Abstract
Hepatocyte nuclear factor (HNF)-1α is a homeodomain-containing transcription factor. Humans heterozygous for mutations in the HNF-1α gene develop maturity-onset diabetes of the young (MODY3), which is associated with reduced insulin secretion. The mechanisms responsible for defective glucose-induced insulin secretion due to HNF-1α deficiency are complex. In order to explore the relationship between HNF-1α and beta-cell proliferation, we have created a novel animal model. Mice lacking one allele of the HNF-1α gene were crossed with transgenic mice expressing the large T antigen driven by the rat insulin II promoter (RIP). The resulting mouse strains allowed us to study the effect of HNF-1α deficiency on the extensive beta-cell proliferation that occurs in these mice. Our results indicate that deficiency of HNF-1α severely constrains the extent of beta-cell proliferation occurring in RIP-Tag mice leading to significant changes in blood glucose concentrations as a result of reduced beta-cell number, insulin content, insulin secretion and intracellular responses in Ca2+. Furthermore expression profiling studies using immortalized cell lines generated from HNF-1α/RIP-Tag mice showed changes in expression of genes involved in cellular growth and proliferation. These results provide insights into the mechanisms whereby HNF-1α affects beta-cell function.
Keywords: hepatocyte nuclear factor-1α, maturity-onset diabetes of the young, pancreatic beta-cells, diabetes, expression profiling
Introduction
The identification of the genes responsible for maturity-onset diabetes of the young (MODY) has yielded important insights into the regulation of normal beta-cell function. MODY is characterized by autosomal dominant inheritance, with onset usually before the age of 25 years, and a primary defect in the function of pancreatic beta-cells (1–3). Mutations in the heterozygous state in the hepatocyte nuclear factor (HNF)-1α gene lead to beta-cell dysfunction, which is associated with MODY3 (4). Previous studies have indicated that expression of the insulin gene as well as genes encoding proteins involved in glucose transport and metabolism (5–10), mitochondrial metabolism (9) and cell-cell adhesion (10) are regulated by HNF-1α. Recently, the reduced expression of Tmem27, a novel target of HNF-1α in pancreatic beta-cells has been shown to result in impaired insulin secretion and beta-cell proliferation (11, 12).
HNF-1α is a homeodomain-containing transcription factor (13), which is enriched in liver, and also expressed in kidney, intestine, and pancreatic islets (14). Mice lacking both alleles of the HNF-1α gene fail to thrive and often die around weaning. The mice exhibit a progressive wasting syndrome, have marked liver enlargement and severe renal Fanconi syndrome with massive glycosuria (15). We have reported that these mice also develop diabetes (16). However, in contrast to humans, heterozygous mice appear normal and are not diabetic (15–17).
In the present study, we further explored the role of HNF-1α in the regulation of beta-cell function by generating a novel mouse model of HNF-1α deficiency by crossing HNF-1α deficient mice with transgenic mice that express the rat insulin II promoter (RIP)-large T antigen (Tag) (18). The latter animals develop hypoglycemia due to beta-cell proliferation leading to an increase in beta-cell mass and insulinomas. Our experimental approach was designed to allow us to test the hypothesis that the level of expression of HNF-1α would have a significant impact on the extent of beta-cell proliferation stimulated by expression of RIP-Tag.
Materials and Methods
Generation of HNF-1α deficient/RIP-Tag mice
HNF-1α (+/−) mice (15) and RIP-Tag mice (Jackson Laboratory, Bar Harbor, ME) were crossbred. Since HNF-1α (−/−) mice are infertile, HNF-1α (−/−)/RIP-Tag mice were generated by intercrossing HNF-1α (+/−)/RIP-Tag mice. Mice were screened by PCR analysis of tail DNA using primers encompassing a promoter region and exon 1 of the mouse HNF-1α gene (forward: 5′-GGCCGCTGGGGCCAGGGTTG-3′ and reverse: 5′-CCAGGGGACCCTCTCCAACC-3′) and an additional reverse primer for the mutated allele designed within the β-galactosidase cassette (5′-GGGAAGGGCGATCGGTGCGG-3′), which yielded PCR products of 300 bp and 400 bp corresponding the wild-type and the mutated allele, respectively (15). The presence of the RIP-Tag transgene was analyzed also by PCR using forward and reverse primers; 5′-CTTAGCAATTCTGAAGGAA-3′ and 5′-GGAGTAAAATATGCTCATCAACCTG-5′, respectively.
All the procedures involving mice were approved by the University of Chicago Institutional Animal Care and Use Committee.
Glucose tolerance testing
Intraperitoneal glucose tolerance tests (IPGTTs) were performed after a 4-h fast. Blood was sampled from the tail vein before and 30, 60, 90 and 120 min after intraperitoneal injection of dextrose (2 mg/g body weight). Glucose levels were measured using a Precision Q.I.D.™ Glucometer (MediSense, Waltham, MA).
Insulin secretion from the in situ-perfused pancreas
The pancreas was perfused in situ in a humidified, temperature-controlled chamber. The perfusate, introduced through the aorta at the level of the celiac artery, consisted of oxygenated Krebs-Ringer bicarbonate solution (KRB) containing 0.25% BSA and a variable concentration of glucose. The perfusion system used three peristaltic pumps (Minipuls 2; Gilson, Middleton, WI), two of which were computer controlled to allow the perfusate glucose concentration to increase progressively from 2 to 26 mM over 100 min, while maintaining a constant total flow rate of 1 ml/min. After 100 min, 20 mM arginine was perfused in the continued presence of 26 mM glucose for an additional 25 min. Before sample collection, the pancreas was perfused with KRB/2 mM glucose for a 30-min equilibration period. Insulin concentrations (picomoles per liter) were measured in the effluent perfusate at 1 and 5 min, and every fifth minute thereafter.
Insulin assays
Insulin concentrations were measured by a double-antibody radioimmunoassay (RIA) using a rat insulin standard in the Radioimmunoassay Core Laboratory of the University of Chicago Diabetes Research and Training Center (DRTC). All samples were assayed in duplicate. Pancreatic insulin content was measured after acid ethanol extraction of the whole pancreas and quantified by RIA.
Intracellular Ca2+ concentration ([Ca2+]i) in pancreatic islets
Islets were isolated by collagenase (Type XI; Sigma-Aldrich, St. Louis, MO) digestion and differential centrifugation through Ficoll gradients described previously (19). Isolated islets from HNF-1α (+/+ and +/−)/RIP-Tag mice were placed on 25-mm coverslips for 48–96 h to allow time for adherence, in RPMI 1640 containing 11.6 mM glucose, supplemented with 10% fetal bovine serum (FBS), 100 μU/ml penicillin, and 100 μg/ml streptomycin (all from Invitrogen, Carlsbad, CA). Before measurement of [Ca2+]i, the islets were loaded with Fura-2 acetoxymethylester (Molecular Probes, Eugene, OR) at 2 μM for 30 min at 37°C and equilibrated in KRB for 30 min at room temperature. and perifused at a flow rate of 2.5 ml/min at 37°C with KRB (without BSA) in a temperature-controlled microperifusion chamber (Medical Systems, Greenvale, NY) mounted onto an inverted microscope (Nikon, Melville, NY) equipped for epifluorescence. Changes in [Ca2+]i were measured in response to changes in the perifusate glucose concentration (a progressive increase from 2 to 26 mM glucose as described above). Results are expressed as the ratio of the emitted light intensity (detected at 510 nm) after excitation at 340 and 380 nm (ratio 340/380).
Assessment of beta-cell mass by point-counting morphometry
Beta-cell mass measurement was performed on pancreata obtained from 10-wk old male HNF-1α (+/+)/RIP-Tag, HNF-1α (+/−)/RIP-Tag and HNF-1α (+/+) mice. The dissected pancreata were cleared of fat and lymph nodes, weighed, fixed in Bouin’s solution, and embedded in paraffin. Beta-cell mass was measured by point-counting morphometry of insulin-immunostained pancreatic sections as previously described (16). Beta-cell mass was calculated by multiplying the pancreatic weight by the relative beta-cell volume. The relative beta-cell volume represents the percentage of total points counted over pancreatic tissue that fall on beta-cells.
Assessment of beta-cell number by fluorescence activated cell sorting (FACS)
HNF-1α deficient/RIP-Tag mice and transgenic mice expressing green fluorescent protein (GFP) under the control of mouse insulin I promoter (MIP) (15) were crossbred. Preparation of single-cell suspensions from pancreata of 10-wk mice were performed using the procedure previously described (15). Briefly, the pancreas was inflated with a solution containing 0.3 mg/ml collagenase in Hanks’ balanced salt solution (Invitrogen), injected via the pancreatic duct. The inflated pancreas was removed, incubated for 10 min at 37°C, and shaken vigorously to disrupt the tissue. The digested pancreas was washed with PBS and incubated in a solution of 0.05% trypsin-EDTA (Invitrogen) at 37°C for 3 min. The digestion was stopped by adding RPMI 1640 supplemented with 10% FBS. The resulting single cells were washed with PBS, resuspended in cold PBS/FBS and filtered using 70 μm mesh. Cells were stained with trypan blue to check viability and preparations showing more than 95% live cells were analyzed. The pancreatic cell suspension was diluted to 2×106 cells/ml with the PBS/FBS solution. We found that the cells could be fixed with 4% paraformaldehyde and gave consistent results up to 4 weeks after the preparation. We tested the same samples, first when they were freshly prepared in PBS/FBS, then fixed in 4% paraformaldehyde, performed FACS analysis after 2 days, 1 week and up to 4 weeks. The results were consistent among the experiments.
Flow cytometric analysis of GFP-labeled beta-cells was performed on a FACScan flow cytometer with Cell Quest software (Becton Dickinson, Franklin Lakes, NJ). GFP-expressing cells were detected using the FL1 channel (absorption spectra 530/30) using non-GFP-expressing pancreas from littermates as a control.
Assessment of beta-cell replication
For the measurement of beta-cell replication rates, 5-bromo-2′-deoxyuridine (BrdU; Sigma-Aldrich) at a dose of 100 ng/g body weight was administered intraperitoneally for 6 h. Pancreatic sections (5 μm thick) were double-stained for BrdU and a mixture of non-beta endocrine cells (antibodies directed against pancreatic polypeptide, somatostatin, and glucagon). BrdU staining was performed by incubation with anti-BrdU mAb followed by a peroxidase-conjugated anti-mouse IgG. Diaminobenzidine with nickel enhancer was used as the chromogen (Cell Proliferation kit; Amersham, Arlington Heights, IL).
Beta cell nuclei were defined as intraislet endocrine cells with a characteristic large round nucleus surrounded by relatively abundant cytoplasm not staining for the red-brown signal from the immunostained mixture of non-beta endocrine cells. All nuclei or the first 1,000 beta- cell nuclei were counted in each section, and data were expressed as the percentage of BrdU-positive beta-cells per total number of beta-cells.
Generation of HNF-1α deficient beta-cell lines
Islets were isolated from HNF-1α (+/+, +/− and −/−)/RIP-Tag mice. Single islets were plated in 96-well plates and were cultured in DMEM (Invitrogen) supplemented with 10% FBS. Beta-cells outgrown from each islet free from fibroblasts were passaged and further cultured.
RNA isolation and quality evaluation
Total RNA was extracted using Trizol reagent (Life Technologies, Gaithersburg, MD) followed by RNeasy Mini column purification (Qiagen, Chatsworth, CA) according to the manufacturers’ instructions. Integrity of RNA was evaluated using an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). The purity/concentration was determined using a GeneSpec III (MiraiBio, Alameda, CA). All RNA samples used for hybridization had an OD260/280 and OD260/230 ratio >1.8 and total RNA concentration >1μg/ml.
Target preparation and microarray hybridization
All gene array hybridizations were performed at the Functional Genomics Facility, University of Chicago. The target preparation protocol followed the Affymetrix GeneChip Expression Analysis Manual (Santa Clara, CA). Briefly, 10 μg of total RNA was used to synthesize double-stranded cDNA using the Superscript Choice System (Life Technologies). First strand cDNA synthesis was primed with a T7-(dT24) oligonucleotide. From the phase-log gel-purified cDNA, biotin-labeled antisense cRNA was synthesized using BioArray High Yield RNA Transcript Labeling Kit (Enzo Diagnostics, Farmingdale, NY). After precipitation with 4 M Lithium Chloride, 20 μg of cRNA was fragmented in fragmentation buffer (40 mM Tris-Acetate, pH 8.1, 100 mM KOAc, 30 mM MgOAc) for 35 min at 94° C and then 12 μg of fragmented cRNA was hybridized to Arrays for 16 hr at 45° C and 60 rpm in an Affymetrix Hybridization Oven 640. The arrays were washed and stained with streptavidin phycoerythrin in Affymetrix Fluidics Station 450 using the Affymetrix GeneChip protocol and scanned using the Affymetrix GeneChip Scanner 3000.
Data acquisition and analysis
The acquisition and initial quantification of array images were performed using the GCOS (Affymetrix). Subsequent data analysis was performed using DNA-Chip Analyzer 1.3 with the CEL files. We used a PM-MM model (PM: a perfect match probe; MM: a single mismatch) to estimate gene expression level and the invariant set approach for data normalization (20, 21). For comparison analyses, thresholds for selecting significant genes were set at a relative difference >2-fold, absolute difference >100 signal intensity and statistical difference at P<0.05.
Pathway analysis
Global functional analyses, network analyses and canonical pathway analyses were performed using Ingenuity Pathway Analysis 3.0 (Ingenuity Systems, Redwood City, CA). A differentially expressed gene list containing gene identifiers and corresponding fold changes was first uploaded as an Excel spreadsheet into the software. Each gene identifier was mapped to its corresponding gene object in the Ingenuity Pathways Knowledge Base. These genes were then used as the starting point for the analyses. The optimal number of genes to be analyzed is approximately 800–1,000 genes.
Quantitative real-time PCR
Total RNA was isolated using the RNeasy Mini Kit (Qiagen, Valencia, CA). First-strand cDNA was prepared from 1 μg of total RNA with the SuperScript II RT kit and random hexamer primers (Invitrogen, Carlsbad, CA). Expression of mRNAs for selected genes was determined by quantitative real-time PCR on an ABI PRISM 7700 Sequence Detector using TaqMan probes (Applied Biosystems, Foster City, CA). The housekeeping gene glyceraldehydes 3-phosphate dehydrogenase (GAPDH) was used as an internal control for cDNA quantity and quality in all assays. The experiments were performed in quadricate.
Statistical analysis
Statistical analyses were performed using Student’s t test. Data are expressed as mean ±SEM. Differences were considered to be significant at P<0.05.
Results
Increased glucose levels and reduced insulin secretion and pancreatic insulin content in HNF-1α (+/−)/RIP-Tag mice
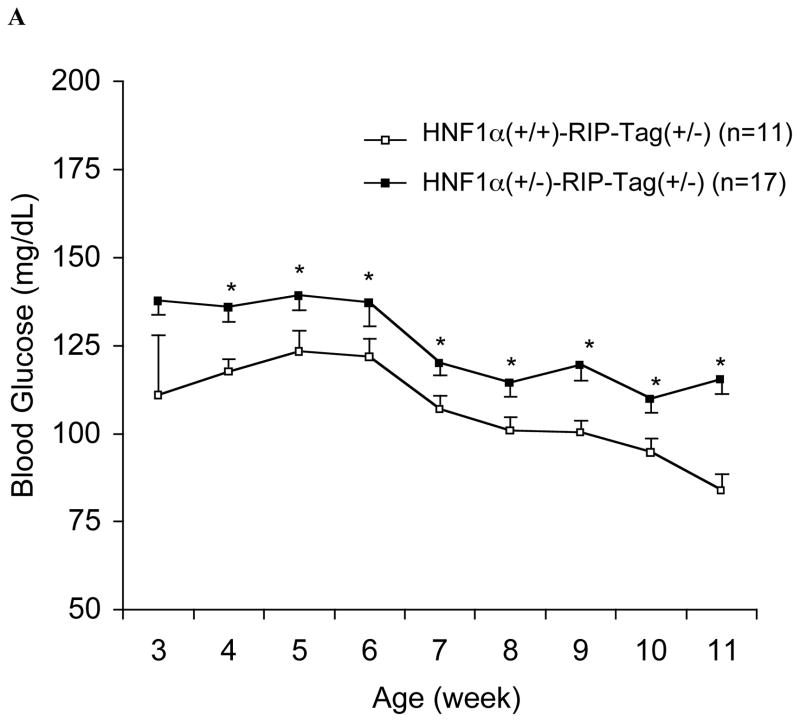
HNF-1α (+/−)/RIP-Tag mice grew normally with no significant difference in body weight compared to HNF-1α (+/+)/RIP-Tag mice. Neither mouse group developed hyperglycemia. However, after 4-wk of age non-fasting blood glucose levels in HNF-1α (+/−)/RIP-Tag mice were significantly and consistently higher than those in HNF-1α (+/+)/RIP-Tag mice (Fig. 1A). HNF-1α (+/−) mice were normoglycemic throughout adult life (16, 17). In contrast, the phenotype of HNF-1α (−/−)/RIP-Tag mice was similar to that reported in HNF-1α (−/−) mice (11) including severe growth retardation, enlarged liver and hyperglycemia despite tumor formation.
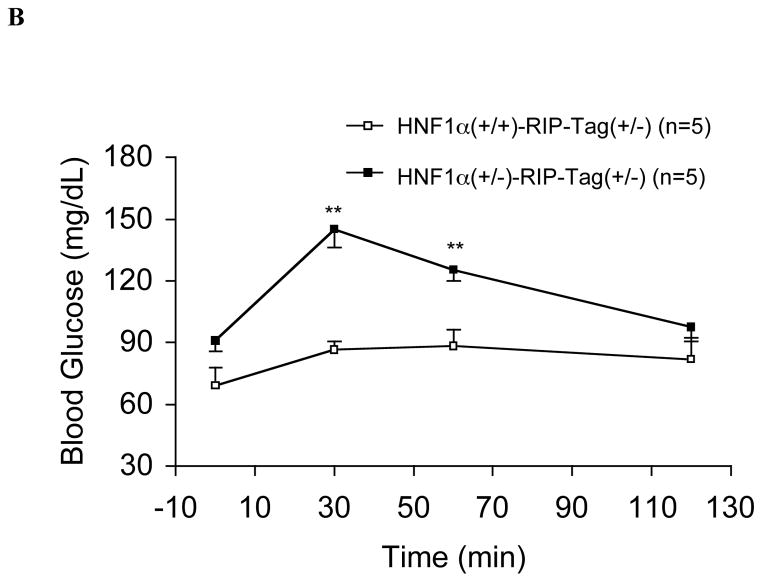
FIG. 1.
A. Time-course of non-fasting blood glucose levels in HNF-1α (+/−)/RIP-Tag mice (H1A (+/−)-Tag) and HNF-1α (+/+)/RIP-Tag mice (H1A (+/+)-Tag). Closed squares; HNF-1α (+/−)/RIP-Tag mice (n = 17); open squares; HNF-1α (+/+)/RIP-Tag mice (n = 11). Statistically significant differences are indicated with an asterisk (*, P<0.05). B. Intraperitoneal glucose tolerance. Intraperitoneal glucose tolerance testing in 10-wk HNF-1α (+/−)/RIP-Tag mice (H1A (+/−)-Tag) and age-matched HNF-1α (+/+)/RIP-Tag mice (H1A (+/+)-Tag). There were significant differences in blood glucose levels at 30-min and 60-min (**, P<0.01 compared with HNF-1α (+/+)/RIP-Tag mice). Closed squares; HNF-1α (+/−)/RIP-Tag mice (n = 5); open squares; HNF-1α (+/+)/RIP-Tag mice (n = 5).
In order to understand the mechanisms responsible for the increase in glucose levels seen with partial deficiency of HNF-1α, subsequent studies focused on analyzing HNF-1α (+/−)/RIP-Tag mice in comparison with HNF-1α (+/+)/RIP -Tag mice. Note that the transgene RIP-Tag induces distinct tumor formation after 3-mo of age, resulting in severe sometimes fatal hypoglylcemia beginning from age 10–11 wks (Fig. 1A). Therefore, 10-wk old mice were used for subsequent studies.
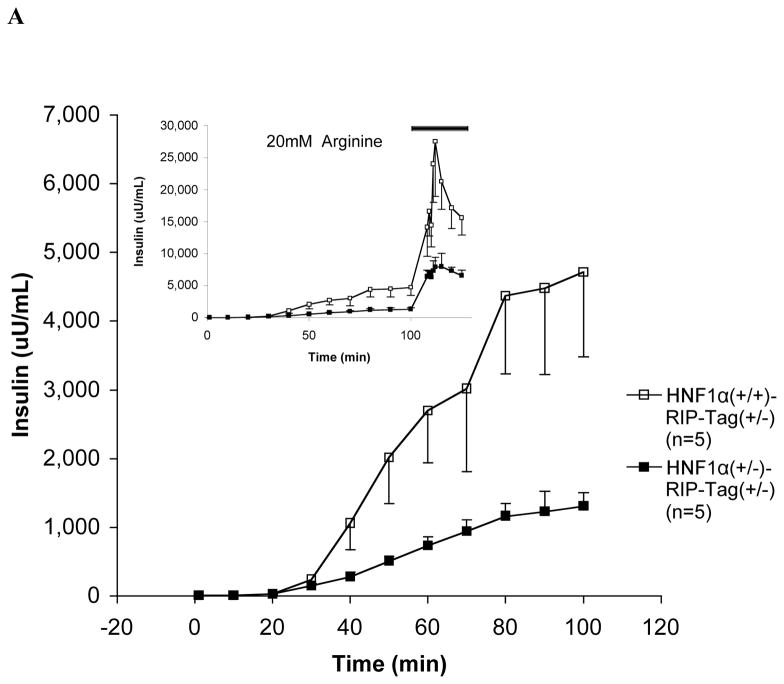
IPGTT showed that blood glucose levels were significantly higher in HNF-1α (+/−)/RIP-Tag mice at 30-min and 60-min after the administration of glucose than in HNF-1α (+/+)/RIP-Tag mice (Fig. 1B). Insulin secretory responses to glucose and arginine in the in situ-perfused pancreas were also attenuated (Fig. 2A). As the glucose concentration in the perfusate was increased from 2 to 26 mM, insulin secretory responses were significantly reduced in HNF-1α (+/−)/RIP-Tag mice compared to HNF-1α (+/+)/RIP-Tag mice. The response to arginine was also significantly lower in the HNF-1α (+/−)/RIP-Tag mice.
FIG. 2.
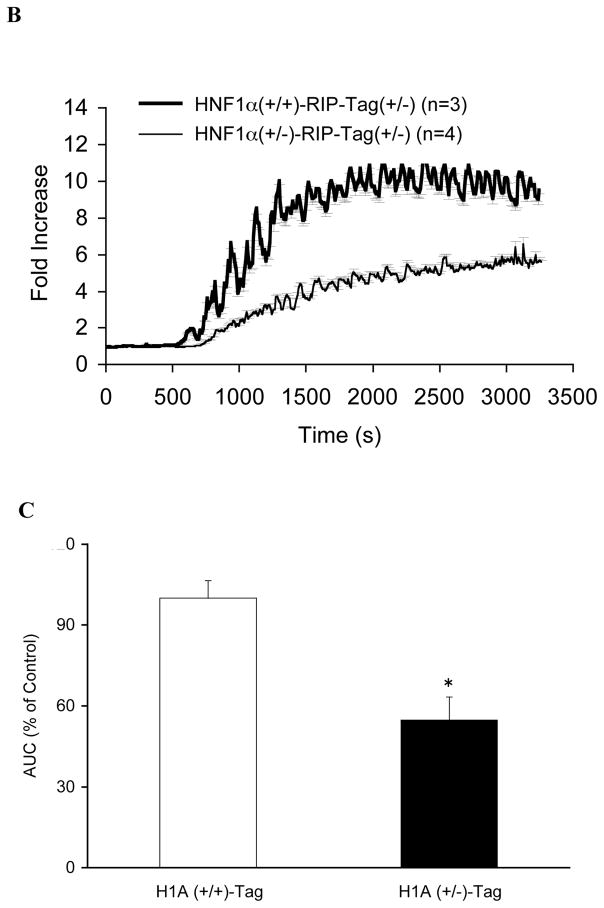
A. Insulin secretion to glucose and arginine in the in situ-perfused pancreas. During the first part of each experiment, the perfusate glucose concentration was increased progressively from 2 to 26 mM. The mean insulin output by pancreata of 10-wk HNF-1α (+/−)/RIP-Tag mice (H1A (+/−)-Tag) (closed squares, n = 5) was significantly less than that from age-matched HNF-1α (+/+)/RIP-Tag mice (H1A (+/+)-Tag) (open squares, n = 5) in response to glucose (P<0.005). (insert) Thereafter, in the continued presence of 26 mM glucose, 20 mM arginine was added to the perfusate (solid black bar). [Inset] The mean insulin output in response to arginine was also significantly less from 10-wk HNF-1α (+/−)/RIP-Tag mice (H1A (+/−)-Tag) compared to age-matched HNF-1α (+/+)/RIP-Tag mice (H1A (+/+)-Tag) (P<0.005). B. Measurements of intracellular calcium mobilization in perifused islets. Changes in intracellular calcium in response to a progressive increase in perfusate glucose from 2 to 26 mM are shown. Thin line: HNF-1α (+/−)/RIP-Tag mice (H1A (+/−)-Tag, n = 4); thick line: HNF-1α (+/+)/RIP-Tag mice (H1A (+/+)-Tag, n = 3). C: Summary of area under the intracellular calcium response curve. Results from HNF-1α (+/+)/RIP-Tag islets (white bars) and HNF-1α (+/−)/RIP-Tag islets (black bars) are depicted. Data are means ± SEM. P<0.05 compared to HNF-1α (+/+)/RIP-Tag islets.
Intracellular calcium mobilization in response to a progressive increase in the concentration of glucose from 2 to 26 mM was also attenuated in islets from HNF-1α (+/−)/RIP-Tag mice (Fig. 2B). The area under the curve (AUC) in the HNF-1α (+/−)/RIP-Tag islets was significantly lower (Fig. 2C; 54.8±8.5% of the value in the HNF-1α (+/+)/RIP-Tag islets, P < 0.05). However, intracellular calcium response to 20 mM KCl was not significantly reduced in HNF-1α (+/−)/RIP-Tag islets (103±16% of the value in the HNF-1α (+/+)/RIP-Tag islets, P > 0.05).
We also measured pancreatic insulin content in male and female mice (Table I). Insulin content of HNF-1α (+/+)/RIP-Tag mice was significantly increased (2-fold in males and 1.6-fold in females) compared to that of HNF-1α (+/+) mice due to insulinoma formation. In contrast, insulin content adjusted for pancreas weight was significantly reduced to ~50% in both male and female HNF-1α (+/−)/RIP-Tag mice compared to age- and sex-matched HNF-1α (+/+)/RIP-Tag controls.
TABLE I.
Pancreatic Insulin Content in 10-wk-old Mice.
| Insulin Content (μg/mg pan creas) |
||
|---|---|---|
| male | female | |
| H1A (+/+)-Tag | 0.31 ± 0.19 (n =13)* | 0.28 ± 0.10 (n =16)* |
| H1A (+/−)-Tag | 0.15 ± 0.05 (n =16) | 0.18 ± 0.06 (n =16) |
| H1A (+/+) | 0.16 ± 0.06 (n =21) | 0.18 ± 0.05 (n =17) |
| H1A (+/−) | 0.11 ± 0.03 (n =20) | 0.13 ± 0.05 (n =28) |
P < 0.05 compared with H1A (+/+) mice.
Alterations in beta-cell mass, number and proliferation in HNF-1α (+/−)/RIP-Tag mice
Beta-cell mass measured by point-counting morphometry showed a tendency to be greater in HNF-1α (+/+)/RIP-Tag mice compared to both HNF-1α (+/−)/RIP-Tag and HNF-1α (+/+) mice (HNF-1α (+/+)/RIP-Tag mice, n=8, 1.8±1.3%). The average values are 8 fold greater than in the controls and were it not for the variability of the measurements this difference should easily be statistically significant; HNF-1α (+/−)/RIP-Tag mice, n=6, 0.22±0.03%; HNF-1α (+/+) mice, n=4, 0.16±0.001%; P=NS vs HNF-1α (+/+)/RIP-Tag mice). However, the differences were not statistically significant. It was difficult to completely exclude the possibility that sampling bias affected these results. Although 3 sections were systematically sampled at relevant intervals from each pancreatic tissue block, extended tumor formation occurred randomly within each individual pancreas resulting in inclusion or exclusion of such large islets for each analysis. To obtain additional insights into the mass of beta-cells present in these animals total beta-cell number was measured. HNF-1α (+/+ and +/−)/RIP-Tag mice were crossbred with MIP-GFP mice and the percentage of GFP-expressing beta-cells present was measured by flow cytometry using single cell suspension from the whole pancreas (19). Beta-cell number of HNF-1α (+/+)/RIP-Tag mice was significantly increased compared to that of HNF-1α (+/+) mice due to tumor formation (Table II). In contrast, HNF-1α (+/−)/RIP-Tag mice had reduced beta-cell number compared to HNF-1α (+/+)/RIP-Tag controls.
TABLE II. Percent of Beta Cell Number in the Pancreas.
Flow cytometric analysis of single cell suspension of whole pancreas.
| Beta-cell number/FACS (10 wk male) | |
|---|---|
| H1A (+/+)-Tag/MIP-GFP (n=3) | 3.6 ± 1.08 %* |
| H1A (+/−)-Tag/MIP-GFP (n=5) | 1.2 ± 0.43 % |
| H1A (+/+)/MI P-GFP (n=8) | 1.0 ± 0.19 % |
| H1A (+/−)/MIP-GFP (n=13) | 0.9 ± 0.13 % |
P < 0.05 compared with H1A (+/+)/MIP-GFP mice.
HNF-1α deficiency led to changes in genes involved in cellular growth and proliferation
To further characterize the molecular events associated with HNF-1α deficiency, transcriptional profiling was performed using immortalized beta-cell lines generated from HNF-1α (+/+, +/− and −/−)/RIP-Tag mice. Initial comparative analysis of expression profiles between HNF-1α (+/−)/RIP-Tag and HNF-1α (+/+)/RIP-Tag beta-cells with the cutoff of > 2-fold changes has identified 1,420 genes, however, the subsequent pathway analysis reported 7 pathways with the same score. To determine the most significant pathway that is affected by a loss of one allele of HNF-1α, we examined genes that were changed > 3-fold, which identified 429 genes (234 upregulated and 195 downregulated; Supplemental Data 1). The pathway most significantly associated with these changes was that of cellular growth and proliferation, cell death, or cancer (score: 60; 35 genes; Supplemental Data 2A). Complete deficiency of HNF-1α resulted in > 2-fold changes in 647 genes (409 upregulated and 238 downregulated; Supplemental Data 1). The pathway analysis identified one specific pathway that was associated with cellular growth and proliferation, cell signaling, or nucleic acid metabolism (score: 49; 33 genes; Supplemental Data 2B). Note that with the cutoff of > 3-fold changes in this comparison, the number of genes to be analyzed was markedly reduced to 237, which was not optimal for the pathway analysis (see Materials and Methods).
Changes of genes involved in cellular growth and proliferation
To identify genes responsible for cellular growth and proliferation, we examined differential expression of genes involved in cell cycle, cellular growth and proliferation (Table III). No significant changes were observed in genes involved in G1 regulation of cell cycle. G2/M regulatory components, such as cell division cycle 2 homolog A (cdc2), cyclin B1, cell division cycle 25 homolog B (cdc25B) and homolog C (cdc25C) mRNAs were reduced (>2 fold) in HNF-1α (+/−)/RIP-Tag beta-cells but not in HNF-1α (−/−)/RIP-Tag beta-cells compared to HNF-1α (+/+)/RIP-Tag controls.
TABLE III.
Changes of genes involved in cell cycle, cellular growth and proliferation in HNF-1α (+/+)/RIP-Tag, HNF-1α (+/−)/RIP-Tag and HNF-1α (−/−)/RIP-Tag beta-cell lines.
| Accession | gene | fold change |
|
|---|---|---|---|
| +/−/+/+ | −/−/+/+ | ||
| cell cycle | |||
| G1 regulatory components | |||
| NM_009870 | cyclin-dependent kinase 4 | 0.97 | 1.21 |
| NM_007631 | cyclin D1 | 1.09 | 1.01 |
| NM_009829 | cyclin D2 | 1.31 | 0.88 |
| NM_007632 | cyclin D3 | 0.54↓ | 0.79 |
| AF059567 | cyclin-dependent kinase inhibitor 2B (p15) | 1.03 | 0.83 |
| NM_009877 | cyclin-dependent kinase inhibitor 2A (p16) | 0.93 | 0.90 |
| BC027026 | cyclin-dependent kinase inhibitor 2C (p18) | 0.83 | 0.94 |
| BC013898 | cyclin-dependent kinase inhibitor 2D (p19) | 0.94 | 1.08 |
| AK007630 | cyclin-dependent kinase inhibitor 1A (P21) | 0.74 | 0.90 |
| NM_009875 | cyclin-dependent kinase inhibitor 1B (P27) | 1.05 | 1.36 |
| NM_009876 | cyclin-dependent kinase inhibitor 1C (P57) | 0.60↓ | 1.72↑ |
| NM_016756 | cyclin-dependent kinase 2 | 0.88 | 1.30 |
| NM_007633 | cyclin E1 | 0.92 | 0.89 |
| AF091432 | cyclin E2 | 0.98 | 1.14 |
| NM_007628 | cyclin A1 | 0.90 | 1.00 |
| X75483 | cyclin A2 | 0.55↓ | 1.09 |
| G2/M regulatory components | |||
| NM_007659 | cell division cycle 2 homolog A (cdc2) | 0.47⇓ | 0.83 |
| AU015121 | cyclin B1 | 0.37⇓ | 0.66 |
| AK013312 | cyclin B2 | 0.90 | 1.08 |
| NM_023117 | cell division cycle 25 homolog B (cdc25B) | 0.41⇓ | 0.67 |
| NM_009860 | cell division cycle 25 homolog C (cdc25C) | 0.42⇓ | 0.79 |
| cellular growth and proliferation | |||
| AI561892 | guanine nucleotide binding protein, alpha stimulating (GNAS) complex locus | 0.33⇓ | 0.24⇓ |
| U03425 | epidermal growth factor receptor (EGFR) | 0.21⇓ | 0.16⇓ |
| NM_009255 | serine (or cysteine) proteinase inhibitor, clade E, member 2 (SERPINE2) | 4.63⇑ | 3.93⇑ |
| BB787243 | insulin-like growth factor binding protein 4 (IGFBP4) | 2.51⇑ | 2.76⇑ |
| BB089170 | vascular endothelial growth factor C (VEGFC) | 0.02⇓ | 0.07⇓ |
| NM_011577 | transforming growth factor, beta 1 | 6.71⇑ | 1.20 |
| AI481026 | insulin-like growth factor binding protein 7 (IGFBP7) | 3.24⇑ | 0.94 |
Abbreviations: +/+, H1A(+/+)/RIP-Tag; +/−, H1A(+/−)/RIP-Tag; −/−, H1A(−/−)/RIP-Tag
A single arrow and a double arrow show changes of 1.5~ 2-fold or > 2-fold, respectively.
We observed a strong differential expression of factors involved in cellular growth and proliferation in both HNF-1α (+/−)/RIP-Tag and HNF-1α (−/−)/RIP-Tag beta-cells such as insulin-like growth factor binding protein 4 (IGFBP4), and serine proteinase inhibitor, clade E, member 2 (SERPINE2), which were listed in both networks described above (Supplemental Data 1). Transforming growth factor-β1 and IGFBP7 were significantly increased in HNF-1α (+/−)/RIP-Tag cells. These results were confirmed by quantitatvie real-time PCR (Supplemental Data 3).
Changes of beta-cell related genes
Lastly, we examined changes in expression of beta-cell related genes, including those that have been reported as downstream targets of HNF-1α in different systems (10–12) (Supplemental Data 4). There was no significant difference in expression levels of insulin I or insulin II genes among HNF-1α (+/+, +/− and −/−)/RIP-Tag beta-cell lines. Glucagon expression was significantly decreased in a dose-dependent manner of HNF-1α. Little change was observed in expression of key transcription factors involved in islet development such as Pdx-1, Foxa-2, Nkx-6.1, Nkx-2.2, Pax-4, Pax-6, Ngn-3, Isl-1 and NeuroD-1. Genes involved in glucose metabolism such as L-Pk, Glut2 and aldolaseB were not significantly changed except that glucokinase was increased in both HNF-1α (+/−)/RIP-Tag and HNF-1α (−/−)/RIP-Tag beta-cells. Genes that have been suggested as direct targets of HNF-1α such as IGF-1 (22), E-cadherin (10) and Tmem27 (11, 12) were not significantly changed in our system except that E-cadherin was slightly elevated in HNF-1α (+/−)/RIP-Tag beta-cells and Tmem 27 was reduced in HNF-1α (−/−)/RIP-Tag beta-cells.
Discussion
The present report describes a new mouse model that was created by crossing HNF-1α (+/−) mice with HNF-1α (+/−)/RIP-Tag mice and compares the HNF-1α (+/−)/RIP-Tag offspring with their HNF-1α (+/+)/RIP-Tag littermates. It is known that expression of the Tag in the pancreatic beta cell driven by the insulin promoter leads to a marked increase in beta cell proliferation, the development of pancreatic beta cell tumors and severe symptomatic hypoglycemia that may be fatal. The creation of RIP-Tag animals that had either both or only one allele of the HNF-1α gene allowed us to define the effects of HNF-1α deficiency on beta cell function and RIP-Tag induced beta cell proliferation.
The results have provided novel insights into the effects of HNF-1α deficiency on pancreatic beta-cell function, growth and proliferation. When compared to their HNF-1α (+/+)/RIP-Tag littermates the HNF-1α (+/−)/RIP-Tag mice demonstrated a number of important changes. Blood glucose concentrations were consistently higher in these animals and they did not develop hypoglycemia. The increase in glucose was due to diminished insulin secretion in the HNF-1α (+/−)/RIP-Tag animals demonstrated very clearly in the perfused mouse pancreas. These animals also demonstrated diminished calcium responses to glucose in pancreatic islets consistent with previously reported alterations in pancreatic beta cell glucose signaling reported with HNF-1α deficiency. Insulin content was also reduced by HNF-1α deficiency and the number of pancreatic islets was reduced in HNF-1α deficient animals.
Pathway analysis of differentially expressed transcriptional profiles based on functional annotation and known molecular interactions is a powerful tool to dissect biological pathway(s) associated with global changes in gene expression. Comparative studies using HNF-1α (+/+, +/− and −/−) RIP-Tag beta-cell lines indicate that the genes whose expression was most significantly reduced lie in pathways relevant to cellular growth and proliferation. These abnormalities in gene expression are undoubtedly relevant to the overall reduction in beta-cell mass observed in HNF-1α (+/−) RIP-Tag mice. Further analysis of transcriptional profiling showed significant upregulation in expression of genes involved in beta-cell proliferation such as SERPINE2, IGFBP4, IGFBP7 and TGF-β1, which were also confirmed by real time PCR. SERPINE2 has been reported to be overexpressed in pancreatic carcinoma and gastric cancer (23). IGFBP4 inhibits IGF-induced cell proliferation and differentiation in many cell types (24). IGFBP7 plays a potential tumor suppressor role in colorectal carcinogenesis. (25). The role of TGF-β1 is a potent inducer of growth inhibition in several cell types in carcinoma development (26). Unexpectedly in light of results from previous reports (7–12), little change was observed in genes involved in beta-cell development (e.g. Pdx-1, Foxa-2, Nkx-6.1, Nkx-2.2, Pax-4, Pax-6, Ngn-3, Isl-1 and NeuroD-1) or in metabolism (e.g. L-PK, Glut2 and aldolaseB). The results differ from those in HNF-1α null mice or isolated islets from these mice that showed no reduction in beta-cell mass (16), but abnormal expression of genes involved in pancreatic islet development such as Pdx-1, HNF-4α and Neuro-D1 (9) and metabolism such as L-PK, Glut2, neutral and basic amino acid transporter, insulin and glucagon (7–10). The phenotype of HNF-1α (+/−) RIP-Tag mice and the pattern of gene expression in HNF-1α (+/−) RIP-Tag beta-cells appears to be most similar to those of transgenic mice that express a dominant-negative form of HNF-1α under the control of rat insulin II promoter (RIP-HNF-1α-P291fsinsC transgenic mice; 10). These mice exhibited progressive hyperglycemia, impaired glucose –stimulated insulin secretion and a reduction in beta-cell number, proliferation rate and pancreatic insulin content accompanied by abnormal islet architecture with reduced expression of Glut2 and E-cadherin. There was no change in expression of transcription factors important for islet development such as Pdx-1, Pax 6, and Nkx2.2. These pleiotropic effects of HNF-1α deficiency in different experimental models could be related to the level of functional HNF-1α.
In summary, the present study used a novel mouse model of HNF-1α diabetes shows that the lack of one allele of the HNF-1α gene resulted in impaired insulin secretion and reduced beta-cell mass. Transcriptional profiling using immortalized beta-cell lines generated from these mice demonstrated abnormal expression of key genes involved in beta-cell proliferation. The results indicate that HNF-1α plays a key role in many facets of the normal regulation of insulin secretion including glucose signaling within the pancreatic beta cell as well as in the regulation of functional beta-cell mass.
Supplementary Material
Supplemental Data 2. Pathway analysis A: A network of genes changed (>3-fold) in HNF -1α (+/−)/RIP-Tag beta-cells compared to HNF-1α (+/+)/RIP-Tag beta-cells. Differentially expressed genes (429) were analyzed using Ingenuity Pathway Analysis 3.0. This network is associated with biological functions including cellular growth and proliferation, cell death, or cancer. B: A network of genes changed (>2-fold) in HNF -1α(−/−)/RIP-Tag beta-cells compared to HNF-1α (+/+)/RIP-Tag beta-cells. Differentially expressed genes (647) were analyzed. This network is associated with biological functions including cellular growth and proliferation, cell signaling, nucleic acid metabolism. The network is displayed graphically as nodes (genes) and edges (the biological relationships between nodes). The node color indicates the expression level of genes (red = upregulated and green = downregulated). Shapes of nodes indicate function as follows: diamond = an enzyme; square = growth factor; triangle = kinase; and circle = other. Asterisks indicate transcripts identified multiple times on the microarray. Solid lines or dashed lines depict direct or indirect interaction, respectively. Arrows imply functional interaction (e.g. activation and inhibition).
Acknowledgments
This research was supported by US Public Health Service Grants DK-20595, DK-61245 and DK-31842.
Abbreviations
- FACS
fluorescent activated cell sorter
- HNF
hepatocyte nuclear factor
- MODY
maturity-onset diabetes of the young
- RIP-Tag
rat insulin II promoter-large T antigen
Footnotes
Conflict of interest
The authors state that they have no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fajans SS, Bell GI, Polonsky KS. Molecular mechanisms and clinical pathophysiology of maturity-onset diabetes of the young. N Engl J Med. 2001;345:971–980. doi: 10.1056/NEJMra002168. [DOI] [PubMed] [Google Scholar]
- 2.Bell GI, Polonsky KS. Diabetes mellitus and genetically programmed defects in β-cell function. Nature. 2001;414:788–791. doi: 10.1038/414788a. [DOI] [PubMed] [Google Scholar]
- 3.Owen K, Hattersley AT. Maturiy-onset diabetes of the young: from clinical description to molecular genetic characterization. Best Pract Res Clin Endo Metab. 2001;15:309–323. doi: 10.1053/beem.2001.0148. [DOI] [PubMed] [Google Scholar]
- 4.Yamagata K, Oda N, Kaisaki PJ, et al. Mutations in the hepatocyte nuclear factor-1alpha gene in maturity-onset diabetes of the young (MODY3) Nature. 1996;384:455–458. doi: 10.1038/384455a0. [DOI] [PubMed] [Google Scholar]
- 5.Ryffel GU. Mutations in the human genes encoding the transpcription factors of the hepatocyte nuclear factor (HNF) 1 and HNF4 families: functional and pathological consequences. J Mol Endocrinol. 2001;27:11–29. doi: 10.1677/jme.0.0270011. [DOI] [PubMed] [Google Scholar]
- 6.Okita K, Yang Q, Yamagata K, et al. Human insulin gene is a target gene of hepatocyte nuclear factor-1α (HNF-1α) and HNF-1β. Biochem Biophys Res Commun. 1999;263:566–569. doi: 10.1006/bbrc.1999.1412. [DOI] [PubMed] [Google Scholar]
- 7.Parrizas M, Maestro MA, Boj SF, et al. Hepatic nuclear factor 1-alpha directs nucleosomal hyperacetylation to its tissue-specific transcriptional targets. Mol Cell Biol. 2001;21:3234–3243. doi: 10.1128/MCB.21.9.3234-3243.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shih DQ, Screenan S, Munoz KN, et al. Loss of HNF-1α function in mice leads to abnormal expression of genes involved in pancreatic islet development and metabolism. Diabetes. 2001;50:2472–2480. doi: 10.2337/diabetes.50.11.2472. [DOI] [PubMed] [Google Scholar]
- 9.Wang H, Antinozzi PA, Hagenfeldt KA, et al. Molecular targets of a human HNF1α mutation responsible for pancreatic β-cell dysfunction. EMBO J. 2000;19:4257–4264. doi: 10.1093/emboj/19.16.4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamagata K, Nammo T, Moriwaki M, et al. Overexpression of dominant-negative mutant hepatocyte nuclear fctor-1 alpha in pancreatic beta-cells causes abnormal islet architecture with decreased expression of E-cadherin, reduced beta-cell proliferation, and diabetes. Diabetes. 2002;51:114–123. doi: 10.2337/diabetes.51.1.114. [DOI] [PubMed] [Google Scholar]
- 11.Fukui K, Yang Q, Cao Y, et al. The HNF-1 target collectrin controls insulin exocytosis by SNARE complex formation. Cell Metab. 2005;2:373–384. doi: 10.1016/j.cmet.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 12.Akpinar P, Kuwajima S, Krutzfeldt J, et al. Tmem27: a cleaved and shed plasma membrane protein that stimulates pancreatic beta cell proliferation. Cell Metab. 2005;2:385–397. doi: 10.1016/j.cmet.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 13.Mendel DB, Crabtree GR. HNF-1, a member of a novel class of dimerizing homeodomain proteins. J Biol Chem. 1991;266:677–680. [PubMed] [Google Scholar]
- 14.Blumenfeld M, Maury M, Chouard T, et al. Hepatic nuclear factor 1 (HNF1) shows a wider distribution than products of its known target genes in developing mouse. Development. 1991;113:589–599. doi: 10.1242/dev.113.2.589. [DOI] [PubMed] [Google Scholar]
- 15.Pontoglio M, Barra J, Hadchouel H, et al. Hepatocyte nuclear factor 1α inactivation results in hepatic dysfunction, phenylketonuria, and renal Fanconi syndrome. Cell. 1996;84:575–585. doi: 10.1016/s0092-8674(00)81033-8. [DOI] [PubMed] [Google Scholar]
- 16.Pontoglio M, Sreenan S, Roe M, et al. Defective insulin secretion in hepatocyte nuclear factor 1α-deficient mice. J Clin Invest. 1998;101:2215–2222. doi: 10.1172/JCI2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee Y, Sauer B, Gonzalez FJ. Laron Dwarfism and non-insulin-dependent diabetes mellitus in the HNF-1α knockout mouse. Mol Cel Biol. 1998;18:3059–3068. doi: 10.1128/mcb.18.5.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanahan D. Heritable formation of pancreatic beta-cell tumours in transgenic mice expressing recombinant insulin/simian virus 40 oncogenes. Nature. 1985;315:115–122. doi: 10.1038/315115a0. [DOI] [PubMed] [Google Scholar]
- 19.Hara M, Wang X, Kawamura T, et al. Transgenic mice with green fluorescent protein-labeled pancreatic β-cells. Am J Physiol Endocrinol Metab. 2003;284:E177–E183. doi: 10.1152/ajpendo.00321.2002. [DOI] [PubMed] [Google Scholar]
- 20.Han ES, Wu Y, McCarter R, et al. Reproducitility, sources of variability, pooling, and sample size: Important considerations for the design of high-density oligonucleotide array experiments. J Gerontol A Biol Sci Med Sci. 2004;59:306–315. doi: 10.1093/gerona/59.4.b306. [DOI] [PubMed] [Google Scholar]
- 21.Naef F, Lim DA, Patil N, et al. DNA hybridization to mismatched templates: A chip study. Phys Rev E Stat Nonlin Soft Matter Phys. 2002;65:040902. doi: 10.1103/PhysRevE.65.040902. [DOI] [PubMed] [Google Scholar]
- 22.Yang Q, Yamagata K, Fukui K, et al. Hepatocyte nuclear factor-1 alpha modulates pancreatic beta-cell growth by regulating the expression of insulin-like growth factor-1 in INS-1 cells. Diabetes. 2002;51:1785–1792. doi: 10.2337/diabetes.51.6.1785. [DOI] [PubMed] [Google Scholar]
- 23.Buchholz M, Biebl A, Neesse A, et al. SERPINE2 (protease nexin I) promotes extracellular matrix production and local invasion of pancreatic tumors in vivo. Cancer Res. 2003;63:4945–51. [PubMed] [Google Scholar]
- 24.Zhou R, Diehl D, Hoeflich A, et al. IGF-binding protein-4: biochemical characteristics and functional consequences. J Endocrinol. 2003;178:177–193. doi: 10.1677/joe.0.1780177. [DOI] [PubMed] [Google Scholar]
- 25.Ruan W, Xu E, Xu F, et al. IGFBP7 plays a potential tumor suppressor role in colorectal carcinogenesis. Cancer Biol Ther. 2007;6:354–359. doi: 10.4161/cbt.6.3.3702. [DOI] [PubMed] [Google Scholar]
- 26.Derynck R, Akhurst RJ, Balmain A. TGF-beta signaling in tumor suppression and cancer progression. Nat Gen. 2001;29:117–129. doi: 10.1038/ng1001-117. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data 2. Pathway analysis A: A network of genes changed (>3-fold) in HNF -1α (+/−)/RIP-Tag beta-cells compared to HNF-1α (+/+)/RIP-Tag beta-cells. Differentially expressed genes (429) were analyzed using Ingenuity Pathway Analysis 3.0. This network is associated with biological functions including cellular growth and proliferation, cell death, or cancer. B: A network of genes changed (>2-fold) in HNF -1α(−/−)/RIP-Tag beta-cells compared to HNF-1α (+/+)/RIP-Tag beta-cells. Differentially expressed genes (647) were analyzed. This network is associated with biological functions including cellular growth and proliferation, cell signaling, nucleic acid metabolism. The network is displayed graphically as nodes (genes) and edges (the biological relationships between nodes). The node color indicates the expression level of genes (red = upregulated and green = downregulated). Shapes of nodes indicate function as follows: diamond = an enzyme; square = growth factor; triangle = kinase; and circle = other. Asterisks indicate transcripts identified multiple times on the microarray. Solid lines or dashed lines depict direct or indirect interaction, respectively. Arrows imply functional interaction (e.g. activation and inhibition).