Abstract
Chloroplasts and bacterial cells divide by binary fission. The key protein in this constriction division is FtsZ, a self-assembling GTPase similar to eukaryotic tubulin. In prokaryotes, FtsZ is almost always encoded by a single gene, whereas plants harbor several nuclear-encoded FtsZ homologs. In seed plants, these proteins group in two families and all are exclusively imported into plastids. In contrast, the basal land plant Physcomitrella patens, a moss, encodes a third FtsZ family with one member. This protein is dually targeted to the plastids and to the cytosol. Here, we report on the targeted gene disruption of all ftsZ genes in P. patens. Subsequent analysis of single and double knockout mutants revealed a complex interaction of the different FtsZ isoforms not only in plastid division, but also in chloroplast shaping, cell patterning, plant development, and gravity sensing. These results support the concept of a plastoskeleton and its functional integration into the cytoskeleton, at least in the moss P. patens.
Keywords: Bryophyte, cell wall, gravitropism, GTPase, chloroplast, plastoskeleton, P. patens, moss
INTRODUCTION
Plants arose from an endosymbiotic event in which a fully differentiated eukaryotic cell engulfed a free-living cyanobacterium, which was subsequently ‘domesticated’ as a chloroplast (Gould et al., 2008; Gray, 1992). This process was not simply a mere merger of two autonomous cells, but a co-evolutionary process at the cellular level, involving gene transfer (e.g. Martin et al., 1998) and rearrangement of cellular structures (Kuroiwa et al., 2008). Consequently, chloroplasts divide by binary fission as bacterial cells do (Kuroiwa et al., 1998; Possingham and Lawrence, 1983). Both processes rely on the protein FtsZ (filamentous temperature sensitive Z), a self-assembling GTPase that can form a contractile ring at the prospective division site (Dai and Lutkenhaus, 1991; de Boer et al., 1992; Gilson and Beech, 2001; Osteryoung et al., 1998; Osteryoung and Vierling, 1995; Strepp et al., 1998; Yoshida et al., 2006). It is now widely accepted that FtsZ is a prokaryotic homolog of the eukaryotic tubulin (Erickson et al., 1996; Lowe and Amos, 1998). In the plastid division complex, FtsZ interacts with ARC6, a descendant of a cyanobacterial cell division protein (Vitha et al., 2003), ARC5, a descendant of a protein involved in eukaryotic cell division (Miyagishima et al., 2008), and with PDV-proteins, which seem to be specific to land plants (Miyagishima et al., 2006).
While most bacteria encode one single FtsZ protein, plants harbor at least three different nuclear-encoded FtsZ proteins in two small protein families, FtsZ1 and FtsZ2. Phylogenetic analyses revealed that all plant FtsZ proteins can be traced back to their cyanobacterial ancestor and that the split into different families occurred in an ancestor of Viridiplantae (Osteryoung et al., 1998; Rensing et al., 2004). The existence of multiple FtsZ isoforms in a single plant immediately suggests that they may have different functions. In fact, FtsZ1 and FtsZ2 proteins can be discriminated by three features: (1) Plant FtsZ1 proteins share the serine residue found in the tubulin signature, whereas all plant FtsZ2 proteins have a threonine residue also found in bacterial FtsZ (El-Shami et al., 2002). (2) FtsZ2 proteins possess a conserved C-terminal domain, which is absent in FtsZ1 proteins. In bacteria, this domain mediates interaction between FtsZ and other cell division proteins (e.g. Ma and Margolin, 1999). (3) FtsZ1 and FtsZ2 proteins differ in their biochemical properties and in vivo localization (El-Kafafi et al., 2005). Evidence for the functional difference of the two families in flowering plants came from studies in Arabidopsis thaliana: while overexpression of AtftsZ1-1 inhibited chloroplast division, slight overexpression of AtftsZ2-1 did not affect the chloroplast division (Stokes et al., 2000) whereas strong overexpression of this gene impaired plastid division (McAndrew et al., 2001). In addition, antisense lines of both genes exhibited strongly reduced chloroplast numbers per cell (Osteryoung et al., 1998). Furthermore, AtFtsZ1::GFP fusion proteins, but not those of the AtFtsZ2-family, were found not only in rings, but also in stromules (Vitha et al., 2001), stroma-filled tubules emanating from the chloroplast (Köhler et al., 1997).
Surprisingly, basal land plants encode a third FtsZ family, making the moss Physcomitrella patens currently the organism with the most different FtsZ proteins, namely five. This finding prompted us to change the nomenclature of the moss FtsZ isoforms (Martin et al., 2009). While the PpFtsZ2-family was not affected, the former FtsZ1-2 is now referred to as FtsZ3, while the most recently found ftsZ gene encodes FtsZ1-2. This new nomenclature is used in the following text and differs from the nomenclature used in previous publications regarding FtsZ1-2 and FtsZ3. Similar to the situation in A. thaliana, both P. patens FtsZ1-isoforms are localized to rings in chloroplasts and surrounding the plastids and to long filaments in stromules. In addition, both moss FtsZ1-isoforms form filamentous networks inside the chloroplasts (Gremillon et al., 2007; Martin et al., 2009; Suppanz et al., 2007). Both FtsZ2-isoforms of P. patens (PpFtsZ2-1, PpFtsZ2-2) are found as network-like structures in the plastids, suggesting that they act as components of a plastoskeleton that may ensure chloroplast shape and integrity (Kiessling et al., 2000; Reski, 2002). Similar to the situation in A. thaliana, however, PpFtsZ2 proteins are not found in stromules. The single member of the third FtsZ-family (PpFtsZ3), which has no ortholog in seed plants, is dually targeted to chloroplasts and to the cytosol in P. patens, localizing to ring-like structures in both compartments. As transient overexpression of this gene hampered cell and chloroplast division, it was suggested that this dually targeted protein may represent a molecular link between cell and organelle division in moss (Kiessling et al., 2004). From studies using fluorescence energy transfer (FRET), it became evident that the different FtsZ isoforms specifically and in a hierarchical order interact in vivo in plastids and in the cytosol of P. patens (Gremillon et al., 2007). It remained elusive, however, what the biological significance of these interactions might be.
In contrast to the situation in A. thaliana, the generation of loss-of-function mutants by targeted gene disruption is a straightforward approach in P. patens (Reski, 1998a). Consequently, targeted disruption of PpftsZ2-1 proved the importance for FtsZ in chloroplast division (Strepp et al., 1998). It was, however, not clear until the complete genome of P. patens was sequenced (Rensing et al., 2008) that this moss encodes not only four, but five different FtsZ proteins (Martin et al., 2009). Here, we report on the creation of a series of FtsZ deletion mutants, including single (ΔPpftsZ1-1, ΔPpftsZ1-2, ΔPpftsZ2-1, ΔPpftsZ2-2, and ΔPpftsZ3), and double mutants (ΔPpftsZ1-1/1–2 and ΔPpftsZ2-1/2–2). Their subsequent analysis revealed specific functions of the different FtsZ families, as well as the individual members of each family, not only in chloroplast division, but also in chloroplast shaping and in different cellular processes. These results are discussed from an evolutionary point of view, supporting the concept of a plastoskeleton and its functional integration into the cytoskeleton, at least in the moss P. patens.
RESULTS
Generation of ftsZ Knockout Mutants
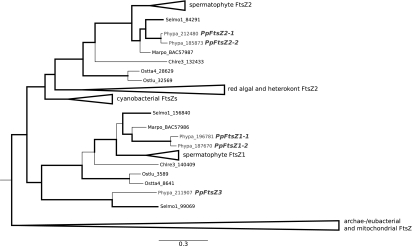
The nuclear genome of P. patens encodes five different ftsZ genes; two closely related paralogs in the FtsZ1 and FtsZ2 family, respectively, and a single member in the basal plant-specific FtsZ3 family (Figure 1; Martin et al., 2009). Targeted gene knockout of ftsZ2-1 was previously reported and proved the function of the encoded protein in chloroplast division (Strepp et al., 1998).
Figure 1.
Phylogenetic Relationships of the Eukaryotic FtsZ Family with a Special Focus on the Gene Family in Physcomitrella patens (Adapted from Tree Described in Martin et al., 2009).
Branch thickness indicates branch support as posterior probabilities. Physcomitrella FtsZ proteins are highlighted in gray. The tree is rooted using the archae-/eubacterial and mitochondrial FtsZs clade as outgroup (shown collapsed). To focus on phylogenetic relationships of the moss gene family, branches of the seed plant (Spermatophyta), red algal (Rhodophyta), and heterokont algal (stramenopiles) FtsZ subfamilies have been collapsed as well. Selmo, Selaginella moellendorffii (Lycopodiophyta); Marpo, Marchantia polymorpha (Marchantiophyta); Chlre, Chlamydomonas reinhardtii (Chlorophyta); Ostta, Ostreococcus tauri (Prasinophyceae); Ostlu, Ostreococcus lucimarinus (Prasinophyceae).
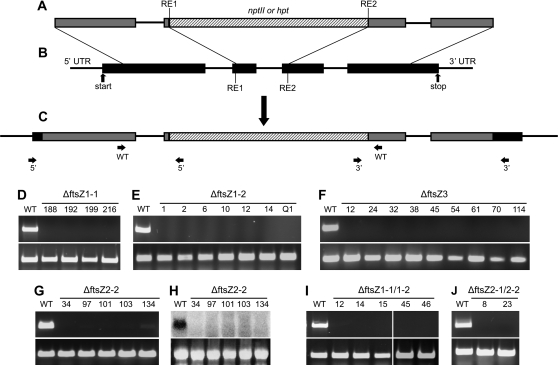
To unravel the functions of the other four FtsZ proteins, we generated single and double deletion mutants making use of P. patens’ high rate of homologous recombination in its nuclear DNA. Knockout constructs comprising genomic ftsZ fragments interrupted by a selection cassette that confers resistance to either hygromycin or G418 were generated for ftsZ1-1, ftsZ1-2, ftsZ3, and ftsZ2-2 (Figure 2A and 2B). The constructs were introduced into P. patens employing PEG-mediated transformation of isolated protoplasts. Several independent single and double knockout mutants were identified (see Table 1 for an overview).
Figure 2.
Generation and Molecular Analysis of P. patens ΔftsZ Mutants.
(A) Exemplary scheme of the generated knockout constructs (RE: restriction enzyme).
(B) The genomic locus.
(C) The altered structure of the genomic locus after integration of the knockout construct by homologous recombination. Primer pairs that were used for direct PCR plant screening are indicated as arrows and are listed in Table 3 for the ftsZ knockouts.
(D–G, I–J) RT–PCR analysis with gene-specific primers (see Table 4). As a control, RT–PCR was performed with primers C45fwd and C45rev, corresponding to the constitutively expressed gene for the ribosomal protein L21. The corresponding PCR-product is always shown in the lower panels.
(H) In addition to RT–PCR, Northern blot analysis was performed for putative ΔftsZ2-2 mutants due to the presence of faint traces of transcript after RT–PCR. As a control for equal loading of RNA ethidium bromide stained 28S rRNA is shown (lower panel).
Table 1.
After Three Rounds of Selection on Medium Containing Antibiotics, Transgenic Plants Were Analyzed Via Direct PCR.
| ΔftsZ1-1 | ΔftsZ1-2 | ΔftsZ3 | ΔftsZ2-2 | ΔftsZ1-1/1–2 | ΔftsZ2-1/2–2 | |
| Direct PCR | 180 | 132 | 238 | 34 | 46 | 192 |
| Correct 5’ and 3’ integration | 22* | 19* | 9 | 5 | 11* | 2 |
| Absence of WT transcript | 4 | 7 | 9 | 5 | 5 | 2 |
All plants with correct 5’ and 3’ integration of the knockout construct into the respective locus were further tested for absence of the ftsZ transcript via RT–PCR. * RNA for subsequent RT–PCR analysis was extracted from only a subset of the plants.
All plants were initially screened via direct PCR (Schween et al., 2002) for disruption of the wild-type ftsZ locus as well as correct 5’ and 3’ integration of the construct with primers that bind outside of the introduced construct and inside the selection cassette (Figure 2C and Supplemental Figure 1). From plants with positive results after direct PCR screening, protonema was grown in liquid cultures to obtain larger amounts of plant material for RNA isolation and subsequent RT–PCR analysis (Figure 2D–2G and 2I–2J). For putative ΔftsZ2-2, an additional Northern blot was performed due to the presence of faint traces of transcript after RT–PCR that was most probably caused by unspecific binding of the primers to ftsZ2-1 (Figure 2H). The confirmed deletion mutants were grown under standard conditions and subsequently compared to wild-type.
Phenotypic Analyses of ΔftsZ1 Mutants
The overall plant morphology of ΔftsZ1-1, ΔftsZ1-2, and ΔftsZ1-1/1–2 mutants did not differ from P. patens wild-type. Likewise, plant growth and development were not obviously different from the control. To investigate a possible involvement of P. patens FtsZ1 proteins in plastid division, chloroplast shape and numbers were compared between the deletion mutants and wild-type. In the latter chloronema cells harbored about 50 lens-shaped chloroplasts (Figure 3A). Interestingly, ΔftsZ1-1 mutants did not show altered chloroplast morphology or number (Figure 3B), while ΔftsZ1-2 mutants had enlarged plastids only in basal chloronema cells (Figure 3C). In contrast, the ΔftsZ1-1/1–2 double mutants displayed chloroplasts with drastically altered shapes in every cell and tissue type. In these mutants, chloroplasts were enlarged and elongated, with pointed poles. Most chloronema cells of these double gene knockout mutants contained three to five chloroplasts per cell only (Figure 3D). When compared to wild-type, the gametophores of these mutants appeared to be smaller (Figure 3H).
Figure 3.
Knockout of ftsZ1 and ftsZ2 Genes Affects Chloroplast Morphology.
Protonema cells of (A) WT, (B) ΔftsZ1-1, (C) ΔftsZ1-2, (D) ΔftsZ1-1/1–2, (E) ΔftsZ2-1, (F) ΔftsZ2-2, and (G) ΔftsZ2-1/2–2 mutants (scale bar = 25 μm).
(H) Gametophores of WT and of ΔftsZ1 and ΔftsZ2 mutants (scale bar = 250 μm).
Sector of a leaf of (I) WT and (J) ΔftsZ2-1/2–2 mutant (scale bar = 50 μm).
Phenotypic Analyses of ΔftsZ2 Mutants
Re-examination of the previously published ΔftsZ2-1 mutants (Strepp et al., 1998) confirmed that they contained only one to three giant chloroplasts per cell (Figure 3E). In contrast, targeted deletion of the ftsZ2-2 gene did not obviously affect shape or number of the plastids (Figure 3F). In contrast, the ΔftsZ2-1/2–2 double mutants harbored one to three macrochloroplasts in every cell (Figure 3G). Gametophores of these mutants were obviously shorter than in the wild-type (Figure 3H). In addition, the double mutant displayed altered leaf morphology, including irregular cell patterning throughout the leaves. While cells in leaves of wild-type, of ΔftsZ2-1 mutants, and of ΔftsZ2-2 mutants were regularly arranged in parallel to the midrib and possessed an approximately rectangular shape, cell shapes in the ΔftsZ2-1/2–2 double mutants were highly diverse, the leaves having also a distorted midrib and a disturbed cell arrangement (Figure 3I and 3J).
Phenotypic Analyses of ΔftsZ3 Mutants
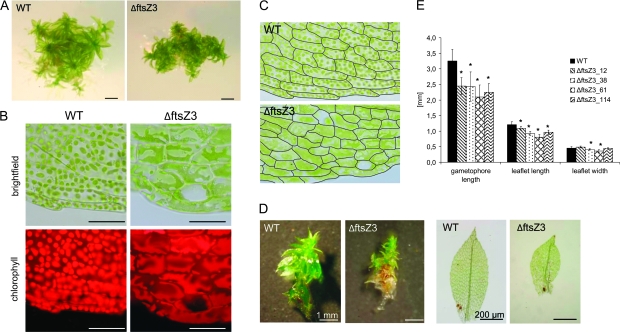
Targeted knockout of the ftsZ3 gene severely affected overall plant morphology. The leafy gametophores (the adult gametophytes) of ΔftsZ3 mutants were shorter, stunted, and more compact than those of wild-type (Figure 4A). The leaf cells of these mutants harbored a variety of unusual chloroplast numbers and shapes. In different cells of a single leaf, macrochloroplasts, enlarged plastids with finger-like outgrowths, and other irregularly shaped chloroplasts were observed (Figure 4B). The ΔftsZ3 mutants also displayed severe perturbations of leaf cell pattern while the overall shape of the leaves was only moderately altered when compared to wild-type (Figures 4C). Under standard growth conditions, the average length of wild-type gametophores was 3.3 mm, while mutant gametophores were reduced to 2.1–2.5 mm in length (Figure 4D and 4E). Further, deletion of the ftsZ3 gene resulted in reduced lengths of the leaves themselves. While in wild-type, the average leaf was 1.2 mm long, the leaves of the deletion mutants were reduced to 0.8–1.1 mm in length (Figure 4D and 4E). In contrast, the gene deletion did not obviously affect the leaf width (Figure 4E).
Figure 4.
Knockout of ftsZ3 Affects Plant and Chloroplast Morphology.
(A) Wild-type and ftsZ3 gametophyte (scale bars = 1 mm).
(B) Micrographs of leaf cells of the wild-type and ΔftsZ3 (scale bar = 50 μm) under brightfield and UV-fluorescence.
(C) Cell patterning in leaf cells of WT (upper panel) and ΔftsZ3 (lower panel). Cell walls were traced using graphical software.
(D) Stereo and light microscope images of gametophores and leaves from WT and ΔftsZ3 grown in parallel under standard conditions for 4 weeks.
(E) Measurement of gametophore and leaf dimensions. Columns represent the average length of gametophores (n = 14) and length and width of leaves (n = 20) grown on Knop medium for 4 weeks. Error bars indicate the absolute average deviation (AAD). Asterisks indicate significant differences between WT and ΔftsZ3 (t-test, P ≤ 0.001).
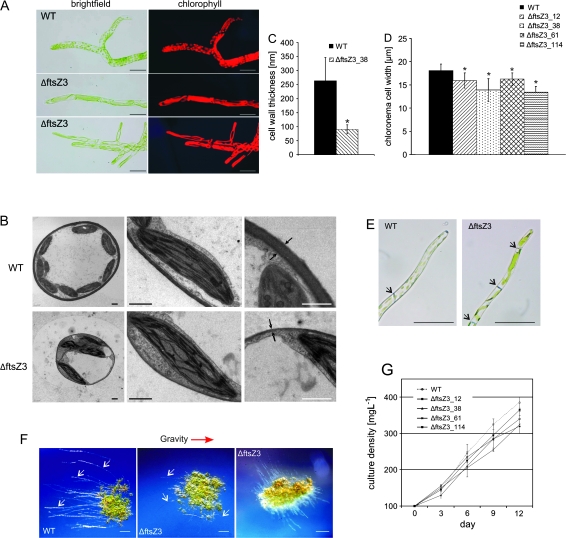
Light microscopy of the filamentous protonema tissues revealed highly unusual plastid morphologies in the ΔftsZ3 mutants: chloroplasts appeared as strongly enlarged or elongated, tubular, and rather thin structures (Figure 5A). However, transmission electron micrographs (TEM) showed that deletion of the ftsZ3 gene did not affect the internal fine structure of the chloroplasts. Surprisingly, cell walls of the mutant were obviously thinner as compared to cell walls of wild-type P. patens (Figure 5B). Subsequently, we measured cell wall thickness at three different positions per thin section. These measurements revealed that cell walls of wild-type were, on average, 263 nm thick, whereas the cell walls in ΔftsZ3 mutants were reduced to about one-third (90 nm). This difference in cell wall thickness was significant, with P ≤ 0.001 (Figure 5C). In addition, chloronema cells of ΔftsZ3 mutants were significantly (P ≤ 0.001) thinner than those of wild-type (Figure 5D).
Figure 5.
Wide Range of Phenotypic Aberrations in ΔftsZ3 Mutants.
(A) Micrographs of chloronema cells of the wild-type and ΔftsZ3 (scale bar = 50 μm).
(B) TEM images of protonema cell cross-sections as well as magnified images of chloroplasts and cell walls are shown. Cell walls are indicated by arrows (scale bar = 1 μm).
(C) Mean cell wall thickness for WT and ΔftsZ3. Cell walls from protonema cells, embedded for TEM analysis 4 d after sub-culturing, were measured (WT: n = 21; KO: n = 24). Asterisks indicate significant differences between WT and mutant (t-test, P ≤ 0.001). Error bars indicate the AAD.
(D) Mean width of chloronema cells 4 d after sub-culturing. Asterisks indicate significant differences between WT and mutants (t-test, P ≤ 0.001). Error bars indicate the AAD (n = 71).
(E) Caulonema cells of WT and ΔftsZ3. Black arrows indicate the position of the cell walls (scale bar = 50 μm).
(F) Comparison of caulonema development in the dark between WT and ΔftsZ3. Stereomicroscope images of protonema colonies, grown in the dark on Knop medium. Caulonema filaments are pointed out by white arrows. The gravity vector is indicated by a red arrow (scale bar = 1 mm).
(G) Biomass accumulation of WT and ΔftsZ3 mutants. Biomass accumulation is displayed as the culture density increasing over time. Liquid cultures were inoculated with 100 mg L−1 dry weight (day 0) and growth was monitored for a period of 12 d. Error bars indicate the AAD (n = 2).
In wild-type P. patens, chloronema cells were cylindrically shaped, with their cross walls perpendicularly orientated with respect to the growth axis. They contained approximately 50 chloroplasts per cell. In the course of normal development, cell differentiation to another cell type, the caulonema, occurred. These caulonema cells originated from chloronema cells and were longer and thinner, with cross walls orientated obliquely to the growth axis. These caulonema cells harbored, on average, 19 chloroplasts, which were smaller than those found in chloronema cells (Figure 5E). Under standard growth conditions, ΔftsZ3 mutants did hardly develop any caulonema cells. Moreover, those few did not show wild-type characteristics (Figure 5E). As it has been reported that the addition of the phytohormone auxin (Decker et al., 2006), as well as growth in the dark (Vandenbussche et al., 2007), induces caulonema formation in P. patens, both treatments were applied separately in this study as well. In accordance with previous findings, wild-type P. patens showed numerous long, negatively gravitropic growing caulonema filaments under inductive conditions. The same conditions induced some caulonema filaments in the ΔftsZ3 mutants. These, however, were much shorter and less numerous than in wild-type and, furthermore, had lost their gravitropic response, as they grew in different directions (Figure 5F). In addition to that, a comparison of biomass accumulation in wild-type and ΔftsZ3 mutants, both inoculated with equal amounts of protonema, revealed growth retardation of the ΔftsZ3 mutants (Figure 5G).
FtsZ Transcript Levels in Wild-Type and ftsZ Deletion Mutants
Our previous studies revealed a complex, well ordered network of FtsZ protein–protein interactions in P. patens (Gremillon et al., 2007). Partial redundancy and co-regulatory effects between ftsZ genes in P. patens have been discussed but not studied at the transcript level by quantitative real-time PCR (qRT–PCR), so far. To this end, measurements of transcript levels from wild-type P. patens and the deletion mutants cultured as protonema under standard conditions for 2 weeks were performed. Dry weights of the cultures were adjusted to 150 mg L−1 each and the cultures were harvested 3 d after the last sub-cultivation at the same time of day. In addition to the five ftsZ genes, transcript abundances of the two constitutively expressed genes l21 and ef1α were analyzed. Use of two reference genes and their mean Cp values for normalization enhanced reliability of qRT–PCR results according to Vandesompele et al. (2002). All transcript levels were measured and compared using ANOVA. Tukey's Honestly Significant Differences (HSD) was used to evaluate potentially significant differences in gene expression (The R Development Core Team, 2009).
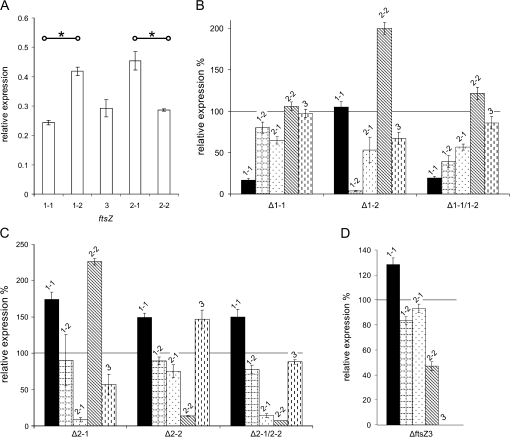
In wild-type P. patens, we found significantly more ftsZ1-2 transcripts than ftsZ1-1 transcripts. Likewise, transcript abundances for ftsZ2-1 were significantly higher than for ftsZ2-2 (Figure 6A). Targeted deletion of one or two ftsZ genes affected transcript abundances of the others when compared to wild-type (Figure 6B–6D). Remarkably, transcript levels of ftsZ1-1 were increased when a gene of the ftsZ2 or ftsZ3 family was deleted, whereas deletion of the ftsZ1-2 gene did not affect ftsZ1-1 transcript abundance. Similarly, transcript abundances of ftsZ2-1 were most affected by deletion of any of the ftsZ1 genes, but nearly not by deletion of ftsZ2-2. However, deletion of the ftsZ1 genes led to reduced transcript levels of ftsZ2-1. In addition, deletion of ftsZ1-2 and of ftsZ2-1, respectively, led to reduced transcript levels of ftsZ3 (Figure 6B–6D).
Figure 6.
FtsZ Transcript Abundance in Wild-Type and in ftsZ Single and Double Knockout Mutants.
(A) Relative expression level of all five ftsZ genes in wild-type. Asterisks indicate significant differences (ANOVA, Tukeys HSD, P < 0.001) between two members of the same family.
(B–D) Wild-type is set to 100% (black line) to depict relative expression levels of the single ftsZ in the mutants. The graphs are grouped by gene family: (B) FtsZ1, (C) FtsZ2, and (D) FtsZ3. Error bars indicate the standard deviation (n = 3). Significant changes in transcript abundance in the wild-type (t-test, P < 0.001) are indicated by an asterisk.
DISCUSSION
The genome of the moss P. patens encodes five ftsZ genes that are members of three distinct subfamilies (Martin et al., 2009). For flowering plants, only genes belonging to two of the ftsZ subfamilies have been found (Osteryoung et al., 1998). To investigate the function of those five moss ftsZ genes, single knockout mutants of all P. patens ftsZ genes and double knockout mutants for the ftsZ1 and ftsZ2 gene families were generated. Since it is puzzling in an evolutionary sense that the morphologically less complex bryophyte P. patens possesses an expanded complement of ftsZ genes as compared to flowering plants, our aim was also to identify functional differences to the previously published A. thaliana FtsZ proteins.
Possible Functions of FtsZ1 Proteins in P. patens
The P. patens proteins FtsZ1-1 and FtsZ1-2 share high sequence identity and are considered as paleologs (Martin et al., 2009), namely they were derived from an ancient large-scale duplication event along the lineage leading to Physcomitrella (Rensing et al., 2007). The targeted deletion of ftsZ1-1 did not lead to an aberrant chloroplast phenotype whereas ΔftsZ1-2 mutants possess enlarged plastids in basal zones of chloronema filaments. In contrast to the lens-shaped wild-type-like plastids observed in the single knockouts, ΔftsZ1-1/1–2 double mutants showed altered plastid shape and size in every cell and tissue type. These findings suggest that PpFtsZ1-1 in interaction with PpFtsZ1-2 fulfils functions related to the maintenance of chloroplast shape. This function could be partially accomplished by FtsZ1-2 in the ΔftsZ1-1 mutants so that loss of the FtsZ1-1 protein did not have visible effects on moss plants. Such partial redundancy between two FtsZ proteins of the same subfamily was previously reported for the FtsZ2 proteins in A. thaliana (McAndrew et al., 2008). From previous studies by Gremillon et al. (2007), it is known that PpFtsZ1-1 is localized to network structures inside the chloroplast and that it participates in the formation of the plastid division apparatus, thus being involved in chloroplast division.
The observed enlargement of chloroplasts in basal chloronema cells of ΔftsZ1-2 mutants revealed a role for PpFtsZ1 in controlling the size of plastids. Localization studies employing an ftsZ1-2:gfp fusion visualized the protein in network structures within the plastids and as a ring surrounding the middle of the organelle (Martin et al., 2009). This ring might exert force on the chloroplast during the plastid division process. It could be shown for bacterial FtsZ that this protein is capable by itself and without the use of any interaction partner to generate a force that is sufficient to constrict liposomes (Osawa et al., 2008). Recently, also a mathematical model describing the FtsZ constriction forces responsible for E. coli cell division has been published (Allard and Cytrynbaum, 2009). Several explanations might account for the enlargement of chloroplasts observed in basal cells of ΔftsZ1-2 protonema filaments but not in younger cells/tip cells. Auxin gradients in P. patens protonema with highest auxin concentration in the tip cell and decreasing auxin levels in the basal cells have been reported previously (Bierfreund et al., 2003). Auxin, like gibberellins and brassinolides, promote preferential cell growth along the longitudinal axis, while other hormones induce expansion along the traverse axis (Shibaoka and Nagai, 1994). Both protonemal cell types, chloronema and caulonema, expand by tip growth (Menand et al., 2007), which is accompanied by substantial changes in the cytoskeleton (Cole and Fowler, 2006; Hussey et al., 2006; Smith and Oppenheimer, 2005). The resulting physiological, biochemical, and structural gradients from dividing apical cells to non-dividing basal filament cells probably influence the formation of complexes between FtsZ and other interaction partners.
In contrast to P. patens, the A. thaliana genome encodes only one FtsZ1 protein. Expression inhibition of AtftsZ1-1 reduced the number of chloroplasts in leaf cells (Osteryoung et al., 1998) while overproduction of the protein led to the formation of either strongly enlarged or long, narrow chloroplasts (Stokes et al., 2000). These observations led to the assumption that AtFtsZ1-1 is involved in chloroplast division. Based on the data obtained for both PpftsZ1 single knockout mutants, both proteins have slightly different functions, but together are needed for chloroplast division. It would be intriguing to analyze plants with a different subset of FtsZ1 proteins, like Nicotiana tabacum, which encodes four FtsZ1 proteins. So far, it was shown that overexpression of NtftsZ1-2 led to the formation of only one to three chloroplasts per mesophyll cell (Jeong et al., 2002).
To date, it is unknown whether, in plants, FtsZ protein amounts correlate to their transcript levels. Here, we found much higher ftsZ1-2 transcript levels compared to ftsZ1-1 transcript levels. If the ratio of transcripts reflects the protein level ratio, a disruption of ftsZ1-2 will unbalance the FtsZ pools to a much larger extent than the disruption of ftsZ1-1, and thus would explain the more severe phenotype of the ΔftsZ1-2 mutants observed in this study. A similar effect of FtsZ pool sizes on the phenotype has been postulated based on the phenotypes of AtftsZ2-1 mutants (McAndrew et al., 2008). In P. patens, targeted deletion of either ftsZ2 genes or the ftsZ3 gene led to increased levels of preferentially the ftsZ1-1 gene, which might reflect a central position of the FtsZ1-1 protein in the protein–protein interaction network.
Possible Functions of FtsZ2 in P. patens
Our re-examination of the ΔftsZ2-1 phenotype confirmed the conclusions from Strepp et al. (1998) that FtsZ2-1 is important for chloroplast division in all gametophytic cells of P. patens. In contrast, targeted deletion of the ftsZ2-2 gene in the present study did not obviously affect chloroplast shape or division. As with the ftsZ1 genes, the double deletion of both ftsZ2 genes gave synergistic effects: the ΔftsZ2-1/2–2 mutants did not only show macrochloroplasts, as described for ΔftsZ2-1, but also had defects not obviously related to chloroplasts, namely aberrant leaf morphology and aberrant cell patterning in the leaves.
Both proteins, FtsZ2-1 and FtsZ2-2, were found in network-like structures in the chloroplast, suggesting that besides a cytoskeleton, a plastoskeleton may exist that helps to maintain plastid shape and integrity (Kiessling et al., 2000). We here found that FtsZ2-2 function in the chloroplast can be substituted by FtsZ2-1. The inability of FtsZ2-2—despite the high sequence similarity of the two proteins—to compensate for loss of FtsZ2-1 might be due to proteins that specifically interact with FtsZ2-1 and that are necessary for the maintenance of plastid integrity. Alternatively, FtsZ2-2 cannot compensate for loss of FtsZ2-1 because the ftsz2-1 gene is more highly expressed (Figure 6). The AtftsZ2-2 mutant showed a relatively mild phenotype in comparison to ΔAtftsZ2-1, although both proteins were reported to have biochemically equivalent functions within the FtsZ2 pool (McAndrew et al., 2008). Hence, partial redundancy in the function of both FtsZ2 proteins seems to exist in P. patens and in A. thaliana. However, a network structure comparable to the plastoskeleton built by FtsZ2-1 and FtsZ2-2 in P. patens has not been described for A. thaliana.
As previously shown for the FtsZ1 family, knockout of the ftsZ2 gene with the highest expression level (ftsZ2-1) resulted in the more dramatic phenotypic aberrations, probably as a result of more dramatic changes in the FtsZ2 protein pools. Nevertheless, cellular defects after double knockout of both ftsZ2 genes were at first glance surprising, as these defects were not seen with other moss mutants with giant chloroplasts, such as in the ΔftsZ2-1 mutants. As plant FtsZ2 and FtsZ3 proteins have a C-terminal motif facilitating protein–protein interactions (Maple et al., 2005; Martin et al., 2009) like bacterial FtsZ have (Haney et al., 2001; Ma and Margolin, 1999; Yan et al., 2000), it seems plausible that depletion of both PpFtsZ2 isoforms in the double mutants affected interaction networks necessary for proper cytosolic functions. The most obvious explanation is a disturbed interaction with PpFtsZ3, which is dually targeted to plastids and to the cytosol in P. patens (Kiessling et al., 2004), and which is interacting with PpFtsZ2-2 in wild-type moss (Gremillon et al., 2007). Consequently, transcript levels of ftsZ2-2 are reduced in the ΔftsZ3 mutants, but not in the other knockout mutants created here.
Possible Functions of FtsZ3 in P. patens
It was previously shown that expression of ftsZ3:gfp labeled plastidic as well as cytosolic rings in P. patens protoplasts, protonema cells, and leaf cells, indicating that this protein may be a molecular link between cell and chloroplast division in moss (Kiessling et al., 2004).
The ΔftsZ3 mutants presented here possess giant chloroplasts with unusual shapes different from those observed in knockout mutants of the other ftsZ genes. However, especially the tubular macrochloroplasts in protonema cells matched the filamentous growth phenotype of E. coli ftsZ mutants (Dai and Lutkenhaus, 1991) very well, demonstrating its function in chloroplast division. Unable to divide, the chloroplasts were growing to enormous lengths and the presence of the other four FtsZ proteins prevented them, in accordance with their function in a plastoskeleton, from ballooning up.
In addition, and in accordance with the dual localization of FtsZ3 in moss, ΔftsZ3 mutants showed numerous phenotypic aberrations, such as retarded plant growth and development, decreased cell wall thickness, altered cell size and shape, and an impaired gravitropic response.
In contrast to our knowledge on cell patterning in A. thaliana epidermal tissue (Larkin et al., 2003), the analysis of cell patterning in moss leaves is just beginning (Harrison et al., 2009). The cellular defects observed here, however, reveal a role of cytosolic FtsZ3 in cell patterning of Physcomitrella. It remains to be elucidated whether PpFtsZ3 interacts with components of the cytoskeleton or is directly involved in cell shaping analogous to the involvement of bacterial FtsZ in Caulobacter crescentus where an FtsZ-dependent mode of peptidoglycan synthesis for cell elongation exists (Aaron et al., 2007). Interestingly, P. patens encodes nine homologs of genes related to the bacterial peptidoglycan synthesis pathway, while A. thaliana encodes only five of them (Machida et al., 2006). Subsequent analyses of some of these genes revealed their functional differences between cyanobacteria, P. patens, and A. thaliana (Garcia et al., 2008), revealing evolutionary differences of these genes during land plant evolution, nicely explaining why β-lactam antibiotics inhibit division of bacteria and moss chloroplasts, but not the division of plastids in seed plants (Kasten and Reski, 1997; Reski, 2009). Another evidence for the ancient function of PpFtsZ3 came from Gremillon et al. (2007), who reported that E. coli FtsZ can substitute PpFtsZ3 in its protein–protein interaction with PpFtsZ2-2 in P. patens cells.
Another striking feature of ΔftsZ3 mutants is their dramatically decreased cell wall thickness, which points to a disturbed transport of building materials (cellulose, hemicelluloses, and pectins) to the cell wall. It is known that cellulose is synthesized at the plasma membrane and that its deposition is guided by cortical microtubules (Paredez et al., 2006). Hemicellulose and pectins are synthesized in the Golgi apparatus and reach the growing cell wall via vesicle transport, which is based on actin filaments (Chen et al., 2007). Thus, the cytoskeleton is crucial for cell wall assembly in plants. Similarly, however, FtsZ in physical interaction with MreB, the prokaryotic actin homolog, directs the insertion of new peptidoglycans into the side walls near the growing poles of E. coli (Varma et al., 2007). To date, it is unclear whether PpFtsZ3 itself, or via interaction with the cytoskeleton, is facilitating cell wall formation in P. patens.
Both cell types of the moss protonema, chloronema and caulonema, grow via tip growth, like pollen tubes of seed plants. While chloronema does not grow in the dark, caulonema grows negatively gravitropic in the absence of light (Cove, 1992). Surprisingly, protonema of ΔftsZ3 mutants seemed to consist of chloronema only. In the dark, however, caulonema started to develop, but, unlike the situation in wild-type, these cells did not follow the gravity vector. Moreover, the cross walls in the mutants did not show the oblique orientation that in wild-type moss is mediated by microtubules (Reski, 1998b). Likewise, gravity sensing in the moss protonema is restricted to the growing tip cells where starch-rich plastids trigger re-orientation of the cytoskeleton to initiate curvature of the growing cell (Blancaflor, 2002; Sack et al., 2001; Schwuchow et al., 2002). Thus, both abnormalities argue in favor of a defective cytoskeleton in the ΔftsZ3 mutants, albeit a putative integration of ancient tubulin FtsZ3 into the classical cytoskeleton of Physcomitrella remains to be elucidated.
Up to now, ftsZ3 genes have only been identified in the moss P. patens and in the lycophyte Selaginella moellendorffii, while flowering plants only encode FtsZ proteins of the FtsZ subfamilies 1 and 2. While PpftsZ1- and PpftsZ2-genes possess six introns, the PpftsZ3 gene possesses only three introns. This ‘more ancient’ gene structure of PpftsZ3 and the importance of its protein for cellular functions reveal its close relationship to its bacterial ancestor. As FtsZ3 not only is dually targeted to plastids and to the cytosol of P. patens, but also exerts specific functions in both compartments, it integrates the FtsZ1/FtsZ2-based plastoskeleton with the cytoskeleton in this species.
Although we here found several FtsZ functions to be similar between P. patens and A. thaliana, there were also striking differences. Given the about 450 million years of separate evolution between both plant species and the basal position mosses retained in land plant evolution (Lang et al., 2008), it is plausible that the chloroplast division apparatus of mosses has retained more bacterial features than those of seed plants. Thus, our results are in good accordance with the elegant works of Machida et al. (2006) and Garcia et al. (2008) demonstrating the importance of homologs of the bacterial Pbp and MurE for plastid division in P. patens (reviewed in Reski, 2009).
Besides that, an involvement of the ‘bacterial tubulin’ FtsZ3 in cellular functions may have resulted in a moss-specific evolution of eukaryotic β-tubulins as described for P. patens by Jost et al. (2004). Thus, the ftsZ gene family may not only serve as a model to study post-duplication evolution and the impact of redundancy, sub- and neofunctionalization, but may also be instructive to understand the evolution of the cytoskeleton in land plants.
METHODS
Plant Material and Growth Conditions
P. patens was cultured as described in Strepp et al. (1998).
For the adjustment of dry weight in P. patens, 10 mL of liquid culture were removed prior to sub-culturing and filtered through gauze (Miracloth, Calbiochem, Schwalbach, Germany). The dry weight was determined after drying the sample for 2 h at 105°C.
Knockout mutants described in this study are deposited in the International Moss Stock Center with the accessions IMSC 40128–40149 (ΔftsZ1-1 (#216), ΔftsZ1-2 (#12, #14, #Q1), ΔftsZ1-1/1–2 (#12, #14, #15, #45, #46), ΔftsZ3 (#12, #32, #38, #61, #114), ΔftsZ2-1, ΔftsZ2-2 (#34, #97, #101, #103, #134), ΔftsZ2-1/2–2 (#8, #23). Mutant numbers as shown in text and figures are indicated in brackets.
Phenotypic Analysis
To investigate the development of negatively gravitropic, dark-grown caulonema filaments, 5 μL droplets from liquid cultures with a dry weight of 100 mg L−1 were placed on solid Knop medium. Plates were sealed and placed in a climate chamber with standard conditions for 6 d. Subsequently, the plates were wrapped in tin foil and positioned vertically. Culturing under standard conditions was carried out for another 7 d. Single protonema colonies were examined with regard to the development of caulonema filaments. Pictures of representative colonies were taken.
For the determination of gametophore, leaf and cell dimensions as well as cell wall thickness, the LSM 5 image browser (Zeiss, Jena, Germany) was employed. Raw data were analyzed in Excel. Statistical evaluation was performed using SigmaPlot 9.0 (Systat Software GmbH, Erkrath, Germany). Statistical evaluation of the results was done via t-test. P-values smaller than 0.001 indicate significant differences between the two groups tested.
Knockout Constructs and Plant Transformation
For generation of knockout constructs, genomic fragments of the ftsZ genes were amplified from P. patens genomic DNA, cloned into pCHR4-TOPO (Invitrogen, Karlsruhe, Germany) and selection cassettes were inserted into the linearized vectors (for details, see Table 2).
Table 2.
Primers for the Amplification of Genomic ftsZ Fragments Are Given as Pairs.
| Knockout construct | Oligonucleotides (5′-3’) | Selection cassette (insertion sites) | |
| ftsZ1-1 | fwd: CTGCAGAAGTGGCGTGTTGAAG | nptII (ClaI-SalI) | |
| rev: AGGCATCCGTCATCCCAGAATTC | |||
| ftsZ1-2 | fwd: ATGGGCTCTACGGCGAGGTT | hpt (HpaI-EcoRV) | |
| rev: GAGGCAGCCTGAATTGCAGCT | |||
| ftsZ3 | fwd: ATGATCACGTGTAGGGTTTGG | nptII (AgeI-HpaI) | |
| rev: CGATGTTGACCTCGTGAAGAG | |||
| ftsZ2-2 | fwd: CAGGGTGAGCTCGCGAGTG | hpt (HindIII-BamHI) | |
| rev: TTTGGTACCCTGATGAAATCAAAACTAATACCAC |
Restriction sites flanking the selection cassettes are listed in brackets. nptII (neomycin phosphotransferase II) was amplified from pRT101neo (R. Strepp); hpt (hygromycin phosphotransferase) was amplified from pCAMBIA1305 (Huether et al., 2005).
Protoplast isolation from P. patens protonema, transformation, and regeneration of stably transformed plants were performed as described previously (Strepp et al., 1998).
For the generation of ftsZ1-1/1–2 double knockouts, a ΔftsZ1-1 mutant was transformed with the knockout construct for ftsZ1-2. For the generation of ftsZ2-1/2–2 double knockouts, the already existing ΔftsZ2-1 mutant was transformed with the knockout construct for ftsZ2-2.
Plant Screening
Plants were screened for disruption of the respective ftsZ locus and correct 5’ and 3’ integration of the knockout construct via direct PCR (Supplemental Figure 1) (Schween et al., 2002). Primers used for the analysis are listed in Table 3.
Table 3.
Primers for PCR Screens of Putative ftsZ Knockout Plants.
| Gene | Screen | Oligonucleotides (5′-3’) |
| ftsZ1-1 | WT | fwd: ATCGCGTTCAAATCGGCGAAG |
| rev: CTTTCAAGCCTGGGCTTAACCA | ||
| 5’ | fwd: AGACGGTTTGTAGATGGCG | |
| rev: TGTCGTGCTCCACCATGTT | ||
| 3’ | fwd: GTTGAGCATATAAGAAAC | |
| rev: CTGTTACAATCTGCATCA | ||
| ftsZ1-2 | WT | fwd: GCGTCGGCACGGTCTGTGTAT |
| rev: CGCGATGTTGGGCTTGTG | ||
| 5’ | fwd: GCGTAGCCTTTGCAGTAC | |
| rev: ATAGTGACCTTAGGCGAC | ||
| 3’ | fwd: TTAACATGTAATGCATGACG | |
| rev: CTGAGATACAGTATTCACTTC | ||
| ftsZ3 | WT | fwd: TTGGACAGGACACGACAGCCGGC |
| rev: GCTCCTGAACCAGTACCGCCACC | ||
| 5’ | fwd: AACCTTGTGGACGACTGTG | |
| rev: TGTCGTGCTCCACCATGTT | ||
| 3’ | fwd: GTTGAGCATATAAGAAACCC | |
| rev: CCAGCTTTGAGTACAGGCTT | ||
| ftsZ2-2 | 5’ | fwd: ATGGCGCTGTTAGGCAGTC |
| rev: ATAGTGACCTTAGGCGAC | ||
| 3’ | fwd: TTAACATGTAATGCATGACG | |
| rev: TCATTAAGTCTGCCACTCC |
WT: PCR product can only be synthesized in the wild-type due to primer binding either in a deleted region or flanking the selection cassette, thus yielding a large product that cannot be amplified. 5’ and 3’: primer pairs bind outside the knockout construct and inside the selection cassette, thus proving correct integration at the genomic locus.
RT–PCR
After RNA-isolation using Trizol reagent (Invitrogen, Karlsruhe, Germany) and cDNA synthesis from 2 μg total RNA using Superscript III Reverse Transcriptase (Invitrogen, Karlsruhe, Germany), RT–PCR was performed to show the absence of the respective ftsZ transcript in the knockout plants. Primers used for the analysis are listed in Table 4. Primers for the constitutively expressed ribosomal gene l21 were used as control.
Table 4.
Primers for RT–PCR Analysis of Putative ftsZ Knockout Plants.
| Gene | Oligonucleotides (5′-3’) |
| ftsZ1-1 | fwd: ATGATGAGCTCCATGGTGAG |
| rev: TCAGACACGGTGTTGACTTC | |
| ftsZ1-2 | fwd: ATGGGCTCTACGGCGAGGTT |
| rev: GAGGCAGCCTGAATTGCAGCT | |
| ftsZ3 | fwd: TTGGACAGGACACGACAGCCGGC |
| rev: CAGTGTCAGCCGCATGGCGCATG | |
| ftsZ2-2 | fwd: TCATTGCTGGTGTAGCGAA |
| rev: ATCGTAGAGCTATTGCCAC | |
| L21 | fwd: GGTTGGTCATGGGTTGCG |
| rev: GAGGTCAACTGTCTCGCC |
Quantitative Real-Time PCR
For qRT–PCR analysis, RNA was extracted using Trizol reagent (Invitrogen, Karlsruhe, Germany) and purified using RNeasy columns (Qiagen, Hilden, Germany). An additional DNase I (Fermentas, St Leon-Rot, Germany) digestion was performed after RNA cleanup. Samples were reverse-transcribed into first-strand cDNA using Taq Man Reverse Transcription Reagents (Applied Biosystems, Darmstadt, Germany). Then, PCR was carried out using gene-specific primers (Table 5) and LightCycler® 480 SYBR Green I Master (Roche, Mannheim, Germany) with a Roche 480 Light Cycler following the manufacturer's instructions. EF1α and L21 were used as reference genes to normalize for variation in the amount of cDNA template. Each real-time PCR experiment contained three technical replicates using 50 ng of cDNA in a total volume of 20 μl. Melting curve analysis for validation of the PCR reaction was carried out routinely. FtsZ expression levels were calculated relative to wild-type transcript abundance employing relative quantification with efficiency correction (Livak and Schmittgen, 2001; Rasmussen, 2001).
Table 5.
Primers for qRT–PCR Analysis of ftsZ Transcript Abundance in Wild-Type and ftsZ Single and Double Knockout Plants.
| Gene | Oligonucleotides (5′-3’) |
| ftsZ1-1 | fwd: CGTTTCTAGGTCGGAGAGGC |
| rev: ACGAAATCTTGATGAGGTGGG | |
| ftsZ1-2 | fwd: ATGGGCTCTACGGCGAGGTT |
| rev: TACCGAGCCTACTCTCCGCTC | |
| ftsZ3 | fwd: GAGCTTTCGATGGCACTTGA |
| rev: TTGTCGAGCTCGGGTGAAC | |
| ftsZ2-1 | fwd: GTTGTTTAGTGGGTGCTCGG |
| rev: GGGGTTCACATCGCAAAGG | |
| ftsZ2-2 | fwd: TGCCATTCAGTCTCCATTGTT |
| rev: TTGCATACTGCATACTCCGTG | |
| EF1α | fwd: CGACGCCCCTGGACATC |
| rev: CCTGCGAGGTTCCCGTAA | |
| L21 | fwd: GGTTGGTCATGGGTTGCG |
| rev: GACCTCAACTGTCTCGCC |
ANOVA of wild-type transcript levels in protonemata was performed using the functions aov, lm, anova, and TukeyHSD implemented in R 2.4.1 (The R Development Core Team, 2009). Significance was inferred assuming a 99% confidence interval.
Northern Blotting
RNA blot hybridization was carried out as described (Frank et al., 2005) with a radioactively labeled ftsZ2-2 cDNA probe that was amplified using the primers 5′-AAGGTAGTACAAATGGGATGGC-3′ and 5′-TCATTAAGTCTGCCACTCCAC-3′.
Microscopic Observations
Bright-field images of cells were taken with a charge-coupled device (CCD) camera (AxioCam MRc5 or ICc1, Zeiss, Jena, Germany) under microscope Axioplan and Stemi 2000C (Zeiss, Jena, Germany). Electron microscopy of protonema cells was carried out at the CM 10 (Philips, Hamburg, Germany) at 60 kV.
Accession Numbers
Sequence data from this article can be found in the GenBank data libraries under accession numbers as follows: FtsZ1-1 XM_001780186 (Phypa_196781); FtsZ2-1 XM_001765901 (Phypa_212480); FtsZ3 XM_001765160 (Phypa_211907); FtsZ2-2 XM_001766723 (Phypa_185873); FtsZ1-2 XM_001769006 (Phypa_187670). P. patens gene models are given in brackets.
SUPPLEMENTARY DATA
Supplementary Data are available at Molecular Plant Online.
FUNDING
This work was supported by the Deutsche Forschungsgemeinschaft (SFB388 and Re837/10) and by the Excellence Initiative of the German Federal and State Governments (GSC-4 and EXC 294).
Supplementary Material
Acknowledgments
We thank Ida Suppanz for designing parts of the ftsZ1-1 knockout construct and Eva L. Decker and Agnes Novakovic for planning the ftsZ2-2 construct and doing the cloning, respectively. We are also grateful to Justine Kiessling for creating the ftsZ3 construct and to Volker Speth for his help with TEM and during the preparation of specimens. No conflict of interest declared.
References
- Aaron M, Charbon G, Lam H, Schwarz H, Vollmer W, Jacobs-Wagner C. The tubulin homologue FtsZ contributes to cell elongation by guiding cell wall precursor synthesis in Caulobacter crescentus. Mol. Microbiol. 2007;64:938–952. doi: 10.1111/j.1365-2958.2007.05720.x. [DOI] [PubMed] [Google Scholar]
- Allard JF, Cytrynbaum EN. Force generation by a dynamic Z-ring in Escherichia coli cell division. Proc. Natl Acad. Sci. U S A. 2009;106:145–150. doi: 10.1073/pnas.0808657106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierfreund NM, Reski R, Decker EL. Use of an inducible reporter gene system for the analysis of auxin distribution in the moss Physcomitrella patens. Plant Cell Rep. 2003;21:1143–1152. doi: 10.1007/s00299-003-0646-1. [DOI] [PubMed] [Google Scholar]
- Blancaflor EB. The cytoskeleton and gravitropism in higher plants. J. Plant Growth Regul. 2002;21:120–136. doi: 10.1007/s003440010041. [DOI] [PubMed] [Google Scholar]
- Chen T, Teng N, Wu X, Wang Y, Tang W, Samaj J, Baluska F, Lin J. Disruption of actin filaments by latrunculin B affects cell wall construction in Picea meyeri pollen tube by disturbing vesicle trafficking. Plant Cell Physiol. 2007;48:19–30. doi: 10.1093/pcp/pcl036. [DOI] [PubMed] [Google Scholar]
- Cole RA, Fowler JE. Polarized growth: maintaining focus on the tip. Curr. Opin. Plant Biol. 2006;9:579–588. doi: 10.1016/j.pbi.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Cove DJ. Chapter 12: Regulation of development in the moss, Physcomitrella patens. In: Russo VEA, Brody S, Cove D, Ottolenghi S, editors. Development: The Molecular Genetic Approach. Berlin: Springer-Verlag; 1992. [Google Scholar]
- Dai K, Lutkenhaus J. ftsZ is an essential cell division gene in Escherichia coli. J. Bacteriol. 1991;173:3500–3506. doi: 10.1128/jb.173.11.3500-3506.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer P, Crossley R, Rothfield L. The essential bacterial cell-division protein FtsZ is a GTPase. Nature. 1992;359:254–256. doi: 10.1038/359254a0. [DOI] [PubMed] [Google Scholar]
- Decker EL, Frank W, Sarnighausen E, Reski R. Moss systems biology en route: phytohormones in Physcomitrella development. Plant Biol. (Stuttg.) 2006;8:397–405. doi: 10.1055/s-2006-923952. [DOI] [PubMed] [Google Scholar]
- El-Kafafi S, Mukherjee S, El-Shami M, Putaux JL, Block MA, Pignot-Paintrand I, Lerbs-Mache S, Falconet D. The plastid division proteins, FtsZ1 and FtsZ2, differ in their biochemical properties and sub-plastidial localization. Biochem J. 2005;387:669–676. doi: 10.1042/BJ20041281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Shami M, El-Kafafi S, Falconet D, Lerbs-Mache S. Cell cycle-dependent modulation of FtsZ expression in synchronized tobacco BY2 cells. Mol. Genet. Genomics. 2002;267:254–261. doi: 10.1007/s00438-002-0660-y. [DOI] [PubMed] [Google Scholar]
- Erickson HP, Taylor DW, Taylor KA, Bramhill D. Bacterial cell division protein FtsZ assembles into protofilament sheets and minirings, structural homologs of tubulin polymers. Proc. Natl Acad. Sci. U S A. 1996;93:519–523. doi: 10.1073/pnas.93.1.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank W, Ratnadewi D, Reski R. Physcomitrella patens is highly tolerant against drought, salt and osmotic stress. Planta. 2005;220:384–394. doi: 10.1007/s00425-004-1351-1. [DOI] [PubMed] [Google Scholar]
- Garcia M, Myouga F, Takechi K, Sato H, Nabeshima K, Nagata N, Takio S, Shinozaki K, Takano H. An Arabidopsis homolog of the bacterial peptidoglycan synthesis enzyme MurE has an essential role in chloroplast development. Plant J. 2008;53:924–934. doi: 10.1111/j.1365-313X.2007.03379.x. [DOI] [PubMed] [Google Scholar]
- Gilson PR, Beech PL. Cell division protein FtsZ: running rings around bacteria, chloroplasts and mitochondria. Res. Microbiol. 2001;152:3–10. doi: 10.1016/s0923-2508(00)01162-1. [DOI] [PubMed] [Google Scholar]
- Gould SB, Waller RF, McFadden GI. Plastid evolution. Annu. Rev. Plant Biol. 2008;59:491–517. doi: 10.1146/annurev.arplant.59.032607.092915. [DOI] [PubMed] [Google Scholar]
- Gray MW. The endosymbiont hypothesis revisited. Int. Rev. Cytol. 1992;141:233–357. doi: 10.1016/s0074-7696(08)62068-9. [DOI] [PubMed] [Google Scholar]
- Gremillon L, Kiessling J, Hause B, Decker EL, Reski R, Sarnighausen E. Filamentous temperature-sensitive Z (FtsZ) isoforms specifically interact in the chloroplasts and in the cytosol of Physcomitrella patens. New Phytologist. 2007;176:299–310. doi: 10.1111/j.1469-8137.2007.02169.x. [DOI] [PubMed] [Google Scholar]
- Haney SA, Glasfeld E, Hale C, Keeney D, He Z, de Boer P. Genetic analysis of the Escherichia coli FtsZ.ZipA interaction in the yeast two-hybrid system: characterization of FtsZ residues essential for the interactions with ZipA and with FtsA. J. Biol. Chem. 2001;276:11980–11987. doi: 10.1074/jbc.M009810200. [DOI] [PubMed] [Google Scholar]
- Harrison CJ, Roeder AHK, Meyerowitz EM, Langdale JA. Local cues and assymmetric cell divisions underpin body plan transitions in the moss Physcomitrella patens. Curr. Biol. 2009;19:461–471. doi: 10.1016/j.cub.2009.02.050. [DOI] [PubMed] [Google Scholar]
- Huether CM, Lienhart O, Baur A, Stemmer C, Gorr G, Reski R, Decker EL. Glyco-engineering of moss lacking plant-specific sugar residues. Plant Biol.(Stuttg.) 2005;7:292–299. doi: 10.1055/s-2005-837653. [DOI] [PubMed] [Google Scholar]
- Hussey PJ, Ketelaar T, Deeks MJ. Control of the actin cytoskeleton in plant cell growth. Annu. Rev. Plant Biol. 2006;57:109–125. doi: 10.1146/annurev.arplant.57.032905.105206. [DOI] [PubMed] [Google Scholar]
- Jeong WJ, Park YI, Suh K, Raven JA, Yoo OJ, Liu JR. A large population of small chloroplasts in tobacco leaf cells allows more effective chloroplast movement than a few enlarged chloroplasts. Plant Physiol. 2002;129:112–121. doi: 10.1104/pp.000588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost W, Baur A, Nick P, Reski R, Gorr G. A large plant beta-tubulin family with minimal C-terminal variation but differences in expression. Gene. 2004;340:151–160. doi: 10.1016/j.gene.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Kasten B, Reski R. β-lactam antibiotics inhibit chloroplast division in a moss (Physcomitrella patens) but not in tomato (Lycopersicon esculentum) J. Plant Physiol. 1997;150:137–140. [Google Scholar]
- Kiessling J, Kruse S, Rensing SA, Harter K, Decker EL, Reski R. Visualization of a cytoskeleton-like FtsZ network in chloroplasts. J. Cell Biol. 2000;151:945–950. doi: 10.1083/jcb.151.4.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiessling J, Martin A, Gremillon L, Rensing SA, Nick P, Sarnighausen E, Decker EL, Reski R. Dual targeting of plastid division protein FtsZ to chloroplasts and the cytoplasm. EMBO Rep. 2004;5:889–894. doi: 10.1038/sj.embor.7400238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler RH, Cao J, Zipfel WR, Webb WW, Hanson MR. Exchange of protein molecules through connections between higher plant plastids. Science. 1997;276:2039–2042. doi: 10.1126/science.276.5321.2039. [DOI] [PubMed] [Google Scholar]
- Kuroiwa T, Kuroiwa H, Sakai A, Takahashi H, Toda K, Itoh R. The division apparatus of plastids and mitochondria. Int. Rev. Cytol. 1998;181:1–41. doi: 10.1016/s0074-7696(08)60415-5. [DOI] [PubMed] [Google Scholar]
- Kuroiwa T, Misumi O, Nishida K, Yagisawa F, Yoshida Y, Fujiwara T, Kuroiwa H. Vesicle, mitochondrial, and plastid division machineries with emphasis on dynamin and electron-dense rings. Int. Rev. Cell Mol. Biol. 2008;271:97–152. doi: 10.1016/S1937-6448(08)01203-3. [DOI] [PubMed] [Google Scholar]
- Lang D, Zimmer AD, Rensing SA, Reski R. Exploring plant biodiversity: the Physcomitrella genome and beyond. Trends Plant Sci. 2008;13:542–549. doi: 10.1016/j.tplants.2008.07.002. [DOI] [PubMed] [Google Scholar]
- Larkin JC, Brown ML, Schiefelbein J. How do cells know what they want to be when they grow up? Lessons from epidermal patterning in Arabidopsis. Annu. Rev. Plant Biol. 2003;54:403–430. doi: 10.1146/annurev.arplant.54.031902.134823. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lowe J, Amos LA. Crystal structure of the bacterial cell-division protein FtsZ. Nature. 1998;391:203–206. doi: 10.1038/34472. [DOI] [PubMed] [Google Scholar]
- Ma X, Margolin W. Genetic and functional analyses of the conserved C-terminal core domain of Escherichia coli FtsZ. J. Bacteriol. 1999;181:7531–7544. doi: 10.1128/jb.181.24.7531-7544.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida M, et al. Genes for the peptidoglycan synthesis pathway are essential for chloroplast division in moss. Proc. Natl Acad. Sci. U S A. 2006;103:6753–6758. doi: 10.1073/pnas.0510693103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maple J, Aldridge C, Moller SG. Plastid division is mediated by combinatorial assembly of plastid division proteins. Plant J. 2005;43:811–823. doi: 10.1111/j.1365-313X.2005.02493.x. [DOI] [PubMed] [Google Scholar]
- Martin A, Lang D, Heckmann J, Zimmer AD, Vervliet-Scheebaum M, Reski R. A uniquely high number of ftsZ genes in the moss Physcomitrella patens. Plant Biology. 2009;11:744–750. doi: 10.1111/j.1438-8677.2008.00174.x. [DOI] [PubMed] [Google Scholar]
- Martin W, Stoebe B, Goremykin V, Hapsmann S, Hasegawa M, Kowallik KV. Gene transfer to the nucleus and the evolution of chloroplasts. Nature. 1998;393:162–165. doi: 10.1038/30234. [DOI] [PubMed] [Google Scholar]
- McAndrew RS, Froehlich JE, Vitha S, Stokes KD, Osteryoung KW. Colocalization of plastid division proteins in the chloroplast stromal compartments establishes a new functional relationship between FtsZ1 and FtsZ2 in higher plants. Plant Phys. 2001;127:1656–1666. [PMC free article] [PubMed] [Google Scholar]
- McAndrew RS, Olson BJ, Kadirjan-Kalbach DK, Chi-Ham CL, Vitha S, Froehlich JE, Osteryoung KW. In vivo quantitative relationship between plastid division proteins FtsZ1 and FtsZ2 and identification of ARC6 and ARC3 in a native FtsZ complex. Biochem. J. 2008;412:367–378. doi: 10.1042/BJ20071354. [DOI] [PubMed] [Google Scholar]
- Menand B, Calder G, Dolan L. Both chloronemal and caulonemal cells expand by tip growth in the moss Physcomitrella patens. J. Exp. Bot. 2007;58:1843–1849. doi: 10.1093/jxb/erm047. [DOI] [PubMed] [Google Scholar]
- Miyagishima SY, Froehlich JE, Osteryoung KW. PDV1 and PDV2 mediate recruitment of the dynamin-related protein ARC5 to the plastid division site. Plant Cell. 2006;18:2517–2530. [Google Scholar]
- Miyagishima SY, Kuwayama H, Urushihara H, Nakanishi H. Evolutionary linkage between eukaryotic cytokinesis and chloroplast division by dynamin proteins. Proc. Natl Acad. Sci. U S A. 2008;105:15202–15207. doi: 10.1073/pnas.0802412105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa M, Anderson DE, Erickson HP. Reconstitution of contractile FtsZ rings in liposomes. Science. 2008;320:792–794. doi: 10.1126/science.1154520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osteryoung KW, Vierling E. Conserved cell and organelle division. Nature. 1995;376:473–474. doi: 10.1038/376473b0. [DOI] [PubMed] [Google Scholar]
- Osteryoung KW, Stokes KD, Rutherford SM, Percival AL, Lee WY. Chloroplast division in higher plants requires members of two functionally divergent gene families with homology to bacterial ftsZ. Plant Cell. 1998;10:1991–2004. doi: 10.1105/tpc.10.12.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredez AR, Somerville CR, Ehrhardt DW. Visualization of cellulose synthase demonstrates functional association with microtubules. Science. 2006;312:1491–1495. doi: 10.1126/science.1126551. [DOI] [PubMed] [Google Scholar]
- Possingham JV, Lawrence ME. Controls to plastid division. Int. Rev. Cytol. 1983;84:1–56. [Google Scholar]
- Rasmussen R. Quantification on the LightCycler instrument. In: Meuer S, Wittwer C, Nakagawara K, editors. Rapid Cycle Real-Time PCR: Methods and Applications. Heidelberg: Springer; 2001. [Google Scholar]
- Rensing SA, et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science. 2008;319:64–69. doi: 10.1126/science.1150646. [DOI] [PubMed] [Google Scholar]
- Rensing SA, Ick J, Fawcett JA, Lang D, Zimmer A, Van de Peer Y, Reski R. An ancient genome duplication contributed to the abundance of metabolic genes in the moss Physcomitrella patens. BMC Evol. Biol. 2007;7:130. doi: 10.1186/1471-2148-7-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing SA, Kiessling J, Reski R, Decker EL. Diversification of ftsZ during early land plant evolution. J. Mol. Evol. 2004;58:154–162. doi: 10.1007/s00239-003-2535-1. [DOI] [PubMed] [Google Scholar]
- Reski R. Physcomitrella and Arabidopsis: the David and Goliath of reverse genetics. Trends Plant Sci. 1998a;3:209–210. [Google Scholar]
- Reski R. Development, genetics and molecular biology of mosses. Botanica Acta. 1998b;111:1–15. [Google Scholar]
- Reski R. Rings and networks: the amazing complexity of FtsZ in chloroplasts. Trends Plant Sci. 2002;7:103–105. doi: 10.1016/s1360-1385(02)02232-x. [DOI] [PubMed] [Google Scholar]
- Reski R. Challenges to our current view on chloroplasts. Biological Chemistry. 2009;390:731–738. doi: 10.1515/BC.2009.089. [DOI] [PubMed] [Google Scholar]
- Sack FD, Schwuchow JM, Wagner T, Kern V. Gravity sensing in moss protonemata. Adv. Space Res. 2001;27:871–876. doi: 10.1016/s0273-1177(01)00151-x. [DOI] [PubMed] [Google Scholar]
- Schween G, Fleig S, Reski R. High-throughput-PCR screen of 15,000 transgenic Physcomitrella plants. Plant Mol. Biol. Rep. 2002;20:43–47. [Google Scholar]
- Schwuchow JM, Kern VD, White NJ, Sack FD. Conservation of the plastid sedimentation zone in all moss genera with known gravitropic protonemata. J. Plant Growth Regul. 2002;21:146–155. doi: 10.1007/s003440010048. [DOI] [PubMed] [Google Scholar]
- Shibaoka H, Nagai R. The plant cytoskeleton. Curr. Opin. Cell Biol. 1994;6:10–15. doi: 10.1016/0955-0674(94)90110-4. [DOI] [PubMed] [Google Scholar]
- Smith LG, Oppenheimer DG. Spatial control of cell expansion by the plant cytoskeleton. Annu. Rev. Cell Dev. Biol. 2005;21:271–295. doi: 10.1146/annurev.cellbio.21.122303.114901. [DOI] [PubMed] [Google Scholar]
- Stokes KD, McAndrew RS, Figueroa R, Vitha S, Osteryoung KW. Chloroplast division and morphology are differentially affected by overexpression of FtsZ1 and FtsZ2 genes in Arabidopsis. Plant Physiol. 2000;124:1668–1677. doi: 10.1104/pp.124.4.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strepp R, Scholz S, Kruse S, Speth V, Reski R. Plant nuclear gene knockout reveals a role in plastid division for the homolog of the bacterial cell division protein FtsZ, an ancestral tubulin. Proc. Natl Acad. Sci. U S A. 1998;95:4368–4373. doi: 10.1073/pnas.95.8.4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suppanz I, Sarnighausen E, Reski R. An integrated physiological and genetic approach to the dynamics of FtsZ targeting and organisation in a moss, Physcomitrella patens. Protoplasma. 2007;232:1–9. doi: 10.1007/s00709-007-0284-5. [DOI] [PubMed] [Google Scholar]
- The R Development Core Team A Language and Environment for Statistical Computing. 2009 V2.4.1. ISSN 3-900051-07-0. [Google Scholar]
- Vandenbussche F, Fierro AC, Wiedemann G, Reski R, Van Der Straeten D. Evolutionary conservation of plant gibberellin signalling pathway components. BMC Plant Biol. 2007;7:65. doi: 10.1186/1471-2229-7-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT–PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma A, de Pedro M, Young KD. FtsZ directs a second mode of peptidoglycan synthesis in Escherichia coli. J. Bacteriol. 2007;189:5692–5704. doi: 10.1128/JB.00455-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitha S, Froehlich JE, Koksharova O, Pyke KA, van Erp H, Osteryoung KW. ARC6 is a J-domain plastid division protein and an evolutionary descendant of the cyanobacterial cell division protein Ftn2. Plant Cell. 2003;15:1918–1933. doi: 10.1105/tpc.013292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitha S, McAndrew RS, Osteryoung KW. FtsZ ring formation at the chloroplast division site in plants. J. Cell Biol. 2001;153:111–120. doi: 10.1083/jcb.153.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan K, Pearce KH, Payne DJ. A conserved residue at the extreme C-terminus of FtsZ is critical for the FtsA–FtsZ interaction in Staphylococcus aureus. Biochem. Biophys. Res. Commun. 2000;270:387–392. doi: 10.1006/bbrc.2000.2439. [DOI] [PubMed] [Google Scholar]
- Yoshida Y, Kuroiwa H, Misumi O, Nishida K, Yagisawa F, Fujiwara T, Nanamiya H, Kawamura F, Kuroiwa T. Isolated chloroplast division machinery can actively constrict after stretching. Science. 2006;313:1435–1438. doi: 10.1126/science.1129689. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.