenin proteins lack these activities [22,24]. Therefore, ANG seems to be the first loss-of-function gene so far identified in ALS [22]. Table 1 lists the genes and genetic factors whose alterations predispose to ALS.
Table 1.
Genes and genetic factors associated with ALS.
| Disease type | Locus | Gene | Inheritance | Clinic features | Ref. |
|---|---|---|---|---|---|
| ALS1 | 21q22.21 | SOD1 | AD | Adult onset, typical ALS | [143] |
| ALS2 | 2q33 | Alsin | AR | Juvenile onset, atypical ALS and PLS, slow progression | [144] |
| ALS3 | 18q21 | Unknown | AD | Adult onset, typical ALS | [145] |
| ALS4 | 9q34 | SETX | AD | Juvenile onset, atypical ALS, slow progression | [146] |
| ALS5 | 15q15.1-21.1 | Unknown | AR | Juvenile onset, atypical ALS, slow progression, no pseudobulbar signs | [147] |
| ALS6 | 16q12 | Unknown | AD | Adult onset, typical ALS, short duration | [148] |
| ALS7 | 20ptel-13 | Unknown | AD | Adult onset, typical ALS, short duration | [149] |
| ALS8 | 20q13.33 | VAPB | AD | Adult onset, atypical ALS, slow progression | [150] |
| ALS-FTD | 9q21-22 | Unknown | AD | Adult onset, ALS with fronto-temporal dementia | [151] |
| ALS-PD | 17q21 | MAPT | AD | Adult onset, ALS with Parkinsonism and dementia | [152] |
| Progressive lower MND | 2p13 | DCTN1 | AD | Adult onset, lower motor neuron disorder | [153] |
| ALS, sporadic | 6q12 | VEGF | Risk factors | Adult onset, typical ALS | [31] |
| 22q12.1-q13.1 | NFHC | [28–30] | |||
| 6q21.3 | HFE | [32] | |||
| 7q21.3 | PON1 | [33] | |||
| 5q13 | SMN | [38] | |||
| 7q36 | DPP6 | [39] | |||
| 12p11.23 | ITPR2 | [40] | |||
| 1p32.1 | FLJ10986 | [41] | |||
| 17q21 | PGRN | [69] | |||
| ALS, familiar and sporadic | 14q11.2 | ANG | AD | Adult onset, typical ALS | [20–22] |
| ALS, familiar and sporadic | 1p36 | TARDBP | AD | Adult onset, typical ALS | [42–46] |
AD: Autosomal dominant; ALS: Amyotropic lateral sclerosis; ALS-FTD: Amyotropic lateral sclerosis–front-temporal dementia; ALS-PD: Amyotropic lateral sclerosis–Parkinsonism and dementia; ANG: Angiogenin; AR: Autosomal recessive; DCTN1: Dynactin p150 subunit; DPP6: Dipeptidyl peptidase 6; FLJ10986: A gene encoding a 48 kDa protein weakly similar to L-ribulokinase; HFE: A hemochromatosis gene involved in iron metabolism; ITPR2: Inositol 1,4,5-triphosphate receptor 2; MAPT: Microtubule-associated protein tau; MND: Motor neuron disease; NFHC: Neurofilament heavy chain; PGRN: Progranulin; PLS: Primary lateral sclerosis; PON1: Paraoxonase 1; SETX: Senataxin; SMN: Survival motor neuron; SOD1: Superoxide dismutase 1; TARDBP: Tar DNA binding protein 43; VAPB: Vesicle-associated membrane protein.
2.2 ALS pathogenesis
Many theories including oxidative stress, excitotoxicity, mitochondrial dysfunction, defective axonal transport, abnormal protein aggregation, and loss of tropic and angiogenic factor support have been proposed as the underlying mechanism of ALS pathogenesis [1,51–53]. Each of these hypotheses is supported by some experimental evidence but at the same time is undermined by contradictory data. For example, motor neuron damage as a result of oxidative stress is a key hypothesis in ALS. It is supported by the findings of elevated oxidative metabolism in ALS, such as the detection of increased biochemical markers of oxidative injury in post mortem examinations of ALS patients [54]. It is also supported by the acquired capacity of some forms of mutant SOD1 to catalyze the production of reactive oxygen species such as superoxide anions (O2-) and peroxynitrite (-ONOO) through either copper catalysis or improper copper and zinc binding [55–57]. However, this hypothesis is undermined by the report that oxidative markers are detected in SOD1G93A mice but not in SOD1G37A mice, although both developed ALS symptoms [58]. It is further undermined by the finding that deletion of copper chaperone for SOD1 diminished the copper load but did not affect the development of ALS [59], and that copper-binding-site-null SOD1 still causes ALS in transgenic mice [60].
The etiology of ALS is likely to be multi-factorial, involving the interplay of several mechanisms to initiate disease and propagate the spread of motor neuron death. A generally accepted hypothesis at present is that several factors, both genetic and environmental, cause mitochondrial dysfunction and excitotoxicity, lead to abnormal protein precipitation and finally apoptosis of motor neurons [52]. Non-neuronal neighboring cells may also play a role in ALS pathogenesis. It has been shown that motor neurons, microglia [61] and astrocytes [62] affect the onset and progress of illness. Astrocytes and microglia harboring SOD1 mutations secrete substances that kill motor neurons. Motor neurons with or without SOD1 mutation showed neuro-degenerative properties when co-cultured with astrocytes that harbor SOD1 mutation, whereas motor neurons with SOD1 mutations showed less neuronal losses when they are surrounded by normal astrocytes [63–65].
Identification of ANG mutations in ALS patients [19–22] and demonstration that these mutations result in loss of angiogenin functions [22–24] provide an alternative viewpoint of ALS pathogenesis. Angiogenin was not the first angiogenic molecule reported to be associated with ALS. In 2001, Oosthuyse et al. have already reported that deletion of the hypoxia-response element in the VEGF promoter caused adult onset motor neuron degeneration similar to ALS [66]. Although mutations in the coding region of VEGF have not been found in ALS patients, a genetic variation in the VEGF promoter that lowers VEGF expression has been shown to be associated with an increased risk of ALS [31]. Reported involvement of both angiogenin and VEGF in ALS suggests a link between angiogenesis and ALS pathogenesis. Further supporting this hypothesis are the findings of null mutations of progranulin (PGRN), another angiogenic protein, in patients with FTD [67,68]. Genetic variations of PGRN have also been recently reported in ALS patients [69]. Many patients with ALS develop dementia, and with FTD develop motor neuron diseases similar to ALS. FTD and ALS share some common neuropathological features such as the accumulation of ubiquitinated neuronal cytoplasmic inclusions containing TDP-43 [3]. Thus, a role of angiogenic factors in ALS pathogenesis has been proposed [53,70,71], and vascular abnormality has been recently demonstrated in SOD1 transgenic mice [72–74]. The findings that the blood spinal cord barrier (BSCB) of SOD1 transgenic mice is impaired and that the BSCB leakage occurs before motor neuron degeneration provide pathological data supporting an active role of blood vessels in ALS pathology [72–74].
2.3 ALS therapy, clinical trials and preclinical studies
There is presently no effective pharmacologic treatment for ALS to halt neuronal death or even slow it appreciably. Riluzole, the only drug approved for ALS since 1995, only extends survival by 2 – 3 months if it is taken for 18 months. Riluzole is thought to act in part by limiting glutamate release. It preferentially blocks tetrodotoxin-sensitive sodium channels, which are associated with damaged neurons [75]. This reduces influx of calcium ions and indirectly prevents stimulation of glutamate receptors. Together with direct glutamate receptor blockade, the effect of the neurotransmitter glutamate on motor neurons is greatly reduced. Riluzole was approved for use in ALS after two independent clinical trials showed a marginal increase in the survival time of ALS patients [8,76]. Unfortunately, patients taking riluzole do not experience any slowing in disease progression or improvement in muscle function. Therefore, riluzole does not present a cure, or even an effective treatment, and the search for better therapeutic agents continues.
Mutant SOD1 transgenic mice have been widely used for ALS drug testing. SOD1 mutations are the cause of ~20% of the familiar ALS and ~3% of the sporadic ALS, and, therefore, ~4% of all ALS cases. More than 100 mutations in SOD1, distributed throughout the gene, have been found in ALS patients [52]. Although it is still unknown why the mutant form of this abundant and ubiquitously expressed enzyme is specifically toxic to motor neurons and causes ALS, it is clear that mice overexpressing the mutant SOD1 genes develop symptoms mimicking that of human ALS patients. Over 70 agents of various categories including tropic factors, antioxidant, antiviral, anti-inflammatory, immunomodulatory, antiapoptosis, antiglutamatergic, calcium regulators, proteasome inhibitors, metal ion regulators, structure proteins as well as energy metabolism-related compounds, have been tested in SOD1G93A mice [51,52,77]. Many of these agents underwent clinical trials. However, the benefits in the mouse have not been translated into clinical efficacy except in the case of riluzole. All the others, including brain-derived neurotrophic factor [78], ciliary neurotrophic factor [79], IGF1 [80,81], and glial-derived neurotrophic factor (GDNF) [82] have failed.
One of the main reasons for the disappointing clinical trials was that the beneficial effect of these agents observed in the SOD1G93A mice was not significant and that the SOD1G93A mice have high noise level in their survival. The ALS Therapy Development Institute (ALSTDI) has conducted a thorough retesting of > 70 drugs in 18,000 ALS mice across 221 studies and failed to reproduce the reported efficacy in SOD1G93A mice [83]. ALSTDI has also reanalyzed the reported data from 5429 mice from 50 published papers and found that the reported beneficial effect in animal survival was actually the noise of the animals. ALSTDI has concluded that 24 ALS mice are needed in each group to reduce the noise and get conclusive results [83]. Moreover, all the SOD1G93A mice in both control and experimental groups should be matched in age, gender, litter size and copy numbers of the transgene.
The reason for a relative minimal effect of exogenous tropic factors and other types of therapeutic proteins could be their failure to cross the blood–brain barrier (BBB) and BSCB. Gene therapy is, therefore, an alternative approach for ALS therapy. Many strategies are under investigation, including the delivery of genes encoding neurotrophic factors, antiapoptotic and antioxidants proteins using viral vectors administered directly into the affected areas of the CNS, or through retrograde transport to motor neurons from intramuscular injection, or through ex vivo gene transfer [84]. AAV (adeno-associated virus)-mediated delivery of IGF-1 [85], GDNF [86] and Bcl-2 [87] gene have been shown to be effective in the SOD1G93A transgenic mice.
VEGF is a prominent angiogenic factor and a new neurotrophic factor linked to ALS. Since the demonstration that deletion of the hypoxia-response element in the promoter of VEGF causes motor neuron degeneration in mice [66] and that polymorphisms in the VEGF promoter that reduce VEGF expression are associated with ALS in the populations of Sweden, Belgium, UK [31] and New England [88], various attempts have been made to target VEGF as a therapeutic approach for ALS. VEGF delivered into SOD1G93A rats intracerebroventricularly [89] or into SOD1G93A mice with a retrogradely transported lentiviral vector [90] has been shown to improve motor neuron function marginally and prolong survival significantly. VEGF overexpression also improves motor muscular function and increases the survival in SOD1G93A transgenic mice [91]. However, intraperitoneal injection of VEGF in SOD1G93A mice had only a modest effect in delaying disease onset and in prolonging survival [92].
3. Angiogenin
3.1 Mechanism of action of angiogenin
Angiogenin was originally isolated from the conditioned medium of HT-29 human colon adenocarcinoma cells based solely on its angiogenic activity in the chicken embryo chorioallantoic membrane angiogenesis assay [17]. Subsequently, it has been found to have a wide tissue distribution with the highest expression in the liver [93]. It is a member of the pancreatic RNase superfamily with a 33% amino-acid identity and an overall homology of 56% to that of RNase A [94]. Angiogenin has a unique ribonucleolytic activity that is several orders of magnitude lower than that of RNase A but is important for its biological activity [95]. The amino-acid residues important for catalysis are conserved in all vertebrate angiogenin from fish to human [96]. Extensive studies on site-directed mutagenesis have shown that angiogenin variants with reduced enzymatic activity also have reduced angiogenic activity [95,97–104]. Structural work indicated that one of the reasons for angiogenin to have a reduced ribonucleolytic activity is that the side chain of Gln 117 occupies part of the enzymatic active site so that substrate binding is compromised [105,106].
Angiogenin is angiogenic, whereas the prototype family member RNase A is not. Two important structural differences between angiogenin and RNase A are responsible for this discrepancy. The first is the segment from amino-acid residues 59 – 68 that forms the receptor binding site in angiogenin [99,107]. Therefore, angiogenin binds to its target cells (including endothelial cells, cancer cells and motor neurons) but RNase A does not. Angiogenin binds to endothelial cells specifically [108] and induces second messenger responses including diacylglycerol and prostacyclin [109,110], and activates MAPK [111] and AKT [112]. Another structural difference between angiogenin and RNase A is that angiogenin has a nuclear localization signal consisting of 29IMRRRGL35, whereas RNase A does not [113]. Therefore, angiogenin undergoes nuclear translocation in endothelial cells where it accumulates in the nucleolus [114,115], binds to the promoter region of ribosomal DNA (rDNA) and stimulates rRNA transcription [116,117], an essential step for ribosome biogenesis, protein translation and cell proliferation.
An angiogenin binding protein has been identified from the surface of endothelial cells [107] and has been characterized to be a type of smooth muscle actin [118,119]. An ~170 kDa angiogenin receptor has also been identified from the endothelial cell surface to mediate nuclear translocation of angiogenin and cell proliferation [120]. Expression of the binding protein and the receptor on endothelial cells seems to be mutually exclusive. The binding protein is expressed on the surface of confluent cells. Binding of angiogenin to the binding protein activates tissue plasminogen activator [121] thereby inducing cell invasion and migration [122]. After the leading cells migrate away, the local cell density decreases, which triggers the expression of an angiogenin receptor. Binding of angiogenin to the receptor stimulates cell proliferation so that the gap created by the migrating cells is filled. Therefore, angiogenin is a multifunctional angiogenic molecule that plays a role in several steps in the angiogenesis process including cell invasion, proliferation and tube formation. Figure 1 shows the current understanding of the mechanism of angiogenin-induced angiogenesis.
Figure 1. Conceptual framework of the interaction between angiogenin and its target cells.
Angiogenin, shown in yellow, can bind to both the receptor and the binding protein, shown in white and orange, respectively. Most the angiogenin and its binding protein complex will dissociate from the cell surface and activates tissue plasminogen activator to produce plasmin, and induce cell invasion into the extracellular matrix. Binding to the 170 kDa receptor induces second messengers and triggers signal transduction. On binding, angiogenin is also internalized and translocated to the nucleus where it accumulates in the nucleolus. All these individual steps are necessary for angiogenesis.
ANG: Angiogenin.
3.2 Role of angiogenin in rRNA transcription
Angiogenin has been shown to undergo nuclear translocation in endothelial [114,115,123] and cancer [124,125] cells. Nuclear translocation of angiogenin in endothelial cells is under tight regulation and is cell density-dependent. It decreases as cell density increases and ceases when cells are confluent [115,120]. Nuclear translocation of angiogenin occurs through receptor-mediated endocytosis [114] and is independent of microtubule system and lysosomal processing [123]. Angiogenin seems to enter the nuclear pore by the classic nuclear pore input route [113]. Nuclear translocation of exogenous angiogenin is very fast. When exogenous angiogenin is added to the cell culture, nuclear angiogenin is detectable in 2 min and is saturated in 30 min [115]. On arriving at the nucleus, angiogenin accumulates in the nucleolus [114] where ribosome biogenesis takes place. Nuclear angiogenin has been shown to bind to the promoter region of rDNA [117] and stimulates rRNA transcription [116,126]. Cell growth requires the production of new ribosomes. Ribosome biogenesis is a process involving rRNA transcription, processing of the prerRNA precursor and assembly of the mature rRNA with ribosomal proteins [127–129]. The rate-limiting step in ribosome biogenesis is the synthesis of rRNA. Therefore, rRNA transcription is an important aspect of growth control. It is also important for maintaining a normal cell function as proteins are required for essentially all cellular activities. It may be particularly relevant for motor neurons to have a robust ribosome biogenesis because of long axonal transport of these cells.
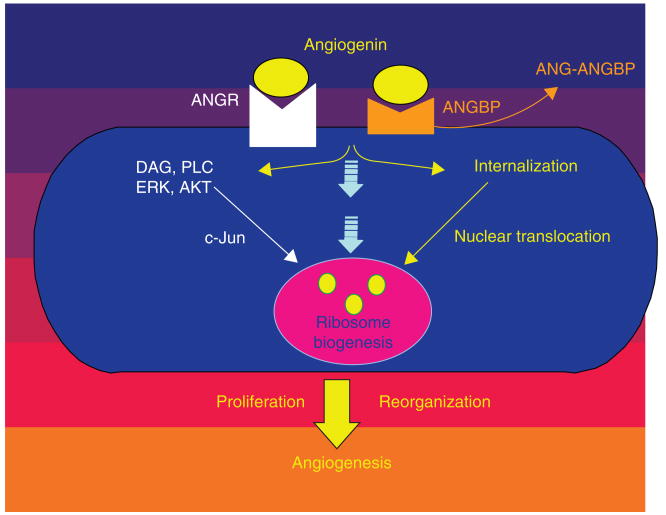
Angiogenin-stimulated rRNA transcription has been demonstrated as a general requirement for angiogenesis [126]. In other words, angiogenin is a permissive factor for other angiogenic factors to induce angiogenesis. Experimental evidence for this contention includes: i) nuclear translocation of endogenous angiogenin in endothelial cells is stimulated by other angiogenic factors including aFGF, bFGF, VEGF and EGF [126]; ii) knocking down ANG expression in endothelial cells inhibits bFGF- and VEGF-induced cell proliferation, accompanied with a decrease in rRNA transcription. Addition of exogenous angiogenin can completely restore the proliferative activity of these angiogenic factors [126]; and iii) angiogenin-specific inhibitors have no effect on binding of VEGF and bFGF to their receptors but inhibit their angiogenic activity [130]. Figure 2 summarizes the function of angiogenin-stimulated rRNA transcription in cell proliferation.
Figure 2. Angiogenin-stimulated rRNA transcription is a general requirement for angiogenesis.
Angiogenin is a permissive factor for other angiogenic proteins to induce cell proliferation. Growth factors such as VEGF activate PI3K–AKT–mTOR pathway to enhance ribosomal protein production. Angiogenin is translocated to the nucleus where it enhances rRNA transcription so that ribosome biogenesis can occur. Angiogenin inhibitors have been shown to abolish cell proliferation stimulated by other angiogenic factors including bFGF and VEGF.
ANG: Angiogenin.
As an angiogenic molecule, angiogenin may thus play a role in maintaining vasculature integrity in the spinal cord. Angiogenin insufficiency, caused by heterozygous missense mutations in the coding region, polymorphisms in the promoter region, or decreased expression owing to other genetic and environmental factors, may result in vascular abnormality and create an unhealthy environment for motor neurons. In addition, angiogenin may play a direct role in motor neuron physiology and its deficiency may accelerate or cause motor neuron degeneration. To this end, angiogenin has been shown to be expressed in motor neurons of both human and mouse spinal cords during development and in adulthood [21,22,24]. Angiogenin deficiency may result in insufficient ribosomal biogenesis and improper mRNA translation either in the entire or in some parts of the cell. Local protein translation of asymmetrically localized mRNA within neurons has been shown to play an important role in neuronal polarity and synaptic plasticity [131]. For example, asymmetrical localization of tau protein and its abnormal metabolism has been linked in both ALS and FTD [132]. Aberrant mRNA formation and RNA processing errors of excitatory amino-acid transporter 2 were found only in neuropathologically affected areas of ALS patients but not in other brain regions [133]. Motor neurons are the longest cells in the body and asymmetrical protein expression may thus be crucial not only during development but also during repair and regeneration. Robust protein translation machinery is essential for motor neuron physiology and a potential role of angiogenin can be predicted from its function in ribosome biogenesis. It is noteworthy that angiogenin is a member of the pancreatic RNase family and that the RNase activity is essential for its biological activity [95]. A link between RNA processing and neurodegeneration has been established [134]. In fact, spinal muscular atrophy, another motor neuron degenerative disease, is caused by mutations in SMN gene that encodes SMN, a protein known to play roles in RNA splicing, ribosome assembly and gene transcription [135].
4. ANG mutations in ALS patients
Recently, linkage analysis in Irish ALS populations identified an association of the G allele of the single nucleotide polymorphism rs11701 in the coding region of ANG (representing the amino-acid residue G86 in the mature protein) [18]. In the same study, a novel missense mutation at position 191 (A to T) in the coding region of ANG, which will result in a substitution of Lys 40 by Ile (K40I), was also found in 2 of the 169 Irish ALS patients but not in 171 control subjects [18]. Subsequently, seven heterozygous missense mutations in the coding region of ANG were identified in 15 patients by sequence screening of 1629 individuals with ALS [21], an overall frequency of ~1% with an overrepresentation of familial ALS (4/259, 1.5%) over sporadic ALS (11/1370, 0.8%). From sequencing 298 ALS patients of a Northern American cohort, four more mutations in ANG gene were identified (1.3% frequency) [22]. More recently, seven more mutations were identified in 9 of the 737 Italian ALS patients [20]. ANG mutations in Italian population also seem to segregate familial ALS (3/132, 2.3%) from sporadic ALS (6/605, 1%) with an overall frequency of 1.2% [20].
A total of 14 missense mutations (at 13-positions) in the coding region of ANG have been identified in 35 of the 3170 ALS patients of the Irish, Scottish, Swedish [21], North American [22] and Italian [20] populations. Among these mutations, 3 occurred in the signal peptide regions and 11 in the mature protein. In the seven sequencing efforts carried out so far, a total of 3003 healthy controls were included and two mutations in the ANG gene were found in non-ALS controls [20,21]. The first is a K17I mutation that was found in an apparent healthy 65-year-old male of European descent. The second is the I46V mutation that was found in 11 of the 1568 Italian healthy controls [19,20,136,137]. Therefore, I46V mutation does not seem to be associated with Italian ALS patients but does seem to be associated with the Scottish ALS patients in whom 3 of the 398 ALS patients but none of the 299 controls harbor this mutation [21]. Table 2 lists the frequencies of ANG mutations that occurred in 3170 ALS patients.
Table 2.
ANG mutations identified in ALS patients.
| Mutations | M(−24)I | F(−13)S | P(−4)L | Q12L | K17I | K17E | S28N |
|---|---|---|---|---|---|---|---|
| Number of cases | 2 | 1 | 2 | 2 | 3 | 2 | 1 |
| Ref. | [19,20] | [19] | [20,22] | [21] | [21,22] | [21] | [22] |
| Mutations | R31K | C39W | K40I | I46V | P112L | V113I | H114R |
| Number of cases | 1 | 2 | 5 | 10 | 1 | 2 | 1 |
| Ref. | [21] | [21] | [18,21] | [20,21,136] | [22] | [20] | [20] |
ALS: Amyotropic lateral sclerosis; ANG: Angiogenin.
5. Properties of mutant angiogenin proteins
Except for the three mutations in the signal peptide region (M-24I, F-13S, P-4S) and the two most recently reported mutations in the mature protein region (V113I, H114R) [20], all the mutant angiogenin proteins have been prepared and characterized by ribonuclease [22,23], nuclear translocation [22] and angiogenesis [22,23] assays. Except for R31K, all of these ALS-associated angiogenin mutant proteins have severely impaired ribonucleolytic activity ranging from < 1% (K40I) to 19% (K17E) of that of the WT angiogenin. R31K has 69% of the enzymatic activity of WT angiogenin [23]. Some of the mutant angiogenin proteins also have reduced thermal stability [23]. Among the three mutant angiogenin proteins (K17I, S28N, P112L) that have been tested in the nuclear translocation assay, S28N and P112L do not undergo nuclear translocation and K17I has a reduced capacity [22]. Two different angiogenesis assays have been used to examine the angiogenic activity of the mutant angiogenin proteins. The endothelial cell tube formation assay on fibrin gel was used to examine the mutants identified from the Northern American ALS patients and the results showed that all three mutants (K17I, S28N, P112L) are inactive [22]. The aorta ring assay was used to test three of the seven mutants identified from the Irish and Scottish ALS populations. All three mutants (Q12L, C39W, K40I) were inactive in the aorta ring angiogenesis assay [23]. Taken together, these results demonstrated that ANG mutations identified in ALS patients are associated with a functional loss of the angiogenic activity of the angiogenin protein.
WT angiogenin has been shown to stimulate neurite outgrowth and pathfinding of motor neurons derived from P19 embryonal carcinoma cells [25]. WT angiogenin also protects P19-derived motor neuron from hypoxia-induced cell death but the ALS-associated mutant angiogenin proteins (Q12L, C39W, K40I) lack this neuroprotective activity [24]. Moreover, these mutant angiogenin proteins are cytotoxic to the P19-derived motor neurons and induce their degeneration, suggesting that ANG mutations may even be causative to ALS [24].
6. Expression of angiogenin in the CNS
Mouse angiogenin is strongly expressed in the developing mouse nervous system both in the brain and in the spinal cord [25]. Immunohistochemistry and immunofluorescence have been used to show that angiogenin expression is the strongest in the brain and spinal cord at 9.5 days postcoitum (pc) [25]. At 11.5 days pc, angiogenin expression remains high in the telencephalon, mesen and mylencephalon as well as in the spinal cord, spinal ganglia and choroids plexus [25]. Until mid-gestation, angiogenin expression is stronger in the nervous system than in any other tissues. Co-staining with peripherin and Islet1 showed that angiogenin is expressed in mouse motor neurons.
Immunohistochemistry was also used to detect expression of human angiogenin in normal spinal cords obtained from fetal (ranging from 15 to 30 weeks gestation) and adult human autopsies. Strong angiogenin staining was observed in the ventral horn motor neurons of both fetal and adult cases [22]. Angiogenin was also detected in the extracellular matrix and interstitial tissues in all cases, consistent with it being a secreted protein. Angiogenin expression in the spinal cord seems to be downregulated as development proceeds but is still strongly expressed in the adulthood. Strong cytoplasmic and nuclear accumulation of angiogenin in motor neurons of both prenatal and adult spinal cords suggests a physiological role of angiogenin, both early in development and later in adulthood, and supports the hypothesis that ANG mutations are relevant to ALS pathology.
Double immunofluorescence with an antiangiogenin mAb 26-2F and antivon Willebrand factor polyclonal antibody showed that angiogenin is also localized in spinal cord endothelial cells, suggesting that angiogenin plays a role in maintaining the integrity of spinal cord vasculature that is important for physiological health of motor neurons [22]. Thus, angiogenin abnormalities may have a dual role in ALS, directly through motor neuron function and indirectly through endothelial cells and aberrant angiogenesis in the spinal cord.
7. Conclusion
Angiogenin is an angiogenic molecule known to play an essential role in angiogenesis by mediating rRNA transcription in endothelial cells [116,117]. The recent discovery of loss-of-function ANG mutations in ALS patients [19–22] and the findings that angiogenin is strongly expressed in the spinal cords both during fetal development and in adulthood [22,25] indicate an important role of angiogenin in motor neuron physiology and pathology. Angiogenin may have a dual role in motor neuron function by acting both on endothelial cells and on motor neurons. Thus, angiogenin may mediate angiogenesis thereby maintaining a normal vasculature, which is essential for motor neuron development, health and survival under various environmental and genetic insults. This is supported by previous findings that angiogenin-stimulated rRNA transcription is required for angiogenesis induced by VEGF [126] and that VEGF is a prominent angiogenic molecule known to be associated with ALS [66,88–92]. Involvement of angiogenin and VEGF, two angiogenic proteins that mediate angiogenesis by different mechanisms, in ALS suggests that angiogenesis insufficiency is linked to ALS pathogenesis [53]. Novel mutations in PGRN gene that encodes progranulin, another angiogenic protein, have recently been reported in ALS patients [69]; adding more evidence that abnormal angiogenesis is associated with ALS.
In addition to a role in angiogenesis, angiogenin may also act on motor neurons directly. This hypothesis is supported by the finding that angiogenin is strongly expressed in the motor neurons of fetal and adult spinal cords [22]. It is also supported by the results that angiogenin undergoes nuclear translocation in motor neurons and stimulates neurite outgrowth and pathfinding [24,25]. The mode of action of angiogenin in both endothelial cells and motor neurons could be related to its activity in mediating ribosome biogenesis. Nuclear angiogenin has been shown to bind to the promoter region of rDNA both in endothelial cells and in cancer cells thereby stimulating rRNA transcription [124,126]. It is conceivable that the role of angiogenin in motor neurons would also be related to rRNA transcription and that a defect in this pathway is likely to result in insufficient synthesis of ribosomes thereby affecting motor neuron viability. The dual role model suggests an essential role of angiogenin in motor neuron physiology. It is consistent with the results that all the angiogenin mutations so far found in ALS patients are heterozygous. Homozygous mutations may be lethal as a complete loss of angiogenin function would be detrimental. This model also implies that a decrease in angiogenin expression, as a result of various environmental and genetic insults, would have a profound effect on motor neuron function. It, at the same time, provides a therapeutic opportunity for ALS treatment by manipulating angiogenin expression levels and activities.
8. Expert opinion
Since the first report in 2004 that missense mutations in the cording region of ANG gene was found in ALS patients [18], 3170 ALS patients and 3003 non-ALS controls have been sequenced in six independent studies [18,19,21,22,136,137] and a total of 14 mutations in 13-positions have been found in 35 patients. Four functional studies have been carried out in which WT angiogenin has been shown to play a direct role in motor neuron physiology and the ALS-associated ANG mutations result in a complete loss of angiogenin activity [22–25]. Although mutations in ANG gene occurred in only 1.1% (ranging from 0.8 to 1% in sporadic ALS and 1.3 to 2.3% in familial ALS), ANG remains to be the second most frequently mutated gene in ALS and is the only loss-of-function gene so far identified in ALS patients. There is a sound rationale for exploring a novel ALS treatment opportunity by manipulating angiogenin levels and/or activities. For this purpose, the efficacy of angiogenin in improving motor muscular function and survival of SOD1G93A mice should be tested. First, WT angiogenin protein, with the ALS-associated mutant angiogenin proteins as controls, could be administered systemically by i.v., i.p., i.m. or s.c. injection. A beneficial effect could be expected from these routes of administration if angiogenin could cross the BBB and BSCB and reaches the CNS. A human angiogenin-specific mAb is available so that the distribution and stability of systemically administered angiogenin could be readily detected. It has been reported that ALS patients and SOD1G93A mice have disrupted BSCB [72–74] so the pharmacokinetic findings of systemically administered angiogenin from the above experiments should be confirmed with WT mice. Even if angiogenin dose not cross the BBB and BSCB, these experiments are still worthy doing because of the possibility that the site of action of angiogenin may be peripheral. The role of axon in ALS has been recognized [138–140] so the possibility that angiogenin acts directly on the neuromuscular junctions or on motor axons directly should not be excluded although there is no direct evidence at present. These experiments are thus both clinically and scientifically significant as they will tell us whether angiogenin is effective and where the site of action might be. If the above routes of administration are ineffective, intrathecal or intracerebroventricular administration of angiogenin protein directly into the CNS may be considered. Alternatively, retrograde delivery of angiogenin-encoding AAV or lentiviral particles could also be used to enhance angiogenin expression in the motor neurons.
Another informative experiment would be to generate and characterize ANG:SOD1G93A double transgenic mice. Both universal and cell-specific promoters should be considered for generating ANG transgenic mice. Characterization of these mice will reveal whether the effect of angiogenin is cell autonomous. Furthermore because the ALS-associated mutant angiogenin proteins are toxic to cultured motor neurons [24], it would be worthwhile to create and characterize transgenic mice overexpressing the mutant forms of ANG. If these mice develop ALS-like symptoms, they will be a valuable animal model, in addition to the SOD1 transgenic mice, to be used for mechanistic study and for screening and testing potential drugs.
Another approach to reveal the role of angiogenin in development in general, and in motor neuron physiology in particular, would be to create and characterize ANG knockout mice. Although humans have only a single ANG gene, mice have six [141]. It is not possible to knockout all of them simultaneously because they are spread out over ~8 million bp. However, mouse ANG1 is clearly the prominent form and the ortholog of the human gene [141,142]. Therefore, it is likely that knocking out mouse ANG1 will suffice for investigating the function of human angiogenin. Because of the possibility that the loss of mouse angiogenin-1 function may be embryonic lethal, it would be advisable to create conditional knockout so that the role of angiogenin-1 in motor neuron can be studied at the different stages during development. If ANG1 deletion results in motor neuron degeneration, ANG1 knockout mice may also be useful for ALS drug screening.
Acknowledgments
This work was supported by a grant from NCI (R01 CA105241). D Wu has received support from a Stanley L Robbins Memorial Research Award.
Footnotes
Declaration of interest
The authors state no conflict of interest and have received no payment in preparation of this manuscript.
Bibliography
Papers of special note have been highlighted as either of interest (•) or of considerable interest (••) to readers.
- 1.Pasinelli P, Brown RH. Molecular biology of amyotrophic lateral sclerosis: insights from genetics. Nat Rev Neurosci. 2006;7:710–23. doi: 10.1038/nrn1971. [DOI] [PubMed] [Google Scholar]
- 2.Massman PJ, Sims J, Cooke N, et al. Prevalence and correlates of neuropsychological deficits in amyotrophic lateral sclerosis. J Neurol Neurosurg Psychiatry. 1996;61:450–5. doi: 10.1136/jnnp.61.5.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neumann M, Sampathu DM, Kwong LK, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–3. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 4.Rippon GA, Scarmeas N, Gordon PH, et al. An observational study of cognitive impairment in amyotrophic lateral sclerosis. Arch Neurol. 2006;63:345–52. doi: 10.1001/archneur.63.3.345. [DOI] [PubMed] [Google Scholar]
- 5.Strong MJ, Lomen-Hoerth C, Caselli RJ, et al. Cognitive impairment, frontotemporal dementia, and the motor neuron diseases. Ann Neurol. 2003;54(Suppl 5):S20–3. doi: 10.1002/ana.10574. [DOI] [PubMed] [Google Scholar]
- 6.Yang W, Sopper MM, Leystra-Lantz C, Strong MJ. Microtubule-associated tau protein positive neuronal and glial inclusions in ALS. Neurology. 2003;61:1766–73. doi: 10.1212/01.wnl.0000099372.75786.f8. [DOI] [PubMed] [Google Scholar]
- 7.Woolley SC, Jonathan SK. Cognitive and behavioral impairment in amyotrophic lateral sclerosis. Phys Med Rehabil Clin N Am. 2008;19:607–17. doi: 10.1016/j.pmr.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 8.Bensimon G, Lacomblez L, Meininger V. A controlled trial of riluzole in amyotrophic lateral sclerosis. ALS/Riluzole Study Group. N Engl J Med. 1994;330:585–91. doi: 10.1056/NEJM199403033300901. [DOI] [PubMed] [Google Scholar]
- 9.Cronin S, Hardiman O, Traynor BJ. Ethnic variation in the incidence of ALS: a systematic review. Neurology. 2007;68:1002–7. doi: 10.1212/01.wnl.0000258551.96893.6f. [DOI] [PubMed] [Google Scholar]
- 10.Boillee S, Vande Velde C, Cleveland DW. ALS: a disease of motor neurons and their nonneuronal neighbors. Neuron. 2006;52:39–59. doi: 10.1016/j.neuron.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 11.Andersen PM, Nilsson P, Keranen ML, et al. Phenotypic heterogeneity in motor neuron disease patients with CuZn-superoxide dismutase mutations in Scandinavia. Brain. 1997;120:1723–37. doi: 10.1093/brain/120.10.1723. [DOI] [PubMed] [Google Scholar]
- 12.Cudkowicz ME, McKenna-Yasek D, Sapp PE, et al. Epidemiology of mutations in superoxide dismutase in amyotrophic lateral sclerosis. Ann Neurol. 1997;41:210–21. doi: 10.1002/ana.410410212. [DOI] [PubMed] [Google Scholar]
- 13.Niemann S, Joos H, Meyer T, et al. Familial ALS in Germany: origin of the R115G SOD1 mutation by a founder effect. J Neurol Neurosurg Psychiatry. 2004;75:1186–8. doi: 10.1136/jnnp.2003.028324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Battistini S, Giannini F, Greco G, et al. SOD1 mutations in amyotrophic lateral sclerosis. Results from a multicenter Italian study. J Neurol. 2005;252:782–8. doi: 10.1007/s00415-005-0742-y. [DOI] [PubMed] [Google Scholar]
- 15.Corrado L, D’Alfonso S, Bergamaschi L, et al. SOD1 gene mutations in Italian patients with Sporadic Amyotrophic Lateral Sclerosis (ALS) Neuromuscul Disord. 2006;16:800–4. doi: 10.1016/j.nmd.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 16.Jones CT, Swingler RJ, Simpson SA, Brock DJ. Superoxide dismutase mutations in an unselected cohort of Scottish amyotrophic lateral sclerosis patients. J Med Genet. 1995;32:290–2. doi: 10.1136/jmg.32.4.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17•.Fett JW, Strydom DJ, Lobb RR, et al. Isolation and characterization of angiogenin, an angiogenic protein from human carcinoma cells. Biochemistry. 1985;24:5480–6. doi: 10.1021/bi00341a030. This paper described the original isolation of angiogenin from the conditioned medium of HT-29 human colon adenocarcinoma cells and characterization of its angiogenic activity in the chick chorioallantoic membrane assay. [DOI] [PubMed] [Google Scholar]
- 18••.Greenway MJ, Alexander MD, Ennis S, et al. A novel candidate region for ALS on chromosome 14q11.2. Neurology. 2004;63:1936–8. doi: 10.1212/01.wnl.0000144344.39103.f6. This was the original paper that, for the first time, connects ANG with ALS. Linkage analysis performed in Irish and Scottish ALS population showed an association of a SNP rs11701 (corresponding to G86 in the mature protein) with ALS. The first ANG mutation (K40I) was reported in this paper. [DOI] [PubMed] [Google Scholar]
- 19••.Conforti FL, Sprovieri T, Mazzei R, et al. A novel Angiogenin gene mutation in a sporadic patient with amyotrophic lateral sclerosis from southern Italy. Neuromuscul Disord. 2008;18:68–70. doi: 10.1016/j.nmd.2007.07.003. Mutation (M-24I) at the start codon in the signal peptide region of ANG was reported in an Italian ALS patient. [DOI] [PubMed] [Google Scholar]
- 20••.Gellera C, Colombrita C, Ticozzi N, et al. Identification of new ANG gene mutations in a large cohort of Italian patients with amyotrophic lateral sclerosis. Neurogenetics. 2008;9:33–40. doi: 10.1007/s10048-007-0111-3. Seven mutations in the coding region of ANG were identified in 9 of the 737 Italian ALS patients. These mutations segregate FALS (3/132, 2.3%) from SALS (6/605, 1%) with an overall frequency of 1.2% in the Italian ALS population. [DOI] [PubMed] [Google Scholar]
- 21••.Greenway MJ, Andersen PM, Russ C, et al. ANG mutations segregate with familial and ‘sporadic’ amyotrophic lateral sclerosis. Nat Genet. 2006;38:411–3. doi: 10.1038/ng1742. This was the original report that ANG mutations were identified in ALS patients. Seven heterozygous missense mutations in ANG were identified in 15 patients by sequence screening of 1,629 individuals with ALS, an overall frequency of ~ 1% with an over representation of familial ALS (4/259, 1.5%) over sporadic ALS (11/1370, 0.8%) [DOI] [PubMed] [Google Scholar]
- 22••.Wu D, Yu W, Kishikawa H, et al. Angiogenin loss-of-function mutations in amyotrophic lateral sclerosis. Ann Neurol. 2007;62:609–17. doi: 10.1002/ana.21221. This was the first report that the mutant angiogenin proteins identified from the North American ALS cohort are inactive in endothelial cell tube formation assay accompanied with a loss of the ribonucleolytic activity and impaired nuclear translocation capacity. Strong expression of ANG protein in the motor neurons of fetal and adult human spinal cords was also reported in this paper. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23••.Crabtree B, Thiyagarajan N, Prior SH, et al. Characterization of human angiogenin variants implicated in amyotrophic lateral sclerosis. Biochemistry. 2007;46:11810–8. doi: 10.1021/bi701333h. This paper characterized the biochemical properties of a subset of reported ANG mutant proteins and showed that they are inactive in ribonuclease assay and in aortic ring angiogenesis assay. [DOI] [PubMed] [Google Scholar]
- 24••.Subramanian V, Crabtree B, Acharya KR. Human angiogenin is a neuroprotective factor and amyotrophic lateral sclerosis associated angiogenin variants affect neurite extension/pathfinding and survival of motor neurons. Hum Mol Genet. 2008;17:130–49. doi: 10.1093/hmg/ddm290. This was the first report that WT ANG protein is neuroprotective but the mutant ANG proteins are not. It also reported, for the first time, that the mutant ANG proteins found in ALS patients are neurotoxic to cultured motor neurons. [DOI] [PubMed] [Google Scholar]
- 25••.Subramanian V, Feng Y. A new role for angiogenin in neurite growth and pathfinding: implications for amyotrophic lateral sclerosis. Hum Mol Genet. 2007;16:1445–53. doi: 10.1093/hmg/ddm095. ANG was reported to stimulate neurite outgrowth and pathfinding of cultured mouse motor neurons. This paper also characterized the ANG expression in the mouse CNS during development. [DOI] [PubMed] [Google Scholar]
- 26.Gros-Louis F, Gaspar C, Rouleau GA. Genetics of familial and sporadic amyotrophic lateral sclerosis. Biochim Biophys Acta. 2006;1762:956–72. doi: 10.1016/j.bbadis.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 27.Chen W, Saeed M, Mao H, et al. Lack of association of VEGF promoter polymorphisms with sporadic ALS. Neurology. 2006;67:508–10. doi: 10.1212/01.wnl.0000227926.42370.04. [DOI] [PubMed] [Google Scholar]
- 28.Al-Chalabi A, Andersen PM, Nilsson P, et al. Deletions of the heavy neurofilament subunit tail in amyotrophic lateral sclerosis. Hum Mol Genet. 1999;8:157–64. doi: 10.1093/hmg/8.2.157. [DOI] [PubMed] [Google Scholar]
- 29.Figlewicz DA, Krizus A, Martinoli MG, et al. Variants of the heavy neurofilament subunit are associated with the development of amyotrophic lateral sclerosis. Hum Mol Genet. 1994;3:1757–61. doi: 10.1093/hmg/3.10.1757. [DOI] [PubMed] [Google Scholar]
- 30.Tomkins J, Usher P, Slade JY, et al. Novel insertion in the KSP region of the neurofilament heavy gene in amyotrophic lateral sclerosis (ALS) Neuroreport. 1998;9:3967–70. doi: 10.1097/00001756-199812010-00036. [DOI] [PubMed] [Google Scholar]
- 31.Lambrechts D, Storkebaum E, Morimoto M, et al. VEGF is a modifier of amyotrophic lateral sclerosis in mice and humans and protects motoneurons against ischemic death. Nat Genet. 2003;34:383–94. doi: 10.1038/ng1211. [DOI] [PubMed] [Google Scholar]
- 32.Goodall EF, Greenway MJ, van Marion I, et al. Association of the H63D polymorphism in the hemochromatosis gene with sporadic ALS. Neurology. 2005;65:934–7. doi: 10.1212/01.wnl.0000176032.94434.d4. [DOI] [PubMed] [Google Scholar]
- 33.Saeed M, Siddique N, Hung WY, et al. Paraoxonase cluster polymorphisms are associated with sporadic ALS. Neurology. 2006;67:771–6. doi: 10.1212/01.wnl.0000227187.52002.88. [DOI] [PubMed] [Google Scholar]
- 34.Comi GP, Bordoni A, Salani S, et al. Cytochrome c oxidase subunit I microdeletion in a patient with motor neuron disease. Ann Neurol. 1998;43:110–6. doi: 10.1002/ana.410430119. [DOI] [PubMed] [Google Scholar]
- 35.Borthwick GM, Taylor RW, Walls TJ, et al. Motor neuron disease in a patient with a mitochondrial tRNAIle mutation. Ann Neurol. 2006;59:570–4. doi: 10.1002/ana.20758. [DOI] [PubMed] [Google Scholar]
- 36.Dhaliwal GK, Grewal RP. Mitochondrial DNA deletion mutation levels are elevated in ALS brains. Neuroreport. 2000;11:2507–9. doi: 10.1097/00001756-200008030-00032. [DOI] [PubMed] [Google Scholar]
- 37.Drory VE, Birnbaum M, Korczyn AD, Chapman J. Association of APOE epsilon4 allele with survival in amyotrophic lateral sclerosis. J Neurol Sci. 2001;190:17–20. doi: 10.1016/s0022-510x(01)00569-x. [DOI] [PubMed] [Google Scholar]
- 38.Veldink JH, Kalmijn S, Van Der Hout AH, et al. SMN genotypes producing less SMN protein increase susceptibility to and severity of sporadic ALS. Neurology. 2005;65:820–5. doi: 10.1212/01.wnl.0000174472.03292.dd. [DOI] [PubMed] [Google Scholar]
- 39.Van Es MA, Van Vught PW, Blauw HM, et al. Genetic variation in DPP6 is associated with susceptibility to amyotrophic lateral sclerosis. Nat Genet. 2008;40:29–31. doi: 10.1038/ng.2007.52. [DOI] [PubMed] [Google Scholar]
- 40.Van Es MA, Van Vught PW, Blauw HM, et al. ITPR2 as a susceptibility gene in sporadic amyotrophic lateral sclerosis: a genome-wide association study. Lancet Neurol. 2007;6:869–77. doi: 10.1016/S1474-4422(07)70222-3. [DOI] [PubMed] [Google Scholar]
- 41.Dunckley T, Huentelman MJ, Craig DW, et al. Whole-genome analysis of sporadic amyotrophic lateral sclerosis. N Engl J Med. 2007;357:775–88. doi: 10.1056/NEJMoa070174. [DOI] [PubMed] [Google Scholar]
- 42.Gitcho MA, Baloh RH, Chakraverty S, et al. TDP-43 A315T mutation in familial motor neuron disease. Ann Neurol. 2008;63:535–8. doi: 10.1002/ana.21344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kabashi E, Valdmanis PN, Dion P, et al. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat Genet. 2008;40:572–4. doi: 10.1038/ng.132. [DOI] [PubMed] [Google Scholar]
- 44.Sreedharan J, Blair IP, Tripathi VB, et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–72. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van Deerlin VM, Leverenz JB, Bekris LM, et al. TARDBP mutations in amyotrophic lateral sclerosis with TDP-43 neuropathology: a genetic and histopathological analysis. Lancet Neurol. 2008;7:409–16. doi: 10.1016/S1474-4422(08)70071-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yokoseki A, Shiga A, Tan CF, et al. TDP-43 mutation in familial amyotrophic lateral sclerosis. Ann Neurol. 2008;63:538–42. doi: 10.1002/ana.21392. [DOI] [PubMed] [Google Scholar]
- 47.Mackenzie IR, Bigio EH, Ince PG, et al. Pathological TDP-43 distinguishes sporadic amyotrophic lateral sclerosis from amyotrophic lateral sclerosis with SOD1 mutations. Ann Neurol. 2007;61:427–34. doi: 10.1002/ana.21147. [DOI] [PubMed] [Google Scholar]
- 48.Robertson J, Sanelli T, Xiao S, et al. Lack of TDP-43 abnormalities in mutant SOD1 transgenic mice shows disparity with ALS. Neurosci Lett. 2007;420:128–32. doi: 10.1016/j.neulet.2007.03.066. [DOI] [PubMed] [Google Scholar]
- 49.Gijselinck I, Sleegers K, Engelborghs S, et al. Neuronal inclusion protein TDP-43 has no primary genetic role in FTD and ALS. Neurobiol Aging. 2007 doi: 10.1016/j.neurobiolaging.2007.11.002. Epup ahead of print. [DOI] [PubMed] [Google Scholar]
- 50.Guerreiro RJ, Schymick JC, Crews C, et al. TDP-43 is not a common cause of sporadic amyotrophic lateral sclerosis. PLoS One. 2008;3:e2450. doi: 10.1371/journal.pone.0002450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cleveland DW, Rothstein JD. From Charcot to Lou Gehrig: deciphering selective motor neuron death in ALS. Nat Rev Neurosci. 2001;2:806–19. doi: 10.1038/35097565. [DOI] [PubMed] [Google Scholar]
- 52.Goodall EF, Morrison KE. Amyotrophic lateral sclerosis (motor neuron disease): proposed mechanisms and pathways to treatment. Expert Rev Mol Med. 2006;8:1–22. doi: 10.1017/S1462399406010854. [DOI] [PubMed] [Google Scholar]
- 53.Lambrechts D, Lafuste P, Carmeliet P, Conway EM. Another angiogenic gene linked to amyotrophic lateral sclerosis. Trends Mol Med. 2006;12:345–7. doi: 10.1016/j.molmed.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 54.Simpson EP, Yen AA, Appel SH. Oxidative stress: a common denominator in the pathogenesis of amyotrophic lateral sclerosis. Curr Opin Rheumatol. 2003;15:730–6. doi: 10.1097/00002281-200311000-00008. [DOI] [PubMed] [Google Scholar]
- 55.Beckman JS, Carson M, Smith CD, Koppenol WH. ALS, SOD and peroxynitrite. Nature. 1993;364:584. doi: 10.1038/364584a0. [DOI] [PubMed] [Google Scholar]
- 56.Estevez AG, Crow JP, Sampson JB, et al. Induction of nitric oxide-dependent apoptosis in motor neurons by zinc-deficient superoxide dismutase. Science. 1999;286:2498–500. doi: 10.1126/science.286.5449.2498. [DOI] [PubMed] [Google Scholar]
- 57.Wiedau-Pazos M, Goto JJ, Rabizadeh S, et al. Altered reactivity of superoxide dismutase in familial amyotrophic lateral sclerosis. Science. 1996;271:515–8. doi: 10.1126/science.271.5248.515. [DOI] [PubMed] [Google Scholar]
- 58.Bruijn LI, Beal MF, Becher MW, et al. Elevated free nitrotyrosine levels, but not protein-bound nitrotyrosine or hydroxyl radicals, throughout amyotrophic lateral sclerosis (ALS)-like disease implicate tyrosine nitration as an aberrant in vivo property of one familial ALS-linked superoxide dismutase 1 mutant. Proc Natl Acad Sci USA. 1997;94:7606–11. doi: 10.1073/pnas.94.14.7606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wong PC, Waggoner D, Subramaniam JR, et al. Copper chaperone for superoxide dismutase is essential to activate mammalian Cu/Zn superoxide dismutase. Proc Natl Acad Sci USA. 2000;97:2886–91. doi: 10.1073/pnas.040461197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang J, Slunt H, Gonzales V, et al. Copper-binding-site-null SOD1 causes ALS in transgenic mice: aggregates of non-native SOD1 delineate a common feature. Hum Mol Genet. 2003;12:2753–64. doi: 10.1093/hmg/ddg312. [DOI] [PubMed] [Google Scholar]
- 61.Boillee S, Yamanaka K, Lobsiger CS, et al. Onset and progression in inherited ALS determined by motor neurons and microglia. Science. 2006;312:1389–92. doi: 10.1126/science.1123511. [DOI] [PubMed] [Google Scholar]
- 62.Yamanaka K, Chun SJ, Boillee S, et al. Astrocytes as determinants of disease progression in inherited amyotrophic lateral sclerosis. Nat Neurosci. 2008;11:251–3. doi: 10.1038/nn2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Di Giorgio FP, Carrasco MA, Siao MC, et al. Non-cell autonomous effect of glia on motor neurons in an embryonic stem cell-based ALS model. Nat Neurosci. 2007;10:608–14. doi: 10.1038/nn1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nagai M, Re DB, Nagata T, et al. Astrocytes expressing ALS-linked mutated SOD1 release factors selectively toxic to motor neurons. Nat Neurosci. 2007;10:615–22. doi: 10.1038/nn1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yamanaka K, Boillee S, Roberts EA, et al. Mutant SOD1 in cell types other than motor neurons and oligodendrocytes accelerates onset of disease in ALS mice. Proc Natl Acad Sci USA. 2008;105:7594–9. doi: 10.1073/pnas.0802556105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66••.Oosthuyse B, Moons L, Storkebaum E, et al. Deletion of the hypoxia-response element in the vascular endothelial growth factor promoter causes motor neuron degeneration. Nat Genet. 2001;28:131–8. doi: 10.1038/88842. This paper generated and characterized mice (VEGF.∂/∂ mice) that lack the hypoxia-response element. VEGF∂/∂ mice have reduced hypoxic VRGF expression in the spinal cord and develop adult onset progressive motor neuron degeneration. [DOI] [PubMed] [Google Scholar]
- 67.Baker M, MacKenzie IR, Pickering-Brown SM, et al. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature. 2006;442:916–9. doi: 10.1038/nature05016. [DOI] [PubMed] [Google Scholar]
- 68.Cruts M, Gijselinck I, Van Der Zee J, et al. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature. 2006;442:920–4. doi: 10.1038/nature05017. [DOI] [PubMed] [Google Scholar]
- 69.Sleegers K, Brouwers N, Maurer-Stroh S, et al. Progranulin genetic variability contributes to amyotrophic lateral sclerosis. Neurology. 2008;71:253–9. doi: 10.1212/01.wnl.0000289191.54852.75. [DOI] [PubMed] [Google Scholar]
- 70.Carmeliet P, Tessier-Lavigne M. Common mechanisms of nerve and blood vessel wiring. Nature. 2005;436:193–200. doi: 10.1038/nature03875. [DOI] [PubMed] [Google Scholar]
- 71.Zacchigna S, Lambrechts D, Carmeliet P. Neurovascular signalling defects in neurodegeneration. Nat Rev Neurosci. 2008;9:169–81. doi: 10.1038/nrn2336. [DOI] [PubMed] [Google Scholar]
- 72.Garbuzova-Davis S, Haller E, Saporta S, et al. Ultrastructure of blood-brain barrier and blood-spinal cord barrier in SOD1 mice modeling ALS. Brain Res. 2007;1157:126–37. doi: 10.1016/j.brainres.2007.04.044. [DOI] [PubMed] [Google Scholar]
- 73.Garbuzova-Davis S, Saporta S, Haller E, et al. Evidence of compromised blood-spinal cord barrier in early and late symptomatic SOD1 mice modeling ALS. PLoS One. 2007;2:e1205. doi: 10.1371/journal.pone.0001205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhong Z, Deane R, Ali Z, et al. ALS-causing SOD1 mutants generate vascular changes prior to motor neuron degeneration. Nat Neurosci. 2008;11:420–2. doi: 10.1038/nn2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Song JH, Huang CS, Nagata K, et al. Differential action of riluzole on tetrodotoxin-sensitive and tetrodotoxin-resistant sodium channels. J Pharmacol Exp Ther. 1997;282:707–14. [PubMed] [Google Scholar]
- 76.Lacomblez L, Bensimon G, Leigh PN, et al. Dose-ranging study of riluzole in amyotrophic lateral sclerosis. Amyotrophic Lateral Sclerosis/Riluzole Study Group II. Lancet. 1996;347:1425–31. doi: 10.1016/s0140-6736(96)91680-3. [DOI] [PubMed] [Google Scholar]
- 77.McGeer EG, McGeer PL. Pharmacologic approaches to the treatment of amyotrophic lateral sclerosis. BioDrugs. 2005;19:31–7. doi: 10.2165/00063030-200519010-00004. [DOI] [PubMed] [Google Scholar]
- 78.Kasarkis EJ, et al. A controlled trial of recombinant methionyl human BDNF in ALS: the BDNF Study Group (Phase III) Neurology. 1999;52:1427–33. doi: 10.1212/wnl.52.7.1427. [DOI] [PubMed] [Google Scholar]
- 79.Akbar MT, Torp R, Danbolt NC, et al. Expression of glial glutamate transporters GLT-1 and GLAST is unchanged in the hippocampus in fully kindled rats. Neuroscience. 1997;78:351–9. doi: 10.1016/s0306-4522(96)00570-2. [DOI] [PubMed] [Google Scholar]
- 80.Borasio GD, Robberecht W, Leigh PN, et al. A placebo-controlled trial of insulin-like growth factor-I in amyotrophic lateral sclerosis. European ALS/IGF-I Study Group. Neurology. 1998;51:583–6. doi: 10.1212/wnl.51.2.583. [DOI] [PubMed] [Google Scholar]
- 81.Lai EC, Felice KJ, Festoff BW, et al. Effect of recombinant human insulin-like growth factor-I on progression of ALS. A placebo-controlled study. The North America ALS/IGF-I Study Group. Neurology. 1997;49:1621–30. doi: 10.1212/wnl.49.6.1621. [DOI] [PubMed] [Google Scholar]
- 82.Manabe Y, Nagano I, Gazi MS, et al. Glial cell line-derived neurotrophic factor protein prevents motor neuron loss of transgenic model mice for amyotrophic lateral sclerosis. Neurol Res. 2003;25:195–200. doi: 10.1179/016164103101201193. [DOI] [PubMed] [Google Scholar]
- 83.Scott S, Kranz JE, Cole J, et al. Design, power, and interpretation of studies in the standard murine model of ALS. Amyotroph Lateral Scler. 2008;9:4–15. doi: 10.1080/17482960701856300. [DOI] [PubMed] [Google Scholar]
- 84.Federici T, Boulis NM. Gene-based treatment of motor neuron diseases. Muscle Nerve. 2006;33:302–23. doi: 10.1002/mus.20439. [DOI] [PubMed] [Google Scholar]
- 85.Kaspar BK, Llado J, Sherkat N, et al. Retrograde viral delivery of IGF-1 prolongs survival in a mouse ALS model. Science. 2003;301:839–42. doi: 10.1126/science.1086137. [DOI] [PubMed] [Google Scholar]
- 86.Wang LJ, Lu YY, Muramatsu S, et al. Neuroprotective effects of glial cell line-derived neurotrophic factor mediated by an adeno-associated virus vector in a transgenic animal model of amyotrophic lateral sclerosis. J Neurosci. 2002;22:6920–8. doi: 10.1523/JNEUROSCI.22-16-06920.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Azzouz M, Hottinger A, Paterna JC, et al. Increased motoneuron survival and improved neuromuscular function in transgenic ALS mice after intraspinal injection of an adeno-associated virus encoding Bcl-2. Hum Mol Genet. 2000;9:803–11. doi: 10.1093/hmg/9.5.803. [DOI] [PubMed] [Google Scholar]
- 88.Terry PD, Kamel F, Umbach DM, et al. VEGF promoter haplotype and amyotrophic lateral sclerosis (ALS) J Neurogenet. 2004;18:429–34. doi: 10.1080/01677060490894450. [DOI] [PubMed] [Google Scholar]
- 89.Storkebaum E, Lambrechts D, Dewerchin M, et al. Treatment of motoneuron degeneration by intracerebroventricular delivery of VEGF in a rat model of ALS. Nat Neurosci. 2005;8:85–92. doi: 10.1038/nn1360. [DOI] [PubMed] [Google Scholar]
- 90.Azzouz M, Ralph GS, Storkebaum E, et al. VEGF delivery with retrogradely transported lentivector prolongs survival in a mouse ALS model. Nature. 2004;429:413–7. doi: 10.1038/nature02544. [DOI] [PubMed] [Google Scholar]
- 91.Wang Y, Mao XO, Xie L, et al. Vascular endothelial growth factor overexpression delays neurodegeneration and prolongs survival in amyotrophic lateral sclerosis mice. J Neurosci. 2007;27:304–7. doi: 10.1523/JNEUROSCI.4433-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zheng C, Nennesmo I, Fadeel B, Henter JI. Vascular endothelial growth factor prolongs survival in a transgenic mouse model of ALS. Ann Neurol. 2004;56:564–7. doi: 10.1002/ana.20223. [DOI] [PubMed] [Google Scholar]
- 93.Weiner HL, Weiner LH, Swain JL. Tissue distribution and developmental expression of the messenger RNA encoding angiogenin. Science. 1987;237:280–2. doi: 10.1126/science.2440105. [DOI] [PubMed] [Google Scholar]
- 94•.Strydom DJ, Fett JW, Lobb RR, et al. Amino acid sequence of human tumor derived angiogenin. Biochemistry. 1985;24:5486–94. doi: 10.1021/bi00341a031. This paper determined the amino-acid sequence of human ANG and identified it as a member of the pancreatic ribonuclease superfamily. [DOI] [PubMed] [Google Scholar]
- 95•.Shapiro R, Riordan JF, Vallee BL. Characteristic ribonucleolytic activity of human angiogenin. Biochemistry. 1986;25:3527–32. doi: 10.1021/bi00360a008. This paper characterized the ribonucleolytic activity of ANG and described the essential role of the enzymatic activity in angiogenesis. [DOI] [PubMed] [Google Scholar]
- 96.Riordan JF. Angiogenin. Methods Enzymol. 2001;341:263–73. doi: 10.1016/s0076-6879(01)41157-8. [DOI] [PubMed] [Google Scholar]
- 97.Curran TP, Shapiro R, Riordan JF, Vallee BL. Modulation of the activity of angiogenin by mutagenesis at Asp-116. Biochim Biophys Acta. 1993;1202:281–6. doi: 10.1016/0167-4838(93)90017-l. [DOI] [PubMed] [Google Scholar]
- 98.Hallahan TW, Shapiro R, Strydom DJ, Vallee BL. Importance of asparagine-61 and asparagine-109 to the angiogenic activity of human angiogenin. Biochemistry. 1992;31:8022–9. doi: 10.1021/bi00149a036. [DOI] [PubMed] [Google Scholar]
- 99.Hallahan TW, Shapiro R, Vallee BL. Dual site model for the organogenic activity of angiogenin. Proc Natl Acad Sci USA. 1991;88:2222–6. doi: 10.1073/pnas.88.6.2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Harper JW, Fox EA, Shapiro R, Vallee BL. Mutagenesis of residues flanking Lys-40 enhances the enzymatic activity and reduces the angiogenic potency of angiogenin. Biochemistry. 1990;29:7297–302. doi: 10.1021/bi00483a020. [DOI] [PubMed] [Google Scholar]
- 101.Shapiro R, Fox EA, Riordan JF. Role of lysines in human angiogenin: chemical modification and site-directed mutagenesis. Biochemistry. 1989;28:1726–32. doi: 10.1021/bi00430a045. [DOI] [PubMed] [Google Scholar]
- 102.Shapiro R, Vallee BL. Site-directed mutagenesis of histidine-13 and histidine-114 of human angiogenin. Alanine derivatives inhibit angiogenin-induced angiogenesis. Biochemistry. 1989;28:7401–8. doi: 10.1021/bi00444a038. [DOI] [PubMed] [Google Scholar]
- 103.Shapiro R, Vallee BL. Identification of functional arginines in human angiogenin by site-directed mutagenesis. Biochemistry. 1992;31:12477–85. doi: 10.1021/bi00164a026. [DOI] [PubMed] [Google Scholar]
- 104.Shapiro R, Weremowicz S, Riordan JF, Vallee BL. Ribonucleolytic activity of angiogenin: essential histidine, lysine, and arginine residues. Proc Natl Acad Sci USA. 1987;84:8783–7. doi: 10.1073/pnas.84.24.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Acharya KR, Shapiro R, Allen SC, et al. Crystal structure of human angiogenin reveals the structural basis for its functional divergence from ribonuclease. Proc Natl Acad Sci USA. 1994;91:2915–9. doi: 10.1073/pnas.91.8.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Russo N, Shapiro R, Acharya KR, et al. Role of glutamine-117 in the ribonucleolytic activity of human angiogenin. Proc Natl Acad Sci USA. 1994;91:2920–4. doi: 10.1073/pnas.91.8.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hu GF, Chang SI, Riordan JF, Vallee BL. An angiogenin-binding protein from endothelial cells. Proc Natl Acad Sci USA. 1991;88:2227–31. doi: 10.1073/pnas.88.6.2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Badet J, Soncin F, Guitton JD, et al. Specific binding of angiogenin to calf pulmonary artery endothelial cells. Proc Natl Acad Sci USA. 1989;86:8427–31. doi: 10.1073/pnas.86.21.8427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bicknell R, Vallee BL. Angiogenin activates endothelial cell phospholipase C. Proc Natl Acad Sci USA. 1988;85:5961–5. doi: 10.1073/pnas.85.16.5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bicknell R, Vallee BL. Angiogenin stimulates endothelial cell prostacyclin secretion by activation of phospholipase A2. Proc Natl Acad Sci USA. 1989;86:1573–7. doi: 10.1073/pnas.86.5.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Liu S, Yu D, Xu ZP, et al. Angiogenin activates Erk1/2 in human umbilical vein endothelial cells. Biochem Biophys Res Commun. 2001;287:305–10. doi: 10.1006/bbrc.2001.5568. [DOI] [PubMed] [Google Scholar]
- 112.Kim HM, Kang DK, Kim HY, et al. Angiogenin-induced protein kinase B/Akt activation is necessary for angiogenesis but is independent of nuclear translocation of angiogenin in HUVE cells. Biochem Biophys Res Commun. 2007;352:509–13. doi: 10.1016/j.bbrc.2006.11.047. [DOI] [PubMed] [Google Scholar]
- 113.Moroianu J, Riordan JF. Identification of the nucleolar targeting signal of human angiogenin. Biochem Biophys Res Commun. 1994;203:1765–72. doi: 10.1006/bbrc.1994.2391. [DOI] [PubMed] [Google Scholar]
- 114.Moroianu J, Riordan JF. Nuclear translocation of angiogenin in proliferating endothelial cells is essential to its angiogenic activity. Proc Natl Acad Sci USA. 1994;91:1677–81. doi: 10.1073/pnas.91.5.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hu G, Xu C, Riordan JF. Human angiogenin is rapidly translocated to the nucleus of human umbilical vein endothelial cells and binds to DNA. J Cell Biochem. 2000;76:452–62. doi: 10.1002/(sici)1097-4644(20000301)76:3<452::aid-jcb12>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 116.Xu ZP, Tsuji T, Riordan JF, Hu GF. The nuclear function of angiogenin in endothelial cells is related to rRNA production. Biochem Biophys Res Commun. 2002;294:287–92. doi: 10.1016/S0006-291X(02)00479-5. [DOI] [PubMed] [Google Scholar]
- 117.Xu ZP, Tsuji T, Riordan JF, Hu GF. Identification and characterization of an angiogenin-binding DNA sequence that stimulates luciferase reporter gene expression. Biochemistry. 2003;42:121–8. doi: 10.1021/bi020465x. [DOI] [PubMed] [Google Scholar]
- 118.Hu GF, Strydom DJ, Fett JW, et al. Actin is a binding protein for angiogenin. Proc Natl Acad Sci USA. 1993;90:1217–21. doi: 10.1073/pnas.90.4.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Moroianu J, Fett JW, Riordan JF, Vallee BL. Actin is a surface component of calf pulmonary artery endothelial cells in culture. Proc Natl Acad Sci USA. 1993;90:3815–9. doi: 10.1073/pnas.90.9.3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hu GF, Riordan JF, Vallee BL. A putative angiogenin receptor in angiogenin-responsive human endothelial cells. Proc Natl Acad Sci USA. 1997;94:2204–9. doi: 10.1073/pnas.94.6.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hu GF, Riordan JF. Angiogenin enhances actin acceleration of plasminogen activation. Biochem Biophys Res Commun. 1993;197:682–7. doi: 10.1006/bbrc.1993.2533. [DOI] [PubMed] [Google Scholar]
- 122.Hu G, Riordan JF, Vallee BL. Angiogenin promotes invasiveness of cultured endothelial cells by stimulation of cell-associated proteolytic activities. Proc Natl Acad Sci USA. 1994;91:12096–100. doi: 10.1073/pnas.91.25.12096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Li R, Riordan JF, Hu G. Nuclear translocation of human angiogenin in cultured human umbilical artery endothelial cells is microtubule and lysosome independent. Biochem Biophys Res Commun. 1997;238:305–12. doi: 10.1006/bbrc.1997.7290. [DOI] [PubMed] [Google Scholar]
- 124.Tsuji T, Sun Y, Kishimoto K, et al. Angiogenin is translocated to the nucleus of HeLa cells and is involved in ribosomal RNA transcription and cell proliferation. Cancer Res. 2005;65:1352–60. doi: 10.1158/0008-5472.CAN-04-2058. [DOI] [PubMed] [Google Scholar]
- 125.Yoshioka N, Wang L, Kishimoto K, et al. A therapeutic target for prostate cancer based on angiogenin-stimulated angiogenesis and cancer cell proliferation. Proc Natl Acad Sci USA. 2006;103:14519–24. doi: 10.1073/pnas.0606708103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126••.Kishimoto K, Liu S, Tsuji T, et al. Endogenous angiogenin in endothelial cells is a general requirement for cell proliferation and angiogenesis. Oncogene. 2005;24:445–56. doi: 10.1038/sj.onc.1208223. This paper reported that ANG-stimulated rRNA transcription is required for angiogenesis stimulated by other angiogenic factor. [DOI] [PubMed] [Google Scholar]
- 127.Comai L. The nucleolus: a paradigm for cell proliferation and aging. Braz J Med Biol Res. 1999;32:1473–8. doi: 10.1590/s0100-879x1999001200004. [DOI] [PubMed] [Google Scholar]
- 128.Melese T, Xue Z. The nucleolus: an organelle formed by the act of building a ribosome. Curr Opin Cell Biol. 1995;7:319–24. doi: 10.1016/0955-0674(95)80085-9. [DOI] [PubMed] [Google Scholar]
- 129.Stoykova AS, Dabeva MD, Dimova RN, Hadjiolov AA. Ribosome biogenesis and nucleolar ultrastructure in neuronal and oligodendroglial rat brain cells. J Neurochem. 1985;45:1667–76. doi: 10.1111/j.1471-4159.1985.tb10521.x. [DOI] [PubMed] [Google Scholar]
- 130.Hirukawa S, Olson KA, Tsuji T, Hu GF. Neamine inhibits xenografic human tumor growth and angiogenesis in athymic mice. Clin Cancer Res. 2005;11:8745–52. doi: 10.1158/1078-0432.CCR-05-1495. [DOI] [PubMed] [Google Scholar]
- 131.Dahm R, Macchi P. Human pathologies associated with defective RNA transport and localization in the nervous system. Biol Cell. 2007;99:649–61. doi: 10.1042/BC20070045. [DOI] [PubMed] [Google Scholar]
- 132.Aronov S, Aranda G, Behar L, Ginzburg I. Axonal tau mRNA localization coincides with tau protein in living neuronal cells and depends on axonal targeting signal. J Neurosci. 2001;21:6577–87. doi: 10.1523/JNEUROSCI.21-17-06577.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lin CL, Bristol LA, Jin L, et al. Aberrant RNA processing in a neurodegenerative disease: the cause for absent EAAT2, a glutamate transporter, in amyotrophic lateral sclerosis. Neuron. 1998;20:589–602. doi: 10.1016/s0896-6273(00)80997-6. [DOI] [PubMed] [Google Scholar]
- 134.Gallo JM, Jin P, Thornton CA, et al. The role of RNA and RNA processing in neurodegeneration. J Neurosci. 2005;25:10372–5. doi: 10.1523/JNEUROSCI.3453-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Monani UR. Spinal muscular atrophy: a deficiency in a ubiquitous protein; a motor neuron-specific disease. Neuron. 2005;48:885–96. doi: 10.1016/j.neuron.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 136.Corrado L, Battistini S, Penco S, et al. Variations in the coding and regulatory sequences of the angiogenin (ANG) gene are not associated to ALS (amyotrophic lateral sclerosis) in the Italian population. J Neurol Sci. 2007;258:123–7. doi: 10.1016/j.jns.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 137.Del Bo R, Scarlato M, Ghezzi S, et al. Absence of angiogenic genes modification in Italian ALS patients. Neurobiol Aging. 2008;29:314–6. doi: 10.1016/j.neurobiolaging.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 138.Fischer LR, Culver DG, Tennant P, et al. Amyotrophic lateral sclerosis is a distal axonopathy: evidence in mice and man. Exp Neurol. 2004;185:232–40. doi: 10.1016/j.expneurol.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 139.Frey D, Schneider C, Xu L, et al. Early and selective loss of neuromuscular synapse subtypes with low sprouting competence in motoneuron diseases. J Neurosci. 2000;20:2534–42. doi: 10.1523/JNEUROSCI.20-07-02534.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kennel PF, Finiels F, Revah F, Mallet J. Neuromuscular function impairment is not caused by motor neurone loss in FALS mice: an electromyographic study. Neuroreport. 1996;7:1427–31. doi: 10.1097/00001756-199605310-00021. [DOI] [PubMed] [Google Scholar]
- 141.Cho S, Zhang J. Ancient expansion of the ribonuclease A superfamily revealed by genomic analysis of placental and marsupial mammals. Gene. 2006;373:116–25. doi: 10.1016/j.gene.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 142.Hooper LV, Stappenbeck TS, Hong CV, Gordon JI. Angiogenins: a new class of microbicidal proteins involved in innate immunity. Nat Immunol. 2003;4:269–73. doi: 10.1038/ni888. [DOI] [PubMed] [Google Scholar]
- 143.Rosen DR, Siddique T, Patterson D, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 144.Yang Y, Hentati A, Deng HX, et al. The gene encoding alsin, a protein with three guanine-nucleotide exchange factor domains, is mutated in a form of recessive amyotrophic lateral sclerosis. Nat Genet. 2001;29:160–5. doi: 10.1038/ng1001-160. [DOI] [PubMed] [Google Scholar]
- 145.Hand CK, Khoris J, Salachas F, et al. A novel locus for familial amyotrophic lateral sclerosis, on chromosome 18q. Am J Hum Genet. 2002;70:251–6. doi: 10.1086/337945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Chen YZ, Bennett CL, Huynh HM, et al. DNA/RNA helicase gene mutations in a form of juvenile amyotrophic lateral sclerosis (ALS4) Am J Hum Genet. 2004;74:1128–35. doi: 10.1086/421054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hentati A, Ouahchi K, Pericak-Vance MA, et al. Linkage of a commoner form of recessive amyotrophic lateral sclerosis to chromosome 15q15-q22 markers. Neurogenetics. 1998;2:55–60. doi: 10.1007/s100480050052. [DOI] [PubMed] [Google Scholar]
- 148.Abalkhail H, Mitchell J, Habgood J, et al. A new familial amyotrophic lateral sclerosis locus on chromosome 16q12.1-16q12.2. Am J Hum Genet. 2003;73:383–9. doi: 10.1086/377156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Sapp PC, Hosler BA, McKenna-Yasek D, et al. Identification of two novel loci for dominantly inherited familial amyotrophic lateral sclerosis. Am J Hum Genet. 2003;73:397–403. doi: 10.1086/377158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Nishimura AL, Mitne-Neto M, Silva HC, et al. A mutation in the vesicle-trafficking protein VAPB causes late-onset spinal muscular atrophy and amyotrophic lateral sclerosis. Am J Hum Genet. 2004;75:822–31. doi: 10.1086/425287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hosler BA, Siddique T, Sapp PC, et al. Linkage of familial amyotrophic lateral sclerosis with frontotemporal dementia to chromosome 9q21–q22. JAMA. 2000;284:1664–9. doi: 10.1001/jama.284.13.1664. [DOI] [PubMed] [Google Scholar]
- 152.Hutton M, Lendon CL, Rizzu P, et al. Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature. 1998;393:702–5. doi: 10.1038/31508. [DOI] [PubMed] [Google Scholar]
- 153.Puls I, Jonnakuty C, LaMonte BH, et al. Mutant dynactin in motor neuron disease. Nat Genet. 2003;33:455–6. doi: 10.1038/ng1123. [DOI] [PubMed] [Google Scholar]