Abstract
Phosphoribulokinase (PRK), a nuclear-encoded plastid-localized enzyme unique to the photosynthetic carbon reduction (Calvin) cycle, was cloned and characterized from the stramenopile alga Vaucheria litorea. This alga is the source of plastids for the mollusc (sea slug) Elysia chlorotica which enable the animal to survive for months solely by photoautotrophic CO2 fixation. The 1633-bp V. litorea prk gene was cloned and the coding region, found to be interrupted by four introns, encodes a 405-amino acid protein. This protein contains the typical bipartite target sequence expected of nuclear-encoded proteins that are directed to complex (i.e. four membrane-bound) algal plastids. De novo synthesis of PRK and enzyme activity were detected in E. chlorotica in spite of having been starved of V. litorea for several months. Unlike the algal enzyme, PRK in the sea slug did not exhibit redox regulation. Two copies of partial PRK-encoding genes were isolated from both sea slug and aposymbiotic sea slug egg DNA using PCR. Each copy contains the nucleotide region spanning exon 1 and part of exon 2 of V. litorea prk, including the bipartite targeting peptide. However, the larger prk fragment also includes intron 1. The exon and intron sequences of prk in E. chlorotica and V. litorea are nearly identical. These data suggest that PRK is differentially regulated in V. litorea and E. chlorotica and at least a portion of the V. litorea nuclear PRK gene is present in sea slugs that have been starved for several months.
Keywords: Alga, Calvin cycle, Elysia chlorotica, kleptoplast, mollusc, phosphoribulokinase, photosynthesis, plastid, redox regulation, stramenopile, symbiosis, Vaucheria litorea
INTRODUCTION
The sacoglossan sea slug Elysia chlorotica maintains a long-term symbiotic (often referred to as kleptoplastic) relationship with photosynthetically active plastids from the stramenopile alga Vaucheria litorea within cells lining the digestive diverticula (Graves et al., 1979; West, 1979; Green et al., 2000; Rumpho et al., 2000). Only the plastids (referred to as kleptoplasts) are retained from the ingested algal cytosol by the sea slug and the kleptoplasts are not vertically transmitted (i.e. they are absent from sea slug eggs; West, 1979; Green et al., 2000). Once the kleptoplastic association is established in each generation, the green sea slug can be removed from its algal prey and will complete its entire lifecycle (about 10 months duration) photoautotrophically, namely with the provision of light and CO2 (West, 1979) (see developmental cycle in Figure 1).
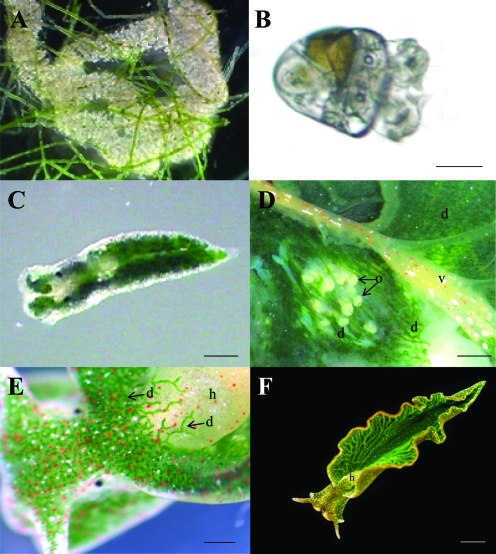
Figure 1.
Lifecycle Stages of Kleptoplastic Elysia chlorotica.
(A) Aposymbiotic sea slug eggs with filaments of Vaucheria litorea in the background. Egg ribbons can vary from 3 to 30 cm in length.
(B) Larval stage. Scale bar = 50 μm.
(C) Juvenile kleptoplastic E. chlorotica having fed on V. litorea for 1 d. Scale bar = 100 μm.
(D) Internal structures of adult E. chlorotica showing the close physical contact of the reproductive ovotestes (o), digestive diverticuli (d) containing green kleptoplasts, and the blood vasculature (v). Scale bar = 250 μm.
(E) Dorsal view of a young, adult E. chlorotica revealing the finely divided digestive diverticuli (d) which distributes the kleptoplasts throughout the body, except within the aposymbiotic heart (h). Scale bar = 500 μm.
(F) Mature E. chlorotica illustrating the uniform green coloring throughout the adult body. Scale bar = 3 mm.
This long-term plastid activity in the sea slug implies that essential photosynthetic proteins, typically encoded by both the algal plastid and nuclear genomes, are present in the animal. Kleptoplast transcriptional and translational activity is evident in E. chlorotica even after being starved for several months of algal prey (Mujer et al., 1996; Pierce et al., 1996; Rumpho et al., 2000). However, the plastid genome of V. litorea encodes only 139 proteins (Rumpho et al., 2008), far fewer than the expected 1000–5000 proteins needed to sustain normal plastid activity (Martin et al., 2002; Richly and Leister, 2004; Bock and Timmis, 2008). Whereas much less is known about the source of kleptoplast proteins that are nuclear-encoded, recently, molecular evidence supports horizontal gene transfer (HGT) of two such nuclear genes or gene families, psbO (Rumpho et al., 2008) and lhc (Pierce et al., 2007), from V. litorea to E. chlorotica. These findings suggest that the likelihood is high of identifying additional examples of HGT of algal nuclear genes essential for photosynthesis in the sea slug.
In this study, phosphoribulokinase (prk), a key nuclear gene encoding an essential chloroplast enzyme unique to the photosynthetic carbon reduction (Calvin) cycle, was targeted for study for both its potential for HGT and regulatory properties. PRK catalyzes the irreversible reaction that generates the substrate ribulose-1,5-bisphosphate (RuBP) for RuBP carboxylase/oxygenase (Rubisco)-dependent CO2 fixation supporting the Calvin cycle. To ameliorate presumed turnover of the enzyme, the PRK pre-protein must be de novo synthesized and imported from the cytosol (see review by Miziorko, 1998). The long-term viability of the association would require E. chlorotica to obtain a prk gene from V. litorea or another photoautotroph via HGT. There is no known alternative pathway to synthesize RuBP in photosynthetic organisms outside of the Archaea, which utilize a special Form III Rubisco (Finn and Tabita, 2004; Sato et al., 2007; Tabita et al., 2008). Hence, all other photosynthetic organisms are dependent on PRK to sustain the cyclic activity.
PRK is also of interest due to its complex regulatory properties and the lack of understanding of this enzyme among Stramenopiles. PRK genes, proteins, and the regulation of enzymatic activity have all been characterized in a number of streptophytes (Wedel and Soll, 1998; Paul et al., 2000; Mouche et al., 2002; Chen et al., 2004; Marri et al., 2005a, 2005b, 2009), chlorophytes (Roesler and Ogren, 1990), cyanobacteria (Su and Bogorad, 1991; Wadano et al., 1995; Tamoi et al., 1998; Wedel and Soll, 1998; Kobayashi et al., 2003; Tamoi et al., 2005), and at least one rhodophyte (Oesterhelt et al., 2007). Among the Stramenopiles, regulation of PRK activity has only been characterized in two diatoms, Odontella sinensis (Michels et al., 2005) and Asterionella formosa (Boggetto et al., 2007), and to a lesser extent in one raphidophyte, Heterosigma carterae (Hariharan et al., 1998). Common to members of all of these phyla is the formation in the dark of a supramolecular complex of PRK with glyceraldehyde-3-P dehydrogenase (GAPDH) and the small (8.5-kDa), non-enzymatic nuclear-encoded protein CP12 (Pohlmeyer et al., 1996; Wedel and Soll, 1998; Graciet et al., 2004). The degree of regulation even within this complex, however, varies considerably among the different phyla. The best characterized are the unicellular green algae and streptophytes in which PRK is activated in the presence of light by dissociation of the inactive complex, via photosynthetic electron flow through thioredoxin which, in turn, reduces the intramolecular disulfides in CP12, and reduction of the intramolecular disulfide of the PRK protein. In contrast, in the rhodophyte (Oesterhelt et al., 2007) and two diatoms (Michels et al., 2005; Boggetto et al., 2007) studied thus far, PRK inactivation by dark complex formation is not complete and the enzyme is not redox regulated. Whereas the addition of DTT to a partially purified PRK extract from the raphidophyte H. carterae stimulated enzyme activity (Hariharan et al., 1998), additional studies are needed to determine whether CP12 is present in this Stramenopile and whether the enzyme is redox regulated in vivo.
Dark inactivation of the Calvin cycle and PRK would prevent futile cycling resulting from the presence of the potentially competing oxidative pentose phosphate pathway (OPPP) in the plastid (Gruber et al., 2009). Interestingly, though, Michels et al. (2005) suggested that the OPPP is not present in the plastids of the diatom Odontella coincident with the absence of dark inactivation of PRK. Bioinformatic analysis supports the absence of a complete OPPP in the plastids of at least three other diatoms (Kroth et al., 2008; Gruber et al., 2009). Hence, this would eliminate futile cycling as a concern in these Stramenopiles. A functional OPPP and dark regulation of PRK is hypothesized to be essential in V. litorea plastids for synthesis of NADPH for fatty acid biosynthesis (the major carbon reserve is lipid and not starch) as well as other carbon substrates for nucleotide and anabolic biosynthesis. This would also be the case for E. chlorotica kleptoplasts; however, regulation would presumably involve redox regulation and formation of the multi-protein complex, all components of which are nuclear-encoded. The presence of CP12 is yet to be demonstrated in V. litorea, and it has never been reported in an animal. Given these observations, the objectives of this study were to advance our understanding of the regulation of a key photosynthetic enzyme, PRK, in Stramenopiles as well as in the unusual case of a mollusc, and determine whether there is any evidence supporting HGT of prk from a stramenopile alga to a mollusc.
RESULTS AND DISCUSSION
PRK Activity and De Novo Synthesis
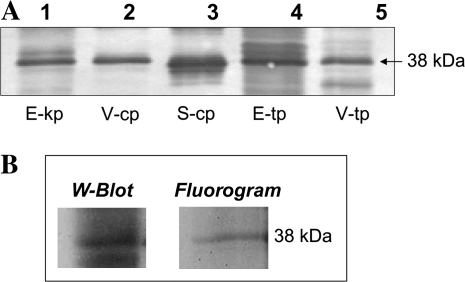
A tobacco PRK-specific antibody decorated a 38-kDa protein in soluble extracts of V. litorea and E. chlorotica (starved 3 months), and in extracts of plastids isolated from V. litorea, E. chlorotica (starved 9 months), and Spinacea oleraceae (streptophyte control) (Figure 2A). PRK enzyme activity from light- and DTTred-treated extracts of E. chlorotica starved 0 months (mean standard error (SE) = 2.3 ± 0.13 μmol min−1 mg protein−1) and 3 months (1.7 ± 0.39 μmol min−1 mg protein−1) were 66 and 49%, respectively, of the corresponding PRK activity measured for V. litorea (3.5 ± 046 μmol min−1 mg protein−1) (Figure 3). PRK activity was not due solely to long-term stability of the protein in the sea slug, because de novo synthesis was detected by immunoprecipitation of a 38-kDa radiolabeled protein after 5 months starvation (Figure 2B). Having shown that the nuclear-encoded PRK protein was synthesized in the sea slug and functional after several months starvation, prk was cloned from V. litorea as a prelude to determining whether the gene was present in E. chlorotica and to characterize and compare regulatory mechanisms in both organisms.
Figure 2.
Detection of PRK in Elysia chlorotica.
(A) PRK Western blot using a tobacco PRK antibody and the alkaline phosphatase detection system. Extracts were loaded on an equal chlorophyll basis as follows: E-kp, E. chlorotica kleptoplasts (from animals starved nine months); V-cp, V. litorea plastids; S-cp, spinach plastids; E-tp, E. chlorotica total proteins (from animals starved for 3 months); V-tp, V. litorea total proteins.
(B) Western blot and fluorogram of immunoprecipitated PRK from 5-month-starved sea slugs, following labeling with [35S]methionine/cysteine for 6 h and separation by SDS–PAGE.
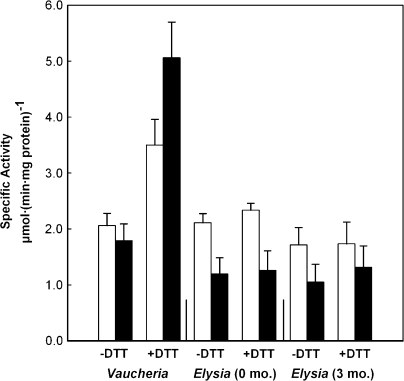
Figure 3.
Redox Activation of PRK in Vaucheria litorea and Elysia chlorotica.
Enzyme activity was measured in crude extracts of algal and sea slug samples collected 5 h into the light (white columns) or dark (black columns) period. The sea slugs had been starved for 0 or 3 months, as indicated. The extracts were prepared in the presence (+DTT) or absence (–DTT) of reducing agent and assayed as described in ‘Methods’. The bars indicate the standard error of the mean with n = 6 for each algal measurement and n = 3 for each sea slug measurement.
cDNA Clone and Plastid Targeting of PRK in V. litorea
A full-length cDNA clone of V. litorea prk was obtained by RT–PCR and 5′- and 3′-RACE and found to be 1299 bp in length, with 1218 bp encoding the protein. The deduced mature protein sequence of 405 amino acid (aa) has a predicted molecular weight of 40.3 kDa and a pI of 4.9 (http://scansite.mit.edu/calc_mw_pi.html). The PRK pre-protein in V. litorea contains a 42-aa bipartite plastid targeting peptide as an N-terminal extension (Figure 4A). This reflects the secondary endosymbiotic origin of V. litorea in which proteins must cross the four membranes that surround the plastids of Stramenopiles, including the outermost chloroplast (i.e. plastid) endoplasmic reticulum (ctER) (Rumpho et al., 2000; McFadden, 2001; Rumpho et al., 2001; Bhattacharya et al., 2004; Lee, 2008). The initial 19 aa of the N-terminal domain constitute the signal sequence as predicted by SignalP v. 3.0 (Nielsen et al., 1999) and HECTAR (a program specific to the Stramenopiles) (Gschloessl et al., 2008). The signal sequence exhibits a slightly positively charged N-terminus followed by a hydrophobic region of at least 10 residues that presumably facilitates binding of the pre-protein to the ctER (Soll and Schleiff, 2004). The positive Lys residue found immediately after the start Met in the closely related stramenopile PRK sequences of diatoms (Kilian and Kroth, 2004) is replaced by a Leu in V. litorea PRK. The motif ASAFAP is frequently found at the border of the ctER signal sequence and plastid transit peptide in Stramenopiles (Kroth, 2002; Kroth et al., 2008). However, the altered motif SFV is present in V. litorea PRK (Figure 4A). Site-directed mutagenesis revealed that the Phe residue in these motifs is relatively important for protein transport into diatom plastids (Kilian and Kroth, 2005).
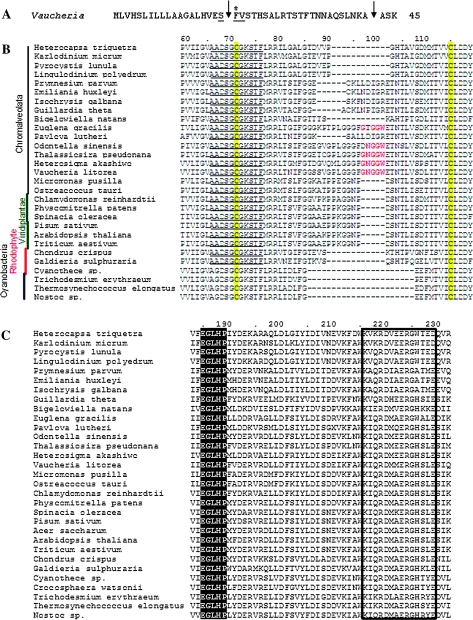
Figure 4.
Signature Motifs of the PRK Protein
(A) N-terminal amino acid sequence of V. litorea PRK pre-protein showing cleavage sites for predicted signal sequence (first arrow; amino acids 1–19) and target peptide sequence (second arrow, amino acids 20–42). The first site is identified by the conserved motif (SFV) at the border of the ER signal sequence (underlined) and a critical Phe (starred).
(B) Clustal W alignment of partial PRK amino acid sequences from different organisms aligned using default parameters. The highly conserved nucleotide binding motif A is shown in bold underline and the two conserved regulatory Cys residues are highlighted in yellow. A 5-amino acid insertion between the Cys residues is shown in red for the Vaucheria sequence and four other chromalveolate sequences.
(C) Clustal W alignment of partial PRK amino acid sequences illustrating the highly conserved Walker B motif (black highlighted) and the PRK signature sequence (boxed). Numbers along the sequences indicate the respective amino acid positions relative to the V. litorea sequence beginning with the start Met.
The plastid transit peptide targeting the pre-protein across the plastid envelope to its final stromal location is made up of the next 23-aa residues as predicted by ChloroP v1.1 (Emanuelsson et al., 2007) (Figure 4A). This sequence is rich in Ser and Thr, similar to that of transit peptides of land plants (Lang et al., 1998; Kroth, 2002; Soll and Schleiff, 2004). The relatively more variable stromal processing peptidase (SPP) cleavage site (NKA↓A) predicted for V. litorea PRK (Figure 4A) differs from both the land plant consensus sequence (I/V-X-A/C↓X) and other stramenopile consensus SPP cleavage sites (V/NM↓A/D/S) (Kilian and Kroth, 2004). This reflects the greater heterogeneity in transit peptide sequences in general, and the low number of non-diatom stramenopile transit peptide sequences currently available.
Regulation of PRK Activity
Three highly conserved regions in PRK protein sequences from cyanobacteria to streptophytes (reviewed by Miziorko, 1998) were also identified in the V. litorea PRK protein as follows. The Walker A motif (AADSGCGKSTF; underlined in Figure 4B) participates in binding the β-phosphate of ATP, possibly through the Ser at the ninth position. The Walker B motif consists of a conserved EGLHP sequence (black highlighted in Figure 4C). Here, Glu (E185) acts as a second ligand for Mg2+-ATP (all amino acid residue numbers refer to the V. litorea sequence unless indicated otherwise). The PRK signature sequence (K-[L/I]-x-R-D-x(3)-R-G-x-[S/T]-x-E) (www.expasy.org/cgi-bin/nicedoc.pl?PDOC00490; boxed in Figure 4C) is predicted to contribute various residues to the active site. In particular, K216 is conserved in all PRKs and binds the sugar-phosphate substrate Ru5P; R224 brings about transition state stabilization. As in other PRK protein sequences, V. litorea PRK has two conserved Cys residues near the N-terminus that correspond to C71 in the Walker A motif and C115 (yellow highlight in Figure 4B). Relative to streptophyte and green algal PRK sequences, there is an insertion of 5 aa between the regulatory Cys residues in V. litorea and other closely related Stramenopiles (Odontella, Thalassiosira, and Heterosigma) (Figure 4B). In contrast, a variable number of amino acid deletions exist between the two Cys in several cyanobacteria and rhodophytes (Figure 4B). These Cys residues are believed to be involved in light regulation of the enzyme in almost all photosynthetic organisms studied thus far (see below).
Under dark oxidizing conditions, it is generally accepted that PRK complexes with GAPDH and the small non-catalytic peptide CP12, inactivating the enzymes and preventing the energy loss that would result if the reductive and OPPP competed for the same intermediates (Pohlmeyer et al., 1996; Wedel et al., 1997; Miziorko, 1998; Wedel and Soll, 1998; Graciet et al., 2004). Dissociation of the complex under light conditions is typically mediated by photosynthetic electron flow through the ferredoxin–thioredoxin system with reduction of the disulfide bridges in CP12 (Wedel and Soll, 1998; Graciet et al., 2004; Marri et al., 2008). In turn, full activation of PRK is similarly driven by thioredoxin reduction of a single disulfide formed by the two conserved Cys residues in PRK (Porter and Hartman, 1986; Porter et al., 1988; Graciet et al., 2004). Despite the presence of the conserved Cys residues in almost all PRKs, and the widespread presence of CP12, light and redox regulation of PRK in vivo is not universal. The diatom O. sinensis (Michels et al., 2005) and the cyanobacterium Synechococcus (Kobayashi et al., 2003) provide two such exceptions. Michels et al. (2005) asserted that this may be due to the insertion of amino acids between the two Cys residues in O. sinensis PRK or deletion in Synechococcus (Kobayashi et al., 2003), physically interfering with formation of the regulatory disulfide bond (Figure 4B). CP12 has been found in all photoautotrophic organisms investigated thus far (Wedel et al., 1997; Petersen et al., 2006a), including a recent report in the diatom Asterionella formosa (Boggetto et al., 2007). Diatoms do not appear to possess a complete plastid OPPP, thereby rendering light regulation of PRK less essential (Armbrust et al., 2004; Michels et al., 2005; Kroth et al., 2008).
Redox regulation of PRK was observed in V. litorea (two-way ANOVA, P = 0.04), but not in E. chlorotica at 0 or 3 months. Tukey's post hoc test of means identified a highly significant (P < 0.001) difference in PRK activity for dark-treated V. litorea samples incubated ± DTT (Figure 3). Although values for PRK activity of light-treated V. litorea samples appeared to increase with the addition of DTT, the difference was marginally non-significant (P = 0.09). A similar observation was made within the +DTT V. litorea treatment; the mean activity of PRK in dark samples was greater than in light, but again was marginally non-significant (P = 0.07). No light/dark treatment effect was observed in the –DTT V. litorea samples (Figure 3). These observations are consistent with reports of more rapid and greater activation of complexed PRK (presumably still partially in the reduced form under dark conditions) compared to ‘free’, already reduced PRK in the light (Howard et al., 2008; Marri et al., 2008). Howard et al. (2008) suggested this may enable a more rapid response to changes in light levels and rapid activation upon complex dissociation. From the initial experiments conducted here on crude extracts, one cannot conclude whether activation of PRK in the dark algal extract is due to reduction of the PRK intramolecular disulfide, which may or may not form due to the presence of the 5-aa insertion between the regulatory Cys (Figure 4B), and/or due to dissociation of the complex possibly through reduction of CP12, or a combination of the two. Additional experiments using partially purified extracts and gel analysis are needed to address these questions.
In contrast to V. litorea PRK, there was an absence of strong support for redox regulation of E. chlorotica PRK measured in extracts from light- or dark-exposed animals, starved for 0 or 3 months, and analyzed by two-way ANOVA (Figure 3). However, t-test analysis of PRK activity values at 0 months revealed a significant light enhancement effect within both DTT treatments (P < 0.05 for each) (Figure 3). This difference was not significant at 3 months starvation. The decreased activity in the dark is most likely due to lower levels of PRK protein. The absence of a DTT effect in the sea slugs would suggest that PRK is not complexed with GAPDH and CP12 in the dark. This is a reasonable hypothesis because CP12 is nuclear-encoded and only present in photoautotrophic organisms. However, the presence of CP12 in V. litorea or E. chlorotica and the formation of the inactive complex in the dark have not yet been studied.
V. litorea PRK Genomic Clone
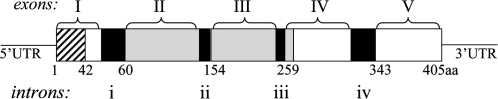
Based on the known cDNA sequence, a genomic prk sequence was amplified from V. litorea DNA using primers (PRK5F and PRK6R) that recognize the 5′ and 3′ untranslated regions, respectively. This primer pair amplified the protein coding region and 31 bp downstream of the cDNA stop codon. The entire 1649-bp amplified fragment was cloned and sequenced. Comparison of the prk cDNA and genomic sequences revealed that the protein-coding region of the gene is split into five exons (Figure 5). The coding sequence for the 42-aa transit peptide is present entirely in exon I, whereas the PRK domain (amino acids 62–266) spans most of exon II, all of exon III, and a small portion of exon IV. The positions of three of the four introns in V. litorea prk are unique; however, the third intron is at a homologous position as a conserved intron in the green alga Chlamydomonas and in the haptophytes Prymnesium (Petersen et al., 2006b) and Emiliania (see recently completed genome at JGI; www.jgi.doe.gov/). This conserved intron is not found in rhodophyte prk, thus lending additional support to the green algal derivation of this gene in V. litorea and other chromalveolates (Li et al., 2006; Petersen et al., 2006b), as discussed below.
Figure 5.
Schematic Representation of Functional Features of the V. litorea PRK Protein Imposed Upon the Corresponding Gene Organization.
Five exons enclosed in brackets (I–V) and four introns represented by black boxes (i–iv) illustrate the genomic structure. Numbered amino acid residues are shown below the corresponding genomic segments. The hatched box represents the plastid transit peptide sequence within exon 1, corresponding to amino acids 1–42. The gray shaded boxes represent the PRK domain sequence, corresponding to amino acids 62–266, and spanning much of exon II, all of exon III, and 24 bp of exon IV.
Prk in E. chlorotica
Primers (PRK5F and PRK5R) were designed to amplify algal prk cDNA between nucleotides –6 and +263, generating a 269-bp algal cDNA fragment and a corresponding 422-bp algal genomic DNA (gDNA) fragment (Figure 6A). Using sea slug genomic DNA (gDNA) as a template, the same primer pair amplified multiple fragments, including two fragments 269 and 421 bp in length (Figure 6A). The sequence of the smaller sea slug gDNA fragment, labeled prkX, was identical to the algal prk cDNA sequence amplified by the same primers (Figure 7). Here, only the protein encoding 263 bp cDNA and corresponding genomic sequences are shown, although the 6-bp upstream of the start codon were identical in all cases. Like the corresponding algal cDNA sequence, sea slug prkX did not contain an intron and the sequence spanned all of exon 1 and 86 bp of exon 2, coding for 87 aa.
Figure 6.
PCR Amplification of prk from V. litorea and E. chlorotica.
(A) PCR amplification of prk from algal and sea slug DNA using primers PRK5F and PRK5R. Lane 1, 1 kb Plus DNA ladder (Invitrogen); lane 2, no DNA (negative control); lane 3, pVA1prk construct (pGEM-T Easy vector with the V. litorea prk cDNA as insert; positive control); lane 4, pVA2prk construct (pGEM-T Easy vector with genomic V. litorea prk as insert); and lane 5, E. chlorotica DNA as template.
(B) PCR amplification of the prk gene from sea slug egg DNA using primers PRK5F and PRK5R. Lane 1, 1 kb plus ladder; lane 2, no DNA (negative control); lane 3, sea slug egg DNA. The arrows in both figures point to prkY (421 bp) and prkX (269 bp) fragments.
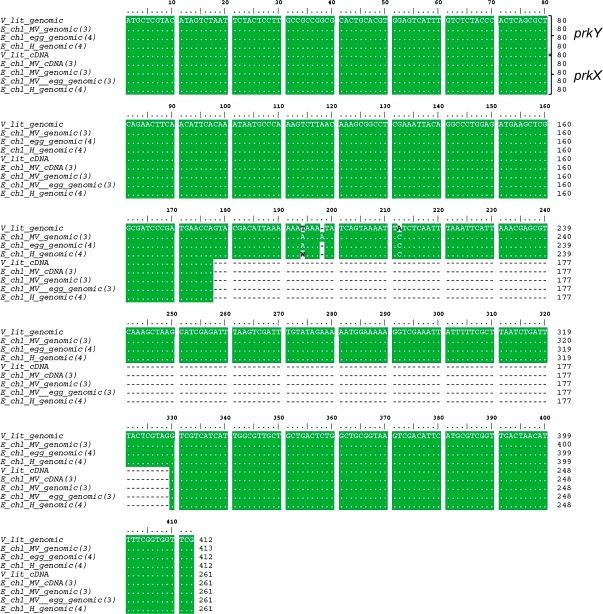
Figure 7.
Alignment of Partial prk Sequences from V. litorea and E. chlorotica.
Clustal W alignment of nucleotide sequences amplified from V. litorea cDNA and genomic DNA, E. chlorotica total DNA from Martha's Vineyard (MV) and Halifax (H) collections, E. chlorotica (MV) egg DNA, and E. chlorotica (MV) cDNA using PRK5F and PRK5R primers (see corresponding PCR bands in Figure 6A and 6B). Numbers in parentheses after the organism name refer to the number of sequences analyzed for each. Numbers at the end of each line indicate the respective nucleotide positions of the amplified product beginning with the start codon ATG. A dot indicates that the residues in that column are identical in all sequences in the alignment. A wavy dash (∼) marks the intron region. W = A or T. The larger, intron-containing sequence was labeled prkY, whereas the shorter sequence was labeled prkX.
The larger sea slug genomic fragment (415 bp), labeled prkY, was nearly identical to prkX except for the presence of the entire first intron (151 bp) of the algal prk genomic clone (Figure 7). Comparing the genomic sequences of prkY from sea slug, sea slug eggs, and the alga revealed only two nucleotide substitutions and one insertion, all within the intron (Figure 7). When translated, prkX and prkY encode the identical first 87 aa of V. litorea PRK, including the entire 42-aa plastid bipartite transit peptide (Figure 4A) and 26 aa of the PRK domain (Figure 5). Evidence for expression of the partial algal prkX sequence in the sea slug was demonstrated through RT–PCR of E. chlorotica cDNA (Figure 7). The sea slug sequence was identical to the corresponding product from V. litorea cDNA as well as all of the other prkX sequences obtained by PCR of gDNA.
The unlikely possibility of algal nuclei remaining in the gut of the sea slug several months after algal feeding, thereby contaminating the total gDNA preparation, was ruled out by the following experiments. First, as discussed above, sea slug egg DNA was separately analyzed; V. litorea plastids are not transmitted in the germline and the eggs never come in contact with the alga, thereby eliminating the possibility of algal nuclear contamination in sea slug egg DNA. Second, DNA samples were analyzed from two or more independent sea slug collections spanning 3 years from Martha's Vineyard (MV), and one collection from Halifax (H), Nova Scotia, with at least three independent isolations of these starved sea slugs. The same products were generated with minimal base pair changes in all cases (Figure 7). Other negative controls included analyzing DNA or cDNA from pufferfish and Dictyostelium with the same reagents; in all cases, no prk PCR products were amplified (results not shown). Complete sequencing of the E. chlorotica mitochondrial genome ruled out the presence of prk in this organellar genome (Rumpho et al., 2008), supporting the possible insertion in the nuclear genome of the sea slug or in an extrachromosomal location.
Evolutionary Origin of PRK in V. litorea and E. chlorotica
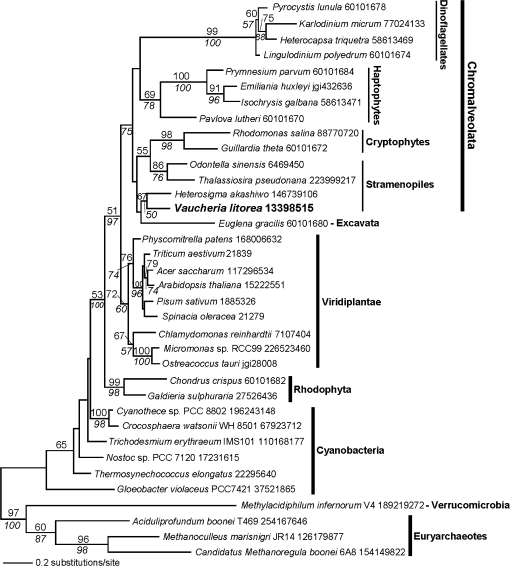
Because we only identified partial prk fragments in the sea slug, they may simply represent pseudogenes or processed pseudogenes that are derived from V. litorea or other phototrophic sources. Focusing only on V. litorea prk gene sequences may not be sufficient and leaves open the possibility of finding other PRK genes in the sea slug from a different source(s), which may also be expressed and functional. Genes for nuclear-encoded plastid-targeted proteins are not always traced back to the dominant endosymbiont. Phylogenetic analysis of the manganese-stabilizing protein PsbO from the fucoxanthin-containing dinoflagellate Karenia brevis indicates that during tertiary symbiosis, the original psbO gene in the dinoflagellate nucleus was replaced by a psbO gene from a haptophyte nuclear genome (Ishida and Green, 2002). In-depth analysis of Karenia ESTs revealed that psbO is only the tip of an iceberg of intracellular gene transfer driven by tertiary endosymbiosis and that HGT has contributed to the plastid proteome in this species (Nosenko et al., 2006). In fact, it was recently postulated that prk in chromalveolates (including Stramenopiles such as V. litorea) that share a red algal-derived plastid gained this gene via HGT from a green alga (Li et al., 2006; Petersen et al., 2006b). The PRK phylogeny shown in Figure 8 supports the cyanobacterial origin of this gene in algae and a specific association (bootstrap support values of 51% PhyML and 97% RAxML) between the chromalveolate and green clades. That the chromalveolate clade includes members of diverse phyla such as haptophytes, cryptophytes, Stramenopiles, and alveolates (i.e. dinoflagellates) suggests that the HGT event occurred in ancient times prior to the radiation of these lineages. The alternative explanation is multiple independent green algal prk gene origins in these taxa. Under the first scenario, the PRK results are consistent with other data showing a large number of green genes of ancient derivation (e.g., involved in carotenoid biosynthesis) in chromalveolates is most likely explained by a cryptic green algal endosymbiosis in this lineage that predates the red algal capture (for details, see Nosenko et al., 2006; Frommolt et al., 2008; Moustafa et al., 2009).
Figure 8.
Maximum Likelihood (PhyML) Phylogenetic Analysis of PRK in Cyanobacteria, Algae, and Plants.
The results of bootstrap analyses are shown with the PhyML bootstrap values above and the RAxML bootstrap values below the branches. The branch lengths in the trees are proportional to the number of substitutions per site (see scale in figure). The branch uniting the cyanobacteria was used to root this phylogeny.
In summary, we cloned and sequenced the nuclear prk gene from the stramenopile alga V. litorea and provided preliminary support for redox regulation of the enzyme. In turn, PRK activity was measureable in E. chlorotica even after 3 months starved of its algal prey, but unlike the algal enzyme, PRK did not exhibit redox regulation in the sea slug. Finally, we amplified two partial prk gene fragments from the sea slug with sequences that are almost identical to the corresponding prk gene in V. litorea. High-throughput genome sequencing (work in progress) will help solidify any claims of HGT and reveal the location and potentially the mechanism of integration of foreign genes in animal nuclear DNA. This work will undoubtedly also identify other algal genes transferred to the sea slug genome and demonstrate expression and targeting of foreign gene products to the kleptoplasts.
METHODS
Culturing of E. chlorotica and V. litorea
Specimens of E. chlorotica were typically collected from a salt marsh pond on Martha's Vineyard, MA. Where indicated, collections were also made in Halifax, Nova Scotia, Canada. Animals were maintained without food in aquaria with aerated artificial seawater (ASW, 925 mosmol kg−1 Instant Ocean, Aquarium Systems, Mentor, OH, USA) at 10°C on a 14-h photoperiod. Months starved refers to the number of months from the time of collection from the field during which the animals were maintained without algal food. Sea slugs were either sampled fresh for PRK activity and plastid isolation, or blotted dry, weighed, frozen with liquid nitrogen, and stored at –80°C for protein, DNA, and RNA isolation. Filaments of Vaucheria litorea were maintained in f/2 media using quarter-strength ASW (250 mosmol kg−1) at 18°C on a 14-h photoperiod. Algal cultures were sub-cultured every 2 weeks.
Plastid Isolation
Sea slugs (∼0.5 g fresh weight) were homogenized in 50 ml ice-cold grinding buffer (GB; 0.1 M 4-[2-hydroxyethyl] piperazine-1-ethanesulfonic acid [HEPES]/KOH, 0.9 M sorbitol, 2 mM MgCl2, 2 mM MnCl2, 4 mM Na2-EDTA, 10 mM Na ascorbate, 2% BSA; pH 7.5) containing 0.5 M N-acetyl-L-cysteine (Sigma, St Louis, MO, USA). The filtered homogenate was centrifuged in a Beckman JS 13.1 rotor at 1200 g for 5 min (4°C) and the plastid pellet re-suspended in 10 ml GB and filtered through a 110-μm nylon mesh. The resulting mucus-reduced extract was then layered on top of a 20%/50% Percoll (Sigma) step gradient prepared in GB and centrifuged for 20 min at 8000 g using a JS 13.1 rotor (4°C). Plastids at the interface were collected by aspiration, washed twice by re-suspending in GB without BSA and N-acetyl-L-cysteine, and centrifuged at 1500 g and 4°C for 5 min. The final plastid pellet was dissolved in re-suspension buffer (0.1 M HEPES/KOH pH 7.5, 0.9 M sorbitol, 10 mM MgCl2).
Plastids were isolated from V. litorea essentially the same way except about 3 g fresh weight filaments were used as the starting material, the GB and final wash buffers contained 0.4 M sorbitol, N-acetyl-L-cysteine was not included in any of the solutions, and a 30%/75% Percoll step gradient was used to separate intact from broken plastids. Chlorophylls a and c were extracted into 90% (v/v) acetone and quantified by measuring the absorbance at 664 and 630 nm using the calculations of Sterman (1988). Spinach plastids were isolated using standard protocols (Rumpho and Edwards, 1984) and chlorophylls a and b extracted and quantified according to Wintermans and de Mots (1965).
Western Blotting
Immunoblotting was performed as previously described (Green et al., 2000). Protein extracts were loaded on an equal chlorophyll basis (270 ng chlorophyll lane−1) into lanes of a 12.5% polyacrylamide gel and the polypeptides separated by SDS–PAGE according to Laemmli (1970). The separated polypeptides were electroblotted onto polyvinylidene fluoride membranes (Immobilon-P). Heterologous, polyclonal antibodies directed against the tobacco PRK protein (Klein and Salvucci, 1995) were used with the alkaline phosphatase detection system.
PRK Enzyme Assay
PRK activity was measured spectrophotometrically at 340 nm and 25°C. The assay coupled the Ru5P-dependent ADP formation by PRK to the oxidation of NADH via pyruvate kinase and lactate dehydrogenase (Hariharan et al., 1998). Algal and sea slug tissue were ground in a liquid nitrogen-cooled mortar at a ratio of 1:1 fresh weight:volume extraction buffer (0.1 M Tris pH 8.0, 10 mM MgCl2, 5 mM 1,4-dithio-D,L-threitol (DTT) (Roesler and Ogren, 1990). DTT was omitted from the grinding media where indicated (–DTT) in the light/dark regulation assays. The homogenate was centrifuged at 13 000 g for 10 min at 4°C and the clarified extract used in the PRK assay and for protein quantification using the BCA method (Pierce Biotechnology, Woburn, MA, USA). The 1-ml PRK assay consisted of 2 mM phosphoenolpyruvate, 0.1 M Tris pH 8.0, 3 mM MgCl2, 5 mM DTT, 2 mM ATP, 10.5 units lactate dehydrogenase, 14 units pyruvate kinase, 0.2 mM NADH, and varying amounts of tissue extract. Background oxidation of NADH in the reaction mixture was monitored for 2 min. This was immediately followed by the addition of 4 mM ribose-5-phosphate (Ri5P), and 2 units of phosphoribose isomerase to initiate the reaction cascade. PRK-dependent oxidation of NADH was monitored for an additional 2 min. All permutations of addition and subtraction of individual components of the reaction mixture were tested to demonstrate both substrate specificity and dependency on PRK. One unit of activity was defined as the amount of enzyme that catalyzed the oxidation of 1 μmol of NADH per minute.
De Novo Synthesis of PRK
Live sea slugs (starved for 5 months) and algal filaments were labeled with 25 μCi ml−1 of [35S] methionine/cysteine (ICN trans [35S] label; specific activity 1175 Ci mmol−1; 1 Ci = 37 GBq) in the light for 6 h at 18°C as described by Mujer et al. (1996). Total soluble proteins were extracted and incubated with tobacco anti-PRK antibody overnight followed by SDS–PAGE (Mujer et al., 1996; Green et al., 2000). Duplicate gels were subjected to either Western blotting using the same PRK antibody, or treated with Amplify Fluorographic Reagent (Amersham Biosciences) and exposed to Kodak Biomax XAR film at –80°C for periods ranging from 48 h to 1 week (Mujer et al., 1996; Green et al., 2000).
Characterization of prk cDNA
Total RNA purified from V. litorea (Mujer et al., 1996) was used to synthesize first strand cDNA with reverse transcriptase (RT) and random primers (Gibco-BRL, Gaithersburg, MD, USA). GenBank sequences of PRK from S. oleracea (Genbank: M21338), Odontella sinensis (Genbank: Y08610), and Chlamydomonas reinhardtii (Genbank: M36123) were aligned to design the degenerate primers 5′-AYWSNGGNTGYGGNAARWSNACNTT and 5′-ATNGTNSWNCCYTCRTCRAANRARTA, for gene-specific PCR amplification. The V. litorea prk sequence thus acquired was used to design specific primers to obtain the 5’ and 3’ ends by rapid amplification of cDNA ends (RACE; following Gibco BRL guidelines). Sequences obtained from these 5’ and 3’ end fragments were used to generate primers (PRK5F = AACAAAATGCTCGTACATAGTC, PRK6R = CTTCTCTAAATGTTGGCAGGA) to amplify the full-length coding region from first strand.
Characterization of prk from E. chlorotica and V. litorea DNA Templates
DNA was isolated from V. litorea (0.5 g fresh weight), adult E. chlorotica (0.1 g fresh weight), and aposymbiotic E. chlorotica eggs (0.1 g fresh weight) using DNAzol ES reagent (Molecular Research Center Inc, Cincinnati, OH, USA). RNase-treated DNA was further purified with a QIAEX II gel extraction kit (Qiagen, Valencia, CA, USA) following the protocol for ‘desalting and concentrating DNA from solutions’. Primers used to amplify prk from algal and sea slug DNA were designed using the V. litorea prk cDNA sequence. Primers included PRK5F (5′-AACAAAATGCTCGTACATAGTC) and PRK5R (5′-CGAACCACCGAAAATGTT). All primers were synthesized by Integrated DNA Technologies (Coralville, IA, USA). PCR conditions used to amplify V. litorea prk included: 1X enzyme reaction buffer, 1.5 mM MgCl2, 0.2 mM deoxynucleotide triphosphate (dNTP) mix (Invitrogen, CA, USA), PRK5F and PRK5R primers (1 μM each), 1–10 ng DNA, and 1.25 U platinum Taq polymerase (Invitrogen, Carlsbad, CA, USA). Cycle conditions were: 95°C for 4 min, 30 cycles of 95°C for 30 s, 50°C for 45 s, and 72°C for 1 min with a final elongation step at 72°C for 10 min. The pGEM-T Easy vector construct (pVA1prk) with the cloned V. litorea prk cDNA sequence was used as a positive control. PCR reactions with DNA from adult E. chlorotica and E. chlorotica eggs were similar to those using algal DNA except the MgCl2 concentration was increased to 3 mM. PCR products were separated using a 1% agarose gel in 1X TBE buffer and visualized by staining with ethidium bromide (Sambrook et al., 1989). Amplified products were cloned into a plasmid vector using the pGEM-T Easy Vector system (Promega). All plasmid constructs containing amplified products were sequenced with an Applied Biosystems ABI 3100 automated sequencer at the University of Maine DNA Sequencing Facility with M13 forward and reverse primers.
Expression of prk Using RT–PCR Analysis
Total RNA was isolated from E. chlorotica starved for 6 months (30 mg fresh weight) and V. litorea (100 mg fresh weight) using an RNeasy mini kit (Qiagen). To remove potential contaminating DNA, the on-column deoxyribonuclease (DNase) treatment was performed during the isolation. First-strand cDNA was synthesized as described previously (Rumpho et al., 2008). A parallel control reaction without RT was run. Reaction conditions were identical for sea slug and algal RNA. First-strand cDNA (2–4 μL) was used as the template in PCR reactions using gene-specific primers.
Phylogenetic Analysis of PRK
A database of PRK proteins was assembled from protein (pblast, tblastn) queries of GenBank (NCBI, Rockville, MD, USA) and genome projects at the Joint Genome Institute (Walnut Creek, CA, USA). The protein data were aligned with Muscle (Edgar, 2004) and manually refined (alignment available upon request from MER). The ML tree (using a 309-aa PRK region) was estimated with PhyML 3.0 (Guindon and Gascuel, 2003) using the WAG substitution model, gamma distribution, with four discrete rate categories, and starting from a random tree. Bootstrap support (100 replications) for nodes in the tree was calculated using PhyML and RAxML BlackBox (Stamatakis et al., 2008) using the same settings as described above.
Accession Numbers
Sequence data from this article can be found in the NCBI/GenBank data libraries under accession numbers AF336986 and DQ388997 and as indicated in Figure 8.
FUNDING
This research was supported by National Science Foundation grants IBN-9808904 (M.R. and J.M.) and IOS-0726178 (M.R. and M.T.); the American Society of Plant Biologists’ Education Foundation (M.R. and M.T.); Ministry for Food, Agriculture, Forestry, and Fisheries, Korean Government, Korea Research Foundation (J.L.); the National Institutes of Health (grant R01ES013679 to D.B.), and the University of Maine (M.R.). This is Maine Agricultural and Forest Experiment Station Publication Number 3079, Hatch Project no. ME08361-08MRF (NC 1168).
Acknowledgments
The authors thank Dr Michael Salvucci for providing antibodies to PRK and Dr Jörn Petersen for analyzing the genomic PRK sequence for introns. No conflict of interest declared.
References
- Armbrust EV, et al. The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science. 2004;306:79–86. doi: 10.1126/science.1101156. [DOI] [PubMed] [Google Scholar]
- Bhattacharya D, Yoon HS, Hackett JD. Photosynthetic eukaryotes unite: endosymbiosis connects the dots. Bioessays. 2004;26:50–60. doi: 10.1002/bies.10376. [DOI] [PubMed] [Google Scholar]
- Bock R, Timmis JN. Reconstructing evolution: gene transfer from plastids to the nucleus. Bioessays. 2008;30:556–566. doi: 10.1002/bies.20761. [DOI] [PubMed] [Google Scholar]
- Boggetto N, Gontero B, Maberly SC. Regulation of phosphoribulokinase and glyceraldehyde 3-phosphate dehydrogenase in a freshwater diatom, Asterionella formosa. J. Phycol. 2007;43:1227–1235. doi: 10.1111/j.1529-8817.2008.00600.x. [DOI] [PubMed] [Google Scholar]
- Chen XF, et al. Molecular cloning and expression analysis of rice phosphoribulokinase gene that is regulated by environmental stresses. Mol. Biol. Rep. 2004;31:249–255. doi: 10.1007/s11033-005-2491-5. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- Finn MW, Tabita FR. Modified pathway to synthesize ribulose 1,5-bisphosphate in methanogenic archaea. J. Bacteriol. 2004;186:6360–6366. doi: 10.1128/JB.186.19.6360-6366.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frommolt R, et al. Ancient recruitment by chromists of green algal genes encoding enzymes for carotenoid biosynthesis. Mol. Biol. Evol. 2008;25:2653–2667. doi: 10.1093/molbev/msn206. [DOI] [PubMed] [Google Scholar]
- Graciet E, Lebreton S, Gontero B. Emergence of new regulatory mechanisms in the Benson–Calvin pathway via protein–protein interactions: a glyceraldehyde-3-phosphate dehydrogenase/CP12/phosphoribulokinase complex. J. Exp. Bot. 2004;55:1245–1254. doi: 10.1093/jxb/erh107. [DOI] [PubMed] [Google Scholar]
- Graves DA, Gibson MA, Bleakney JS. Digestive diverticula of Alderia modesta and Elysia chlorotica (Opisthobranchia, Sacoglossa) Veliger. 1979;21:415–422. [Google Scholar]
- Green BJ, et al. Mollusc–algal chloroplast endosymbiosis: photosynthesis, thylakoid protein maintenance, and chloroplast gene expression continue for many months in the absence of the algal nucleus. Plant Physiol. 2000;124:331–342. doi: 10.1104/pp.124.1.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber A, Weber T, Bartulos CR, Vugrinec S, Kroth PG. Intracellular distribution of the reductive and oxidative pentose phosphate pathways in two diatoms. J. Basic Microbiol. 2009;49:58–72. doi: 10.1002/jobm.200800339. [DOI] [PubMed] [Google Scholar]
- Gschloessl B, Guermeur Y, Cock JM. HECTAR: A method to predict subcellular targeting in heterokonts. BMC Bioinformatics. 2008;9:393. doi: 10.1186/1471-2105-9-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hariharan T, Johnson PJ, Cattolico RA. Purification and characterization of phosphoribulokinase from the marine chromophytic alga Heterosigma carterae. Plant Physiol. 1998;117:321–329. doi: 10.1104/pp.117.1.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard TP, Metodiev M, Lloyd JC, Raines CA. Thioredoxin-mediated reversible dissociation of a stromal multiprotein complex in response to changes in light availability. Proc. Natl. Acad. Sci. U S A. 2008;105:4056–4061. doi: 10.1073/pnas.0710518105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida K, Green BR. Second- and third-hand chloroplasts in dinoflagellates: phylogeny of oxygen-evolving enhancer 1 (PsbO) protein reveals replacement of a nuclear-encoded plastid gene by that of a haptophyte tertiary endosymbiont. Proc. Natl. Acad. Sci. U S A. 2002;99:9294–9299. doi: 10.1073/pnas.142091799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian O, Kroth PG. Presequence acquisition during secondary endocytobiosis and the possible role of introns. J. Mol. Evol. 2004;58:712–721. doi: 10.1007/s00239-004-2593-z. [DOI] [PubMed] [Google Scholar]
- Kilian O, Kroth PG. Identification and characterization of a new conserved motif within the presequence of proteins targeted into complex diatom plastids. Plant J. 2005;41:175–183. doi: 10.1111/j.1365-313X.2004.02294.x. [DOI] [PubMed] [Google Scholar]
- Klein RR, Salvucci ME. Rubisco, rubisco activase and ribulose-5-phosphate kinase gene expression and polypeptide accumulation in a tobacco mutant defective in chloroplast protein synthesis. Photosynth. Res. 1995;43:213–223. doi: 10.1007/BF00029934. [DOI] [PubMed] [Google Scholar]
- Kobayashi D, Tamoi M, Iwaki T, Shigeoka S, Wadano A. Molecular characterization and redox regulation of phosphoribulokinase from the cyanobacterium Synechococcus sp PCC 7942. Plant Cell Physiol. 2003;44:269–276. doi: 10.1093/pcp/pcg048. [DOI] [PubMed] [Google Scholar]
- Kroth PG. Protein transport into secondary plastids and the evolution of primary and secondary plastids. International Review of Cytology—A Survey of Cell Biology. 2002;221:191–255. doi: 10.1016/s0074-7696(02)21013-x. [DOI] [PubMed] [Google Scholar]
- Kroth PG, et al. A model for carbohydrate metabolism in the diatom Phaeodactylum tricornutum deduced from comparative whole genome analysis. PLoS ONE. 2008;3:e1426. doi: 10.1371/journal.pone.0001426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lang M, Apt KE, Kroth PG. Protein transport into ‘complex’ diatom plastids utilizes two different targeting signals. J. Biol. Chem. 1998;273:30973–30978. doi: 10.1074/jbc.273.47.30973. [DOI] [PubMed] [Google Scholar]
- Lee RE. In Phycology. Cambridge: Cambridge University Press; 2008. Heterokontophyta, Xanthophyceae; pp. 413–423. [Google Scholar]
- Li S, Nosenko T, Hackett JD, Bhattacharya D. Phylogenomic analysis identifies red algal genes of endosymbiotic origin in the chromalveolates. Mol. Biol. Evol. 2006;23:663–674. doi: 10.1093/molbev/msj075. [DOI] [PubMed] [Google Scholar]
- Marri L, Sparla F, Pupillo P, Trost P. Co-ordinated gene expression of photosynthetic glyceraldehyde-3-phosphate dehydrogenase, phosphoribulokinase, and CP12 in Arabidopsis thaliana. J. Exp. Bot. 2005a;56:73–80. doi: 10.1093/jxb/eri020. [DOI] [PubMed] [Google Scholar]
- Marri L, Trost P, Pupillo P, Sparla F. Reconstitution and properties of the recombinant glyceraldehyde-3-phosphate dehydrogenase/CP12/phosphoribulokinase supramolecular complex of Arabidopsis. Plant Physiol. 2005b;139:1433–1443. doi: 10.1104/pp.105.068445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marri L, Trost P, Trivelli X, Gonnelli L, Pupillo P, Sparla F. Spontaneous assembly of photosynthetic supramolecular complexes as mediated by the intrinsically unstructured protein CP12. J. Biol. Chem. 2008;283:1831–1838. doi: 10.1074/jbc.M705650200. [DOI] [PubMed] [Google Scholar]
- Marri L, Zaffagnini M, Collin V, Issakidis-Bourguet E, Lemaire SD, Pupillo P, Sparla F, Miginiac-Maslow M, Trost P. Prompt and easy activation by specific thioredoxins of Calvin cycle enzymes of Arabidopsis thaliana associated in the GAPDH/CP12/PRK supramolecular complex. Mol. Plant. 2009;2:259–269. doi: 10.1093/mp/ssn061. [DOI] [PubMed] [Google Scholar]
- Martin W, et al. Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc. Natl. Acad. Sci. U S A. 2002;99:12246–12251. doi: 10.1073/pnas.182432999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden GI. Primary and secondary endosymbiosis and the origin of plastids. J. Phycol. 2001;37:951–959. [Google Scholar]
- Michels AK, Wedel N, Kroth PG. Diatom plastids possess a phosphoribulokinase with an altered regulation and no oxidative pentose phosphate pathway. Plant Physiol. 2005;137:911–920. doi: 10.1104/pp.104.055285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miziorko HM. Phosphoribulokinase: current perspectives on the structure/function basis for regulation and catalysis. In: Purich DL, editor. Advances in Enzymology and Related Areas of Molecular Biology. Chichester: John Wiley & Sons, Inc.; 1998. pp. 95–127. [DOI] [PubMed] [Google Scholar]
- Mouche F, Gontero B, Callebaut I, Mornon JP, Boisset N. Striking conformational change suspected within the phosphoribulokinase dimer induced by interaction with GAPDH. J. Biol. Chem. 2002;277:6743–6749. doi: 10.1074/jbc.M106401200. [DOI] [PubMed] [Google Scholar]
- Moustafa A, Beszteri B, Maier UG, Bowler C, Valentin K, Bhattacharya D. Genomic footprints of a cryptic plastid endosymbiosis in diatoms. Science. 2009;324:1724–1726. doi: 10.1126/science.1172983. [DOI] [PubMed] [Google Scholar]
- Mujer CV, Andrews DL, Manhart JR, Pierce SK, Rumpho ME. Chloroplast genes are expressed during intracellular symbiotic association of Vaucheria litorea plastids with the sea slug Elysia chlorotica. Proc. Natl. Acad. Sci. U S A. 1996;93:12333–12338. doi: 10.1073/pnas.93.22.12333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H, Brunak S, von Heijne G. Machine learning approaches for the prediction of signal peptides and other protein sorting signals. Protein Eng. 1999;12:3–9. doi: 10.1093/protein/12.1.3. [DOI] [PubMed] [Google Scholar]
- Nosenko T, Lidie KL, Van Dolah FM, Lindquist E, Cheng JF, Bhattacharya D. Chimeric plastid proteome in the florida ‘red tide’ dinoflagellate Karenia brevis. Mol. Biol. Evol. 2006;23:2026–2038. doi: 10.1093/molbev/msl074. [DOI] [PubMed] [Google Scholar]
- Oesterhelt C, Klocke S, Holtgrefe S, Linke V, Weber AP, Scheibe R. Redox regulation of chloroplast enzymes in Galdieria sulphuraria in view of eukaryotic evolution. Plant Cell Physiol. 2007;48:1359–1373. doi: 10.1093/pcp/pcm108. [DOI] [PubMed] [Google Scholar]
- Paul MJ, Driscoll SP, Andralojc PJ, Knight JS, Gray JC, Lawlor DW. Decrease of phosphoribulokinase activity by antisense RNA in transgenic tobacco: definition of the light environment under which phosphoribulokinase is not in large excess. Planta. 2000;211:112–119. doi: 10.1007/s004250000269. [DOI] [PubMed] [Google Scholar]
- Petersen J, Teich R, Becker B, Cerff R, Brinkmann H. The GapA/B gene duplication marks the origin of streptophyta (charophytes and land plants) Mol. Biol. Evol. 2006a;23:1109–1118. doi: 10.1093/molbev/msj123. [DOI] [PubMed] [Google Scholar]
- Petersen J, Teich R, Brinkmann H, Cerff R. A ‘green’ phosphoribulokinase in complex algae with red plastids: evidence for a single secondary endosymbiosis leading to haptophytes, cryptophytes, heterokonts, and dinoflagellates. J. Mol. Evol. 2006b;62:143–157. doi: 10.1007/s00239-004-0305-3. [DOI] [PubMed] [Google Scholar]
- Pierce S, Biron R, Rumpho M. Endosymbiotic chloroplasts in molluscan cells contain proteins synthesized after plastid capture. J. Exp. Biol. 1996;199:2323–2330. doi: 10.1242/jeb.199.10.2323. [DOI] [PubMed] [Google Scholar]
- Pierce SK, Curtis NE, Hanten JJ, Boerner SL, Schwartz JA. Transfer, integration and expression of functional nuclear genes between multicellular species. Symbiosis. 2007;43:57–64. [Google Scholar]
- Pohlmeyer K, Paap BK, Soll J, Wedel N. CP12: a small nuclear-encoded chloroplast protein provides novel insights into higher-plant GAPDH evolution. Plant Mol. Biol. 1996;32:969–978. doi: 10.1007/BF00020493. [DOI] [PubMed] [Google Scholar]
- Porter MA, Hartman FC. Commonality of catalytic and regulatory sites of spinach phosphoribulokinase: characterization of a tryptic peptide that contains an essential cysteinyl residue. Biochemistry. 1986;25:7314–7318. [Google Scholar]
- Porter MA, Stringer CD, Hartman FC. Characterization of the regulatory thioredoxin site of phosphoribulokinase. J. Biol. Chem. 1988;263:123–129. [PubMed] [Google Scholar]
- Richly E, Leister D. An improved prediction of chloroplast proteins reveals diversities and commonalities in the chloroplast proteomes of Arabidopsis and rice. Gene. 2004;329:11–16. doi: 10.1016/j.gene.2004.01.008. [DOI] [PubMed] [Google Scholar]
- Roesler KR, Ogren WL. Chlamydomonas reinhardtii phosphoribulokinase: sequence, purification, and kinetics. Plant Physiol. 1990;93:188–193. doi: 10.1104/pp.93.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumpho ME, Edwards GE. Inhibition of 3-phosphoglycerate-dependent O2 evolution by phosphoenolpyruvate in C4 mesophyll chloroplasts of Digitaria sanguinalis (L) Scop. Plant Physiol. 1984;76:711–718. doi: 10.1104/pp.76.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumpho ME, et al. Horizontal gene transfer of the algal nuclear gene psbO to the photosynthetic sea slug Elysia chlorotica. Proc. Natl. Acad. Sci. U S A. 2008;105:17867–17871. doi: 10.1073/pnas.0804968105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumpho ME, Summer EJ, Manhart JR. Solar-powered sea slugs: mollusc/algal chloroplast symbiosis. Plant Physiol. 2000;123:29–38. doi: 10.1104/pp.123.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumpho ME, Summer EJ, Green BJ, Fox TC, Manhart JR. Mollusc/algal chloroplast symbiosis: how can isolated chloroplasts continue to function for months in the cytosol of a sea slug in the absence of an algal nucleus? Zoology—Analysis of Complex Systems. 2001;104:303–312. doi: 10.1078/0944-2006-00036. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- Sato T, Atomi H, Imanaka T. Archaeal type III RuBisCOs function in a pathway for AMP metabolism. Science. 2007;315:1003–1006. doi: 10.1126/science.1135999. [DOI] [PubMed] [Google Scholar]
- Soll J, Schleiff E. Protein import into chloroplasts. Nature Rev. Mol. Cell Biol. 2004;5:198–208. doi: 10.1038/nrm1333. [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- Sterman NT. Spectrophotometric and fluorometric chlorophyll analysis. In: Lobban CS, Chapman DJ, Kramer BP, editors. Experimental Phycology. Cambridge: Cambridge University Press; 1988. pp. 35–46. [Google Scholar]
- Su X, Bogorad L. A residue substitution in phosphoribulokinase of Synechocystis PCC 6803 renders the mutant light sensitive. J. Biol. Chem. 1991;266:23698–23705. [PubMed] [Google Scholar]
- Tabita FR, Satagopan S, Hanson TE, Kreel NE, Scott SS. Distinct form I, II, III, and IV Rubisco proteins from the three kingdoms of life provide clues about Rubisco evolution and structure/function relationships. J. Exp. Bot. 2008;59:1515–1524. doi: 10.1093/jxb/erm361. [DOI] [PubMed] [Google Scholar]
- Tamoi M, Miyazaki T, Fukamizo T, Shigeoka S. The Calvin cycle in cyanobacteria is regulated by CP12 via the NAD(H)/NADP(H) ratio under light/dark conditions. Plant J. 2005;42:504–513. doi: 10.1111/j.1365-313X.2005.02391.x. [DOI] [PubMed] [Google Scholar]
- Tamoi M, Murakami A, Takeda T, Shigeoka S. Acquisition of a new type of fructose-1,6-bisphosphatase with resistance to hydrogen peroxide in cyanobacteria: molecular characterization of the enzyme from Synechocystis PCC 6803 Biochim. Biophys. Acta Struct. Mol. Enzymol. 1998;1383:232–244. doi: 10.1016/s0167-4838(97)00208-2. [DOI] [PubMed] [Google Scholar]
- Wadano A, Kamata Y, Iwaki T, Nishikawa K, Hirahashi T. Purification and characterization of phosphoribulokinase from the cyanobacterium Synechococcus PCC7942. Plant Cell Physiol. 1995;36:1381–1385. [PubMed] [Google Scholar]
- Wedel N, Soll J. Evolutionary conserved light regulation of Calvin cycle activity by NADPH-mediated reversible phosphoribulokinase/CP12/glyceraldehyde-3-phosphate dehydrogenase complex dissociation. Proc. Natl. Acad. Sci. U S A. 1998;95:9699–9704. doi: 10.1073/pnas.95.16.9699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedel N, Soll J, Paap BK. CP12 provides a new mode of light regulation of Calvin cycle activity in higher plants. Proc. Natl. Acad. Sci. U S A. 1997;94:10479–10484. doi: 10.1073/pnas.94.19.10479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West HH. Northeastern University: 1979. Chloroplast symbiosis and development of the ascoglossan opisthobranch Elysia chlorotica. PhD Thesis; p. 161 pp. [Google Scholar]
- Wintermans JF, de Mots A. Spectrophotometric characteristics of chlorophylls a and b and their pheophytins in ethanol. Biochim. Biophys. Acta. 1965;109:448–453. doi: 10.1016/0926-6585(65)90170-6. [DOI] [PubMed] [Google Scholar]