Abstract
Background
The electrocardiographic QT interval is associated with risk of sudden cardiac death (SCD). A previous genome-wide association study demonstrated that allelic variants (rs10494366 and rs4657139) in NOS1AP, which encodes a carboxy-terminal PDZ ligand of neuronal nitric oxide synthase, are associated with the QT interval in white adults. The present analysis was conducted to validate the association between NOS1AP variants and the QT interval and to examine the association with SCD in a combined population of 19,295 black and white adults from the Atherosclerosis Risk in Communities (ARIC) study and the Cardiovascular Health Study (CHS).
Methods and Results
We examined 19 tagging SNPs in the genomic blocks containing rs10494366 and rs4657139 in NOS1AP. SCD was defined as a sudden pulseless condition of cardiac origin in a previously stable individual. General linear models and Cox proportional hazards regression models were used. Multiple SNPs in NOS1AP, including rs10494366, rs4657139, and rs16847548 were significantly associated with adjusted QT interval in whites (P<0.0001). In whites, after adjusting for age, sex, and study, the relative hazard (RH) of SCD associated with each C allele at rs16847548 was 1.31 (95% CI: 1.10 to 1.56, P=0.002), assuming an additive model. In addition, a downstream neighboring SNP, rs12567209, not correlated with rs16847548 or QT interval, was also independently associated with SCD in whites (RH = 0.57, 95%CI: 0.39, 0.83; P = 0.003). Adjustment for QT interval and CHD risk factors attenuated, but did not eliminate, the association between rs16847548 and SCD, and such adjustment had no effect on the association between rs12567209 and SCD. No significant associations between tagging SNPs in NOS1AP and either QT interval or SCD were observed in blacks.
Conclusions
In a combined analysis of two population-based prospective cohort studies, sequence variations in NOS1AP were associated with baseline QT interval and the risk of SCD in white U.S. adults.
Keywords: death, sudden, QT interval, genetics, epidemiology
Sudden cardiac death (SCD) and cardiac arrhythmias remain a daunting public health problem. It is estimated that there are between 250,000 and 400,000 sudden cardiac deaths in the United States each year1, 2. Nearly half of all coronary heart disease (CHD) deaths are sudden and approximately 1/3 of these deaths are the first clinical manifestation of disease3. Thus it is important to identify risk factors, both genetic and environmental, for SCD.
Previous studies have identified family history of SCD as a powerful risk factor for SCD that is independent of traditional risk factors for CHD or a family history of myocardial infarction4–6. Moreover, a number of genes have been linked to rare, heritable arrhythmias that predispose to SCD. However, the genetic factors underlying SCD in the general population are largely unknown.
SCD is a complex phenotype of heterogeneous etiology with multiple factors contributing to risk that can be broadly classified into three categories: 1) atherosclerosis and thrombosis; 2) electrogenesis and propagation; and 3) initiating influences and triggers7. Indeed, SCD is a multi-factorial disorder involving the interaction of multiple genes in conjunction with environmental influences. A number of pathways modulate the electrophysiology of the heart and have been associated with enhanced risk of SCD. Altered ventricular repolarization reflected in abnormalities of the electrocardiographic QT interval is an intermediate phenotype that is not only associated with an increased risk of SCD in both the presence and absence of structural heart disease8 but also heritable within families9 and in population-based studies10. Using a genome-wide association study (GWAS) we have previously demonstrated that allelic variants in NOS1AP (nitric oxide synthase 1 adaptor protein), which encodes a cytosolic ligand of neuronal nitric oxide synthase (nNOS), are associated with altered QT intervals in white adults11. This association has subsequently been replicated in additional white populations12–17. The objectives of the present study were to validate the association of NOS1AP variants with QT interval prolongation in a large U.S. population-based cohort of white and black adults and, more importantly, to establish the association between NOS1AP variants and the risk of SCD in these community-based individuals. We hypothesized that at least one of the NOS1AP SNPs examined would be associated with the QT interval; moreover, at least one such SNP would also be associated with the risk of SCD.
Methods
The Atherosclerosis Risk in Communities (ARIC) study and the Cardiovascular Health Study (CHS)
The ARIC study and CHS are both population-based prospective cohort studies of cardiovascular disease. The ARIC Study includes 15,792 persons aged 45–64 years at baseline (1987–89), randomly chosen from four US communities18. ARIC cohort members completed four clinic examinations, conducted approximately three years apart between 1987 and 1998. CHS includes 5,888 participants > 65 years of age identified from four U.S. communities using Medicare eligibility lists. The original cohort included 5201 participants recruited in 1989–1990 and 687 additional subjects were recruited in 1992–1993 to enhance the racial/ethnic diversity of the cohort19.
Clinic examinations for both ARIC and CHS participants included assessment of cardiovascular risk factors, self-reported medical family history, employment and educational status, diet, physical activity, comorbidities, and clinical and laboratory measurements. ARIC and CHS participants were contacted annually by telephone for identification of all hospitalizations and deaths, and lists of discharges from local hospitals were scanned for events. Deaths were identified from death certificates, and potential out-of-hospital fatal coronary heart disease events were investigated by interviewing one or more next of kin and by the completion of a questionnaire by the patient’s physician. ARIC and CHS staff abstract discharge diagnoses on all hospitalizations, as well as conduct standardized committee review of all CHD, stroke and cardiovascular death events18, 20, 21. Comprehensive data were gathered on cardiovascular events and deaths from hospital records, interviews with physicians, next of kin and/or witnesses, death certificates, and autopsy reports22. In addition to similar study protocols between ARIC and CHS, extensive review of the data definitions and study sources was carried out with review of files by an independent set of investigators so that only comparable clinical variables were included in the current analysis of the combined cohorts.
The following exclusion criteria, based on missing exposure or outcome data, were applied to obtain the final sample for the present analysis: 1) self-reported race/ethnicity other than black or white (48 in ARIC, 39 in CHS), 2) samples not genotyped due to lack of DNA or consent for genetic research (103 in ARIC; 432 in CHS), 3) samples with < 75% of genotypes called (1052 in ARIC, 305 in CHS), 4) missing electrocardiograms (not performed or not transmitted) or poor quality data (missing leads or artifacts) for either QT or heart rate (189 in ARIC, 124 in CHS), and 5) unconfirmed SCD (82 in ARIC, 2 in CHS). After these exclusions, 14,309 of 15,783 ARIC participants (91%) and 4,986 of 5,888 CHS participants (85%) were included in the present analysis for a combined sample size of 19,295 individuals.
Assessment of Sudden Cardiac Death
Each parent study classified all cases of fatal CHD according to standard protocols. To identify cases of SCD in ARIC and CHS for the present study, all cases of fatal CHD that occurred by July 31, 2002 in CHS and December 31, 2002 in ARIC were reviewed and adjudicated by a committee of physicians. SCD was operationally defined as a sudden pulseless condition from a cardiac origin in a previously stable individual. After review of data available from death certificates, informant interviews, physician questionnaires, coroner reports, and hospital discharge summaries, the reviewers classified each CHD death as definite sudden arrhythmic death, possible sudden arrhythmic death, definite non-sudden death, or unclassifiable. We a priori sought to exclude cases with non-arrhythmic characteristics including those with evidence of progressive hypotension or advanced congestive heart failure prior to death. We also excluded those cases with advanced dementia or terminal illness such as end stage cancer or liver disease. Each event was independently adjudicated by two investigators. If disagreement existed between the first two reviewers, a third investigator independently reviewed the event to provide final classification. As part of event review, information was systematically abstracted regarding duration of symptoms, whether the event was witnessed, other circumstances of the event, and medical co-morbidities of the patient in order to help classify whether the subject had experienced SCD. Those classified as “definite sudden arrhythmic death” were either confirmed by evidence of “instantaneous death” or in the case of unwitnessed deaths, there was descriptive information regarding the position of the body that indicated a sudden event had occurred. All suspected SCD, defined as a sudden pulseless condition from a cardiac origin in a previously stable individual, that we could not classify as “definite” were classified as “possible SCD”. Cases were identified as either in or out of hospital deaths. The primary outcome of SCD described in the present study combines both definite and possible sudden arrhythmic death. For the present analysis, participants were censored at time of loss to follow up or death if the cause of death was other than SCD. The administrative censoring date was July 31, 2002 for CHS and December 31, 2002 for ARIC, based on the study’s adjudication schedules.
Assessment of QT interval at the baseline examinations of each study
At the baseline visit of the ARIC study, participants were asked not to smoke or ingest caffeine for at least 1 hour prior to the electrocardiogram. After resting for 5–10 minutes while the electrodes were being placed, a standard supine 12-lead electrocardiogram and a 2-minute paper recording of a three-lead (leads V1, II, and V5) rhythm strip were made. The ECGs were digitally recorded, and identical methods (MAC personal computer, Marquette Electronics, Milwaukee, Wisconsin) were used in all clinical centers. A similar protocol was used at the baseline visit of CHS. MAC PC-DT ECG acquisition units (Marquette Electronics, Inc., Milwaukee, WI) were used to record a 10-second 12-lead simultaneous ECG at a sample rate of 250 per second per lead. All tracings from the baseline visits of both CHS and ARIC were transmitted over analogue phone lines to a central ECG Reading Center in Edmonton, Alberta, Canada for analysis. The QT interval from the digital 12-lead ECG was determined using the Novacode ECG measurement and classification program23.
Assessment of Covariates
Both the ARIC study and CHS have extensive data on behavioral, clinical, and serologic factors relevant to selected cardiovascular phenotypes and outcomes. At each visit, demographic, anthropometric, and cardiovascular risk factor data were collected. Data from the baseline visits of both ARIC and CHS were used for the present analyses. Participants described themselves as white or black in response to an interviewer-administered questionnaire, which also contained questions on highest education attained, smoking status, and marital status. Collection of fasting blood samples and processing for total cholesterol followed standard study protocols19, 24. Hypertension was defined as systolic blood pressure (SBP) ≥ 140 mmHg, diastolic blood pressure (DBP) ≥ 90 mmHg, or use of antihypertensive medications. Diabetes was defined as fasting glucose ≥ 126 mg/dL, non-fasting glucose ≥ 200 mg/dL, or history/treatment of diabetes. History of myocardial infarction (MI) at the baseline examination was defined by either a self-reported heart attack requiring hospitalization, self-reported history of physician-diagnosed MI, or a history of MI identified on the baseline electrocardiogram, which was verified in CHS25. A positive self-reported family history of CVD was defined as having at least one parent with CHD or stroke in ARIC and having at least one full sibling with heart attack or stroke in CHS.
SNP Selection and Genotyping
Since over 80% of the present study subjects were whites, SNPs were selected to tag the linkage disequilibrium (LD) block containing rs10494366 and rs4657139 (the most significant SNPs from fine mapping in our initial study in whites) and its neighboring LD blocks in the thirty trio samples of U.S. residents with northern and western European ancestry (CEU population) used in the HapMap Project26, 27. Twenty-one SNPs were selected using the computer program Tagger with criteria of r2 >0.65 and minor allele frequency (MAF) >0.05 in CEU (data from 19 SNPs passed QC tests). SNPs from coding regions were not specifically interrogated.
Genotyping was performed using TaqMan assays (Applied Biosystems) in conjunction with the BioTrove OpenArray SNP genotyping platform28. Data completeness (reported as the percentage of samples that have a genotype call at a given SNP) across all SNPs and samples was 93.7%, and the accuracy of genotyping, determined by comparison to concordance calls generated for 58 samples genotyped multiple times (range 2–19 times, median 6, resulting in ~350 comparison per SNP), was 99.1%. Two SNPs were dropped from further analysis due to significantly lower accuracy (96.5% and 96.1%). Individual samples with <75% complete data were also removed from further analysis, as low data completeness was strongly associated with higher genotyping error rates. The overall accuracy and data completeness were 99.4% and 97.8%, respectively, after removing poor quality DNA samples and SNPs.
Statistical Analysis
Differences in baseline characteristics by subsequent SCD status were assessed using t-tests and χ2 tests within each self-reported race/ethnicity. All analyses were stratified by self-reported ethnicity. Because the study protocols were similar across ARIC and CHS, the number of SCD events was modest within each study, and initial genotype-phenotype analyses indicated that the associations did not differ between the two studies, all analyses were pooled across the two studies to increase statistical power. The rare allele of each SNP in whites was designated as the minor allele. Deviations from Hardy-Weinberg proportions were assessed using the chi-squared goodness of fit test within each ethnicity group.
To analyze the QT-interval, a Z score for QT interval was created for each individual by standardizing an individual’s QT-interval to the study-specific population mean and standard error. A generalized linear model was then used to assess the association between SNPs and these Z scores representing QT intervals assuming the additive genetic model while adjusting for age, sex, and heart rate. All results presented in the present paper were generated using the study-specific age-, sex-, and heart-rate-adjusted QT intervals. Parallel analyses were also performed using Bazett’s heart rate-corrected QT duration (QTc)29, and similar results and inferences were obtained (data not shown). Analysis of the QT interval included both individuals who later experienced SCD and those who did not. Since 19 SNPs were tested, we used a Bonferroni-corrected-alpha of 0.003 (0.05/19) as the threshold for statistical significance in the age, sex, and study-adjusted model.
For SCD risk, single SNP genotype--based analyses were performed. Cumulative incidence of SCD in the presence of competing events (deaths due to other causes) was estimated overall and by SNP genotype30. To estimate the relative hazards and the significance of the association between each SNP genotype and SCD risk while adjusting for covariates, Cox proportional hazards models were constructed, and a Bonferroni-corrected-alpha of 0.003 was used to declare statistical significance in the age, sex, and study adjusted analysis. An additive model was assumed for each SNP. For significant SNPs, a model assuming 3 genotypic risks was also constructed to confirm the use of the additive model. The proportional hazards assumption was checked with Schoenfeld’s residual31.
As exploratory analyses, the role of genotypic effects across various high-risk subgroups was examined both with stratified analyses and by fitting interaction terms into the regression models. All analyses were performed with either SAS (version 9.0) or STATA (version 9.2).
Statement of Responsibility
The authors had full access to and take full responsibility for the integrity of the data. All authors have read agree to the manuscript as written.
Results
Clinical characteristics of ARIC and CHS participants at the baseline examinations
Baseline demographic and cardiovascular risk factors are shown by SCD status and by self-reported race in Table 1. As expected, many of the well established cardiovascular risk factors were significantly associated with SCD risk. Among both whites and blacks, individuals who died from SCD were significantly older, had higher systolic blood pressure, and fibrinogen and lower HDL cholesterol. They were also more likely to be male, smokers, less well educated, and have a history of diabetes, hypertension, and myocardial infarction at the baseline examination. Individuals who experienced SCD had significantly longer mean QT intervals at baseline, prior to the event. In whites, the age-, sex-, heart-rate, and study-adjusted mean QT duration was 403 ms for those who did not ultimately have SCD and 411 for those who did (P<0.001); and the corresponding values were 402 and 410 ms, respectively, in blacks (P<0.001).
Table 1.
Baseline characteristics of 14,737 white and 4,558 black ARIC and CHS participants by SCD status and race
| White |
Blacks |
|||
|---|---|---|---|---|
| No SCD | SCD | No SCD | SCD | |
| N | 14,403 | 334 | 4,394 | 164 |
| Age, yr | 59.4±10.0 | 66.2±10.3 | 56.7±9.3 | 60.7±9.4 |
| Male (%) | 6,540 (45.4) | 227 (68.0) | 1,645 (37.4) | 81 (49.4) |
| Not currently married (%) | 2.633 (19.1) | 80 (27.2) | 1,618 (42.0) | 63 (49.6) |
| Education (%) | ||||
| < high school | 2,804 (19.5) | 111 (33.2) | 1,766 (40.3) | 93 (57.1) |
| High school graduate | 6,214 (43.2) | 130 (38.9) | 1,269 (29.0) | 38 (23.3) |
| > college | 5,364 (37.3) | 93 (27.9) | 1,346 (30.7) | 32 (19.6) |
| Smoking (%) | ||||
| Never | 5,748 (39.9) | 107 (27.2) | 2,006 (45.8) | 63 (38.4) |
| Former | 5,403 (37.6) | 136 (40.7) | 1,127 (25.7) | 46 (28.1) |
| Current | 3,240 (22.5) | 91 (32.0) | 1,248 (28.5) | 55 (33.5) |
| BMI, kg/m2 | 26.8±4.7 | 27.5±5.0 | 29.4±6.0 | 29.3±6.1 |
| Cholesterol, mg/dL | 214±40 | 214±39 | 213±44 | 220±47 |
| HDL-cholesterol, mg/dL | 52±17 | 45±13 | 56±17 | 51±14 |
| LDL-cholesterol, mg/dL | 135±37 | 138±35 | 135±41 | 140±43 |
| SBP, mmHg | 123±20 | 134±22 | 130±22 | 142±26 |
| DBP, mmHg | 71±10 | 72±12 | 79±12 | 80±14 |
| Fibrinogen | 303±63 | 331±68 | 323±73 | 350±79 |
| Hypertension (%) | 4,974 (34.7) | 191 (57.3) | 2,519 (57.6) | 136 (83.4) |
| Diabetes (%) | 1,440 (10.0) | 81 (24.2) | 834 (19.4) | 70 (43.2) |
| History of MI (%) | 715 (5.0) | 93 (27.8) | 154 (3.5) | 30 (18.3) |
| Heart rate (bpm) | 66± 10 | 66±12 | 66± 11 | 70±12 |
| Adjusted QT*, ms | 403±0.1 | 411±1.0 | 402±0.3 | 410±1.7 |
Results presented as mean±S.D. or N(percent);
Age-, sex-, heart-rate-, and study-adjusted mean QT and standard error All within-race comparisons between SCD and non-SCD are significantly different (P<0.05) except for current marital status (P=0.09 in blacks), BMI (P=0.83), total cholesterol (P=0.85 in whites), LDL-cholesterol (P=0.22 in whites and 0.15 in blacks), DBP (P=0.07 in whites and 0.05 in blacks), and heart rate (P=0.14 in whites)
While the two studies are largely comparable, there were modest differences in the significance and magnitude of associations between cardiovascular risk factors and SCD risk between the two studies. For example, smoking, BMI, and total cholesterol were not significantly associated with SCD in CHS but were in ARIC (Table 2).
Table 2.
Baseline characteristics of 14,309 ARIC and 4,986 CHS participants by SCD status
| ARIC |
CHS |
Combined |
||||
|---|---|---|---|---|---|---|
| No SCD | SCD | No SCD | SCD | No SCD | SCD | |
| N (%) | 14,033 (98.1) | 276 (1.9) | 4,764 (95.6) | 222 (4.4) | 18,797 (97.4) | 498 (2.6) |
| Age, yr | 54.1±5.8 | 56.7±5.7 | 72.7±5.5 | 74.0±5.8 | 58.8±9.9 | 64.4±10.4 |
| Black (%) | 3,649 (26.0) | 120 (43.5) | 745 (15.6) | 44 (19.8) | 4,394 (23.4) | 164 (32.9) |
| Male (%) | 6,208 (44.2) | 176 (63.8) | 1,977 (41.5) | 132 (59.5) | 8,185 (43.4) | 308 (61.8) |
| Not currently married (%) | 2,668 (20.8) | 59 (29.4) | 1,583 (33.2) | 84 (38.2) | 4,251 (24.1) | 143 (34.0) |
| Education (%) | ||||||
| < high school | 3,206 (22.9) | 120 (43.6) | 1,364 (28.7) | 84 (37.8) | 4,570 (24.4) | 204 (41.0) |
| High school graduate | 5,757 (41.1) | 93 (33.8) | 1,726 (36.3) | 75 (33.8) | 7,483 (39.9) | 168 (33.8) |
| > college | 5,048 (36.0) | 62 (22.6) | 1,662 (35.0) | 63 (28.4) | 6,710 (35.8) | 125 (25.2) |
| Smoking (%) | ||||||
| Never | 5,534 (39.5) | 77 (27.9) | 2,220 (46.7) | 93 (41.9) | 7,754 (41.3) | 170 (34.1) |
| Former | 4,535 (32.4) | 87 (31.5) | 1,995 (41.9) | 95 (42.8) | 6,530 (34.8) | 182 (36.6) |
| Current | 3,945 (28.1) | 112 (40.6) | 543 (11.4) | 34 (15.3) | 4,488 (23.9) | 146 (29.3) |
| BMI, kg/m2 | 27.7±5.3 | 28.9±5.7 | 26.6±4.7 | 27.1±5.1 | 27.4±5.2 | 28.1±5.5 |
| Cholesterol, mg/dL | 214.3±41.6 | 223.1±44.0 | 211.6±39.2 | 207.6±38.5 | 213.6±41.0 | 216.2±42.3 |
| SBP, mmHg | 121±18 | 132.2±23.4 | 136.1±21.7 | 142.6±22.8 | 124.7±20.4 | 136.8±23.7 |
| DBP, mmHg | 74±11 | 76.6±13.6 | 70.6±11.3 | 72.7±12.3 | 72.8±11.2 | 74.9±13.2 |
| Fibrinogen | 302±65 | 337±72 | 323±67 | 337±72 | 307±66 | 337±72 |
| Hypertension (%) | 4,735 (33.9) | 170 (62.0) | 2,758 (58.0) | 157 (70.7) | 7,493 (40.0) | 327 (65.9) |
| Diabetes (%) | 1,542 (11.1) | 89 (32.4) | 732 (15.5) | 62 (28.0) | 2,274 (12.2) | 151 (30.4) |
| History of MI (%) | 444 (3.2) | 76 (27.5) | 425 (8.9) | 47 (21.2) | 869 (4.6) | 123 (24.7) |
| Adjusted QT*, ms | 400±0.1 | 410±1.1 | 411±0.3 | 417±1.5 | 403±0.1 | 411±0.9 |
Age-, sex- and heart-rate-adjusted mean QT and standard error; with additional adjustment for study for the combined analysis
Association between QT interval and SCD
Over a median follow up of 14.1 years in ARIC and 12.2 years in CHS, 334 whites and 164 blacks experienced sudden cardiac death (cumulative incidence rate per 1,000 person-years: 2.0 overall; 1.4 in ARIC and 4.5 in CHS; 1.8 in whites and 2.9 in blacks). Given the older age of the CHS cohort, 222 of the 498 SCD events (45%) occurred in CHS while 276 events occurred in ARIC. Of all CHD mortality adjudicated (N = 985), 40.2% were classified as definite SCD and 10.4% as possible SCD. The majority of cases of SCD occurred out of hospital (90%).
Among whites, longer QT interval was associated with the development of SCD after adjust for age, sex, heart rate, and study. Compared to whites in the first quintile of QT interval, those in the 2nd, 3rd, 4th, and 5th quintiles were 1.43 (95% CI 0.97 to 2.13), 1.77 (95% CI 1.16 to 2.69), 2.33 (95% CI 1.49 to 3.64), and 3.58 (95% CI 2.20 to 5.81) times more likely to have suffered SCD, respectively (P for trend <0.0001). Similar dose response relationship was observed when the analysis was repeated using QT deciles (P for trend <0.0001). Among blacks, longer QT interval was also associated with SCD risk; however, the dose response relationship was less apparent, possibly due to the smaller number of cases. Compared to blacks in the first quintile of QT interval, those in the 2nd, 3rd, 4th, and 5th quintiles were about 1.75 (95% CI 1.07 to 2.88), 2.04 (95% CI 1.19 to 3.49), 1.62 (95% CI 0.86 to 3.05), and 2.54 (95% CI 1.34 to 4.82) times more likely to have suffered SCD (P for trend =0.02).
Association between 19 SNPs in NOS1AP and QT interval
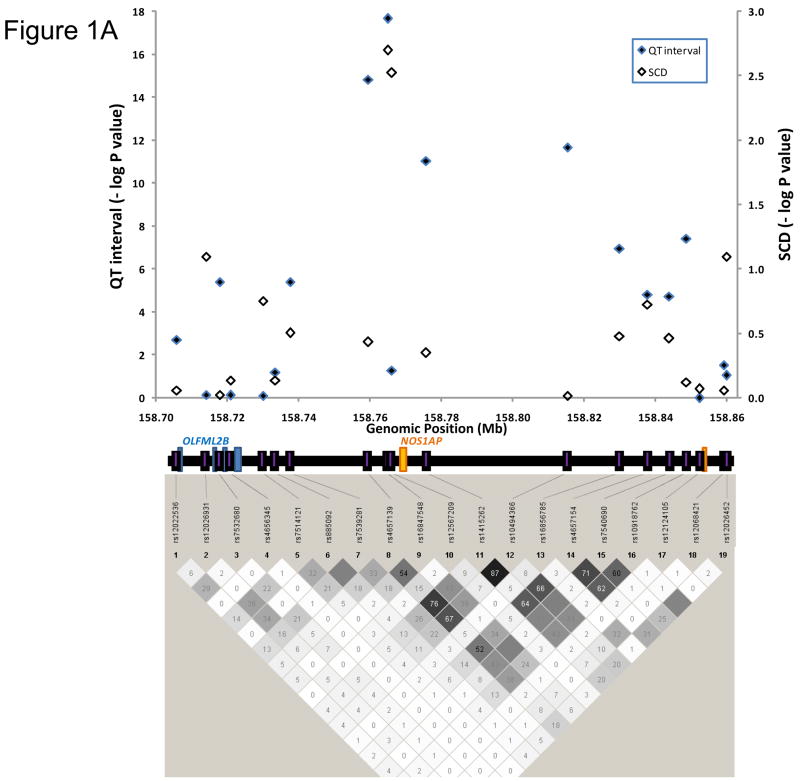
Allele frequencies of the 19 SNPs examined are shown in Table 3 by ethnicity. The LD pattern of these 19 SNPs were quite different in blacks as compared with whites, with considerably less LD among SNPs in blacks (Figure 1). Eleven of 19 SNPs examined were significantly associated with age-, sex-, and heart rate-adjusted QT interval in whites with P≤0.003. However, no SNPs were significantly associated with QT interval in blacks.
Table 3.
Genotypic association between 19 SNPs in NOS1AP and QT interval and SCD by self-reported race
| Whites | Blacks | |||||||
|---|---|---|---|---|---|---|---|---|
| Distance from neighboring SNP (Kb) | SNP | Alleles† | MAF | QT-interval P Value | SCD P Value | MAF | QT-interval P Value | SCD P Value |
| rs12022536 | A/G | 0.19 | 0.002 | 0.87 | 0.50 | 0.86 | 0.98 | |
| 8.3 | rs12026931 | C/T | 0.10 | 0.74 | 0.09 | 0.03 | 0.32 | 0.64 |
| 3.7 | rs7532680* | T/C | 0.19 | <0.001 | 0.95 | 0.73 | 0.51 | 0.68 |
| 3.3 | rs4656345 | A/G | 0.04 | 0.76 | 0.74 | 0.01 | 0.43 | 0.02 |
| 9.2 | rs7514121 | G/A | 0.22 | 0.81 | 0.18 | 0.43 | 0.78 | 0.60 |
| 3.4 | rs885092 | A/G | 0.15 | 0.06 | 0.73 | 0.26 | 0.53 | 0.41 |
| 4.2 | rs7539281 | A/G | 0.26 | <0.001 | 0.31 | 0.60 | 0.18 | 0.21 |
| 21.9 | rs4657139 | A/T | 0.33 | <0.001 | 0.36 | 0.85 | 0.30 | 0.41 |
| 5.4 | rs16847548 | C/T | 0.22 | <0.001 | 0.002 | 0.19 | 0.74 | 0.64 |
| 1.2 | rs12567209 | A/G | 0.07 | 0.05 | 0.003 | 0.07 | 0.92 | 0.07 |
| 9.6 | rs1415262* | C/G | 0.35 | <0.001 | 0.80 | 0.80 | 0.22 | 0.22 |
| 39.6 | rs10494366 | G/T | 0.35 | <0.001 | 0.44 | 0.61 | 0.68 | 0.87 |
| 0.99 | rs16856785 | C/G | 0.10 | <0.001 | 0.33 | 0.60 | 0.82 | 0.72 |
| 7.8 | rs4657154 | A/G | 0.27 | <0.001 | 0.174 | 0.28 | 0.62 | 0.54 |
| 6.1 | rs7540690 | A/G | 0.22 | <0.001 | 0.33 | 0.10 | 0.76 | 0.72 |
| 4.8 | rs10918762 | G/A | 0.22 | <0.001 | 0.19 | 0.25 | 0.78 | 0.37 |
| 3.6 | rs12124105 | T/A | 0.06 | 0.99 | 0.34 | 0.01 | 0.34 | 0.30 |
| 6.9 | rs12068421 | T/C | 0.14 | 0.03 | 0.76 | 0.30 | 0.92 | 0.05 |
| 0.74 | rs12026452** | A/G | 0.15 | 0.09 | 0.85 | 0.04 | 0.32 | 0.14 |
QT interval refers to an age-, sex-, heart rate-, and study- corrected QT interval; SCD analysis adjusted for age, sex, and study. Highlighted rows indicate most significant SNPs from the current study
Allele listed first represents the minor allele in Whites
Genotypes out of HWE, P<0.01, in Blacks
Genotypes out of HWE, P<0.01, in Whites
Figure 1.
Plots showing the linkage disequilibrium (LD) pattern and association results for both QT interval and SCD in whites (A) and blacks (B) for 19 SNPs genotyped to tag the NOS1AP locus and surrounding region that exhibited the strongest association with QT interval in previous studies11. The bottom panel is a plot showing the pairwise LD between SNPs. The value within each diamond represents the pair-wise correlation between SNPs (measured as R-square) defined by the top left and the top right sides of the diamond. Shading represents the magnitude and significance of the pair-wise LD, with a black to white gradient reflecting higher to lower LD values; see http://www.broad.mit.edu/mpg/haploview/ for further details. NOS1AP exons 1 and 2 are shown in orange. The top panel is a plot showing the significance for each SNP, with genomic position on the X-axis and the negative base-10 logarithm of the p-value on the Y-axis. Information regarding genomic position was taken from Human Genome Build 35.
In whites, the most significant SNP in the present study was rs16847548 (P=2.2 × 10−18), which is in LD with rs4657139 but was not typed in previous studies11–15, 17. The frequency of the C allele of rs16847548 was 0.22 in whites. After adjusting for age, sex, and heart rate, the mean QT interval of individuals with TT, TC, and CC genotypes at rs16847548 were 399, 401, and 403 ms respectively in ARIC; 411, 414, and 416 ms respectively in CHS; and 402, 404, and 407 ms respectively in the combined dataset. This effect size of approximately 5 ms difference between the two homozygous groups is consistent with our previous observations. In whites, the percent variation (R2) in the QT interval distribution (uncorrected QT interval) that was explained by rs16847548 was 0.2% in both the individual studies and the combined dataset. In comparison, the R2 associated with other variables was: 0.5% for age, 0.1% for sex, 0.2% for diabetes, 0.4% for history of MI at baseline, and 67% for heart rate in white ARIC participants. Among white CHS participants, the R2 associated with age, sex, diabetes, history of MI at baseline, and heart rate were 0.03%, 1.1%, 0.4%, 0.5%, and 63%, respectively.
Associations between NOS1AP genotypes and SCD
Consistent with the observation of longer mean QT interval associated with the C allele of rs16847548, this allele was also significantly associated with increased risk of SCD in whites. Indeed, only 179 of the 8,905 individuals (2%) carrying the TT genotype at rs16847548 suffered from SCD (Table 4), whereas, 25 of the 706 (3.5%) individuals with the CC genotype experienced SCD. In whites, the crude relative hazards that were estimated using a co-dominant model suggested a dose-response relationship between copies of the C allele at rs16847548 and SCD. Using an additive model, the age-, sex-, and study-adjusted RH for each C allele was 1.31, 95% CI 1.10 to 1.56; P=0.002 (Table 4).
Table 4.
Unadjusted and adjusted relative hazard (RH) of SCD by rs16847548 and rs12567209 genotypes in whites from ARIC and CHS
| rs16847548 | rs12567209 | |||||
|---|---|---|---|---|---|---|
| TT | TC | CC | GG | AG | AA | |
| No SCD | N=8,726 | N=4,885 | N=681 | N=12,197 | N=1,932 | N=69 |
| 61.1% | 34.2% | 4.8% | 85.9% | 13.6% | 0.5% | |
| SCD | N=179 | N=127 | N=25 | N=303 | N=25 | N=2 |
| 54.1% | 38.4% | 7.6% | 91.8% | 7.6% | 0.6% | |
| RH (95% CI) | 1.00 (ref.) | 1.26 (1.00–1.58) | 1.79 (1.18–2.72) | 1.00 (ref.) | 0.51 (0.34–0.76) | 1.31 (0.32–5.25) |
| Model 1 | 1.00 (ref) | 1.31 (1.10–1.56) | 1.00 (ref.) | 0.57 (0.39–0.83) | ||
| P value | 0.002 | P value | 0.003 | |||
| Model 2 | 1.00 | 1.27 (1.06, 1.51) | 1.00 | 0.62 (0.42, 0.90) | ||
| P value | 0.008 | P value | 0.012 | |||
| Model 3 | 1.00 | 1.22 (1.03–1.46) | 1.00 | 0.63 (0.43, 0.92) | ||
| P value | 0.02 | P value | 0.02 | |||
| Model 4 | 1.00 | 1.17 (0.97–1.42) | 1.00 | 0.60 (0.40, 0.91) | ||
| P value | 0.09 | P value | 0.02 | |||
P Value obtained from regression model assuming additive genetic model
Model 1 included age, sex, and study
Model 2 included model 1 + both rs16847548 and rs12567209
Model 3 = model 2 + heart rate (continuous) and QT-interval (quintiles)
Model 4 = model 3+ current marital and smoking status, education, BMI, total cholesterol and fibrinogen levels, hypertension, diabetes, and history of MI, heart rate (continuous) and QT-interval (quintiles)
In addition, a downstream neighboring SNP, rs12567209, not correlated with rs16847548 (r2 = 0.02), was also associated with SCD in whites (age-, sex-, and study-adjusted RH for each A allele =0.57 assuming an additive model, 95% CI 0.39 to 0.83; P=0.003). Due to the low frequency of the A allele (MAF=0.07), a dominant model was also used for the analysis of rs12567209. The age-, sex-, and study-adjusted relative hazard of SCD comparing those with at least one copy of the A allele to those with the GG genotype was 0.53 (95% CI 0.36 to 0.79; P=0.002), thus both the additive and the dominant models were consistent with the data. The present study is not able to distinguish whether one model was a better fit than the other (additive model shown in Table 4). Surprisingly, rs12567209 was only modestly associated with QT interval (Table 3), suggesting that the effect on risk for SCD was not necessarily conveyed through modulation of QT interval. The mean age-, sex-, heart-rate, and study-adjusted QT interval for GG, AG, and AA genotypes were 403, 403, and 401 ms respectively (P for additive =0.05; P for dominant model =0.08).
To demonstrate the independent effect on risk of SCD for rs16847548 and rs12567209, we included both SNPs in the same model and found that both SNPs remained associated with SCD. Moreover, there was no significant interaction between these two SNPs (P for interaction=0.68 adjusted for age, sex, and study). Assuming an additive model for both SNPs and after adjusting for age, sex, and study, the RH for each copy of the C allele of rs16847548 was 1.27 (95% CI 1.06 to 1.51; P=0.008) and 0.62 (95% CI 0.42 to 0.90; P=0.012) for each copy of the A allele of rs12567209 (Table 4). The associations between both SNPs and SCD risk were relatively consistent across the two studies although the study-specific p values did not reach the Bonferroni-corrected significance level due to their smaller sample sizes. The corresponding RH for rs16847548 was 1.32 (95% CI 1.01 to 1.70; P=0.04) for ARIC and 1.24 (95% CI 0.98 to 1.57; P=0.08) for CHS. The corresponding RH for rs12567209 was 0.84 (95% CI 0.52 to 1.39; P=0.51) for ARIC and 0.44 (95% CI 0.25 to 0.79; P=0.006).
No SNPs were significantly associated with SCD in blacks at the α=0.003 level (Table 3). The RH for each C allele of rs16847548 was 1.07 (95% CI 0.81 to 1.41; P=0.64) after adjusting for age, sex, and study. On the other hand, the RH for each A allele of rs12567209 was 1.40 (opposite direction as the association in whites; 95% CI 0.97 to 2.03; P=0.07) after adjusting for age, sex, and study (Supplementary Table 1).
Multivariate genotype association analyses with SCD
To explore whether the effect of NOS1AP SNPs on the risk of SCD is entirely mediated through modulation of the QT interval, we added both QT interval and heart rate as variables into the Cox proportional hazards model in whites, after adjusting for age, sex, and study. The relative hazard of SCD associated with each additional copy of the C allele at rs16847548 decreased from 1.27 (model 2 in Table 4) to 1.22 (model 3 in Table 4), and the relative hazard for each additional A allele at rs12567209 changed from 0.62 to 0.63. Further adjustment for existing cardiovascular risk factors that were associated with SCD modestly attenuated the significance of the associations for rs16847548 (RH=1.17; 95% CI 0.97 to 1.42).
Additional analyses were performed to examine the impact of either co-morbidities or other SNPs on the robustness of the association between rs16847548 and rs12567209 and SCD risk in whites. First, exclusion of whites with a previous history of MI strengthened both associations (for each C allele of rs16847548 RH=1.39 for model 1, 95% CI 1.13 to 1.69; P=0.002; for each A allele of rs12567209 RH=0.49 for model 1; 95% CI 0.30 to 0.78; P=0.003). Second, exclusion of 623 individuals with electrocardiographic QRS complex >120 ms, which is indicative of a bundle branch block or other conduction defect, also resulted in a stronger association between rs16847548 and SCD risk (RH=1.40 for model 1; 95% CI 1.16 to 1.68; P<0.001). However, when QRS duration was included in the fully adjusted model (model 3), the relative hazard changed minimally from 1.23 to 1.25 (95% CI 1.04 to 1.51). The age-, sex-, and study-adjusted RH for rs12567209 changed from 0.57 to 0.62 (95% CI 0.42 to 0.92; P=0.02) upon exclusion of QRS complex >120 ms. Finally, among whites, none of the tests of interaction between either SNP, SCD, and known cardiovascular risk factors (study, history of MI, sex, diabetes, age at last follow up, diabetes, hypertension, family history of CVD, obesity, dyslipdemia, and smoking) was statistically significant (Supplemental Tables S2 and S3).
Lack of association between rs16847548 and rs12567209 and non-sudden cardiac mortality in whites
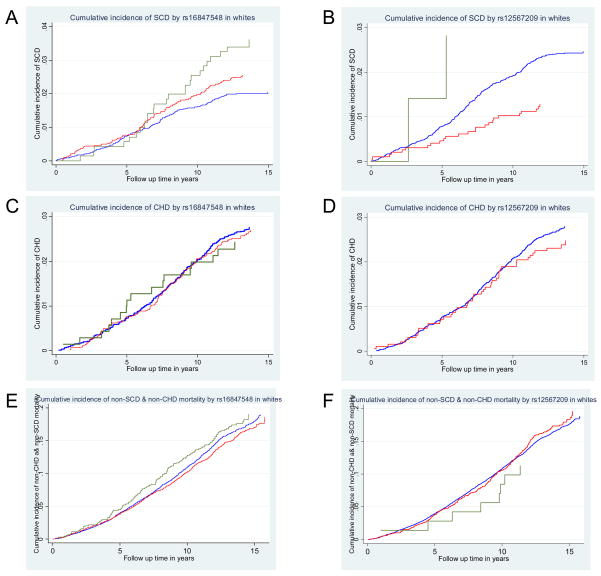
It is possible that the association between rs16847548 and/or rs12567209 with SCD is due to an association with overall CHD mortality. Therefore, survival analyses were also conducted for CHD mortality that was not coded as SCD (non-SCD CHD mortality) and all other mortality that were neither SCD nor CHD (non-SCD & non-CHD) mortality. Figure 2 shows the cumulative incidences of SCD, non-SCD CHD, and non-SCD & non-CHD mortality, while accounting for each other as competing case of death, by rs16847548 or rs12567209 in whites. The cumulative incidence of SCD per 1,000 person-years of whites with the TT, TC, and CC genotypes at rs16847548 were 1.5, 2.0, and 2.8, respectively (Figure 2A). On the other hand, rs16847548 was not associated with non- sudden CHD mortality (age-, sex-, and study-adjusted RH=0.98, 95% CI 0.83 to 1.17; P=0.86; Figure 2C) nor with non-SCD & non-CHD mortality (age-, sex-, and study-adjusted RH=1.00, 95% CI 0.94 to 1.07, P=0.94; Figure 2E). For rs12567209, the cumulative incidence of SCD per 1,000 person-years of whites with the GG, AG, and AA genotypes were 1.9, 0.9, and 2.2, respectively (Figure 2B). As for rs16847548, no association was observed with non- sudden CHD mortality (age-, sex-, and study-adjusted RH=0.83, 95% CI 0.62 to 1.11); P=0.21; Figure 2D) nor with non-SCD & non-CHD mortality (age-, sex-, and study-adjusted RH=99 95% CI 0.89 to 1.10, P=0.86; Figure 2F).
Figure 2.
Cumulative incidence curves, accounting for competing cause of death, in whites. Kaplan-Meier survival curves in whites by rs16847548 (A,C,E) and by rs1267209 (B,D,F). (A,B) SCD, (C,D) non-SCD CHD mortality, and (E,F) non-SCD & non-CHD mortality. For rs16847548, green lines represent CC, red lines represent CT, and blue lines represent TT. For rs12567209, green lines represent AA, red lines represent AG, and blue lines represent GG. Only 1 CHD death was observed in rs12567209 AA individuals (D), and hence no curve is shown for that genotype.
Discussion
In this study, common sequence variations in NOS1AP are associated with both inter-individual variation in the QT interval and risk of SCD in whites from two large cohorts of adults in the U.S. The at-risk allele (C at rs16847548) is common, with 39% of the general white population carrying one copy of the C allele and 5% carrying two copies. More specifically, in the combined population, white U.S. adults carrying the CC genotype at rs16847548 of NOS1AP were about 72% more likely to die of SCD and had a mean QT interval that was approximately 5 ms longer than their counterparts with the TT genotype, even after accounting for age, sex, and heart rate. On the other hand, the less common A allele of rs12567209 (~13% of the general white population are carriers) was independently associated with a decreased risk of SCD (RH=0.57) in whites. Notably, both of the genetic effects were specific for SCD rather than other forms of death from CHD. Finally, in spite of demographic differences between ARIC and CHS whites, the genetic effect estimates were comparable in the two populations separately and there was no evidence of significant heterogeneity (Tables S2 and S3).
The present study identifies a novel gene, NOS1AP, along with a new set of cellular interactions, which can potentially affect SCD risk in the general population. The multivariate analyses shows that even after adjusting for QT interval and heart rate a significant association still remains between both rs16847548 and rs12567209 and SCD risk. This result was somewhat unanticipated given that our initial hypothesis evolved from a model in which SNPs influence QT interval, and that increasing QT interval would increase risk for SCD. However, given that adjusting for QT interval largely does not attenuate the risk for SCD associated with these SNPs, this suggests an alternate model, in which these SNPs modulate an unmeasured, or hidden, factor, which itself modulates both QT interval and risk for SCD, and need not do so equivalently. However, the possibility that the remaining significant association between these SNPs and SCD risk is due to the QT interval as assessed by ECG representing an imperfect measure of cardiac repolarization, the potential misclassification of the actual QT interval measurements and SCD definition, or the presence of additional genetic variation (i.e. not having identified the causal SNPs) cannot be excluded. Although a recent study reported no association between NOS1AP and SCD in the Rotterdam Study, it is important to note that rs16847548 and rs12567209 were not directly studied nor efficiently tagged and that the number of SCD events was small12. In addition, the positive association between rs16847548 of NOS1AP and SCD risk supports the approach of using either precursors or intermediate phenotypes in genetic studies of complex diseases7, 32, 33 as NOS1AP was first identified to be a candidate gene for SCD through a previous GWAS of QT interval11.
The results of the present study, together with previous reports of associations between NOS1AP and the QT interval in multiple populations of European descent11, 13–17, suggest novel and potentially causal mechanisms linking NOS1AP and SCD risk. As an adapter protein, the gene product of NOS1AP (CAPON) serves to physically bridge neuronal nitric oxide synthase (nNOS) and its targets and modulator proteins. In guinea pig ventricular myocytes, CAPON is localized near ryanodine receptors, and the over expression of CAPON results in shortening of the cardiac action potential, a decrease in L-type Ca current and a smaller increase in the delayed rectifier potassium current, IKr, resulting in prolongation of the QT interval34.
Several limitations are warranted in the interpretation of these findings. First, although we have identified the association of sequence variation at the NOS1AP locus with QT interval and SCD risk, it is not known whether we have identified the functional variants. For example, it is likely that rs12567209 is only in linkage disequilibrium with the causal SNP since its association with SCD in whites was in the opposite direction as its association in blacks. Even though rs16847548 had the strongest association (judging by p-values of all 19 SNPs), with both QT interval and SCD in both the ARIC and CHS cohorts, simulation studies have shown that the causative SNP may not necessarily have the smallest P-value since P-values fluctuate by chance due to the nature of random sampling, dependent on the sample size and allele frequency35. Thus, it is possible that rs16847548 is also in linkage disequilibrium with another causal SNP. Second, no significant association between the 19 NOS1AP SNPs and QT interval or SCD was observed in the black participants at a conservative α=0.003. The discordance in the associations between blacks and whites may represent the result of lower statistical power (due to inappropriate tagging SNPs and smaller numbers of events) in the blacks or it may represent a real genetic difference. Given the observed carrier frequency for rs16847548 of 34% (based on allele frequency of 0.19 in blacks) and assuming an overall genotypic relative hazard of 1.37, as observed in whites, at least 649 SCD cases in the blacks would be necessary for the study to have 80% power with an alpha of 0.00331. If the correlation between our genotyped SNPs and the putative ungenotyped functional variant is lower in blacks, then we would have less power to detect an effect in blacks. On the other hand, it is possible that the causal allele in blacks is not rs16847548 or that there exists only one causal allele in NOS1AP but the pattern of linkage disequilibrium between the causal variant and rs16847548 differs between blacks and whites. Third, despite corroborating functional data from guinea pig cardiomyocytes34, it is still possible that the associated variants in the NOS1AP locus actually influence (or are in LD with) a distant gene rather than NOS1AP, as this “action at a distance” has on rare occasions been observed at other rare diseases36. Fourth, s with all genetic association studies of complex traits, an independent replication study of comparable size and phenotype is not only the best defense against possible false positive reporting in our study but is necessary before certainty of the observed associations can be established. Lastly, the present study was not ideal for assessing the utility of genetic risk prediction among those already at high risk as the number of high-risk individuals is relatively modest in these studies.
In summary, in this study, we report that sequence variations in NOS1AP, a novel candidate gene that was previously identified through GWAS of the QT interval11, are associated with both QT interval and the subsequent risk of SCD in a large cohort of 14,737 white U.S. adults. As expected, individuals carrying the at-risk allele at rs16847548 had a modest increased risk of SCD, with each allele increasing SCD risk by about 30% compared to those who did not carry the risk allele. On the other hand, the A minor allele of rs12567209 was associated with a reduced risk of SCD, and the associations of these two SNPs were independent of each of other. Although the genetic effects described here are modest, if replicated in other populations, this effort may be an important step towards the identification of a panel of susceptibility alleles that may potentially used for risk assessment in the general population. Future studies that explore the pathways mediating the association between variations of NOS1AP and SCD risk will also be crucial and may shed light on both targeted prevention strategies and novel therapeutic targets for abnormal cardiac repolarization and SCD risk.
Supplementary Material
Acknowledgments
The authors thank the staff and participants of the ARIC study and CHS for their important contributions. The authors would like to thank Ms. Ashley O’Connor for her effort in genotyping theses two large cohorts.
Funding Sources
This work was supported by the Donald W. Reynolds Foundation (WHLK, DEA, WP, BB, BD, GT, EM, PMS, AC), NHGRI HG02757 (AC), the Leducq Foundation (EM, PMS, AC), and K01DK067207 (WHLK).
The Atherosclerosis Risk in Communities Study is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute contracts N01-HC-55015, N01-HC-55016, N01-HC-55018, N01-HC-55019, N01-HC-55020, N01-HC-55021, and N01-HC-55022.
The Cardiovascular Heart Study is supported by contracts N01-HC-35129, N01-HC-45133, N01-HC-75150, N01-HC-85079 through N01-HC-85086, N01 HC-15103, N01 HC-55222, and U01 HL080295 from the National Heart, Lung, and Blood Institute, with additional contribution from the National Institute of Neurological Disorders and Stroke.
Footnotes
Short summary of potential clinical impact
Nearly half of all coronary heart disease (CHD) deaths are sudden. Family history of sudden cardiac death (SCD) is a powerful risk factor for SCD; however, the genetic factors underlying SCD in the general population are largely unknown. The electrocardiographic QT interval is associated with risk of SCD. A previous genome-wide association study reported that allelic variants in NOS1AP, which encodes a ligand of neuronal nitric oxide synthase, are associated with the QT interval in white adults. The present analysis was conducted to validate the association between NOS1AP variants and the QT interval and to further examine the association with SCD in a combined population of 19,295 black and white adults from two population-based cohort studies. Among whites, we found that multiple SNPs in NOS1AP were associated with adjusted QT interval in whites (P<0.0001), and two SNPs were independently associated with SCD. One SNP, with a minor allele frequency (MAF) of 22%, was associated with a 31% greater risk of SCD for each copy of the variant allele, while a neighboring SNP (MAF 7%) was associated with a 43% lower risk for SCD. No associations between SNPs in NOS1AP and either QT interval or SCD were observed in blacks. Although the genetic effects described here are modest, if replicated in other populations, this effort may represent one step towards using genetic risk markers, along with other risk factors, to help identify patients who warrant our most aggressive SCD preventive strategies.
Disclosure Statement
None
References
- 1.Chugh SS, Jui J, Gunson K, Stecker EC, John BT, Thompson B, Ilias N, Vickers C, Dogra V, Daya M, Kron J, Zheng ZJ, Mensah G, McAnulty J. Current burden of sudden cardiac death: multiple source surveillance versus retrospective death certificate-based review in a large U.S. community. J Am Coll Cardiol. 2004;44:1268–1275. doi: 10.1016/j.jacc.2004.06.029. [DOI] [PubMed] [Google Scholar]
- 2.Zheng ZJ, Croft JB, Giles WH, Mensah GA. Sudden cardiac death in the United States, 1989 to 1998. Circulation. 2001;104:2158–2163. doi: 10.1161/hc4301.098254. [DOI] [PubMed] [Google Scholar]
- 3.Myerburg RJ, Castellanos A. Emerging paradigms of the epidemiology and demographics of sudden cardiac arrest. Heart Rhythm. 2006;3:235–239. doi: 10.1016/j.hrthm.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 4.Dekker LR, Bezzina CR, Henriques JP, Tanck MW, Koch KT, Alings MW, Arnold AE, de Boer MJ, Gorgels AP, Michels HR, Verkerk A, Verheugt FW, Zijlstra F, Wilde AA. Familial sudden death is an important risk factor for primary ventricular fibrillation: a case-control study in acute myocardial infarction patients. Circulation. 2006;114:1140–1145. doi: 10.1161/CIRCULATIONAHA.105.606145. [DOI] [PubMed] [Google Scholar]
- 5.Friedlander Y, Siscovick DS, Weinmann S, Austin MA, Psaty BM, Lemaitre RN, Arbogast P, Raghunathan TE, Cobb LA. Family history as a risk factor for primary cardiac arrest. Circulation. 1998;97:155–160. doi: 10.1161/01.cir.97.2.155. [DOI] [PubMed] [Google Scholar]
- 6.Jouven X, Desnos M, Guerot C, Ducimetiere P. Predicting sudden death in the population: the Paris Prospective Study I. Circulation. 1999;99:1978–1983. doi: 10.1161/01.cir.99.15.1978. [DOI] [PubMed] [Google Scholar]
- 7.Arking DE, Chugh SS, Chakravarti A, Spooner PM. Genomics in sudden cardiac death. Circ Res. 2004;94:712–723. doi: 10.1161/01.RES.0000123861.16082.95. [DOI] [PubMed] [Google Scholar]
- 8.Vrtovec B, Delgado R, Zewail A, Thomas CD, Richartz BM, Radovancevic B. Prolonged QTc interval and high B-type natriuretic peptide levels together predict mortality in patients with advanced heart failure. Circulation. 2003;107:1764–1769. doi: 10.1161/01.CIR.0000057980.84624.95. [DOI] [PubMed] [Google Scholar]
- 9.Newton-Cheh C, Larson MG, Corey DC, Benjamin EJ, Herbert AG, Levy D, D’Agostino RB, O’Donnell CJ. QT interval is a heritable quantitative trait with evidence of linkage to chromosome 3 in a genome-wide linkage analysis: The Framingham Heart Study. Heart Rhythm. 2005;2:277–284. doi: 10.1016/j.hrthm.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 10.Straus SM, Kors JA, De Bruin ML, van der Hooft CS, Hofman A, Heeringa J, Deckers JW, Kingma JH, Sturkenboom MC, Stricker BH, Witteman JC. Prolonged QTc interval and risk of sudden cardiac death in a population of older adults. J Am Coll Cardiol. 2006;47:362–367. doi: 10.1016/j.jacc.2005.08.067. [DOI] [PubMed] [Google Scholar]
- 11.Arking DE, Pfeufer A, Post W, Kao WH, Newton-Cheh C, Ikeda M, West K, Kashuk C, Akyol M, Perz S, Jalilzadeh S, Illig T, Gieger C, Guo CY, Larson MG, Wichmann HE, Marban E, O’Donnell CJ, Hirschhorn JN, Kaab S, Spooner PM, Meitinger T, Chakravarti A. A common genetic variant in the NOS1 regulator NOS1AP modulates cardiac repolarization. Nat Genet. 2006;38:644–651. doi: 10.1038/ng1790. [DOI] [PubMed] [Google Scholar]
- 12.Aarnoudse AJ, Newton-Cheh C, de Bakker PI, Straus SM, Kors JA, Hofman A, Uitterlinden AG, Witteman JC, Stricker BH. Common NOS1AP variants are associated with a prolonged QTc interval in the Rotterdam Study. Circulation. 2007;116:10–16. doi: 10.1161/CIRCULATIONAHA.106.676783. [DOI] [PubMed] [Google Scholar]
- 13.Eijgelsheim M, Aarnoudse AL, Rivadeneira F, Kors JA, Witteman JC, Hofman A, van Duijn CM, Uitterlinden AG, Stricker BH. Identification of a Common Variant at the NOS1AP Locus Strongly Associated to QT - Interval Duration. Hum Mol Genet. 2008 doi: 10.1093/hmg/ddn341. [DOI] [PubMed] [Google Scholar]
- 14.Lehtinen AB, Newton-Cheh C, Ziegler JT, Langefeld CD, Freedman BI, Daniel KR, Herrington DM, Bowden DW. Association of NOS1AP genetic variants with QT interval duration in families from the Diabetes Heart Study. Diabetes. 2008;57:1108–1114. doi: 10.2337/db07-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Post W, Shen H, Damcott C, Arking DE, Kao WH, Sack PA, Ryan KA, Chakravarti A, Mitchell BD, Shuldiner AR. Associations between genetic variants in the NOS1AP (CAPON) gene and cardiac repolarization in the old order Amish. Hum Hered. 2007;64:214–219. doi: 10.1159/000103630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raitakari OT, Blom-Nyholm J, Koskinen TA, Kahonen M, Viikari JS, Lehtimaki T. Common variation in NOS1AP and KCNH2 genes and QT interval duration in young adults. The Cardiovascular Risk in Young Finns Study. Ann Med. 2008:1–8. doi: 10.1080/07853890802392529. [DOI] [PubMed] [Google Scholar]
- 17.Tobin MD, Kahonen M, Braund P, Nieminen T, Hajat C, Tomaszewski M, Viik J, Lehtinen R, Ng GA, Macfarlane PW, Burton PR, Lehtimaki T, Samani NJ. Gender and effects of a common genetic variant in the NOS1 regulator NOS1AP on cardiac repolarization in 3761 individuals from two independent populations. Int J Epidemiol. 2008;37:1132–1141. doi: 10.1093/ije/dyn091. [DOI] [PubMed] [Google Scholar]
- 18.The Atherosclerosis Risk in Communities (ARIC) Study: design and objectives. The ARIC investigators. Am J Epidemiol. 1989;129:687–702. [PubMed] [Google Scholar]
- 19.Fried LP, Borhani NO, Enright P, Furberg CD, Gardin JM, Kronmal RA, Kuller LH, Manolio TA, Mittelmark MB, Newman A. The Cardiovascular Health Study: design and rationale. Ann Epidemiol. 1991;1:263–276. doi: 10.1016/1047-2797(91)90005-w. [DOI] [PubMed] [Google Scholar]
- 20.Ives DG, Fitzpatrick AL, Bild DE, Psaty BM, Kuller LH, Crowley PM, Cruise RG, Theroux S. Surveillance and ascertainment of cardiovascular events. The Cardiovascular Health Study. Ann Epidemiol. 1995;5:278–285. doi: 10.1016/1047-2797(94)00093-9. [DOI] [PubMed] [Google Scholar]
- 21.Price TR, Psaty B, O’Leary D, Burke G, Gardin J. Assessment of cerebrovascular disease in the Cardiovascular Health Study. Ann Epidemiol. 1993;3:504–507. doi: 10.1016/1047-2797(93)90105-d. [DOI] [PubMed] [Google Scholar]
- 22.Sotoodehnia N, Siscovick DS, Vatta M, Psaty BM, Tracy RP, Towbin JA, Lemaitre RN, Rea TD, Durda JP, Chang JM, Lumley TS, Kuller LH, Burke GL, Heckbert SR. Beta2-adrenergic receptor genetic variants and risk of sudden cardiac death. Circulation. 2006;113:1842–1848. doi: 10.1161/CIRCULATIONAHA.105.582833. [DOI] [PubMed] [Google Scholar]
- 23.Rautaharju PM, Park LP, Chaitman BR, Rautaharju F, Zhang ZM. The Novacode criteria for classification of ECG abnormalities and their clinically significant progression and regression. J Electrocardiol. 1998;31:157–187. [PubMed] [Google Scholar]
- 24.General Description and Study Management. ARIC Protocol. 1987 [Google Scholar]
- 25.Psaty BM, Kuller LH, Bild D, Burke GL, Kittner SJ, Mittelmark M, Price TR, Rautaharju PM, Robbins J. Methods of assessing prevalent cardiovascular disease in the Cardiovascular Health Study. Ann Epidemiol. 1995;5:270–277. doi: 10.1016/1047-2797(94)00092-8. [DOI] [PubMed] [Google Scholar]
- 26.A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Bakker PI, Yelensky R, Pe’er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 28.www.biotrove.com. 2007.
- 29.Bazett H. An analysis of the time-relations of electrocardiograms. Heart-A Journal for the Study of the Circulation. 1920;7:353–370. [Google Scholar]
- 30.Gooley TA, Leisenring W, Crowley J, Storer BE. Estimation of failure probabilities in the presence of competing risks: new representations of old estimators. Stat Med. 1999;18:695–706. doi: 10.1002/(sici)1097-0258(19990330)18:6<695::aid-sim60>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 31.Schoenfeld DA. Sample-size formula for the proportional-hazards regression model. Biometrics. 1983;39:499–503. [PubMed] [Google Scholar]
- 32.Cohen JC, Boerwinkle E, Mosley TH, Jr, Hobbs HH. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med. 2006;354:1264–1272. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 33.Lange LA, Carlson CS, Hindorff LA, Lange EM, Walston J, Durda JP, Cushman M, Bis JC, Zeng D, Lin D, Kuller LH, Nickerson DA, Psaty BM, Tracy RP, Reiner AP. Association of polymorphisms in the CRP gene with circulating C-reactive protein levels and cardiovascular events. JAMA. 2006;296:2703–2711. doi: 10.1001/jama.296.22.2703. [DOI] [PubMed] [Google Scholar]
- 34.Chang KC, Barth AS, Sasano T, Kizana E, Kashiwakura Y, Zhang Y, Foster DB, Marban E. CAPON modulates cardiac repolarization via neuronal nitric oxide synthase signaling in the heart. Proc Natl Acad Sci U S A. 2008;105:4477–4482. doi: 10.1073/pnas.0709118105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang Q, Morrison AC, Boerwinkle E. Linkage disequilibrium structure and its impact on the localization of a candidate functional mutation. Genet Epidemiol. 2001;21 (Suppl 1):S620–625. doi: 10.1002/gepi.2001.21.s1.s620. [DOI] [PubMed] [Google Scholar]
- 36.Lettice LA, Horikoshi T, Heaney SJ, van Baren MJ, van der Linde HC, Breedveld GJ, Joosse M, Akarsu N, Oostra BA, Endo N, Shibata M, Suzuki M, Takahashi E, Shinka T, Nakahori Y, Ayusawa D, Nakabayashi K, Scherer SW, Heutink P, Hill RE, Noji S. Disruption of a long-range cis-acting regulator for Shh causes preaxial polydactyly. Proc Natl Acad Sci U S A. 2002;99:7548–7553. doi: 10.1073/pnas.112212199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.