Abstract
The I-TASSER algorithm for protein 3D structure prediction was tested in CASP8, with the procedure fully automated in both the Server and Human sections. The quality of the server models is close to that of human ones but incorporating more diverse templates from other servers improves the results of human predictions in the distant homology category. For the first time, the sequence-based contact predictions from machine learning techniques are found helpful for both template-based modeling (TBM) and template-free modeling (FM). In TBM, although the average accuracy of the sequence-based contact predictions is lower than that from template-based ones, the novel contacts in the sequence-based predictions, which are complementary to the threading templates in the weakly or unaligned regions, are important to improve the global and local packing of these regions. Moreover, the newly developed atomic structural refinement algorithm was tested in CASP8 and found to improve the hydrogen-bonding networks and the overall TM-score, which is mainly due to its ability of removing steric clashes so that the models can be generated from cluster centroids. Nevertheless, one of the major issues of the I-TASSER pipeline is the model selection where the best models could not be appropriately recognized when the correct templates are detected only by the minority of the threading algorithms. There are also problems related with domain-splitting and mirror image recognition which mainly influences the performance of I-TASSER modeling in the FM-based structure predictions.
Keywords: Protein structure prediction, threading, I-TASSER, CASP8, contact prediction, free modeling
INTRODUCTION
When will computers beat humans in protein structure prediction? Or are there still any human insights that cannot be reproduced in automated approaches? During the CASP experiments, several groups 1–3 demonstrated that the interventions by human experts, who made use of biochemical information (function, family characteristics, mutagenesis, catalytic residues, etc.), can indeed help with template recognition, structural assembly, and final model selection. Nevertheless, fully automated algorithms have an advantage in genome-wide structure prediction4–6; they also allow non-experts to generate structural models on their own or through Internet services7–9. Undoubtedly, with the expeditious accumulation of genome-wide sequences, the development of fully automated computer-based structure prediction methods becomes unprecedentedly demanding10.
Recent years have witnessed significant progress in automated structure prediction6,11. In CASP7, for example, it was stated in assessors’ reports 12–14 that “the best prediction server (Zhang-Server) was ranked third overall, i.e. it outperformed all but two of the human participating groups”. Actually, in the current framework of CASP, it is difficult to have an entirely fair assessment of the performance of automated vs. human prediction because human predictors can use all the models generated by servers and therefore have a better pool of initial templates to start with.
In CASP8, we participated in both human (as ‘Zhang’) and server (as ‘Zhang-Server’) predictions. For the purpose of the development and testing of automated structure prediction approaches, both Zhang and Zhang-Server used identical I-TASSER approach15. Compared with CASP7, new developments in I-TASSER include the employment of de novo sequence-based contact predictions16, and atomic-level hydrogen-bonding (H-bond) optimization17. Because the only difference between Zhang and Zhang-Server is that the ‘human’ prediction uses more templates (including those generated by other groups in the Server section), the difference between their performances may be viewed as a measure of the effect due to the different template pools used in human and server predictions.
MATERIALS AND METHODS
The I-TASSER prediction pipeline includes four general steps: template identification, structure reassembly, atomic model construction, and final model selection.
Template identification
Target sequences are threaded through a non-redundant PDB structure library for identifying appropriate global-structure templates (for TBM targets) or local fragments (for FM targets). Threading is done by MUSTER18, which uses an extended sequence profile-profile alignment algorithm with the alignment score assisted by secondary structure match, fragment structure profile, solvent accessibility, backbone torsion angle, and hydrophobic scoring matrix. For hard targets, additional templates identified by LOMETS19, a local meta-threading server including FUGUE20, HHSEARCH21, PROSPECT22, PPA15 and SP323, are used. In human prediction, we additionally include the models generated by other groups in the Server Section in the template pool. Having more threading templates is the only source of differences between Zhang and Zhang-Server predictions.
Structure assembly
Continuous fragments excised from the threading templates are used to assemble full-length models15,24 with unaligned loop regions built by ab initio modeling in a lattice system25. The structure assembly process consists of two sets of simulations15. The first set uses the threading templates as initial structures. In the second set, the simulations start from the cluster centroids generated by SPICKER26, which clusters all the trajectories from the first set of simulations. Spatial restraints collected from the PDB structures hit by TM-align27 using the cluster centroids as query structures are also incorporated in the I-TASSER simulations. The purpose of the second stage is to refine the local geometry as well as the global topology of the SPICKER centroids.
Energy force field
The structure assembly simulations (for both the threading-aligned and the ab initio modeled regions) are guided by a unified knowledge-based force field, which includes four components: (1) general knowledge-based statistics terms from the PDB (C-alpha/side-chain correlations25, H-bonds28 and hydrophobicity29); (2) spatial restraints from threading templates19; (3) sequence-based contact predictions from SVMSEQ16.
The last energy term is relatively new in comparison with the force field used in the previous CASP experiment30. SVMSEQ is a support-vector-machine (SVM) based residue-residue contact predictor that only uses sequence information16. It was trained using local window features (position-specific scoring matrices, secondary structure and solvent accessibility predictions) and in-between segment features (residue separations, secondary structure of the contacting residues, and state distributions of the contacting residues). Nine sets of predictions are generated, based Cα, Cβ and side-chain center positions, each with contact cutoffs 6 A, 7 A, and 8 A. All nine predictions are used in I-TASSER simulation as restraints, with weights proportional to their confidence.
Atomic model construction
The SPICKER cluster centroids from I-TASSER are reduced models, with each residue represented by its Cα and side-chain center. The full-atomic models are built by REMO17, a new protocol we developed for constructing full-atomic models from C-alpha traces by optimizing the H-bond networks. The basic backbone fragments (Cα, C, N, O) are matched from a secondary structure specific backbone isomer library which consist of a total of 68,206 non-redundant isomers from high-resolution PDB structures. The driving force in the REMO refinement protocol includes H-bonding, clash/break-amendment, I-TASSER restraints, and the CHARMM22 potential. Based on a test set of 230 non-homologous proteins, REMO has the ability of removing steric clashes while retaining a topology score (e.g. TM-score) similar to that of cluster centroids. Moreover, the H-bond network was improved in more than 80% (187/230) of test proteins by REMO17.
Model selection
The reduced models from I-TASSER are ranked based on the structure density in SPICKER clusters26. For each reduced model, atomic models from REMO are selected based on an empirical scoring function which is equal to the sum of the number of H-bonds divided by the target length, the TM-score31 of the model with the SPICKER cluster centroid, and the average TM-score of the model with the initial templates (used for easy target only). The weights of the empirical score have been trained in benchmark tests. The highest scoring models are finally submitted.
Multiple-domain proteins
The procedure to deal with multiple-domain proteins is similar to what we used in CASP730. If a segment of the target sequence with >80 residues has no aligned residues in the top two threading templates, the target is judged a multiple domain protein, and domain boundaries are automatically assigned based on the boundaries of the large gaps. The I-TASSER simulations will be run for the full chain as well as the separate domains. The final full-length models are generated by docking the models of all domains together through a quick Metropolis Monte Carlo simulation where energy is defined as the RMSD of the domain models to the full-chain models plus the reciprocal of the number of inter-domain steric clashes. This procedure is only applied to proteins that have some domains not aligned in the top-scoring templates. If multiple-domain templates are available with all domains aligned, the whole-chain will be modeled in I-TASSER simultaneously.
RESULTS AND DISCUSSION
A total of 164 domains from 121 protein targets were eventually assessed in the Server Section, and 71 domains in the Human Section. Among the 164 domains, 50 are high-accuracy (HA), 102 are template-based modeling (TBM) and only 12 are free-modeling (FM, including TBM/FM) targets. Because more targets were tested in the server section, and the methods used in our server and human predictions are essentially identical, our report will mainly focus on the server predictions. In particular, we summarize what went right and what were the major problems with our approach.
What went right?
I-TASSER pulls templates closer to the native conformation
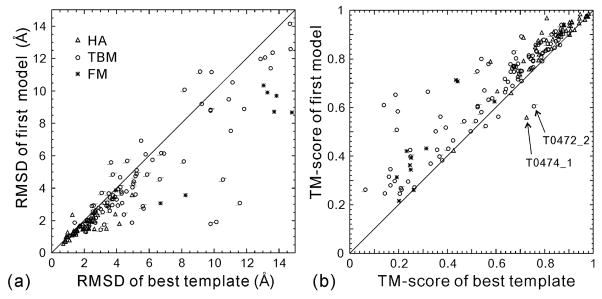
As observed in both benchmark tests15 and previous CASP experiments30, one of the most important advantages of I-TASSER is that the fragment assembly procedure can consistently drive the initial template structures closer to the native states. In Figure 1a, we present the RMSD of the first I-TASSER server models versus the RMSD of the best threading templates used in I-TASSER for all 164 domains, with both RMSDs calculated for the aligned regions of threading alignments. Although FM targets are supposed to have no appropriate templates, we show them in the plot because the I-TASSER procedure always starts from the top scoring templates obtained by threading no matter how weak the alignment scores are. In fact, even when the global topology of the templates is incorrect, the super-secondary structure segments are useful as structural building blocks. Apparently, I-TASSER simulations improve the template structure in the majority of test cases as measured by RMSD. For 139 out of 164 domains, the RMSD of the final models is lower than that of the templates. In the remaining 22 (3) cases, the RMSD of the I-TASSER models is higher (equal to) that of the templates. Overall, the average RMSD of the best threading template is 5.54 A for the aligned regions, with an average alignment coverage of 91%; this RMSD is reduced to 4.24 A by I-TASSER.
Figure 1.
Comparison of the best threading templates with the first model predicted by the I-TASSER server. RMSD for models is calculated in the same aligned region as the threading template. The highlights in (b) are two domains where I-TASSER deteriorates the best templates.
Because some threading alignments are very short, and may consist of only a small piece of structure, a TM-score comparison should reflect more appropriately the adding of I-TASSER in full-chain model construction from the templates. Figure 1b is a comparison of final models versus the best threading templates in terms of TM-score. Now, 150 targets have a final model with a higher TM-score than the templates, and 10 (4) have a final model with a lower (equal) TM-score than the templates. Noticeably, there are two domains, T0472_2 and T0474, where the first submitted models are significantly worse than the best templates. T0472 has a duplicated β3α two-domain structure, with its closest structural template being 3bid, a domain-swapped dimer. Because our threading library includes only single-chain proteins, most of the whole-chain threading templates have only the N-terminal domain aligned. The first submitted model by our I-TASSER server is based on the whole-chain modeling, and has a reasonably good quality for the N-terminal domain (RMSD=1.54 A and TM-score=0.731) but a low-quality C-terminal domain (TM-score=0.605 for T0472_2). The second submitted model by the server for T0472 was built by modeling the domains separately, followed by domain docking as described in Methods, and has a TM-score of 0.767 for T0472_2, slightly higher than that of the template (TM-score=0.755).
T0474 is small protein of 80 residues solved by Structural Genomics Consortium, and has a very extended structure (85.3A from N to C terminus). All the three closest templates (2ay0, 2bj1, 2hza) are dimers, with the “necks” of the chains intertwined with each other. The individual chains are apparently unstable on their own, but our server attempted to fold the chain as an individual compact domain, which resulted in a much less extended structural model with a TM-score=0.560. The second submitted model has a more extended structure with a TM-score=0.683, which is still lower than the best template withTM-score=0.726.
Restraints from multiple templates cover a larger portion of the structure than those from the best single templates
One of the major driving forces of the structure refinement in I-TASSER are the high-quality consensus restraints taken from multiple templates by MUSTER18 or LOMETS19. Five types of template-based restraints are used in I-TASSER: (1) side-chain contact restraints taken from the top N templates (N=20 for easy targets, 30 for medium and 50 for hard targets); (2) Cα contact restraints from the top N templates; (3) long-range Cα distance-map from the top 4 templates (i.e. |i-j|>6, each residue pair having up to 4 different distance restraints); (4) short-range Cα distance-map for |i-j|≤6 with the average distance from the top N templates; (5) pair-wise contact potential based on the frequency of the side-chain contacts appearing in the top N templates32.
Although there has been a long-time belief that consensus restraints should have a better accuracy than those from single templates, there is no systematic comparison of the two based on the same set of templates. In Table I, we present a detailed list of the accuracy and coverage of four restraint types taken either from multiple templates or from the best single threading template that has the highest TM-score to the native in the top N templates. In all categories of targets (i.e. HA, TMB and FM), the consensus contact predictions have a higher coverage, i.e. more correct contacts are predicted. However, somewhat contrary to expectation, the accuracy of the contacts based on single templates is slightly higher than that of the consensus ones, which is probably due to the fact that we are using the best individual template from threading. In fact, if we use the first template (as ranked by threading rather than TM-score), the accuracy of the contact prediction is similar to that of consensus contacts, but the coverage is lower than when the best threading template (i.e. highest TM-score) is used. Here, we compare consensus restraints to the best templates because we try to highlight the possible reason that I-TASSER improves the quality of the best templates as shown in Figure 1. Overall, the average accuracy/coverage for side-chain and Cα contact predictions are 0.34/0.55 and 0.59/0.55 from the best single template, compared to 0.31/0.64 and 0.56/0.64 from multiple templates. One reason for the apparently higher accuracy of Cα contacts in comparison with side-chain contacts is that side-chain contacts are more variable due to rotamer conformations, and therefore are more difficult to predict.
Table I.
Comparison of spatial restraints taken from multiple templates and from the single best threading template (the latter shown in parentheses).
| Side-chain contact restraints |
Cα contact restraints |
Short distance mapd | Long distance mape | RMf | TMg | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Na | Accb | Covc | Na | Accb | Covc | |||||
| HA-targets | ||||||||||
| T0388_1 | 163 | 0.42(0.51) | 0.96(0.92) | 103 | 0.82(0.88) | 0.96(0.95) | 0.26(0.24) | 0.59(0.53) | 1.2 | 0.950 |
| T0390_1 | 107 | 0.37(0.45) | 0.93(0.90) | 105 | 0.68(0.81) | 0.93(0.87) | 0.61(0.48) | 0.55(0.71) | 1.6 | 0.919 |
| T0392_1 | 59 | 0.36(0.38) | 0.95(0.78) | 75 | 0.69(0.78) | 0.87(0.67) | 0.38(0.57) | 0.48(0.91) | 1.4 | 0.905 |
| T0396_1 | 68 | 0.38(0.43) | 0.88(0.81) | 15 | 0.48(0.60) | 0.93(0.80) | 0.31(0.37) | 0.91(1.01) | 2.1 | 0.895 |
| T0398_1 | 117 | 0.33(0.44) | 0.93(0.91) | 100 | 0.65(0.88) | 0.91(0.89) | 0.76(0.31) | 0.46(0.49) | 0.7 | 0.977 |
| T0398_2 | 120 | 0.36(0.52) | 0.95(0.89) | 99 | 0.66(0.97) | 0.93(0.88) | 0.43(0.19) | 0.37(0.35) | 0.6 | 0.985 |
| T0400_1 | 158 | 0.38(0.53) | 0.85(0.83) | 99 | 0.68(0.86) | 0.91(0.87) | 0.52(0.38) | 0.58(0.89) | 1.5 | 0.921 |
| T0402_1 | 95 | 0.34(0.40) | 0.84(0.68) | 97 | 0.75(0.82) | 0.92(0.75) | 0.56(0.55) | 0.74(1.06) | 1.9 | 0.867 |
| T0404_1 | 56 | 0.37(0.45) | 0.89(0.84) | 58 | 0.75(0.83) | 0.88(0.86) | 0.34(0.27) | 0.53(0.61) | 1.0 | 0.918 |
| T0416_1 | 196 | 0.37(0.44) | 0.87(0.73) | 105 | 0.71(0.85) | 0.82(0.75) | 0.43(0.32) | 0.56(0.94) | 1.5 | 0.940 |
| T0418_1 | 146 | 0.42(0.47) | 0.92(0.76) | 92 | 0.70(0.74) | 0.90(0.79) | 0.41(0.49) | 0.44(0.95) | 1.6 | 0.917 |
| T0418_2 | 47 | 0.38(0.34) | 0.83(0.64) | 13 | 0.42(0.36) | 0.62(0.38) | 0.46(0.81) | 0.70(1.29) | 1.8 | 0.791 |
| T0422_2 | 58 | 0.47(0.52) | 0.93(0.90) | 14 | 0.46(0.42) | 0.79(0.36) | 0.57(0.52) | 0.70(0.93) | 1.7 | 0.845 |
| T0423_1 | 138 | 0.40(0.50) | 0.88(0.84) | 125 | 0.81(0.90) | 0.80(0.72) | 0.36(0.25) | 0.77(0.63) | 1.5 | 0.941 |
| T0426_1 | 287 | 0.45(0.50) | 0.98(0.87) | 263 | 0.85(0.92) | 0.96(0.91) | 0.20(0.20) | 0.18(0.41) | 0.7 | 0.987 |
| T0428_1 | 228 | 0.43(0.54) | 0.98(0.95) | 157 | 0.81(0.89) | 0.95(0.90) | 0.23(0.24) | 0.27(0.47) | 1.0 | 0.974 |
| T0432_1 | 93 | 0.41(0.57) | 0.92(0.83) | 21 | 0.62(0.67) | 0.86(0.57) | 0.41(0.51) | 0.76(1.00) | 1.8 | 0.911 |
| T0435_1 | 116 | 0.39(0.49) | 0.84(0.79) | 128 | 0.69(0.89) | 0.84(0.80) | 0.59(0.69) | 1.25(1.80) | 3.8 | 0.842 |
| T0437_1 | 48 | 0.33(0.42) | 0.73(0.67) | 48 | 0.53(0.62) | 0.67(0.48) | 0.49(0.45) | 1.10(1.20) | 1.6 | 0.849 |
| T0438_1 | 144 | 0.36(0.39) | 0.81(0.74) | 127 | 0.67(0.77) | 0.86(0.76) | 0.60(0.44) | 0.84(0.79) | 1.5 | 0.926 |
| T0438_2 | 188 | 0.41(0.45) | 0.90(0.86) | 180 | 0.79(0.89) | 0.87(0.78) | 0.35(0.28) | 0.58(0.56) | 1.2 | 0.967 |
| T0441_2 | 131 | 0.33(0.34) | 0.83(0.73) | 123 | 0.67(0.70) | 0.83(0.76) | 0.63(0.72) | 0.74(1.23) | 2.0 | 0.902 |
| T0442_1 | 134 | 0.45(0.48) | 0.90(0.90) | 123 | 0.80(0.82) | 0.84(0.80) | 0.52(0.30) | 0.89(0.59) | 1.2 | 0.950 |
| T0442_2 | 46 | 0.37(0.36) | 0.76(0.70) | 51 | 0.75(0.79) | 0.75(0.65) | 0.58(0.26) | 0.77(0.64) | 0.8 | 0.939 |
| T0444_1 | 265 | 0.43(0.53) | 0.95(0.91) | 49 | 0.66(0.73) | 0.86(0.73) | 0.26(0.26) | 0.33(0.52) | 1.3 | 0.962 |
| T0445_1 | 158 | 0.40(0.47) | 0.92(0.80) | 118 | 0.71(0.82) | 0.86(0.78) | 0.36(0.37) | 0.55(0.96) | 1.6 | 0.913 |
| T0447_1 | 576 | 0.42(0.47) | 0.90(0.84) | 400 | 0.79(0.82) | 0.81(0.81) | 0.40(0.31) | 0.67(0.65) | 1.4 | 0.975 |
| T0450_1 | 469 | 0.34(0.42) | 0.90(0.80) | 353 | 0.68(0.77) | 0.83(0.69) | 0.45(0.37) | 0.68(0.90) | 1.5 | 0.968 |
| T0452_1 | 140 | 0.37(0.47) | 0.75(0.81) | 98 | 0.63(0.74) | 0.78(0.74) | 0.64(0.39) | 0.71(1.16) | 1.9 | 0.888 |
| T0452_2 | 131 | 0.39(0.45) | 0.91(0.83) | 109 | 0.76(0.81) | 0.88(0.79) | 0.35(0.29) | 0.51(0.72) | 1.2 | 0.953 |
| T0453_1 | 79 | 0.37(0.43) | 0.82(0.72) | 67 | 0.84(0.92) | 0.96(0.90) | 0.50(0.39) | 0.45(1.01) | 1.6 | 0.872 |
| T0454_1 | 31 | 0.43(0.46) | 0.94(0.94) | 9 | 0.36(0.36) | 0.56(0.56) | 0.32(0.38) | 0.46(0.69) | 1.1 | 0.86 |
| T0455_1 | 108 | 0.33(0.39) | 0.92(0.80) | 129 | 0.82(0.92) | 0.96(0.90) | 0.36(0.39) | 0.48(0.95) | 1.6 | 0.909 |
| T0456_2 | 143 | 0.36(0.41) | 0.92(0.76) | 73 | 0.62(0.62) | 0.81(0.45) | 0.68(0.87) | 0.85(1.70) | 5.1 | 0.872 |
| T0458_1 | 62 | 0.43(0.51) | 0.97(0.94) | 48 | 0.90(0.89) | 0.90(0.88) | 0.27(0.23) | 0.37(0.50) | 0.8 | 0.947 |
| T0459_1 | 66 | 0.51(0.54) | 0.95(0.77) | 28 | 0.62(0.69) | 0.93(0.79) | 0.49(0.44) | 0.63(1.05) | 1.6 | 0.877 |
| T0461_1 | 118 | 0.37(0.41) | 0.92(0.91) | 114 | 0.70(0.78) | 0.92(0.89) | 0.32(0.29) | 0.89(0.81) | 1.8 | 0.911 |
| T0470_1 | 78 | 0.40(0.48) | 0.87(0.78) | 23 | 0.59(0.75) | 0.74(0.65) | 0.56(0.62) | 0.85(1.11) | 2.1 | 0.877 |
| T0470_2 | 46 | 0.41(0.44) | 0.78(0.57) | 20 | 0.80(0.85) | 0.80(0.55) | 0.34(0.41) | 0.42(0.78) | 1.3 | 0.909 |
| T0472_2 | 21 | 0.42(0.42) | 0.90(0.90) | 26 | 0.68(0.68) | 0.81(0.81) | 0.61(0.36) | 1.19(0.93) | 2.9 | 0.605 |
| T0474_1 | 0 | 0.00(0.00) | 0.00(0.00) | 1 | 0.25(1.00) | 1.00(1.00) | 0.26(0.27) | 0.66(0.94) | 2.4 | 0.559 |
| T0479_1 | 103 | 0.39(0.37) | 0.86(0.71) | 119 | 0.83(0.88) | 0.87(0.81) | 0.44(0.51) | 0.65(1.19) | 2.0 | 0.892 |
| T0486_1 | 181 | 0.39(0.40) | 0.88(0.77) | 151 | 0.79(0.83) | 0.89(0.79) | 0.40(0.41) | 0.54(1.14) | 1.5 | 0.937 |
| T0488_1 | 58 | 0.34(0.46) | 0.95(0.93) | 71 | 0.77(0.90) | 0.94(0.93) | 0.32(0.27) | 0.40(0.64) | 1.3 | 0.899 |
| T0491_1 | 66 | 0.39(0.41) | 0.89(0.80) | 104 | 0.87(0.88) | 0.95(0.88) | 0.56(0.56) | 1.04(1.26) | 2.0 | 0.839 |
| T0499_1 | 43 | 0.44(0.48) | 0.88(0.81) | 41 | 0.83(0.90) | 0.95(0.93) | 0.46(0.49) | 0.72(0.99) | 1.4 | 0.795 |
| T0504_3 | 53 | 0.59(0.50) | 0.66(0.60) | 51 | 0.81(0.79) | 0.76(0.73) | 0.67(0.64) | 0.62(2.94) | 1.8 | 0.749 |
| T0505_1 | 154 | 0.42(0.51) | 0.93(0.88) | 114 | 0.78(0.84) | 0.92(0.86) | 0.38(0.26) | 0.61(0.73) | 1.5 | 0.940 |
| T0506_1 | 108 | 0.41(0.44) | 0.92(0.89) | 108 | 0.81(0.87) | 0.84(0.83) | 0.46(0.47) | 0.88(0.91) | 1.7 | 0.904 |
| T0508_1 | 200 | 0.44(0.52) | 0.84(0.78) | 122 | 0.80(0.86) | 0.84(0.78) | 0.43(0.36) | 0.64(0.90) | 1.4 | 0.936 |
| Average (HA) | 128.0 | 0.39(0.45) | 0.87(0.79) | 97.3 | 0.70(0.79) | 0.86(0.77) | 0.45(0.41) | 0.65(0.92) | 1.6 | 0.895 |
| TBM targets | ||||||||||
| T0389_1 | 111 | 0.34(0.40) | 0.79(0.69) | 74 | 0.71(0.75) | 0.86(0.69) | 0.87(0.52) | 0.93(1.50) | 3.2 | 0.822 |
| T0391_1 | 133 | 0.34(0.37) | 0.70(0.62) | 128 | 0.68(0.69) | 0.69(0.60) | 0.77(0.78) | 2.44(3.38) | 11.2 | 0.708 |
| T0393_1 | 160 | 0.28(0.33) | 0.64(0.46) | 102 | 0.34(0.54) | 0.58(0.51) | 0.86(0.71) | 0.92(1.53) | 3.6 | 0.802 |
| T0393_2 | 34 | 0.23(0.40) | 0.68(0.65) | 10 | 0.27(0.43) | 0.40(0.30) | 0.63(0.60) | 0.90(1.19) | 2.1 | 0.789 |
| T0394_1 | 258 | 0.30(0.30) | 0.51(0.43) | 175 | 0.56(0.48) | 0.60(0.46) | 0.72(0.78) | 2.46(4.56) | 10.9 | 0.638 |
| T0395_1 | 212 | 0.30(0.32) | 0.53(0.34) | 106 | 0.51(0.61) | 0.53(0.36) | 0.91(0.78) | 2.43(2.83) | 14.9 | 0.545 |
| T0397_2 | 52 | 0.17(0.17) | 0.63(0.35) | 79 | 0.48(0.52) | 0.75(0.56) | 1.10(1.10) | 1.79(2.36) | 3.9 | 0.623 |
| T0399_1 | 141 | 0.22(0.26) | 0.40(0.42) | 150 | 0.45(0.49) | 0.53(0.49) | 1.16(0.96) | 2.94(3.09) | 8.1 | 0.524 |
| T0401_1 | 115 | 0.26(0.26) | 0.52(0.45) | 100 | 0.53(0.46) | 0.62(0.44) | 0.98(0.90) | 1.42(2.23) | 4.2 | 0.716 |
| T0406_1 | 119 | 0.27(0.31) | 0.67(0.52) | 41 | 0.34(0.32) | 0.32(0.22) | 0.59(0.54) | 1.15(2.35) | 3.3 | 0.778 |
| T0407_1 | 280 | 0.33(0.34) | 0.56(0.49) | 186 | 0.71(0.75) | 0.76(0.70) | 0.93(0.78) | 1.39(2.04) | 4.2 | 0.768 |
| T0407_2 | 86 | 0.13(0.15) | 0.20(0.22) | 96 | 0.27(0.36) | 0.26(0.36) | 1.57(1.52) | 1.80(5.02) | 11.2 | 0.315 |
| T0408_1 | 51 | 0.48(0.35) | 0.88(0.45) | 15 | 0.45(0.46) | 0.67(0.40) | 0.36(0.44) | 1.00(4.80) | 1.8 | 0.827 |
| T0409_1 | 43 | 0.25(0.36) | 0.74(0.58) | 59 | 0.57(0.79) | 0.83(0.63) | 0.67(0.34) | 1.03(1.34) | 3.0 | 0.651 |
| T0411_1 | 110 | 0.32(0.44) | 0.76(0.70) | 64 | 0.41(0.56) | 0.77(0.72) | 0.75(0.56) | 0.76(1.33) | 3.3 | 0.794 |
| T0412_1 | 143 | 0.36(0.40) | 0.80(0.73) | 94 | 0.59(0.61) | 0.83(0.82) | 0.73(0.72) | 1.07(1.76) | 3.1 | 0.837 |
| T0413_1 | 295 | 0.24(0.20) | 0.36(0.26) | 197 | 0.45(0.38) | 0.49(0.38) | 1.08(1.17) | 2.14(4.71) | 9.2 | 0.602 |
| T0414_1 | 127 | 0.44(0.37) | 0.56(0.39) | 131 | 0.69(0.76) | 0.72(0.62) | 0.89(0.83) | 1.80(1.47) | 8.0 | 0.632 |
| T0415_1 | 95 | 0.38(0.43) | 0.76(0.74) | 92 | 0.72(0.81) | 0.80(0.80) | 0.85(0.38) | 1.29(1.03) | 2.2 | 0.814 |
| T0417_1 | 125 | 0.27(0.27) | 0.69(0.51) | 105 | 0.47(0.57) | 0.70(0.56) | 0.92(0.96) | 1.15(2.37) | 4.3 | 0.751 |
| T0419_1 | 208 | 0.30(0.25) | 0.40(0.32) | 88 | 0.41(0.33) | 0.43(0.38) | 0.77(0.60) | 2.53(3.48) | 11.8 | 0.584 |
| T0419_2 | 216 | 0.27(0.29) | 0.42(0.36) | 92 | 0.42(0.53) | 0.41(0.45) | 0.78(0.62) | 2.66(2.24) | 10.0 | 0.610 |
| T0420_1 | 152 | 0.22(0.23) | 0.58(0.43) | 110 | 0.41(0.53) | 0.53(0.50) | 1.01(0.98) | 1.52(2.26) | 3.4 | 0.751 |
| T0421_1 | 187 | 0.31(0.22) | 0.45(0.34) | 73 | 0.42(0.26) | 0.47(0.33) | 0.82(1.00) | 1.91(3.18) | 7.4 | 0.665 |
| T0422_1 | 160 | 0.38(0.47) | 0.86(0.83) | 103 | 0.74(0.84) | 0.81(0.70) | 0.58(0.46) | 1.58(0.94) | 4.0 | 0.881 |
| T0424_1 | 164 | 0.32(0.37) | 0.75(0.65) | 195 | 0.68(0.77) | 0.85(0.81) | 0.48(0.51) | 0.96(1.13) | 2.3 | 0.862 |
| T0424_2 | 75 | 0.36(0.39) | 0.71(0.63) | 53 | 0.63(0.65) | 0.77(0.60) | 0.65(0.60) | 0.96(1.33) | 2.3 | 0.766 |
| T0424_3 | 28 | 0.22(0.24) | 0.68(0.54) | 36 | 0.57(0.67) | 0.78(0.67) | 0.29(0.26) | 1.17(1.17) | 1.9 | 0.718 |
| T0425_1 | 180 | 0.36(0.38) | 0.73(0.69) | 127 | 0.54(0.58) | 0.63(0.61) | 0.71(0.67) | 1.44(1.50) | 2.9 | 0.833 |
| T0427_1 | 195 | 0.35(0.36) | 0.69(0.54) | 126 | 0.50(0.55) | 0.62(0.56) | 0.74(0.78) | 1.91(1.93) | 3.2 | 0.83 |
| T0427_2 | 158 | 0.33(0.39) | 0.70(0.61) | 113 | 0.43(0.45) | 0.58(0.42) | 0.71(0.68) | 1.60(2.08) | 3.9 | 0.807 |
| T0429_1 | 42 | 0.37(0.42) | 0.79(0.52) | 60 | 0.70(0.85) | 0.82(0.47) | 0.86(0.89) | 2.40(0.99) | 9.0 | 0.342 |
| T0429_2 | 57 | 0.19(0.21) | 0.25(0.25) | 63 | 0.34(0.26) | 0.32(0.22) | 1.32(0.98) | 3.03(3.23) | 11.4 | 0.296 |
| T0430_1 | 108 | 0.22(0.25) | 0.23(0.26) | 97 | 0.40(0.43) | 0.34(0.31) | 0.91(1.09) | 3.29(2.63) | 8.5 | 0.517 |
| T0430_2 | 167 | 0.11(0.12) | 0.22(0.20) | 91 | 0.20(0.19) | 0.16(0.15) | 1.54(1.71) | 4.57(7.57) | 15.2 | 0.430 |
| T0431_1 | 75 | 0.30(0.40) | 0.84(0.83) | 71 | 0.53(0.75) | 0.79(0.77) | 1.02(0.64) | 1.34(1.87) | 3.6 | 0.779 |
| T0431_2 | 324 | 0.36(0.43) | 0.81(0.79) | 136 | 0.63(0.78) | 0.67(0.74) | 0.58(0.39) | 0.89(1.33) | 2.9 | 0.892 |
| T0433_1 | 199 | 0.34(0.40) | 0.64(0.53) | 135 | 0.69(0.79) | 0.78(0.68) | 0.80(0.71) | 0.78(1.55) | 2.4 | 0.879 |
| T0434_1 | 162 | 0.39(0.47) | 0.61(0.57) | 152 | 0.72(0.90) | 0.74(0.68) | 0.74(0.74) | 2.58(3.20) | 12.3 | 0.689 |
| T0436_1 | 414 | 0.29(0.32) | 0.66(0.57) | 247 | 0.57(0.59) | 0.68(0.58) | 0.71(0.69) | 2.34(2.47) | 6.2 | 0.833 |
| T0440_1 | 291 | 0.36(0.39) | 0.68(0.63) | 184 | 0.62(0.67) | 0.69(0.64) | 0.73(0.60) | 1.81(1.59) | 3.4 | 0.858 |
| T0441_1 | 72 | 0.25(0.30) | 0.83(0.82) | 72 | 0.60(0.68) | 0.74(0.64) | 0.55(0.56) | 0.85(1.21) | 2.3 | 0.818 |
| T0443_3 | 39 | 0.32(0.11) | 0.51(0.10) | 30 | 0.50(0.09) | 0.10(0.03) | 0.83(1.24) | 2.25(7.17) | 10.3 | 0.39 |
| T0445_2 | 89 | 0.28(0.33) | 0.75(0.60) | 77 | 0.59(0.54) | 0.75(0.47) | 0.75(0.79) | 0.88(1.68) | 2.4 | 0.788 |
| T0446_1 | 31 | 0.28(0.29) | 0.77(0.74) | 44 | 0.70(0.71) | 0.75(0.73) | 0.96(0.86) | 1.85(2.10) | 3.6 | 0.663 |
| T0446_2 | 23 | 0.19(0.22) | 0.57(0.48) | 37 | 0.53(0.47) | 0.81(0.46) | 0.96(1.13) | 2.55(2.66) | 3.0 | 0.543 |
| T0448_1 | 227 | 0.30(0.34) | 0.57(0.54) | 141 | 0.50(0.64) | 0.50(0.58) | 0.84(0.78) | 1.07(1.77) | 4.6 | 0.769 |
| T0449_1 | 344 | 0.27(0.29) | 0.58(0.47) | 345 | 0.57(0.65) | 0.70(0.60) | 0.98(1.07) | 1.52(2.58) | 4.8 | 0.780 |
| T0451_1 | 105 | 0.25(0.29) | 0.66(0.52) | 116 | 0.66(0.78) | 0.74(0.69) | 0.66(0.59) | 0.96(1.73) | 2.7 | 0.813 |
| T0454_2 | 94 | 0.29(0.29) | 0.69(0.55) | 22 | 0.31(0.31) | 0.45(0.36) | 0.61(0.76) | 0.98(2.32) | 3.4 | 0.736 |
| T0456_1 | 53 | 0.33(0.36) | 0.92(0.75) | 68 | 0.74(0.81) | 0.93(0.79) | 0.44(0.54) | 0.62(1.24) | 2.7 | 0.757 |
| T0457_1 | 194 | 0.30(0.34) | 0.57(0.53) | 108 | 0.48(0.49) | 0.56(0.54) | 0.88(0.83) | 1.51(1.86) | 4.2 | 0.767 |
| T0457_2 | 92 | 0.19(0.19) | 0.47(0.36) | 73 | 0.41(0.47) | 0.56(0.47) | 1.22(1.30) | 1.73(2.88) | 5.7 | 0.606 |
| T0460_1 | 62 | 0.11(0.05) | 0.11(0.05) | 48 | 0.21(0.08) | 0.12(0.04) | 1.56(1.20) | 2.71(7.27) | 12.3 | 0.262 |
| T0462_1 | 62 | 0.34(0.44) | 0.66(0.55) | 61 | 0.64(0.74) | 0.75(0.70) | 0.56(0.63) | 2.24(2.38) | 2.2 | 0.760 |
| T0462_2 | 56 | 0.34(0.44) | 0.79(0.54) | 50 | 0.51(0.75) | 0.70(0.54) | 0.85(0.68) | 1.65(1.80) | 2.0 | 0.721 |
| T0463_1 | 185 | 0.25(0.30) | 0.60(0.57) | 148 | 0.60(0.65) | 0.69(0.65) | 0.87(0.74) | 1.36(2.00) | 6.2 | 0.762 |
| T0464_1 | 50 | 0.42(0.45) | 0.40(0.36) | 43 | 0.57(0.62) | 0.53(0.42) | 0.80(0.68) | 2.64(1.57) | 4.1 | 0.561 |
| T0466_1 | 63 | 0.18(0.05) | 0.17(0.06) | 86 | 0.67(0.17) | 0.21(0.13) | 1.28(1.41) | 3.08(6.40) | 10.1 | 0.297 |
| T0468_1 | 49 | 0.30(0.30) | 0.49(0.35) | 52 | 0.33(0.36) | 0.52(0.31) | 1.07(1.07) | 1.95(3.02) | 5.7 | 0.396 |
| T0469_1 | 45 | 0.47(0.47) | 0.67(0.64) | 21 | 0.65(0.65) | 0.52(0.52) | 0.69(0.54) | 1.34(1.35) | 2.2 | 0.737 |
| T0471_1 | 96 | 0.35(0.50) | 0.78(0.55) | 71 | 0.54(0.63) | 0.66(0.59) | 0.59(0.39) | 1.65(1.60) | 1.9 | 0.800 |
| T0472_1 | 45 | 0.59(0.59) | 0.60(0.58) | 46 | 0.94(0.93) | 0.65(0.61) | 0.46(0.42) | 1.37(0.85) | 5.0 | 0.660 |
| T0473_1 | 51 | 0.40(0.40) | 0.61(0.55) | 19 | 0.52(0.48) | 0.63(0.63) | 0.51(0.64) | 1.59(1.92) | 1.9 | 0.705 |
| T0475_1 | 109 | 0.43(0.51) | 0.87(0.74) | 114 | 0.79(0.81) | 0.78(0.68) | 0.69(0.75) | 0.70(1.33) | 2.5 | 0.839 |
| T0477_1 | 213 | 0.32(0.36) | 0.80(0.63) | 136 | 0.60(0.62) | 0.76(0.59) | 0.62(0.64) | 1.14(1.88) | 4.8 | 0.857 |
| T0478_1 | 95 | 0.14(0.09) | 0.06(0.07) | 27 | 0.00(0.00) | 0.00(0.00) | 0.50(0.40) | 2.27(3.40) | 8.1 | 0.426 |
| T0478_2 | 97 | 0.24(0.26) | 0.13(0.16) | 25 | 0.00(0.24) | 0.00(0.24) | 0.62(0.38) | 2.67(2.55) | 9.3 | 0.425 |
| T0480_1 | 19 | 0.20(0.29) | 0.79(0.47) | 20 | 0.33(0.50) | 0.70(0.35) | 1.23(0.80) | 1.38(4.95) | 2.7 | 0.368 |
| T0481_1 | 110 | 0.32(0.35) | 0.65(0.54) | 32 | 0.50(0.52) | 0.44(0.34) | 0.56(0.64) | 2.20(2.40) | 3.4 | 0.746 |
| T0483_1 | 267 | 0.40(0.43) | 0.86(0.82) | 144 | 0.59(0.66) | 0.86(0.78) | 0.56(0.57) | 0.71(1.28) | 4.5 | 0.857 |
| T0485_1 | 201 | 0.37(0.34) | 0.56(0.42) | 135 | 0.70(0.77) | 0.68(0.47) | 0.80(0.86) | 1.37(3.82) | 5.6 | 0.747 |
| T0487_1 | 137 | 0.20(0.23) | 0.58(0.52) | 143 | 0.51(0.64) | 0.62(0.62) | 0.93(0.91) | 1.44(2.03) | 3.2 | 0.790 |
| T0487_2 | 84 | 0.17(0.17) | 0.31(0.36) | 94 | 0.66(0.60) | 0.40(0.41) | 1.26(1.33) | 2.16(3.84) | 6.1 | 0.503 |
| T0487_3 | 44 | 0.12(0.09) | 0.20(0.18) | 58 | 0.40(0.20) | 0.24(0.14) | 1.48(1.01) | 2.34(3.59) | 6.2 | 0.383 |
| T0487_4 | 60 | 0.02(0.03) | 0.03(0.05) | 79 | 0.15(0.19) | 0.09(0.10) | 1.94(2.01) | 2.81(5.46) | 12.0 | 0.246 |
| T0487_5 | 103 | 0.16(0.20) | 0.38(0.36) | 85 | 0.28(0.37) | 0.36(0.34) | 0.96(1.08) | 1.83(2.93) | 4.8 | 0.580 |
| T0489_1 | 213 | 0.26(0.20) | 0.31(0.21) | 83 | 0.44(0.17) | 0.29(0.10) | 0.96(1.35) | 2.60(6.28) | 10.6 | 0.502 |
| T0490_1 | 370 | 0.32(0.33) | 0.71(0.60) | 280 | 0.58(0.61) | 0.72(0.64) | 0.67(0.66) | 1.06(1.59) | 2.6 | 0.897 |
| T0492_1 | 57 | 0.36(0.42) | 0.74(0.53) | 61 | 0.69(0.75) | 0.75(0.66) | 0.67(0.77) | 2.32(2.44) | 6.3 | 0.725 |
| T0493_1 | 138 | 0.40(0.42) | 0.83(0.77) | 104 | 0.70(0.69) | 0.78(0.69) | 0.59(0.51) | 0.86(1.18) | 2.0 | 0.878 |
| T0494_1 | 319 | 0.38(0.41) | 0.80(0.76) | 213 | 0.79(0.82) | 0.75(0.73) | 0.62(0.60) | 1.27(1.65) | 3.5 | 0.900 |
| T0495_1 | 117 | 0.20(0.24) | 0.34(0.26) | 95 | 0.44(0.44) | 0.39(0.28) | 1.10(1.30) | 2.56(3.77) | 13.7 | 0.465 |
| T0496_2 | 11 | 0.25(0.20) | 0.82(0.36) | 3 | 0.38(0.25) | 1.00(0.33) | 0.48(1.12) | 1.12(3.77) | 3.1 | 0.707 |
| T0497_1 | 102 | 0.32(0.32) | 0.83(0.58) | 100 | 0.75(0.81) | 0.86(0.72) | 0.60(0.60) | 0.68(1.30) | 2.1 | 0.862 |
| T0498_1 | 26 | 0.15(0.13) | 0.19(0.15) | 10 | 0.12(0.00) | 0.10(0.00) | 3.19(3.24) | 3.60(3.92) | 9.2 | 0.272 |
| T0501_1 | 223 | 0.30(0.35) | 0.57(0.52) | 113 | 0.47(0.52) | 0.59(0.58) | 0.84(0.82) | 1.82(2.03) | 3.8 | 0.771 |
| T0501_2 | 109 | 0.25(0.27) | 0.51(0.44) | 89 | 0.54(0.63) | 0.56(0.54) | 1.17(1.26) | 1.56(2.54) | 4.8 | 0.67 |
| T0502_1 | 78 | 0.29(0.32) | 0.64(0.60) | 113 | 0.64(0.64) | 0.69(0.63) | 0.66(0.69) | 1.96(2.00) | 3.4 | 0.75 |
| T0503_1 | 133 | 0.36(0.37) | 0.65(0.55) | 111 | 0.88(0.92) | 0.90(0.87) | 0.48(0.58) | 1.15(1.97) | 2.7 | 0.800 |
| T0504_1 | 52 | 0.50(0.32) | 0.50(0.40) | 63 | 0.91(0.78) | 0.62(0.60) | 0.90(1.22) | 1.95(5.50) | 15.0 | 0.423 |
| T0504_2 | 74 | 0.45(0.22) | 0.45(0.34) | 65 | 0.80(0.54) | 0.72(0.60) | 1.29(1.38) | 1.95(4.44) | 16.3 | 0.279 |
| T0505_2 | 88 | 0.22(0.27) | 0.48(0.44) | 79 | 0.53(0.52) | 0.58(0.44) | 0.94(0.70) | 1.04(2.07) | 3.7 | 0.666 |
| T0506_2 | 60 | 0.34(0.36) | 0.52(0.52) | 48 | 0.86(0.91) | 0.65(0.65) | 0.56(0.51) | 1.33(1.45) | 2.8 | 0.757 |
| T0507_1 | 111 | 0.26(0.31) | 0.57(0.51) | 62 | 0.39(0.49) | 0.60(0.56) | 0.93(0.76) | 1.23(2.10) | 5.1 | 0.676 |
| T0509_1 | 168 | 0.34(0.37) | 0.82(0.73) | 132 | 0.68(0.72) | 0.81(0.65) | 0.62(0.56) | 0.91(1.28) | 2.1 | 0.891 |
| T0510_1 | 130 | 0.24(0.07) | 0.25(0.08) | 147 | 0.55(0.32) | 0.33(0.17) | 1.12(1.15) | 2.40(5.64) | 14.8 | 0.431 |
| T0510_2 | 51 | 0.29(0.46) | 0.65(0.65) | 19 | 0.29(0.30) | 0.53(0.42) | 0.93(0.88) | 1.24(1.96) | 4.7 | 0.562 |
| T0511_1 | 215 | 0.35(0.39) | 0.80(0.70) | 179 | 0.61(0.61) | 0.68(0.56) | 0.75(0.69) | 1.29(2.12) | 4.8 | 0.825 |
| T0512_1 | 387 | 0.25(0.25) | 0.52(0.36) | 396 | 0.48(0.52) | 0.60(0.52) | 1.15(0.89) | 1.67(2.42) | 4.1 | 0.808 |
| T0513_1 | 186 | 0.26(0.26) | 0.63(0.53) | 181 | 0.51(0.51) | 0.57(0.49) | 0.80(0.73) | 1.83(2.60) | 9.5 | 0.713 |
| T0514_1 | 126 | 0.16(0.08) | 0.22(0.10) | 135 | 0.38(0.03) | 0.37(0.02) | 1.34(1.78) | 2.46(8.02) | 15.0 | 0.316 |
| Average (TBM) | 132.0 | 0.29(0.31) | 0.58(0.48) | 99.1 | 0.53(0.55) | 0.59(0.50) | 0.87(0.84) | 1.72(2.66) | 5.7 | 0.668 |
| FM targets | ||||||||||
| T0397_1 | 62 | 0.16(0.05) | 0.23(0.05) | 66 | 0.35(0.09) | 0.27(0.08) | 1.18(1.19) | 2.23(6.19) | 10.2 | 0.262 |
| T0405_1 | 30 | 0.11(0.02) | 0.07(0.03) | 6 | 0.00(0.07) | 0.00(0.17) | 1.03(1.32) | 2.14(8.03) | 9.1 | 0.373 |
| T0405_2 | 167 | 0.08(0.01) | 0.03(0.02) | 112 | 0.00(0.00) | 0.00(0.00) | 1.73(2.49) | 3.17(7.77) | 14.9 | 0.300 |
| T0416_2 | 28 | 0.00(0.00) | 0.00(0.00) | 9 | 0.00(0.00) | 0.00(0.00) | 1.05(0.95) | 4.57(6.46) | 4.1 | 0.528 |
| T0443_1 | 37 | 0.44(0.60) | 0.11(0.08) | 5 | 0.00(0.00) | 0.00(0.00) | 0.70(0.47) | 1.86(5.70) | 8.3 | 0.468 |
| T0443_2 | 39 | 0.27(0.00) | 0.08(0.00) | 46 | 1.00(0.00) | 0.22(0.00) | 1.78(0.33) | 3.94(4.09) | 8.0 | 0.351 |
| T0465_1 | 81 | 0.29(0.11) | 0.19(0.16) | 43 | 0.43(0.15) | 0.07(0.19) | 1.03(1.23) | 2.83(6.08) | 10.4 | 0.363 |
| T0476_1 | 44 | 0.14(0.15) | 0.23(0.16) | 22 | 0.55(0.29) | 0.50(0.23) | 1.27(1.37) | 2.57(12.77) | 6.7 | 0.398 |
| T0482_1 | 51 | 0.42(0.09) | 0.35(0.08) | 52 | 0.59(0.00) | 0.50(0.00) | 1.26(1.28) | 2.21(8.99) | 8.1 | 0.446 |
| T0496_1 | 111 | 0.13(0.03) | 0.04(0.04) | 60 | 0.20(0.00) | 0.02(0.00) | 1.34(2.55) | 3.31(6.93) | 12.5 | 0.317 |
| T0510_3 | 22 | 0.00(0.07) | 0.00(0.09) | 24 | 0.00(0.00) | 0.00(0.00) | 1.67(2.32) | 3.72(9.15) | 10.9 | 0.249 |
| T0513_2 | 48 | 0.00(0.04) | 0.00(0.02) | 38 | 0.00(0.00) | 0.00(0.00) | 1.75(1.33) | 5.13(5.93) | 4.3 | 0.507 |
| Average (FM) | 60.0 | 0.17(0.10) | 0.11(0.06) | 40.2 | 0.26(0.05) | 0.13(0.05) | 1.32(1.40) | 3.14(7.34) | 9.0 | 0.380 |
| Average (All) | 125.5 | 0.31(0.34) | 0.64(0.55) | 94.2 | 0.56(0.59) | 0.64(0.55) | 0.77(0.75) | 1.50(2.47) | 4.7 | 0.712 |
Number of contacts appearing in the native structure.
Accuracy of contact predictions: the number of correctly predicted contacts divided by the total number of contact predictions.
Coverage of contact predictions: the number of correctly predicted contacts divided by the number of contacts in the native structure.
Error of short-range distance predictions (|i–j|≤6) relative to the native structure.
Error of long-range distance predictions (|i–j|>6) relative to the native structure.
RMSD (A) of the first submitted model by Zhang-Server (best in top 5 shown for FM).
TM-score of the first submitted model by Zhang-Server (best in top 5 shown for FM).
The 8th and 9th columns of Table I show the errors of short- and long-range Cα distance predictions, respectively. For short-range distance prediction, single-template based prediction has a slightly smaller average error than the multiple-template based one. But for the long-range distance prediction, the distant error from multiple templates (i.e. the best in top 4 predictions) is much smaller than that from the best single template. Moreover, as the major advantage of using multiple templates, multiple-template based predictions cover again a larger portion of the structure. Overall, the multiple-template based prediction produces on average 1,302/2,563 short/long-range distance predictions while single-template prediction produces only 1,099/2,243 short/long-range predictions.
Interestingly, there are some targets for which the accuracy and coverage of contact predictions is apparently high but the quality of the final models is still poor. For example, two FM targets (T0476_1 and T0482_1) have Cα contact predictions with an accuracy and coverage both >0.5 (see Table I). But all the 11 correctly predicted contacts in T0476_1 are concentrated in two beta-hairpins (one at the tail and another in the middle, both being short-range), and are actually not helpful for assembling the global topology. On the contrary, the side-chain contact predictions have a lower accuracy but cover a larger portion of the structure. A similar situation is seen with T0482_1 as well. In fact, the correlation coefficient (calculated for all 164 domains) between the TM-score of the final models and the product of accuracy and coverage of side-chain contacts is 0.87, while the same quantity for Cα contacts is 0.79, which indicates that side-chain contact predictions are more important for the structure assembly.
Sequence-based contact predictions help both FM and TBM modeling
In addition to the consensus restraints from multiple templates, the second important contribution to the I-TASSER template structural refinement is the sequence-based contact prediction from SVMSEQ16. Our original purpose when developing SVMSEQ was to improve the I-TASSER structure assembly only for FM targets, because for TBM/HA targets, the overall accuracy of SVMSEQ is lower than that of the template-based contact prediction16. However, we found that the SVMSEQ prediction also improves the quality of models for the TBM targets.
In Table II, we present a summary of the SVMSEQ contact prediction for both side-chain and Cα contacts. As expected, the sequence-based contact predictions have the highest impact on FM targets. For these targets, the average accuracy of the side-chain contacts by LOMETS is only 17%, covering 11% of all native contacts. But the SVMSEQ prediction on side-chain contacts (with a 8 A cutoff distance) has an accuracy of 38.1%, with a coverage of 29.9% of all contacts in the native structure; out of this coverage, 21.8% are newly predicted contacts that are not generated by LOMETS. If we look at Cα contacts, the average accuracy of SVMSEQ predictions is 44.8%, compared with 26% by LOMETS. This covers 35.3% of all native contacts, with 29.3% being new. The Cβ predictions have similar results to Cα. These sequence-based ‘de novo’ predictions are of great value for I-TASSER in the case of FM target predictions.
Table II.
Summary of sequence-based contact predictions compared with the template-based contact predictions.
| Side-chain contacts | Cα contacts | Cβ contacts | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tema | S6b | S7c | S8d | Tema | S6b | S7c | S8d | S6a | S7b | S8c | Come | ||
| HA | NPf | 114.9 | 15.2 | 21.9 | 28.3 | 84.7 | 17.6 | 28.6 | 35.5 | 11.2 | 22.4 | 31.9 | 10.8 |
| ACCg | 0.39 | 0.219 | 0.32 | 0.403 | 0.7 | 0.26 | 0.397 | 0.475 | 0.162 | 0.328 | 0.442 | 0.228 | |
| COVh | 0.87 | 0.136 | 0.199 | 0.25 | 0.86 | 0.162 | 0.265 | 0.333 | 0.099 | 0.202 | 0.292 | 0.381 | |
| NNi | 0.7 | 1.8 | 5.4 | 1.1 | 9 | 15.6 | 0.7 | 7.2 | 14.4 | 4.7 | |||
| CONj | 0.007 | 0.017 | 0.049 | 0.013 | 0.082 | 0.144 | 0.007 | 0.062 | 0.129 | 0.153 | |||
|
| |||||||||||||
| TBM | NPf | 78.5 | 15.5 | 22.7 | 28.6 | 61.8 | 20.4 | 33.3 | 42.1 | 14.5 | 26.5 | 36.9 | 20.3 |
| ACCg | 0.29 | 0.216 | 0.322 | 0.389 | 0.53 | 0.274 | 0.419 | 0.494 | 0.209 | 0.366 | 0.472 | 0.325 | |
| COVh | 0.58 | 0.135 | 0.203 | 0.247 | 0.59 | 0.167 | 0.276 | 0.352 | 0.128 | 0.225 | 0.307 | 0.253 | |
| NNi | 2.7 | 4.8 | 8.5 | 3.6 | 11.9 | 19.1 | 2.5 | 9.5 | 16.6 | 8.4 | |||
| CONj | 0.024 | 0.045 | 0.072 | 0.033 | 0.102 | 0.163 | 0.024 | 0.082 | 0.143 | 0.105 | |||
|
| |||||||||||||
| FM | NPf | 6.2 | 9 | 13 | 15.9 | 5.8 | 9.9 | 15.1 | 18.8 | 7.8 | 13.3 | 17.8 | 14.5 |
| ACCg | 0.17 | 0.211 | 0.312 | 0.381 | 0.26 | 0.238 | 0.363 | 0.448 | 0.199 | 0.32 | 0.422 | 0.448 | |
| COVh | 0.11 | 0.164 | 0.245 | 0.299 | 0.13 | 0.179 | 0.277 | 0.353 | 0.151 | 0.24 | 0.332 | 0.267 | |
| NNi | 6.8 | 9.5 | 11.5 | 7.6 | 11.8 | 15.5 | 5.3 | 10.1 | 14.5 | 10.2 | |||
| CONj | 0.124 | 0.176 | 0.218 | 0.133 | 0.215 | 0.293 | 0.101 | 0.178 | 0.271 | 0.187 | |||
|
| |||||||||||||
| All | NPf | 84.3 | 15 | 21.8 | 27.6 | 64.7 | 18.8 | 30.5 | 38.4 | 13 | 24.3 | 34 | 17 |
| ACCg | 0.31 | 0.216 | 0.321 | 0.392 | 0.56 | 0.267 | 0.408 | 0.485 | 0.194 | 0.351 | 0.459 | 0.305 | |
| COVh | 0.64 | 0.138 | 0.205 | 0.252 | 0.64 | 0.167 | 0.273 | 0.347 | 0.121 | 0.219 | 0.304 | 0.311 | |
| NNi | 2.4 | 4.2 | 7.8 | 3.1 | 11 | 17.8 | 2.2 | 8.9 | 15.7 | 7.4 | |||
| CONj | 0.026 | 0.046 | 0.075 | 0.034 | 0.104 | 0.167 | 0.024 | 0.083 | 0.148 | 0.132 | |||
Contact predictions from multiple threading templates by LOMETS19.
Contact prediction from SVMSEQ16 with a cutoff of 6 A.
Contact prediction from SVMSEQ with a cutoff of 7 A.
Contact prediction from SVMSEQ with a cutoff of 8 A.
Contact prediction by taking consensus of predictions from CASP8 servers.
Total number of predictions
Accuracy of contact predictions: the number of correctly predicted contacts divided by the total number of contact predictions.
Coverage of contact predictions: the number of correctly predicted contacts divided by the number of contacts in the native structure.
Number of true-positive predictions which are not generated by the template-based predictions.
Coverage of novel predictions: NN divided by the number of contacts in the native structure.
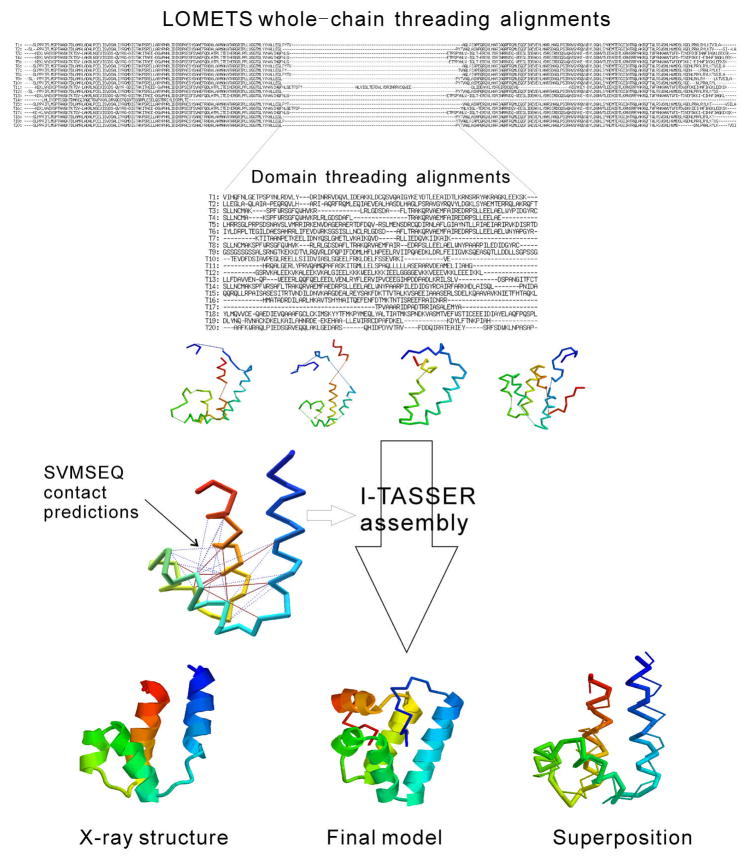
In Figure 2, we show one example of successful modeling on an FM target, T0416_2, by the I-TASSER server. I-TASSER first runs LOMETS on the whole chain (332 residues), which yields alignments dominated by 3crmA and 2qgnA. However, there is a middle region spanning 87 residues (L112-T198) that has no alignment with any of the top 20 templates. The server then automatically defines this region as a new domain and runs LOMETS again on the domain, which results in a number of weakly scoring hits. Although none of these templates for the small domain has a correct fold, some have close fragments, which provides building blocks for I-TASSER assembly (Row 3 of Figure 2). Out of the top 29 side-chain contact predictions by SVMSEQ, 13 (45%) are correct, covering 46% of all native contacts (Row 4 of Figure 2). Under the guidance of these restraints, I-TASSER finally assembles a model for T0416_2 (S124-K180, as defined by the assessors) with a RMSD=3.4 A and a TM-score=0.53.
Figure 2.
The procedure of the I-TASSER server in modeling a FM target of T0416_2. The upper part shows the top 20 alignments by LOMETS19 for the whole-chain sequence followed by the subsequent threading on the domain which was missed in the whole-chain threading. The examples of 4 templates closest to the target are shown in the third row. The fourth row shows the native backbone structure with inter-residue lines indicating the side-chain contact predictions by SVMSEQ16 (red solid lines are true-positive and green dashed lines are false-positive predictions). The domain modeling was done in the sequence (L112-T198) but the tails (L112-E125 and F192-T198 shown as backbones in the final models) are trimmed during docking with other parts of the structures. The superposition is made on S124-K180 according to the assessor’s definition of T0416_2. The image is generated by MVP40.
The accuracy of SVMSEQ predictions for HA/TBM targets is similar to that for FM ones. However, the coverage and accuracy of the contacts by LOMETS are much higher than SVMSEQ predictions for these targets. Nevertheless, SVMSEQ still generates a considerable number of correct contacts which cannot be generated by template-based predictions. The SVMSEQ-based Cα contact predictions with a 8 A cutoff, for example, provide 14.4% and 16.3% of new true-positive contact predictions for HA and TBM targets, respectively. These restraints are useful in modeling the regions lacking threading alignments as well as improving the global topology. It is worth mentioning that when we use the SVMSEQ-predicted contacts in the I-TASSER assembly, a large percentage of them are false positive. However, these false positive predictions do not necessarily affect the modeling of the regions with good templates because the consensus restraints from LOMETS are strong and dominating in those regions compared with the weak noise from SVMSEQ predictions. For the weakly aligned regions, however, the false-positive rate of SVMSEQ is lower than that of LOMETS, and therefore becomes helpful.
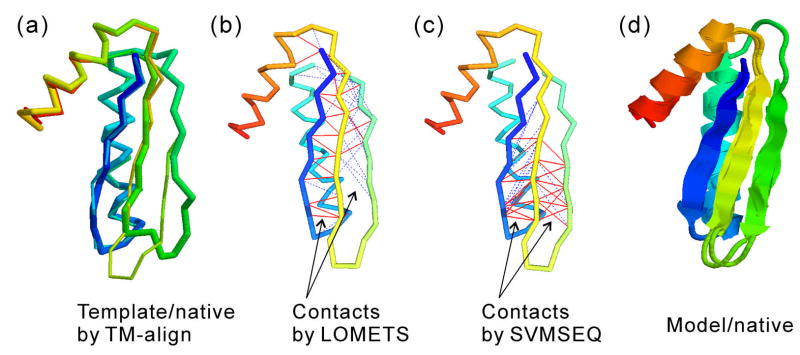
Figure 3 is one such example of a TBM-HA target, T0437_1, demonstrating the positive contribution of SVMSEQ to homology-based modeling. The LOMETS threading alignments are dominated by the template 2jz5A, which has a sequence identity of 32% to the target. The best threading alignment generated by HHsearch21 has an RMSD =2.30 A and TM-score =0.778. If we structurally align 2jz5A to the experimental structure by TM-align27, the RMSD is 1.34 A with TM-score=0.838 (Figure 3a). Although the global topology of 2jz5A matches the target well, there is a major mismatch in the region V49-T60 (the lower part of the second beta-sheet, Figure 3a). Correspondingly, there is no correct contact prediction from LOMETS in this region (Figure 3b). The sequence-based SVMSEQ contact prediction, however, generates 10 correct Cα contact predictions in this region (2 others are false positive, Figure 3c). These restraints help I-TASSER generate models with a correct beta-sheet structure in this region. The RMSD of the overall model is 1.13 A, which is even closer than the best structural alignment (Figure 3d). In this example, although the overall accuracy of the SVMSEQ prediction is still lower than LOMETS, the novel contacts from the sequence-based prediction improve the quality of local structures. In other regions (e.g. the N-terminal beta-sheet), SVMSEQ generates a number of false positive contact predictions. Since the LOMETS predictions provide strong consensus restraints, these weak false-positive predictions did not reduce the modeling accuracy in those regions.
Figure 3.
SVMSEQ contact predictions improve the modeling of T0437_1. (a) Structural superposition of the target (thin backbone) on the best template 2jz5A (thick backbone) with structural alignment generated by TM-align27 (RMSD =1.34A, TM-score =0.838). (b) Backbone structure of the native with lines between residues indicating Cα contact prediction from LOMETS19. Red solid lines are true-positive and green dashed ones are false-positive. There is no true-positive contact in the lower part of the second beta-hairpin. (c) Same as (b) but contacts are from SVMSEQ16 with 10 true-positive predictions in the lower part of the second beta-hairpin. (d) Superposition of the I-TASSER server model on the native with a RMSD =1.13 A and a TM-score=0.885. The image is generated by MVP40.
In the last column of Table II, we also list a consensus prediction taken from 6 CASP8 servers including LEE-SERVER, MULTICON-CMFR, MUProt, SAM-T08-2stage, RR_FANG_1, and Parings. A consensus contact is collected if it is predicted by more than half of the servers. These contacts were used in our human predictions. Somewhat unexpectedly, the consensus prediction from multiple servers does not outperform the prediction from the single program SVMSEQ. For FM targets, the consensus prediction has a slightly higher accuracy than SVMSEQ but a lower coverage. The overall accuracy of consensus contact prediction for all targets is lower than SVMSEQ but the coverage is similar. The SVMSEQ server also participated in CASP8 contact prediction33, but it submitted predictions obtained by combining results from SVMSEQ and LOMETS. Although this combination helps increase the accuracy for TBM/HA targets, it substantially decreases the accuracy of the original SVMSEQ predictions for FM targets, which was eventually assessed in the contact prediction section of CASP8.
Atomic-level structure refinement improves hydrogen-bonding networks
The SPICKER program26 clusters the structure decoys from I-TASSER and generates two types of reduced models: the cluster centroid (as ‘combo’) obtained by averaging the coordinates of all clustered decoys and the decoy closest to the centroid (as ‘closc’). Combo structures are usually closer to the native but have more structural clashes than the closc models. When constructing the full-atomic models, REMO17 has the advantage to eliminate clashes from combo and optimize the hydrogen-bonding network, over a number of other similar algorithms 34–36.
In Table III, we compare the REMO models of 149 domains (corresponding to 117 targets) with the full-atom models regenerated by Pulchra34 based on the same set of closc and combo models. The models of these 149 domains have been generated by the I-TASSER server without domain splitting, and we selected them for these comparisons so that we can eliminate the possible influence of the domain docking procedure. Clearly, the models by Pulchra based on combo have a better TM-score and HBscore compared with that on closc. But Pulchra could not remove the steric clashes in the combo models. Here, HBscore is defined as the number of H-bonds appearing in both model and native divided by that in the native structure, with H-bonds defined by HBPLUS 3.037. The final models generated by REMO have on average a better TM-score and HBscore than both the Pulchra models. The average number of steric clashes of the REMO models is 1.6, which is close to the average in the experimental structures in the PDB17.
Table III.
| RMSD | TM-score | HBscore (all-atom) | HBscore (backbone) | Nclash | |
|---|---|---|---|---|---|
| REMO+combo | 4.50 A | 0.725 | 0.496 | 0.643 | 1.6 |
| Pulchra+closc | 4.75 A | 0.708 | 0.380 | 0.520 | 3.5 |
| Pulchra+combo | 4.51 A | 0.716 | 0.390 | 0.531 | 34.3 |
Human and automated server predictions are consistent
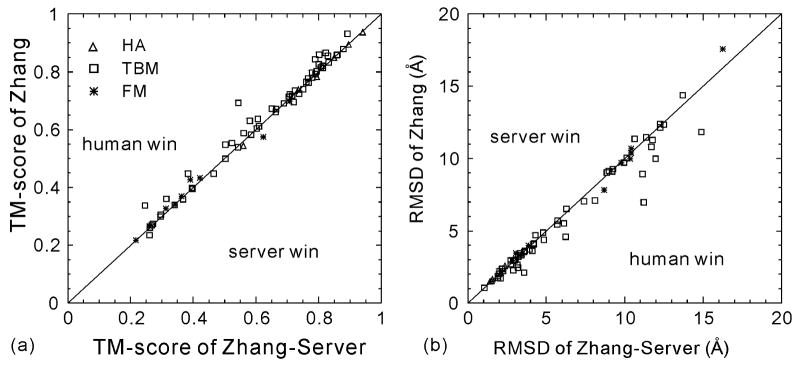
Figure 4 is a head-to-head comparison of Zhang-Server and Zhang in terms of TM-score and RMSD for the first models of 71 domains that have been tested in both the Server and the Human sections. There are slightly more targets with the human model having a higher TM-score than the server prediction, which results in a 1.8% overall increase in TM-score. Because the strategies of human and server predictions are identical, this difference reflects the gain from using multiple threading programs from other servers in addition to LOMETS. However, the “human-won” targets are mainly in the TBM and FM categories. For HA targets, the average TM-score of the server models is actually 0.6% higher than that of human-predicted models. This shows that at least for the easy targets, human interventions are not necessary.
Figure 4.
Comparison of the first models predicted by human (as “Zhang”) and server (as “Zhang-Server”) for all 164 domains.
What went wrong?
I-TASSER fails to select non-consensus correct folds
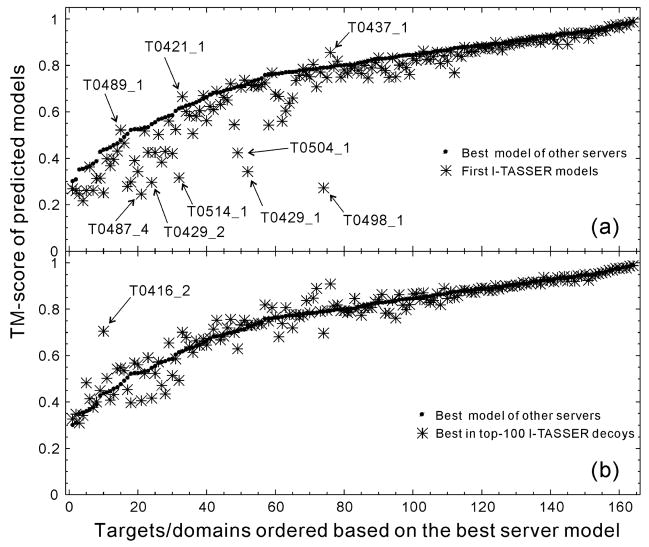
To help highlight the problems of the I-TASSER structure modeling and especially to identify the targets which I-TASSER failed to generate good models for, we use the best model generated by the servers in CASP8 other than Zhang-Server as the reference. All models were downloaded from http://predictioncenter.gc.ucdavis.edu/download_area/CASP8/server_predictions. In Figure 5a, we compare, for each target, the TM-score of the first model predicted by the I-TASSER server with that of the best model generated by other servers. Although there are several targets where I-TASSER generates better models than all others, the I-TASSER models are worse than the best models from other servers for most targets in the TBM/FM categories. The average TM-score of the I-TASSER models, calculated for all 164 domains, is 0.712 versus 0.765 for the best of other servers.
Figure 5.
TM-score of the I-TASSER server prediction (stars) in control with the best model (solid spheres) predicted by other servers in CASP8. (a) The first model by I-TASSER. (b) The best in top 100 models in I-TASSER simulation.
In Figure 5b, we list the best (by TM-score) of the top 100 (as ranked by SPICKER) models generated by the I-TASSER simulations with reference to the best models from other servers. These models were generated by I-TASSER but many of them were ranked low by SPICKER and not selected for submission. The average TM-score of these models is 0.765, equal to that of the best models by other servers. This difference highlights a major problem of the I-TASSER pipeline: the model selection. The top 100 I-TASSER models for each target are available at http://zhang.bioinformatics.ku.edu/casp8/decoys; these will serve as a benchmark set for the next stage model selection development.
I-TASSER builds models as guided by the consensus restraints from multiple threading templates. The consensus information is reinforced in the final step when the structures are clustered by SPICKER. These procedures are based on the assumption that a consensus template structure, ranked high by different scores of multiple threading programs, should be of better quality than those hit only by individual threading algorithms because there are much more ways for a threading program to pick up a wrong alignment than a right one6. For some targets, this assumption does not hold, and the selection based on consensus usually fails to select the correct fold. This turns out to be the major reason for the failure of I-TASSER model selection, especially for most of the cases highlighted in Figure 5a.
For example, T0498_1 is a designed protein which was designed to have a high sequence similarity (95%) with T0499_1, but to have a different fold, i.e. T0498_1 has a 3α fold while T0499_1 has an αβ fold38. Among all LOMETS programs, only MUSTER18 has a correct but weakly scoring hit on the template 2fs1A with a 3α conformation and a TM-score =0.67. However, because of the high sequence and profile similarity, the majority of the high-scoring alignments are with the αβ fold templates from 2igd, 1zxhA, 1mhxA, and 2i2yA. Thus, although I-TASSER did generate models with TM-score>0.70 in this case, the correct 3α fold was ranked low, and the selection preferred the incorrect αβ fold.
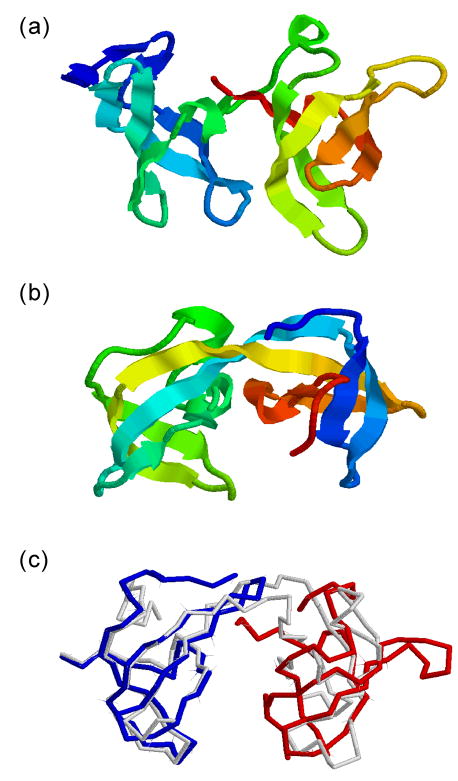
While T0498_1 is a special challenge for modeling and ranking which probably occurs very rarely in nature, T0504_1 is another example of a similar ranking problem. T0504 is a three-domain protein but I-TASSER modeled T0504_1 and T0504_2 together because these regions were aligned simultaneously. T0504_3 was successfully modeled, with the first model having an RMSD =1.77A. The best template for T0504_1 and T0504_2 is 2g3r which is hit only by HHsearch21, with a low rank. The majority of LOMETS programs detect 2gf7A as a template, which has a similar architecture of two domains, both having a two-beta-hairpin wound structure (Figure 6b). Interestingly, domains in 2gf7A swap one beta-hairpin with each other, which results in a different topology from T0504 (Figure 6a). This situation is similar to oligomer domain swapping39 but the swap here occurs within a single protein chain. This may reflect a new evolution mechanism where oligomer domain swapping is followed by gene fusion. Correspondingly, the first I-TASSER model has a similar architecture to the target (Figure 6c) but the TM-scores of both T0504_1 and T0504_2 are low because of the different orientation of the beta-hairpins.
Figure 6.
Structural modeling for T0504. (a) The experimental structure of the first two domains of T0504. (b) The template structure of 2gf7A detected by LOMETS which has the beta-hairpin swapped and may reflect a new evolution mechanism from the target. (c) Superposition of the native on the I-TASSER model (white backbone). The native structures of T0504_1 and T0504_2 are in blue and red. The architecture of the model and the native is similar but with different orientation of beta-hairpins.
T0514_1 is another example of I-TASSER ranking. The difference from T0499_1 and T0504_1 is that LOMETS has no strong hit on any of the templates. I-TASSER is usually good at assembling fragments from multiple weakly hit templates15. But in this example, the I-TASSER server failed to rank the best model as the first. The third submitted model has a TM-score =0.490 while the first model is a mirror image of the third model and has a TM-score =0.316 (see below).
Problem in domain splitting
Inappropriate domain assignment is the second major reason for the failure of I-TASSER modeling. This can happen in two scenarios. The first is when each individual domain has good templates from different proteins but the threading programs fail to detect them when whole-chain sequences are used. The difficulty in this scenario is that we do not have an efficient algorithm for domain prediction. One such case is T0429, which is a two-domain protein. The first domain T0429_1 has an alignment with template 2f5kA hit by HHsearch with a TM-score=0.85, and the second domain T0429_2 has a hit from 1oi1A by MUSTER with a TM-score=0.47. However, because of the failure of domain splitting, I-TASSER attempted to fold the target based on ab initio modeling, which resulted in models significantly worse than the best model by other servers which was based on the correct templates (Figure 5a).
The second scenario occurs when one of multiple domains has no strong alignment while other domains have strong templates. If we model the target as a whole chain, the final clustering will be dominated by the well-aligned regions, which will result in the weakly-aligned domains having insufficient sampling because the structures of those domains are more diverse. One such example is T0487 which is a 685-residue target consisting of 5 domains. The sequences of all 5 domains are strongly aligned with the template 1yvuA, except for T0487_4 which is a 87-residue domain (S178-V264) with no correct alignment with 1yvuA. Because the target is big, I-TASSER does not have sufficient sampling in this region, and the SPICKER clustering is dominated by the other well-aligned regions. As a result, the model of T0487_4 has a much worse quality than the best of other servers which obviously split the target into domains and hit the correct templates (1r4kA and 1si2A) for this domain (information obtained from the head of the models). This problem was noticed in the CASP7 experiment30 and we have attempted to split the sequence into domains and model the domains separately. However, this does not always work better than folding the whole-chain sequence because the corresponding chain connectivity restraints and interactions with partner domains are lost in the individual domain modeling. One solution to the problem may be to fold the easy domains first and then fold the remaining domains while keeping the structures of the other domains frozen.
Potential function fails to recognize mirror image fold for FM targets
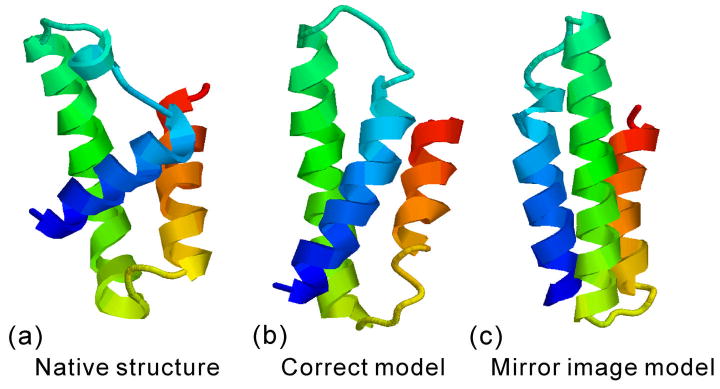
The predicted distance map and contact restraints have no ability to distinguish mirror image structures because both the right model and the mirror can satisfy the restraints equally well. This is one of the problems of I-TASSER in free modeling when the models are generated from scratch and no template can be used to guide the model selection. T0405_1 is one such example, which is the first domain (N2-E73) of a two-domain target T0405 (Figure 7). The I-TASSER server correctly recognized the target as having two domains but incorrectly split the first domain as M1-L101. As expected, the accuracy of the contact predictions from LOMETS is low (11% for side-chain and 0% for Cα contacts, see Table I); but SVMSEQ predictions have an accuracy of 25% for side-chain contacts and 20% for Cα contacts. The I-TASSER server generated two types of models for T0405_1 which are mirror images of each other with a distance-RMSD=2.1 A (Figures 7b and 7c). But the incorrect mirror image was finally picked up by SPICKER (Figure 7c). There are several other big, hard targets where the mirror image structure was also ranked higher than the correct one. For example, in the above-mentioned target T0514, which is a 154-residue protein with a beta-sandwich topology, I-TASSER ranks the mirror image structure as the first model and the one with the correct image as the third.
Figure 7.
The I-TASSER modeling for T0405_1 (a), where the mirror image structure (c) is ranked higher than the correct model (b).
CONCLUSIONS
The I-TASSER pipeline was tested in the CASP8 experiment. The success mainly comes from the fact that the algorithm manages to make use of information from multiple templates to assemble models with an optimized knowledge-based potential25 to accommodate the global and local structural packing. The multiple template information is represented in I-TASSER as consensus spatial restraints and rigid structural fragments. The consensus restraints have a similar accuracy to those from the top individual templates but cover a larger portion of the structure and a larger fraction of native contacts. The rigid structure fragments excised from the PDB template structures help reduce the entropy of the conformational search and increase the fidelity of local structures. Encouragingly, the procedure has been made fully automated and generates models with a quality close to the human predictions for at least close homology modeling.
For the first time, the sequence-based contact predictions from machine-learning techniques16 are found helpful in both TBM and FM 3D structure assembly. In TBM, although the overall accuracy is most desirable, the key factor that determines the usefulness of the de novo contact predictions is the complementarity to the template-based predictions, that is, only those contacts that are novel relative to the templates are essential. The false-positive predictions in the well-aligned regions are mostly neutralized by the strong template-based restraints. However, special treatment of the false-positive predictions, e.g. removing the sequence-based contacts involving the well-aligned regions while keeping those in weakly aligned or unaligned regions, may further eliminate possible side effects of the de novo contact predictions in TBM. Progress has also been made in atomic-level structural refinement which optimizes the hydrogen-bonding network and improves local structural packing17.
Nevertheless, one of the major issues of the current I-TASSER approach lies in the selection of correct models. This is especially the case when the best templates are hit only by a minority of threading algorithms and ranked low in the scoring function. External statistical and physics-based atomic potentials may be borrowed to deal with this issue in combination with the I-TASSER potentials and SPICKER clustering. Another related issue is the mirror image recognition for free modeling, for which chirality-dependent energy terms need to be introduced in I-TASSER. Finally, incorrect domain splitting turns out to be the major issue influencing the quality of the I-TASSER models for multiple-domain targets. Since both separate domain modeling and simultaneous modeling of multiple domains have defects, i.e. individual domain modeling misses the restraint information from partners while simultaneous modeling suffers from insufficient sampling for small and weakly aligned domains, one solution may be to model the domain structures in a sequential order while keeping the other domains frozen. All these issues highlighted in the CASP8 experiment will be of highest priorityin the development of the next generation of I-TASSER.
Acknowledgments
The author thanks Drs. S. Wu, Y. Li and A. Roy for assistance in CASP8, Dr. A. Szilagyi for reading the manuscript. The project is supported in part by the Alfred P. Sloan Foundation, NSF Career Award (DBI 0746198), and the National Institute of General Medical Sciences (R01GM083107).
References
- 1.Murzin AG, Bateman A. CASP2 knowledge-based approach to distant homology recognition and fold prediction in CASP4. Proteins. 2001;(Suppl 5):76–85. doi: 10.1002/prot.10037. [DOI] [PubMed] [Google Scholar]
- 2.Ginalski K, Rychlewski L. Protein structure prediction of CASP5 comparative modeling and fold recognition targets using consensus alignment approach and 3D assessment. Proteins. 2003;53 (Suppl 6):410–417. doi: 10.1002/prot.10548. [DOI] [PubMed] [Google Scholar]
- 3.Das R, Qian B, Raman S, Vernon R, Thompson J, Bradley P, Khare S, Tyka MD, Bhat D, Chivian D, Kim DE, Sheffler WH, Malmstrom L, Wollacott AM, Wang C, Andre I, Baker D. Structure prediction for CASP7 targets using extensive all-atom refinement with Rosetta@home. Proteins. 2007;69(S8):118–128. doi: 10.1002/prot.21636. [DOI] [PubMed] [Google Scholar]
- 4.Baker D, Sali A. Protein structure prediction and structural genomics. Science. 2001;294(5540):93–96. doi: 10.1126/science.1065659. [DOI] [PubMed] [Google Scholar]
- 5.Skolnick J, Fetrow JS, Kolinski A. Structural genomics and its importance for gene function analysis. Nat Biotechnol. 2000;18(3):283–287. doi: 10.1038/73723. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y. Progress and challenges in protein structure prediction. Current opinion in structural biology. 2008;18(3):342–348. doi: 10.1016/j.sbi.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic acids research. 2005;33(Web Server issue):W244–248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nature protocols. 2009;4(3):363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Y. Protein structure prediction: When is it useful? Corr Opin Struct Biol. 2009 doi: 10.1016/j.sbi.2009.02.005. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kryshtafovych A, Fidelis K, Moult J. Progress from CASP6 to CASP7. Proteins. 2007;69 (Suppl 8):194–207. doi: 10.1002/prot.21769. [DOI] [PubMed] [Google Scholar]
- 12.Battey JN, Kopp J, Bordoli L, Read RJ, Clarke ND, Schwede T. Automated server predictions in CASP7. Proteins. 2007;69(S8):68–82. doi: 10.1002/prot.21761. [DOI] [PubMed] [Google Scholar]
- 13.Kopp J, Bordoli L, Battey JN, Kiefer F, Schwede T. Assessment of CASP7 predictions for template-based modeling targets. Proteins. 2007;69(S8):38–56. doi: 10.1002/prot.21753. [DOI] [PubMed] [Google Scholar]
- 14.Jauch R, Yeo HC, Kolatkar PR, Clarke ND. Assessment of CASP7 structure predictions for template free targets. Proteins. 2007;69(S8):57–67. doi: 10.1002/prot.21771. [DOI] [PubMed] [Google Scholar]
- 15.Wu S, Skolnick J, Zhang Y. Ab initio modeling of small proteins by iterative TASSER simulations. BMC biology. 2007;5:17. doi: 10.1186/1741-7007-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu S, Zhang Y. A comprehensive assessment of sequence-based and template-based methods for protein contact prediction. Bioinformatics (Oxford, England) 2008;24(7):924–931. doi: 10.1093/bioinformatics/btn069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li YQ, Zhang Y. REMO: A new protocol to refine full atomic protein models from C-alpha traces by optimizing hydrogen-bonding networks. Proteins. 2009 doi: 10.1002/prot.22380. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu ST, Zhang Y. MUSTER: Improving Protein Sequence Profile-Profile Alignments by Using Multiple Sources of Structure Information. Proteins. 2008 doi: 10.1002/prot.21945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu ST, Zhang Y. LOMETS: A local meta-threading-server for protein structure prediction. Nucl Acids Res. 2007;35:3375–3382. doi: 10.1093/nar/gkm251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi J, Blundell TL, Mizuguchi K. FUGUE: sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. Journal of molecular biology. 2001;310(1):243–257. doi: 10.1006/jmbi.2001.4762. [DOI] [PubMed] [Google Scholar]
- 21.Soding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21(7):951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 22.Xu Y, Xu D. Protein threading using PROSPECT: design and evaluation. Proteins. 2000;40(3):343–354. [PubMed] [Google Scholar]
- 23.Zhou H, Zhou Y. Fold recognition by combining sequence profiles derived from evolution and from depth-dependent structural alignment of fragments. Proteins. 2005;58(2):321–328. doi: 10.1002/prot.20308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y, Skolnick J. Automated structure prediction of weakly homologous proteins on a genomic scale. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:7594–7599. doi: 10.1073/pnas.0305695101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, Kolinski A, Skolnick J. TOUCHSTONE II: A new approach to ab initio protein structure prediction. Biophysical journal. 2003;85:1145–1164. doi: 10.1016/S0006-3495(03)74551-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, Skolnick J. SPICKER: A clustering approach to identify near-native protein folds. Journal of computational chemistry. 2004;25(6):865–871. doi: 10.1002/jcc.20011. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y, Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic acids research. 2005;33(7):2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y, Hubner I, Arakaki A, Shakhnovich E, Skolnick J. On the origin and completeness of highly likely single domain protein structures. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:2605–2610. doi: 10.1073/pnas.0509379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen H, Zhou HX. Prediction of solvent accessibility and sites of deleterious mutations from protein sequence. Nucleic acids research. 2005;33(10):3193–3199. doi: 10.1093/nar/gki633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y. Template-based modeling and free modeling by I-TASSER in CASP7. Proteins. 2007;69(S8):108–117. doi: 10.1002/prot.21702. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y, Skolnick J. Scoring function for automated assessment of protein structure template quality. Proteins. 2004;57:702–710. doi: 10.1002/prot.20264. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y, Skolnick J. The protein structure prediction problem could be solved using the current PDB library. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:1029–1034. doi: 10.1073/pnas.0407152101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu ST, Zhang Y. Protein residue contact prediction by SVMSEQ and LOMETS servers. CASP8 Abstract. 2008:114. [Google Scholar]
- 34.Rotkiewicz P, Skolnick J. Fast procedure for reconstruction of full-atom protein models from reduced representations. Journal of computational chemistry. 2008;29(9):1460–1465. doi: 10.1002/jcc.20906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petrey D, Xiang Z, Tang CL, Xie L, Gimpelev M, Mitros T, Soto CS, Goldsmith-Fischman S, Kernytsky A, Schlessinger A, Koh IY, Alexov E, Honig B. Using multiple structure alignments, fast model building, and energetic analysis in fold recognition and homology modeling. Proteins. 2003;53 (Suppl 6):430–435. doi: 10.1002/prot.10550. [DOI] [PubMed] [Google Scholar]
- 36.Holm L, Sander C. Database algorithm for generating protein backbone and side-chain co-ordinates from a C alpha trace application to model building and detection of co-ordinate errors. Journal of molecular biology. 1991;218(1):183–194. doi: 10.1016/0022-2836(91)90883-8. [DOI] [PubMed] [Google Scholar]
- 37.McDonald IK, Thornton JM. Satisfying hydrogen bonding potential in proteins. Journal of molecular biology. 1994;238(5):777–793. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
- 38.He Y, Chen Y, Alexander P, Bryan PN, Orban J. NMR structures of two designed proteins with high sequence identity but different fold and function. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(38):14412–14417. doi: 10.1073/pnas.0805857105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bennett MJ, Schlunegger MP, Eisenberg D. 3D domain swapping: a mechanism for oligomer assembly. Protein Sci. 1995;4(12):2455–2468. doi: 10.1002/pro.5560041202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu D, Zhang Y. MVP: Macromolecular Visualization and Processing. http://zhang.bioinformatics.ku.edu/MVP.