Introduction
During mitosis, kinetochores form persistent attachments to microtubule tips and undergo corrective detachment in response to phosphorylation by Ipl1 (Aurora B) kinase1. The Dam1 complex is required to establish and maintain bioriented attachment to microtubule tips in vivo, and it contains multiple sites phosphorylated by Ipl12-10. Moreover, a number of kinetochore-like functions can be reconstituted in vitro with pure Dam1 complex11-14. These functions are believed to derive from the ability of the complex to self-assemble into rings12, 13, 15-17. Here we show that rings are not necessary for dynamic microtubule attachment, Ipl1-dependent modulation of microtubule affinity, or the ability of Dam1 to move processively with disassembling microtubule tips. Using two fluorescence-based assays, we found that the complex exhibited a high affinity for microtubules (Kd ∼6 nM) that was reduced by phosphorylation at Ser 20, a single Ipl1 target residue in Dam1. Moreover, individual complexes undergo one-dimensional diffusion along microtubules and detach 2.5-fold more frequently after phosphorylation by Ipl1. Particles consisting of 1 to 4 Dam1 complexes – too few to surround a microtubule – were captured and carried by disassembling tips. Thus, even a small number of binding elements could provide a dynamic, phosphoregulated microtubule attachment and thereby facilitate accurate chromosome segregation.
Results and Discussion
Purified recombinant Dam1 complex possesses many properties important for kinetochore-microtubule attachment. Similarly to kinetochores, the complex tracks with the tips of both growing and shortening microtubules, even under tensile forces up to 3 pN11-13. It also modulates filament dynamics in a tension-dependent manner14. When mixed with stabilized microtubules the complex self-assembles into rings, each composed of 16 to 25 individual heterodecamers encircling a filament12, 13, 15-17. This discovery has led to an attractive model, now widely embraced18-20, in which rings function as sliding clamps that allow kinetochores to maintain dynamic attachments to microtubule tips. Theoretical considerations suggest that the ring mechanism is an ideal way to harness microtubule disassembly for the production of force and movement21-24. However, direct evidence that kinetochores in vivo contain rings25, 26, or even that the in vitro kinetochore-like functions of the Dam1 complex require rings, is lacking.
We have developed two methods using fluorescence microscopy to quantify the interaction between the Dam1 kinetochore complex and microtubule filaments. First we constructed a fluorescent version of the Dam1 complex. Previous work showed that a carboxy-terminal GFP tag on Dad1 of the Dam1 complex has no detectable phenotype27. We added this GFP tag into the polycistronic vector expressing the Dam1 complex components15 and purified the complex. In the resulting complex, Dad1-GFP is present at stoichiometric levels (Fig. 1a). The GFP-tagged complex migrates as a single peak on gel filtration with a Stoke's radius of 78 Å (Fig. 1b), which is slightly larger than that reported for unlabelled complex (76 Å)15. The purified complex forms rings on microtubules and tracks the tips of depolymerizing microtubules (T. Gonen and C. L. Asbury, unpublished data).
Figure 1. Purification of the GFP-tagged Dam1 Complex.
(a) The relative abundance of each component of the Dam1 complex was similar for the wild type (WT) and GFP-tagged complexes. The components of the complexes were separated by SDS-PAGE and visualized by Coomassie brilliant blue staining. The left gel (14% acrylamide) shows separation of the small components of the Dam1 complex. The right gel (8% acrylamide) shows separation of Dad1-GFP from Dam1. (b) During gel filtration, the GFP-tagged Dam1 complex elutes in a single peak with an apparent Stoke's radius of 78 Å (solid trace). The elution profile for a mixture of standard proteins is shown for comparison (dotted trace: ferritin, 61 Å; aldolase, 48 Å; RNAse A, 18 Å).
Previously, the affinity of the Dam1 complex for microtubules was estimated using western blot analysis to measure the amount of complex that co-pellets with microtubules. These assays tested concentrations of microtubules ranging from 0.1−5 μM12, 28. We measured the binding of the GFP-tagged complex to microtubules using quantitative fluorescence microscopy and detected binding at 2.5 nM microtubules (Fig. 2a-c), well below the detection limit of the previous analysis. The increased sensitivity of our assay allows us to measure binding at low microtubule concentrations, thereby avoiding inhibition by unpolymerized tubulin.
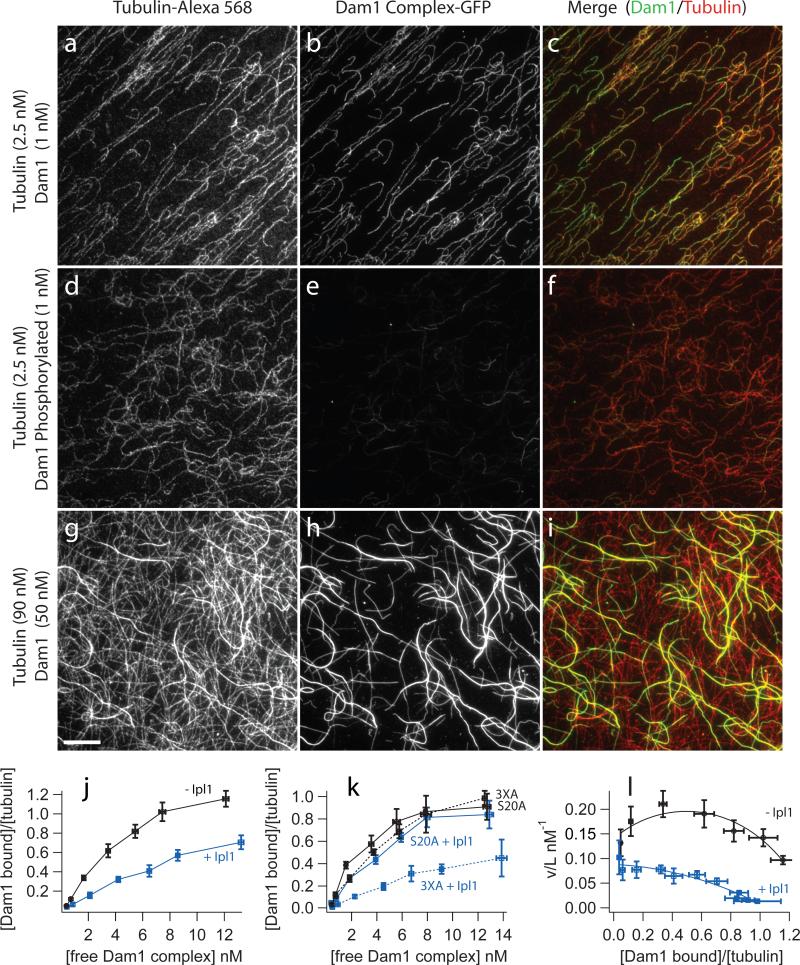
Figure 2. Phosphorylation of Dam1 Ser 20 by Ipl1 decreases microtubule affinity of the Dam1 complex.
Binding of GFP-tagged Dam1 complex to microtubules assembled from Alexa-568 labeled-tubulin was recorded by fluorescence microscopy as described in the Methods. (a-f) Representative images of Dam1 complex (1 nM) either unphosphorylated (a-c) or phosphorylated by Ipl1 (d-f) bound to taxol-stabilized microtubules (2.5 nM tubulin dimer). Thousands of such images (n>2000) were analyzed to generate the binding curves shown below (j-l). (g-i) Images of higher concentrations of the Dam1 complex (50 nM) binding to microtubules (at 90 nM tubulin dimer) highlight the cooperativity of binding. The scale bar in (g) applies to panels (a-i) and represents 10 μm. The intensities of images in a given column were scaled the same. (j) Plot of binding data with binding density, ν, on the vertical axis plotted against free ligand concentration, L. Unphosphorylated Dam1 complex (black symbols), Dam1 complex phosphorylated by Ipl1 (blue symbols). Data are mean ± s.e.m. from 8−11 (unphosphorylated) and 4−10 (phosphorylated). (k) Plot comparing microtubule binding of mutant Dam1 complexes with Ala substitutions at residues targeted by Ipl1. Even when phosphorylation was blocked at three of four target residues (3XA), the affinity of the complex for microtubules is severely reduced after Ipl1 treatment (blue dotted curve), compared with the untreated control (black dotted curve). Conversely, when phosphorylation is blocked at a single Ser residue (S20A), Ipl1 treatment caused no significant reduction in binding (blue solid curve) relative to an untreated control (black solid curve). Data are mean ± s.e.m. from three replicates. (l) Scatchard plot of the same data shown in (j), unphosphorylated Dam1 complex (black symbols), Dam1 complex phosphorylated by Ipl1 (blue symbols). The curves represent the fit to the McGhee and von Hippel model30.
Increasing amounts of Dam1 complex were bound to a constant concentration of taxol-stabilized microtubules. A standard plot of binding density (Dam1 complex bound/tubulin dimer) versus the concentration of free Dam1 complex (Fig. 2j) shows half-maximal binding occurs at 3.5 nM Dam1 complex. A Scatchard plot of the same data (Fig. 2l) was non-linear with a downward curvature, which indicates positive cooperativity29.
A model to describe cooperative binding of a ligand to a linear lattice was developed previously30. This approach provides a measure of the apparent affinity (Kd) of the ligand for the lattice, the number of lattice subunits bound per ligand (n), and a measure (w) of the cooperativity between ligands. Fitting our binding data to this model gave a Kd of 6.2 ± 0.9 nM, an n of 0.74 ± 0.045 tubulin dimers per Dam1 complex and w of 2.3 ± 0.35 (Fig. 2l). The value for n suggests 18 Dam1 complexes would complete a ring around a 13 protofilament microtubule, consistent with that previously reported (16−25)13, 16. The parameter w is the increase in affinity gained by a Dam1 complex binding to the lattice next to another Dam1 complex, compared with binding to a bare lattice; a value of 2.3 indicates positive cooperativity. Images of the Dam1 complex bound at high microtubule concentrations highlight its cooperative-binding characteristics; some microtubules seemed to be completely decorated, whereas others in the same field were only sparsely labeled (Fig. 2g-i). Together, these results show that the Dam1 complex binds to microtubules cooperatively, with a Kd 30-fold higher than previously reported12.
In vivo, phosphorylation by Ipl1 causes detachment of aberrantly attached kinetochores, but the mechanism for this corrective detachment is unknown31. Using our quantitative epifluorescence assay, we found that phosphorylation of the wild-type Dam1 complex reduced its Kd for microtubules (Fig. 2a-f,j). A Scatchard plot (Fig. 2l) of this data showed a nearly linear trend, indicating reduced cooperativity, compared with the unphosphorylated complex. Application of the McGhee and von Hippel model yields a Kd of 10.8 ± 1.4 nM Dam1 complex, an n of 0.96 ± 0.05 and w of 1.4 ± 0.22, indicating some positive cooperativity. Thus, phosphorylation decreases affinity and cooperativity.
Mutation of the four Ipl1 phosphorylation sites on Dam1 is lethal and three of these sites cluster on the C-terminal end of Dam1 (S257, S265 S292)9. Previous work showed that replacing all four Ipl1 target residues with Asp, a phosphomimetic amino acid, decreases affinity for the Ndc80 complex, whereas replacing just the C-terminal targets decreases the tendency for self-assembly with no apparent affect on microtubule affinity17, 32. We used two different mutant Dam1 complexes to determine which sites were required for the decreased affinity and cooperativity of binding that we observed with Ipl1 phosphorylation. In the first mutant, the three C-terminal Ipl1 target sites in Dam1 were mutated to alanine (Dam1 3XA) to block phosphorylation. In the second mutant, Ser 20 alone (the fourth target site) was mutated to Ala (Dam1 S20A). The binding affinities of these mutant complexes were determined in the absence and presence of Ipl1 phosphorylation. Surprisingly, preventing phosphorylation of S20 blocked Ipl1 regulation of microtubule binding, whereas preventing phosphorylation of all three C-terminal sites has no effect on Ipl1 regulation (Fig. 2k). Thus, the binding of the Dam1 complex to microtubules is regulated by Ipl1 phosphorylation of a single residue, Ser 20, in the aminoterminus of Dam1.
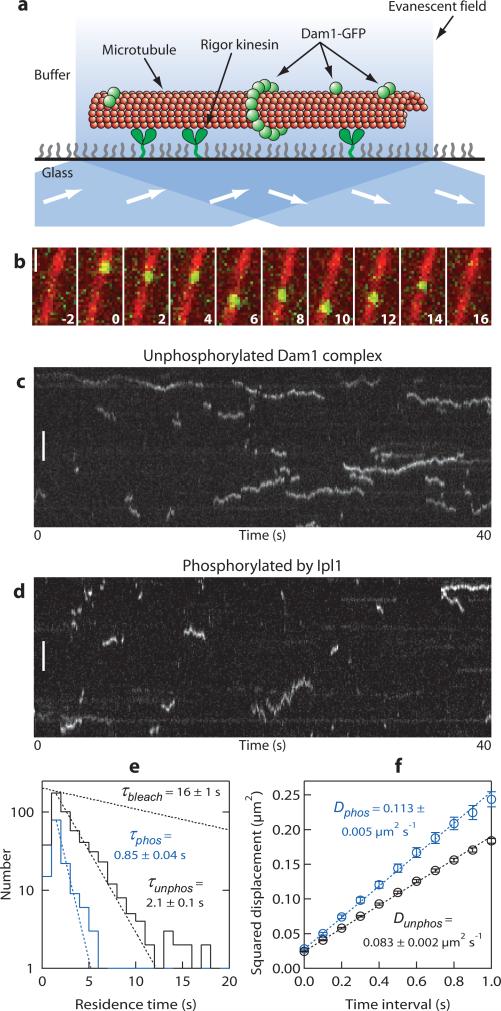
To investigate the effects of phosphorylation on microtubule binding of the Dam1 complex at concentrations below those accessible by epifluorescence, we developed an assay using total internal reflection fluorescence (TIRF) microscopy (Fig. 3a). The TIRF assay allows binding and movement of single GFP-tagged molecules on individual microtubules to be observed at high temporal resolution (here, 10 frames per second). At a concentration of 40 pM, individual particles composed of either phosphorylated or unphosphorylated Dam1 complexes bound transiently to taxol-stabilized microtubules. The particles diffused along the microtubule lattice and occasionally coalesced into brighter diffusing oligomers (Fig. 3b-d; Supplemental Information, Movies 1,2), consistent with previous observations at higher concentrations of the complex13. Occasionally, diffusing particles appeared to transfer from one microtubule to another at locations where the filaments overlapped (for example, see Supplemental Information, Movie 3). Long-duration events (> 5 s) were more common with the unphosphorylated Dam1 complex than with the phosphorylated complex (compare Fig. 3c with 3d). Residence times for the particles were distributed exponentially (Fig. 3e). The mean residence time for unphosphorylated particles (τunphos) was 2.1 ± 0.1 s (n = 514), which is significantly longer than that for Ipl1-phosphorylated complex ( τphos of 0.85 ± 0.04 s n = 145), indicating that phosphorylation decreases their affinity for microtubules. The mean squared displacement for both types of particles increased linearly with time (Fig. 3f), as expected for a random diffusive process. From the slope of each plot, the average diffusion coefficient for unphosphorylated particles (Dunphos) was 0.083 ± 0.001 μm2 s−1, which is significantly lower than that for phosphorylated particles (Dphos of 0.113 ± 0.002 μm2 s−1). These data show that Ipl1 phosphorylation directly weakens the interaction between the Dam1 complex and microtubules.
Figure 3. Ipl1 phosphorylation speeds lattice diffusion and decreases the residence time of the Dam1 complex on microtubules.
(a) Schematic representation of the TIRF assay developed to observe the dynamics of GFP-tagged Dam1 complexes (green spheres) interacting with individual microtubules (red). Microtubules were immobilized above the glass surface by an engineered ‘rigor’ kinesin mutant that binds tightly to microtubules38. Excitation by total internal reflection, in combination with a surface-blocking treatment that prevents non-specific adsorption, allowed movement of individual Dam1 complexes to be recorded in the evanescent field. (b) Selected frames from a Supplemental Information, Movie 4 showing one-dimensional diffusion of the Dam1 complex (green) along a taxol-stabilized microtubule (red). Time, s; scale bar, 1 μm. (c, d) Kymographs showing the effect of Ipl1 phosphorylation on microtubule-binding and one-dimensional diffusion of the Dam1 complex (total concentration, 40 pM). Position along the microtubule is depicted on the vertical axis while time changes along the horizontal axis. Long-duration events (> 5 s) are more common with unphosphorylated complex (c), compared with the phosphorylated complex (d). Scale bars, 3 μm. (e) Residence-time distributions for unphosphorylated (black histogram, n = 514 binding events on 14 microtubules, 8 recordings) and Ipl1 phosphorylated complex (blue histogram, n = 145 events, 10 microtubules, 5 recordings). Dotted lines show exponential fits used to determine mean residence times, τphos and τunphos. For comparison, the exponential distribution of bleach times for single GFP molecules recorded under identical experimental conditions is also depicted (τbleach, from Supplementary Information, Fig. S1). (f) While attached to microtubules, the phosphorylated complex diffused more rapidly than the unphosphorylated complex. Mean squared displacement is plotted against time for unphosphorylated (black symbols) and phosphorylated (blue symbols) Dam1 complex. Dotted lines show linear fits used to determine diffusion coefficients, Dphos and Dunphos. Data are mean ± s.e.m., computed from 514 and 145 records of unphosphorylated and phosphorylated complex, respectively.
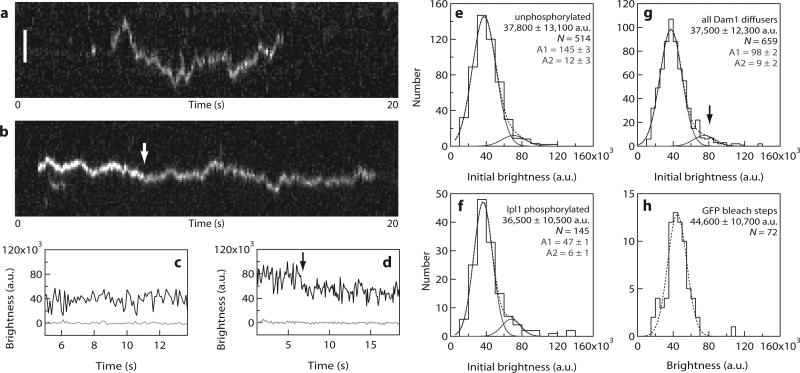
Fluorescence intensities of the individual particles diffusing on stabilized microtubules (Fig. 4) were quite dim and inconsistent with the brightness that would be expected for rings composed of 16 or more GFP-tagged Dam1 complexes. Most particles disappeared in a single step, which probably represents detachment of an individual Dam1 complex (Fig. 4a, c). However, many of the brighter particles showed stepwise decreases in intensity while they remained attached to the microtubule (Fig. 4b, d), consistent with photobleaching of two GFP molecules sequentially. At the low concentrations used here (40 pM) the distributions of initial brightness values for unphosphorylated and phosphorylated particles were not significantly different (Fig. 4e, f). This indicates that the measured phosphorylation-dependent differences in residence time and diffusion coefficient (Fig. 3e, f) are not caused by differences in oligomeric state. The combined initial brightness-distribution for all particles was fitted by the sum of two Gaussians (Fig. 4g), corresponding to a large population (90%) with a unitary brightness of 37,500 ± 12,300 arbitrary units (a.u., mean ± s.d.) and a small population (10%) with twice the brightness (75,000 ± 12,300 a.u.). The unitary brightness is consistent with that of single GFP molecules (44,500 ± 10,900 a.u.), measured in control experiments where GFP-MCAK was bound tightly to microtubules under otherwise identical conditions (Fig. 4h). These data establish that a single Dam1 complex is sufficient to form a dynamic attachment to the microtubule lattice.
Figure 4. Single Dam1 complexes are sufficient to form a dynamic microtubule attachment.
(a, b) Kymographs showing the binding and one-dimensional diffusion of individual particles, each composed of one or more Dam1 complexes, along taxol-stabilized microtubules. Position along the microtubule is depicted on the vertical axis while time changes along the horizontal axis. Scale bar 2 μm. (c, d) Records of brightness versus time for the particles shown in a, b. The brightness of most diffusing particles was roughly constant over time (as in a, c). However, some particles exhibited stepwise reductions in intensity during the interval when they remained attached to the microtubule (b, d, arrows), consistent with photobleaching of individual GFP molecules. (e-g) Distributions of initial brightness values for Dam1 particles diffusing on taxol-stabilized microtubules, each fit by the sum of two Gaussians (dotted lines). Distributions for unphosphorylated (e) and Ipl1 phosphorylated (f) Dam1 particles were not significantly different. Both datasets are pooled in (g), which shows a large population (90% of particles) with a unitary brightness of 37,500 ± 12,300 a.u. (mean ± s.d.) plus a small population (10%) with twice the brightness, 75,000 ± 12,300 a.u. (n = 659 events, 24 microtubules, 13 recordings). The unitary brightness exhibited by most diffusing particles was close to that of single GFP molecules, indicating that oligomerization of many Dam1 complexes (for example, into a ring encircling the microtubule) is not required for binding and diffusion on the microtubule lattice. (h) Control data showing the distribution of brightness values for single GFP molecules (n = 72 bleach events from 5 recordings) fused to the kinesin-family motor, MCAK, which was bound tightly to microtubules (see also Supplementary Information, Fig. S1). The experimental conditions were otherwise identical to those in (a-g). The dotted line shows a single Gaussian fit, which indicates that under these conditions one GFP molecule corresponds to a brightness of 44,600 ± 10,700 a.u.
The ability of single Dam1 complexes to diffuse along a microtubule suggests that rings may also be unnecessary for processive movement driven by microtubule depolymerization. To test this hypothesis, we modified our TIRF assay to study interactions with dynamic rather than stabilized microtubules. Short, GMPCPP-stabilized microtubule seeds were anchored to a coverslip and the Dam1 complex was then introduced along with free tubulin and GTP, to allow microtubule growth. The affinity of the complex for microtubules was decreased significantly by the free tubulin; a 25-fold higher concentration was required to achieve binding levels similar to those seen with taxol-stabilized microtubules (1 nM versus 40 pM). Notably, the short seeds were decorated much more brightly with Dam1 than the dynamic extensions grown from them (Fig. 5a and Supplemental Information, Movies 5-7), confirming earlier observations showing a higher affinity for GMPCPP-containing microtubule segments12. After microtubule-extensions of several micrometers were grown, the free tubulin level was lowered by buffer-exchange to induce disassembly.
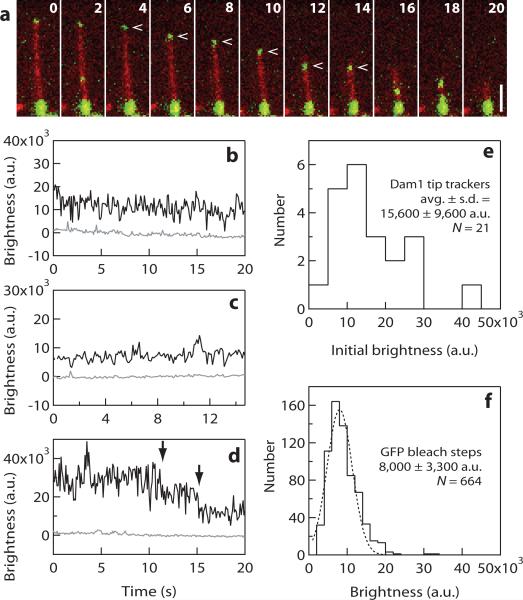
Figure 5. Rings are not required for processive disassembly-driven movement of the Dam1 complex.
(a) Selected frames from Supplemental Information, Movie 6 showing processive movement of the Dam1 complex (upper green particle, indicated by arrow) driven by the disassembling tip of an individual microtubule (red). The lower portion of the microtubule, which corresponds to a GMPCPP-stabilized segment of the filament, is also brightly decorated with Dam1-GFP. Elapsed times are in seconds. Scale bar, 2 μm. (b-d) Records of brightness versus time for several particles that exhibited processive disassembly-driven movement. The brightness of some particles was roughly constant over time (as in b and c), whereas others showed a stepwise loss of brightness consistent with photobleaching of single GFP molecules (arrows in d). (e) Distribution of initial brightness values for individual particles that exhibited disassembly-driven movement before additional particles accumulated at the disassembling microtubule tip. Initial brightness values for most particles (19 out of 21) were between 5,000 and 30,000 a.u., a range corresponding to 1−4 GFP molecules. This result indicates that oligomerization into a ring is not required for dissasembly-driven movement of the Dam1 complex, as many more than 4 complexes are required to form a complete ring encircling the microtubule12, 16. (f) Control data showing the distribution of brightness values for single GFP molecules (histogram, n = 664 bleach events from 38 recordings) fused to MCAK, which was bound tightly to microtubules. The experimental conditions were otherwise identical to those in (a-e). The dotted line shows a Gaussian fit, which indicates that under these conditions one GFP molecule corresponds to a brightness of 8,000 ± 3,300 a.u. (mean ± s.d.). Note that the mean value corresponding to a single GFP is different here than in Fig. 4, primarily because a different camera was used for these experiments.
When Dam1 particles initially diffusing on the microtubule lattice encountered a disassembling tip, they began to move with the tip in the direction of shortening, sometimes encountering and coalescing with additional particles (Supplemental Information, Movies 5-7). To determine the minimum number of complexes required for disassembly-driven movement, we measured the initial brightness values for a subset of tip-tracking particles that fulfilled two criteria: they were closest to the tip at the onset of filming and they began tracking before encountering any other particles. These criteria excluded particles that could have tracked because of oligomerization with other particles. The initial brightness values for 19 out of 21 particles fell between 5,000 and 30,000 a.u. (Fig. 5b–e), which, under the conditions of these experiments, corresponds to 1−4 GFP molecules (Fig. 5f). This result indicates that oligomerization into a ring is not required for disassembly-driven movement, as many more than four Dam1 complexes are required to form a complete ring encircling a microtubule.
The simplest explanation for processive tip tracking of Dam1 particles is that their diffusion is biased at the microtubule tip. As described previously21, the diffusion of a particle with multiple microtubule-binding elements is expected to become biased at the filament tip because fluctuations toward the lattice allow more binding interactions to be made and therefore lower the free energy of the system. In principle, particles with as few as 2−4 microtubule-binding elements can track shortening tips through this mechanism if the free energy change associated with each binding interaction is large enough and the stepping rate is fast enough for the biased diffusion to outrun microtubule disassembly. The equilibrium Kd and diffusion coefficient measured for the Dam1 complex suggest a binding energy of approximately19 kBT and a stepping rate of approximately 2500 s−1 (assuming a step distance of 8 nm, the spacing of tubulin dimers in the microtubule lattice), which are well within the range of parameters that can produce tip tracking by the Hill mechanism21. This biased diffusion model differs fundamentally from the purest form of the ring model, in which peeling protofilaments push directly against the ring (that is, the ‘conformational wave’ model or ‘forced walk’ model)22, 24. Although previous observations of microtubule-driven Dam1 movement were consistent with either mechanism, the tip-tracking of small oligomers is incompatible with the conformational wave mechanism. Thus, the microtubule-driven movement of the Dam1 complex is probably driven, at least in part, by a biased diffusion mechanism. The contribution these small oligomers can make to the load-bearing capacity of the kinetochore remains a subject of future study.
Accurate chromosome segregation during cell division requires kinetochores to make regulated attachments to the tips of dynamic microtubules. We found that small particles containing as few as one to four individual Dam1 complexes were sufficient to reconstitute the features required for dynamic attachment to a microtubule, for phosphoregulation and for disassembly-driven movement. This finding shows that rings are not necessary for these activities. The Dam1 complex is one of several kinetochore components known to bind microtubules33, 34. Future work will show how the other microtubule-binding components contribute to the floating grip of kinetochores.
Methods
Construction of the Dad1-GFP Dam1 Complex Plasmid
PCR was used to amplify a region of PC4 SPC34H15 to include the unique Pci1 site 5’ of the Dad1 gene, and to add a Pac1 site on the 3’ end of Dad1 (5’ primer TTTGTGATGCTCGTCAGGG, 3’ primer TTAATTAACTTCGTTTTCGATTGA), GFP was amplified by PCR out of pFA6a, which has a native Pac1 site 5’ of GFP and a BstX1 site was added to the 3’ end (5’ primer AGAGTCAAGCCTGAAAATAATATGTCAGAAACATTCGCAACTCCCACTCCCAATAATCGAGGTCGACGGATCCCCGGG; 3’ primer CCAATTCACTGGGCTTATTTAGAAGTGGCGCGCC). Both fragments were ligated into Topo Zero Blunt (Invitrogen). A triple ligation was then performed between PC4 SPC34H cleaved with Pci1 and BstX1, the Dad1 fragment cleaved with Pci1 and Pac1, and the GFP fragment cleaved with Pac1 and BstX1. Ser to Ala mutations were made by site-directed mutagenesis of Dam1 in a separate vector followed by replacement of the wild type Dam1 with the mutated Dam1 in the polycistronic vector.
Purification of the Dam1 Complex
Dam1 complex polycistronic vectors were transformed into BL21 Rosetta (Novagen). Cultures were grown to about 30 Klett units, and the cultures were induced overnight at 23° C. Pellets were lysed using a French press in 50 mM sodium phosphate buffer (pH 6.9) containing 350 mM NaCl and protease inhibitors (0.01 mg/mL chymostatin, 0.01 mg/mL aprotinin, 0.01 mg/mL leupeptin, 0.01 mg/mL pepstatin 0.002 mg/mL benzamadine, 1 mM phenylmethylsulfonyl fluoride). Proteins were purified by affinity chromatography using talon resin, according to the manufacturer's instructions (BD Biosciences). Peak elutions were concentrated to approximately 1 ml (using an Amicon Ultra centrifugal filter with a moleculat weight cut-off of 50,000, Millipore) and then subjected to gel filtration chromatography on an SDX200 16/60 column (GE Healthcare). Peak fractions were concentrated and cleared at 13,000g. Glycerol (10% final concentration) was added and aliquots were snap frozen and stored at −80 °C.
Phosphorylation of the Dam1 complex
GST-Ipl1 was expressed from plasmid pSB196 (a gift of Sue Biggins, Fred Hutchinson Cancer Research Center, Seattle, WA) at 21 °C and purified using glutathione-Sepharose 4B according to the manufacturer's instructions (GE Healthcare Biosciences) except that the elution buffer was 50 mM Tris buffer, pH 8.0, containing 100 mM NaCl and 5 mM glutathione. GST-Sli15 (residues 554−698) was expressed from plasmid pSB503 (a gift from Sue Biggins) at 37 °C and purified using glutathione-Sepharose 4B resin, according to the manufacturer's instructions except that the elution buffer was 20 mM Tris buffer, pH 8.0, containing 200 mM NaCl, 1 mM β-mercaptoethanol, 1 mM EDTA and 10 mM glutathione.
Purified recombinant Dam1 complex was phosphorylated in a reaction (50 μL) containing 4 μM Dam1 complex, 0.5 μM GST-Ipl1, 0.5 μM GST-Sli15 (residues 554−698), 200 mM NaCl, 10 mM ATP, 25 mM MgCl2 and 50 mM HEPES buffer, pH 7.2. Reactions were incubated for 90 minutes at 30 °C. Glycerol (5%) was added, aliquots were snap frozen and stored at −80 °C . The conditions were optimized to obtain an average stoichiometry of 3.0 phosphate molecules per Dam1 molecule. Using mass spectrometry, we detected phosphorylation on residues Ser 20, Ser 265 and Ser 292 in Dam110. The peptide containing Ser 257 is difficult to detect by mass spectrometry, and we could not assess the phosphorylation state of the Ser 257 Ipl1 consensus site.
In our comparisons of the unphosphorylated complex and the phosphorylated complex, controls with either no kinase (Fig. 2) or no ATP (data not shown) were performed and gave similar results.
Microtubule Binding Assays
The binding assay was adapted from an assay designed to measure microtubule nucleation35. Briefly, increasing concentrations of GFP-tagged Dam1 complex were incubated with taxol-stabilized microtubules. The Dam1 complex bound to microtubules was pelleted onto a coverslip and quantified by fluorescence microscopy. See Supplementary Information for details of the assay.
Plots were generated and curves were fit using IGOR software (Wavemetrics). Confidence intervals were computed by maximum likelihood estimation, as implemented by IGOR and described36. In addition to the fit using the McGhee and von Hippel model30 described earlier, we also fitted the data to the Hill model37. Both the McGhee and von Hippel model and the Hill model supported the conclusions that Ipl1 phosphorylation of Dam1 complex reduces its affinity for microtubules and reduces the cooperativity of binding.
TIRF Microscopy
A TIRF illumination system was custom built on a Nikon inverted stage microscope (to be described in detail elsewhere). The coverslips used to construct flow cells were acid-washed followed by silanization with a short chain PEG-silane (Gelest). Holes were drilled in the slides allowing attachment of tubing to adapters such that solutions could be flowed in and out of the flow cells using a peristaltic pump. Flow cells were first washed with double-distilled water. To secure microtubules to the coverslip, kinesin with a mutation (G234A) that causes rigor-like microtubule-binding was used38. The rigor kinesin was allowed to adhere to the coverslip surface for approximately 15 minutes and then washed in BRB80 containing 70 mM KCl and 1 mg mL−1 κ-casein. For comparison of phosphorylated and unphosphorylated complex, taxol-stabilized Alexa-647 labeled microtubules were flowed in and allowed to bind for approximately 5 min. The microtubules were then washed with BRB80 containing 70 mM KCl, 1 mg mL−1 κ-casein and 10 μM taxol. Finally, GFP-tagged Dam1 complex in BRB80 containing 70 mM KCl, 1 mg mL−1 κ-casein, 10 μM taxol and an oxygen scavenger was flowed in at reported concentrations. The oxygen scavenger was 200 μg mL−1 glucose oxidase, 35 μg mL−1 catalase, 25 mM glucose, and 5 mM DTT.
For live microtubules, Alexa-647 labeled GMPCPP seeds were flowed in and allowed to bind for approximately 5 min. Bound microtubules were washed with BRB80 containing 70 mM KCl, 1 mg mL−1 κ-casein and 1 mM GTP. GFP-tagged Dam1 complex was flowed in at reported concentrations in BRB80 containing 70 mM KCl, 1 mg mL−1 κ-casein, 1 mM GTP, 2 mg mL−1 tubulin (Alexa-647 labeled 1:100) and oxygen scavenger. Finally, depolymerization was induced by flowing in the same buffer without tubulin or Dam1 complex. Flow cells were constructed such that each coverslip contained two separate, side-by-side chambers. Experiments were performed in the first chamber and in the second MCAK-GFP was always used as a control so that the brightness of a single GFP could be determined.
TIRF Data Analysis
Software was developed to analyze the TIRF movies using Labview (National Instruments). This software allowed the tracking of the position and brightness of individual fluorescent particles. Only isolated particles that did not cross paths with other particles were tracked. For calculating detachment rates and diffusion coefficients, only events lasting longer than one second were included. We note that the non-zero y-intercepts in our squared displacement versus time plots (Fig. 3) reflect localization uncertainty inherent in the position measurements due to background noise, limited photostatistics, and any errors introduced by our particle tracking software. The offset was approximately 0.026 μm2, which corresponds to a localization uncertainty of 160 nm.
Supplementary Material
Supplementary Movies
Movie S1: Unphosphorylated Dam1 complex binding and diffusing along MTs
Movie S2: Ipl1 phosphorylated Dam1 complex binding and diffusing along MTs
Movie S3: Dam1 complex particles transferring between MTs
Movie S4: Dam1 complex diffusing on an MT
Movie S5: Dam1 complex tracking depolymerizing MTs
Movie S6: Dam1 complex tracking a single depolymerizing MT
Movie S7: Dam1 complex particle jumping from one MT onto another, getting caught by the depolymerizing tip and following it back to the seed
In all movies, red represents MTs (tubulin tagged with Alexa-647) while green represents the Dam1 complex (Dad1-GFP). Movies S1-S4 show the Dam1 complex (40 pM) interacting with taxol stabilized MTs. Movies S5-S7 show the Dam1 complex (1 nM) interacting with dynamic MTs grown from GMPCPP MT seeds. Depolymerization is induced by flowing in buffer lacking tubulin and the Dam1 complex. Experiments are further described in Methods.
Supplementary Figure Legends
Figure S1. Quantifying the brightness of individual GFP molecules (a) Kymograph showing binding of individual GFP-MCAK molecules to a taxol-stabilized MT. Position along the MT is depicted on the vertical axis while time changes along the horizontal axis. MCAK is a dimeric, kinesin-family protein that binds tightly to MTs in the absence of nucleotides. At the low GFP-MCAK concentration used here (∼20 pM), the MT is sparsely decorated so that individual dimers remain visible until the GFP is photobleached. Scale bar, 3 μm. (b and c) Records of brightness versus time for individual, MT-bound GFP-MCAK molecules. As expected for dimers of GFP, many molecules (26 of 46 records, or 56%) photobleached in two steps (arrows in b). However, due to partial proteolysis during purification (see e, below), a significant fraction (44%) bleached in a single step (arrow in c). In either case the step amplitudes were similar, and well-fit by a Gaussian distribution centered at ∼45,000 a.u., so step amplitudes from both double- and single-bleach traces were pooled to generate the histogram of Figure 4f. (d) Photobleach times were distributed exponentially, as expected for bleaching of single fluorophores. The dotted curve shows an exponential fit used to determine the mean bleach time (τbleach) of 16 ± 1 (n = 72). (e) Coomassie brilliant blue-stained SDS-PAGE gel (left, false color) showing full-length MCAK-GFP plus degradation products, which represent 37% of the total MCAK (by gel densitometry). Immunoblot analyses using antibody to GFP (Santa Cruz Biotechnology, Santa Cruz, CA) (scanned blot in the middle) and antibody to MCAK1 (scanned film on the right) indicates that the degradation products lack GFP and contain MCAK. Assuming all the GFP-free MCAK is competent for dimerization and MT binding, the expected fraction of fluorescent molecules with two GFP tags is 46%, reasonably close to our measured fraction (56%). We observed no significant degradation of Dad1-GFP (See Figure 1a).
Supplementary Methods
Microtubule binding assay
Taxol-stabilized MTs were made as follows. Alexa-568 labeled bovine brain tubulin was mixed with unlabeled tubulin at a ratio of 1:41 to a final concentration of 52 μM in 34 μL BRB80 containing 1 mM GTP. BRB80 was made as described http://mitchison.med.harvard.edu/protocols/poly.html. The mixture was cleared at 4 °C at 90,000 × g for 10 min. The tubulin in the supernatant was then polymerized in a total volume of 40 μL BRB80 containing 5 mM MgCl2, 1.75 mM GTP and 3.5% DMSO at 37 °C for 30 min. MTs were diluted 6-fold in BRB80 containing 10 μM taxol. A small sample was used to determine the concentration of polymerized tubulin. We find between 80% to 90% of the tubulin is polymerized by this method.
Varying concentrations (0−50 nM) of phosphorylated and unphosphorylated complex were added to bovine brain taxol-stabilized MTs (2.5 nM, 1:41 Alexa-568-labeled tubulin/unlabeled tubulin) in BRB80 containing 45 mM NaCl, 4 mM phosphate buffer and 9 μM taxol in a total volume of 250 μL. The mixture was incubated for 10 minutes at 21 °C. The reaction was fixed by addition of 750 μL of 2% glutaraldehyde in BRB80 and incubated for 15 minutes at 20 °C. An aliquot (250 μL) of the reaction mixture was layered onto a 15% glycerol cushion (1 mL) on top of a polylysine glass coverslip in a Beckman TLS-55 centrifuge tube fitted with a custom spacer (Ellard Instrumentation, Monroe, WA). The MTs were pelleted onto the coverslip at 135,000 × g for 10 minutes at 21 °C in a Beckman Optima TLX ultracentrifuge. The glycerol cushion was removed by aspiration and the coverslip was taken out of the tube, placed MT side down on a drop of Citifluor (Ted Pella, Redding CA) on a slide and sealed with nail polish.
1. Moore, A.T. et al. MCAK associates with the tips of polymerizing microtubules. J Cell Biol 169, 391−397 (2005).
Acknowledgments
We thank J. J. Miranda and S. C. Harrison (Harvard Medical School) for providing the expression plasmid for the Dam1 complex and M. Wagenbach for providing MCAK-GFP. We also thank A. Franck, A. Powers, S. Biggins and J. Stumpff for helpful scientific discussion. This work was supported by an NSF IGERT traineeship to J.C., Searle Scholar Award 06-L-111 (to C.L.A.), Packard Fellowship for Science and Engineering #2006-30521 (to C.L.A.), and by grants R01GM40506, R01GM79373 and R01GM69429 from the National Institute of General Medical Sciences (to T.N.D., C.L.A. and L.W., respectively)
References
- 1.Pinsky BA, Kotwaliwale CV, Tatsutani SY, Breed CA, Biggins S. Glc7/protein phosphatase 1 regulatory subunits can oppose the Ipl1/aurora protein kinase by redistributing Glc7. Mol Cell Biol. 2006;26:2648–2660. doi: 10.1128/MCB.26.7.2648-2660.2006. PMCD: PMC1430313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tanaka K, Kitamura E, Kitamura Y, Tanaka TU. Molecular mechanisms of microtubule-dependent kinetochore transport toward spindle poles. J Cell Biol. 2007;178:269–281. doi: 10.1083/jcb.200702141. PMCID: PMC2064446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Franco A, Meadows JC, Millar JB. The Dam1/DASH complex is required for the retrieval of unclustered kinetochores in fission yeast. J Cell Sci. 2007;120:3345–3351. doi: 10.1242/jcs.013698. PMID: 17881496. [DOI] [PubMed] [Google Scholar]
- 4.Cheeseman IM, Enquist-Newman M, Muller-Reichert T, Drubin DG, Barnes G. Mitotic spindle integrity and kinetochore function linked by the Duo1p/Dam1p complex. J Cell Biol. 2001;152:197–212. doi: 10.1083/jcb.152.1.197. PMID: 11149931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He X, Rines DR, Espelin CW, Sorger PK. Molecular analysis of kinetochoremicrotubule attachment in budding yeast. Cell. 2001;106:195–206. doi: 10.1016/s0092-8674(01)00438-x. PMID: 11511347. [DOI] [PubMed] [Google Scholar]
- 6.Janke C, Ortiz J, Tanaka TU, Lechner J, Schiebel E. Four new subunits of the Dam1-Duo1 complex reveal novel functions in sister kinetochore biorientation. Embo J. 2002;21:181–193. doi: 10.1093/emboj/21.1.181. PMID: 11782438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones MH, He X, Giddings TH, Winey M. Yeast Dam1p has a role at the kinetochore in assembly of the mitotic spindle. Proc Natl Acad Sci U S A. 2001;98:13675–13680. doi: 10.1073/pnas.241417098. PMID: 11698664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y, et al. The mitotic spindle is required for loading of the DASH complex onto the kinetochore. Genes Dev. 2002;16:183–197. doi: 10.1101/gad.959402. PMID: 11799062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheeseman IM, et al. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell. 2002;111:163–172. doi: 10.1016/s0092-8674(02)00973-x. PMID: 12408861. [DOI] [PubMed] [Google Scholar]
- 10.Shimogawa MM, et al. Mps1 phosphorylation of Dam1 couples kinetochores to microtubule plus ends at metaphase. Curr Biol. 2006;16:1489–1501. doi: 10.1016/j.cub.2006.06.063. PMID: 16890524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Asbury CL, Gestaut DR, Powers AF, Franck AD, Davis TN. The Dam1 kinetochore complex harnesses microtubule dynamics to produce force and movement. Proc Natl Acad Sci U S A. 2006;103:9873–9878. doi: 10.1073/pnas.0602249103. PMID: 16777964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Westermann S, et al. Formation of a dynamic kinetochore- microtubule interface through assembly of the Dam1 ring complex. Mol Cell. 2005;17:277–290. doi: 10.1016/j.molcel.2004.12.019. PMID: 15664196. [DOI] [PubMed] [Google Scholar]
- 13.Westermann S, et al. The Dam1 kinetochore ring complex moves processively on depolymerizing microtubule ends. Nature. 2006;440:565–569. doi: 10.1038/nature04409. PMID: 16415853. [DOI] [PubMed] [Google Scholar]
- 14.Franck AD, et al. Tension applied through the Dam1 complex promotes microtubule elongation providing a direct mechanism for length control in mitosis. Nat Cell Biol. 2007;9:832–837. doi: 10.1038/ncb1609. PMID: 175726669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miranda JJ, De Wulf P, Sorger PK, Harrison SC. The yeast DASH complex forms closed rings on microtubules. Nat Struct Mol Biol. 2005;12:138–143. doi: 10.1038/nsmb896. PMID: 15640796. [DOI] [PubMed] [Google Scholar]
- 16.Miranda JJ, King DS, Harrison SC. Protein arms in the kinetochore-microtubule interface of the yeast DASH complex. Mol Biol Cell. 2007;18:2503–2510. doi: 10.1091/mbc.E07-02-0135. PMID: 17460120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang HW, et al. Architecture of the Dam1 kinetochore ring complex and implications for microtubule-driven assembly and force-coupling mechanisms. Nat Struct Mol Biol. 2007 doi: 10.1038/nsmb1274. PMID: 17643123. [DOI] [PubMed] [Google Scholar]
- 18.Howard J, Hyman AA. Microtubule polymerases and depolymerases. Curr Opin Cell Biol. 2007;19:31–35. doi: 10.1016/j.ceb.2006.12.009. PMID: 17184986. [DOI] [PubMed] [Google Scholar]
- 19.Westermann S, Drubin DG, Barnes G. Structures and functions of yeast kinetochore complexes. Annual review of biochemistry. 2007;76:563–591. doi: 10.1146/annurev.biochem.76.052705.160607. PMID: 17362199. [DOI] [PubMed] [Google Scholar]
- 20.Karp G. Cell and Molecular Biology. Edn. 5 John Wiley & Sons, Inc.; Hoboken, NJ: 2007. [Google Scholar]
- 21.Hill TL. Theoretical problems related to the attachment of microtubules to kinetochores. Proc Natl Acad Sci U S A. 1985;82:4404–4408. doi: 10.1073/pnas.82.13.4404. PMID: 3859869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koshland DE, Mitchison TJ, Kirschner MW. Polewards chromosome movement driven by microtubule depolymerization in vitro. Nature. 1988;331:499–504. doi: 10.1038/331499a0. PMID: 3340202. [DOI] [PubMed] [Google Scholar]
- 23.Molodtsov MI, Grishchuk EL, Efremov AK, McIntosh JR, Ataullakhanov FI. Force production by depolymerizing microtubules: a theoretical study. Proc Natl Acad Sci U S A. 2005;102:4353–4358. doi: 10.1073/pnas.0501142102. PMID: 15767580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Efremov A, Grishchuk EL, McIntosh JR, Ataullakhanov FI. In search of an optimal ring to couple microtubule depolymerization to processive chromosome motions. Proc Natl Acad Sci U S A. 2007;104:19017–19022. doi: 10.1073/pnas.0709524104. PMID: 18029449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dong Y, Vanden Beldt KJ, Meng X, Khodjakov A, McEwen BF. The outer plate in vertebrate kinetochores is a flexible network with multiple microtubule interactions. Nat Cell Biol. 2007;9:516–522. doi: 10.1038/ncb1576. PMID: 17435749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McIntosh JR. Rings around kinetochore microtubules in yeast. Nat Struct Mol Biol. 2005;12:210–212. doi: 10.1038/nsmb0305-210. PMID: 15744320. [DOI] [PubMed] [Google Scholar]
- 27.Enquist-Newman M, et al. Dad1p, third component of the Duo1p/Dam1p complex involved in kinetochore function and mitotic spindle integrity. Mol Biol Cell. 2001;12:2601–2613. doi: 10.1091/mbc.12.9.2601. PMID: 11553702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheeseman IM, et al. Implication of a novel multiprotein Dam1p complex in outer kinetochore function. J Cell Biol. 2001;155:1137–1145. doi: 10.1083/jcb.200109063. PMID: 11756468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cantor CR, Schimmel PR. Biophysical Chemistry. I. II. III. W. H. Freeman & Company; San Francisco: 1980. [Google Scholar]
- 30.McGhee JD, von Hippel PH. Theoretical aspects of DNA-protein interactions: cooperative and non-co-operative binding of large ligands to a one-dimensional homogeneous lattice. J Mol Biol. 1974;86:469–489. doi: 10.1016/0022-2836(74)90031-x. PMID: 4416620. [DOI] [PubMed] [Google Scholar]
- 31.Pinsky BA, Kung C, Shokat KM, Biggins S. The Ipl1-Aurora protein kinase activates the spindle checkpoint by creating unattached kinetochores. Nat Cell Biol. 2006;8:78–83. doi: 10.1038/ncb1341. PMID: 16327780. [DOI] [PubMed] [Google Scholar]
- 32.Shang C, et al. Kinetochore protein interactions and their regulation by the Aurora kinase Ipl1p. Mol Biol Cell. 2003;14:3342–3355. doi: 10.1091/mbc.E02-11-0765. PMID: 12925767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. PMID: 17129783. [DOI] [PubMed] [Google Scholar]
- 34.Davis TN, Wordeman L. Rings, bracelets, sleeves, and chevrons: new structures of kinetochore proteins. Trends in cell biology. 2007;17:377–382. doi: 10.1016/j.tcb.2007.08.001. PMID: 17766118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gunawardane RN, Zheng Y, Oegema K, Wiese C. Purification and Reconstitution of Drosophilia γ-Tubulin Complexes. In: Palazzo RE, Davis TN, editors. Methods in Cell Biology. Vol. 67. Academic Press; San Diego: 2001. pp. 1–26. PMID: 11550462. [DOI] [PubMed] [Google Scholar]
- 36.Press WH, Vetterling WT, Teukolsky SA, Flannery BP. Numerical Recipes in C: The Art of Scientific Computing. Edn. 2nd Cambridge University Press; Cambridge, UK: 1996. [Google Scholar]
- 37.Hill A. The possible effects of the aggregation of the molecules of haemoglobin on its dissociation curves. J. Physiol. 1910;40:iv–vii. [Google Scholar]
- 38.Rice S, et al. A structural change in the kinesin motor protein that drives motility. Nature. 1999;402:778–784. doi: 10.1038/45483. PMID: 10617199. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Movies
Movie S1: Unphosphorylated Dam1 complex binding and diffusing along MTs
Movie S2: Ipl1 phosphorylated Dam1 complex binding and diffusing along MTs
Movie S3: Dam1 complex particles transferring between MTs
Movie S4: Dam1 complex diffusing on an MT
Movie S5: Dam1 complex tracking depolymerizing MTs
Movie S6: Dam1 complex tracking a single depolymerizing MT
Movie S7: Dam1 complex particle jumping from one MT onto another, getting caught by the depolymerizing tip and following it back to the seed
In all movies, red represents MTs (tubulin tagged with Alexa-647) while green represents the Dam1 complex (Dad1-GFP). Movies S1-S4 show the Dam1 complex (40 pM) interacting with taxol stabilized MTs. Movies S5-S7 show the Dam1 complex (1 nM) interacting with dynamic MTs grown from GMPCPP MT seeds. Depolymerization is induced by flowing in buffer lacking tubulin and the Dam1 complex. Experiments are further described in Methods.
Supplementary Figure Legends
Figure S1. Quantifying the brightness of individual GFP molecules (a) Kymograph showing binding of individual GFP-MCAK molecules to a taxol-stabilized MT. Position along the MT is depicted on the vertical axis while time changes along the horizontal axis. MCAK is a dimeric, kinesin-family protein that binds tightly to MTs in the absence of nucleotides. At the low GFP-MCAK concentration used here (∼20 pM), the MT is sparsely decorated so that individual dimers remain visible until the GFP is photobleached. Scale bar, 3 μm. (b and c) Records of brightness versus time for individual, MT-bound GFP-MCAK molecules. As expected for dimers of GFP, many molecules (26 of 46 records, or 56%) photobleached in two steps (arrows in b). However, due to partial proteolysis during purification (see e, below), a significant fraction (44%) bleached in a single step (arrow in c). In either case the step amplitudes were similar, and well-fit by a Gaussian distribution centered at ∼45,000 a.u., so step amplitudes from both double- and single-bleach traces were pooled to generate the histogram of Figure 4f. (d) Photobleach times were distributed exponentially, as expected for bleaching of single fluorophores. The dotted curve shows an exponential fit used to determine the mean bleach time (τbleach) of 16 ± 1 (n = 72). (e) Coomassie brilliant blue-stained SDS-PAGE gel (left, false color) showing full-length MCAK-GFP plus degradation products, which represent 37% of the total MCAK (by gel densitometry). Immunoblot analyses using antibody to GFP (Santa Cruz Biotechnology, Santa Cruz, CA) (scanned blot in the middle) and antibody to MCAK1 (scanned film on the right) indicates that the degradation products lack GFP and contain MCAK. Assuming all the GFP-free MCAK is competent for dimerization and MT binding, the expected fraction of fluorescent molecules with two GFP tags is 46%, reasonably close to our measured fraction (56%). We observed no significant degradation of Dad1-GFP (See Figure 1a).
Supplementary Methods
Microtubule binding assay
Taxol-stabilized MTs were made as follows. Alexa-568 labeled bovine brain tubulin was mixed with unlabeled tubulin at a ratio of 1:41 to a final concentration of 52 μM in 34 μL BRB80 containing 1 mM GTP. BRB80 was made as described http://mitchison.med.harvard.edu/protocols/poly.html. The mixture was cleared at 4 °C at 90,000 × g for 10 min. The tubulin in the supernatant was then polymerized in a total volume of 40 μL BRB80 containing 5 mM MgCl2, 1.75 mM GTP and 3.5% DMSO at 37 °C for 30 min. MTs were diluted 6-fold in BRB80 containing 10 μM taxol. A small sample was used to determine the concentration of polymerized tubulin. We find between 80% to 90% of the tubulin is polymerized by this method.
Varying concentrations (0−50 nM) of phosphorylated and unphosphorylated complex were added to bovine brain taxol-stabilized MTs (2.5 nM, 1:41 Alexa-568-labeled tubulin/unlabeled tubulin) in BRB80 containing 45 mM NaCl, 4 mM phosphate buffer and 9 μM taxol in a total volume of 250 μL. The mixture was incubated for 10 minutes at 21 °C. The reaction was fixed by addition of 750 μL of 2% glutaraldehyde in BRB80 and incubated for 15 minutes at 20 °C. An aliquot (250 μL) of the reaction mixture was layered onto a 15% glycerol cushion (1 mL) on top of a polylysine glass coverslip in a Beckman TLS-55 centrifuge tube fitted with a custom spacer (Ellard Instrumentation, Monroe, WA). The MTs were pelleted onto the coverslip at 135,000 × g for 10 minutes at 21 °C in a Beckman Optima TLX ultracentrifuge. The glycerol cushion was removed by aspiration and the coverslip was taken out of the tube, placed MT side down on a drop of Citifluor (Ted Pella, Redding CA) on a slide and sealed with nail polish.
1. Moore, A.T. et al. MCAK associates with the tips of polymerizing microtubules. J Cell Biol 169, 391−397 (2005).