Abstract
In most plants, a large fraction of photo-assimilated carbon is stored in the chloroplasts during the day as starch and remobilized during the subsequent night to support metabolism. Mutations blocking either starch synthesis or starch breakdown in Arabidopsis thaliana reduce plant growth. Maltose is the major product of starch breakdown exported from the chloroplast at night. The maltose excess 1 mutant (mex1), which lacks the chloroplast envelope maltose transporter, accumulates high levels of maltose and starch in chloroplasts and develops a distinctive but previously unexplained chlorotic phenotype as leaves mature. The introduction of additional mutations that prevent starch synthesis, or that block maltose production from starch, also prevent chlorosis of mex1. In contrast, introduction of mutations in disproportionating enzyme (DPE1) results in the accumulation of maltotriose in addition to maltose, and greatly increases chlorosis. These data suggest a link between maltose accumulation and chloroplast homeostasis. Microscopic analyses show that the mesophyll cells in chlorotic mex1 leaves have fewer than half the number of chloroplasts than wild-type cells. Transmission electron microscopy reveals autophagy-like chloroplast degradation in both mex1 and the dpe1/mex1 double mutant. Microarray analyses reveal substantial reprogramming of metabolic and cellular processes, suggesting that organellar protein turnover is increased in mex1, though leaf senescence and senescence-related chlorophyll catabolism are not induced. We propose that the accumulation of maltose and malto-oligosaccharides causes chloroplast dysfunction, which may by signaled via a form of retrograde signaling and trigger chloroplast degradation.
Keywords: Carbohydrate metabolism, photosynthesis, senescence, chloroplast biology, Arabidopsis, autophagy
INTRODUCTION
Starch is one of the major products of photosynthesis in most higher plants. It is synthesized in chloroplasts during the day and broken down at night to sustain metabolism and growth. The major component of starch is amylopectin, a polymer in which chains of α-1,4-linked glucose are connected via α-1,6-bonds (branch points) to form a tree-like structure (Buléon et al., 1998; Zeeman et al., 2007b). Amylopectin is able to form a semi-crystalline matrix as neighboring chains form double helices, which pack in an ordered manner. The extent to which starch accumulates in leaves varies between species. Arabidopsis partitions 40–50% of newly assimilated carbon into starch (Zeeman and ap Rees, 1999), and mutants unable to synthesize or degrade starch are compromised in their growth rate in a diurnal cycle (Caspar et al., 1985; Zeeman et al., 1998).
The pathway of starch breakdown inside chloroplasts is relatively complex and requires the coordinated actions of a suite of enzymes (Zeeman et al., 2007a). The initial steps involve the phosphorylation of the starch granule surface by enzymes of the glucan, water dikinase class (GWD; Lorberth et al., 1998; Yu et al., 2001). This is proposed to disrupt the semi-crystalline packing of amylopectin (Edner et al., 2007; Hejazi et al., 2008). A set of glucan hydrolases then degrades the amylopectin via a network of reactions to maltose and glucose, both of which can exit the chloroplast via distinct transporters (MEX1 and pGlcT, respectively) that facilitate their diffusion across the inner chloroplast envelope (Schäfer et al., 1977; Rost et al., 1996; Weber et al., 2000; Niittylä et al., 2004).
Weise et al. (2004) demonstrated that for several species, maltose is the major form of carbohydrate exported from isolated starch-containing chloroplasts that were incubated in the dark. Maltose is produced by chloroplastic isoforms of β-amylases (Scheidig et al., 2002; Kaplan and Guy, 2005; Fulton et al., 2008), which act at the granule surface or on malto-oligosaccharide intermediates released from the granule by α-amylases or de-branching enzymes (Delatte et al., 2006; Kötting et al., 2009). Glucose can be produced via the metabolism of maltotriose or other malto-oligosaccharides by disproportionating enzyme (DPE1 or D-enzyme; Lin and Preiss, 1988; Critchley et al., 2001). When acting on maltotriose (its preferred substrate), DPE1 transfers a maltosyl unit from one molecule to another, resulting in glucose and maltopentaose. The latter can then be further metabolized to maltose and maltotriose by β-amylase.
Mutating important enzymes of starch breakdown cause starch excess (sex) phenotypes, due to an imbalance between the synthesis rate during the day and the breakdown rate during the night. This results in the gradual accretion of starch in the leaves as they age (Zeeman and ap Rees, 1999). The most severe sex phenotype observed to date is caused by mutations affecting GWD, reflecting its key position at the start of the pathway (Yu et al., 2001). Mutations affecting steps in the network of reactions downstream of GWD (e.g. β-amylase isoforms) result in less severe sex phenotypes. In some sex mutants, intermediates of the starch breakdown pathway accumulate. In the mex1 mutant, maltose cannot be exported from the chloroplast and accumulates in the stroma during starch breakdown at night, reaching levels more than 40 times that of wild-type plants (Niittylä et al., 2004; Lu et al., 2006). In the dpe1 mutant, maltotriose accumulates during the night, as it cannot be disproportionated and is a poor substrate for β-amylases (Critchley et al., 2001).
Unlike most sex mutants, mex1 is exceptional, as it displays an as yet unexplained chlorotic phenotype, with pale green or yellowish leaves (Niittylä et al., 2004). Different mechanisms could account for this observed chlorosis. For example, chlorophyll biosynthesis could be specifically down-regulated. Alternatively, chlorophyll catabolism, which would result in the accumulation of chlorophyll catabolites (Hörtensteiner, 2006) could be induced. It has also been reported that whole chloroplasts can be degraded inside lytic vacuoles (Wittenbach et al., 1982; Ono et al., 1995; Minamikawa et al., 2001). Recent evidence suggests that this occurs via autophagy (Wada et al., 2009)—a process by which cellular components are degraded to recycle nutrients. Both chlorophyll catabolism and chloroplast autophagy are processes that occur during the senescence of leaves. Autophagy can also be induced by nutrient limitation (Chen et al., 1994; Aubert et al., 1996; Rose et al., 2006).
This work aimed to understand the cellular processes resulting in the chlorotic phenotype of mex1. The results suggest that whole-chloroplast degradation via an autophagy-like process is the cause.
RESULTS
Chlorosis in mex1 Is Associated with the Production of Starch Breakdown Intermediates
The mex1 mutant has a previously unexplained chlorotic phenotype (Figure 1). When grown in either a 16-h or a 12-h photoperiod, the chlorophyll content of mex1 plants was lower than the wild-type on a leaf area basis (measured either with a chlorophyll meter on intact plants; Table 1). The reduction in mex1 was most significant in older leaves. Compared with the wild-type, young leaves had similar or only slightly reduced chlorophyll content.
Figure 1.
The Chlorotic Phenotype of mex1 Mutants.
Photographs of 5-week-old wild-type and mex1 plants grown in a 12-h photoperiod. Note the green young leaves and pale mature leaves of mex1.
Table 1.
Chlorophyll Content and Chloroplast Number in Differently Aged Leaves of mex1 and Wild-Type.
| Mean chlorophyll content (SPAD units) |
Mean chloroplast number per mesophyll cell | |||
| Leaf age | 16-h photoperiod | 12-h photoperiod | 12-h photoperiod | |
| Wild-type | Young | 33.1 ± 0.3 | 35.1 ± 0.4 | 11.1 ± 0.4 |
| Mature | 30.6 ± 0.5 | 33.1 ± 0.6 | 73.1 ± 2.0 | |
| Old | 23.7 ± 0.4 | 29.4 ± 0.6 | n.d. | |
| mex1 | Young | 26.7 ± 1.7 | 29.9 ± 1.6* | 9.4 ± 0.3* |
| Mature | 20.1 ± 0.6* | 25.8 ± 0.2* | 35.0 ± 1.6* | |
| Old | 12.3 ± 0.7* | 21.7 ± 0.5* | n.d. | |
Plants were grown and analyzed in independent experiments. For chlorophyll contents, the values are the means ± SE of three to six measurements on different plants. Measurements were made using a SPAD chlorophyll meter and expressed on a leaf area basis. For the 16-h photoperiod, old, mature, and young leaves (leaf numbers 1, 3–4, 7–8, respectively) of 3-week-old plants were analyzed. For the 12-h photoperiod, 4-week-old plants were analyzed (leaf numbers 1, 6–7, 10–11). For chloroplast numbers, values are the mean chloroplast count ± SE of 50 mesophyll cells prepared from three leaves (mature leaf, 7–8; young leaf, 14) from wild-type and mex1 plants (4 and 5 weeks old, respectively). Values for mex1 marked with an asterisk are significantly different from the corresponding wild-type values (student's t-test, p ≤ 0.05). n.d., not determined.
Niittylä et al. (2004) reported that blocking starch biosynthesis in mex1 by introducing a mutation in the plastidial isoform of phosphoglucomutase (the pgm mutant; Caspar et al., 1985) appeared to rescue the chlorotic phenotype, resulting in green, pgm-like plants. In contrast, mutation of disproportionating enzyme to prevent the metabolism of malto-oligosaccharides derived from starch (the dpe1 mutant; Critchley et al., 2001) made the chlorotic phenotype much worse (Niittylä et al., 2004). These data imply that the accumulation of starch breakdown products may trigger the chlorotic phenotype. To investigate this possibility further, we created additional multiple mutant lines in which starch breakdown was affected upstream of maltose production and analyzed the starch, chlorophyll, and malto-oligosaccharide contents. In addition, we recreated the dpe1/mex1 double mutant in an all-Columbia background (to exclude ecotype effects; the original double mutant was in a mixed Columbia and Wassilewskija background). For this, we isolated a new mutant allele of dpe1 in the Columbia background from the GABI-KAT collection (line GK_339B11), which harbors a T-DNA insertion in the seventh intron of the DPE1 gene (Supplemental Figure 1A). Homozygous mutants (designated dpe1-2) lacked the DPE1 protein (Supplemental Figure 1B).
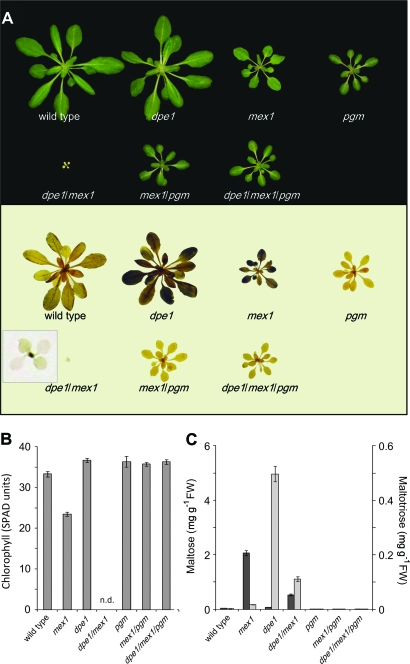
The visible phenotype of the mex1/pgm and dpe1/mex1 double mutants were consistent with those previously reported (Figure 2A and 2B; Niittylä et al., 2004). The mex1/pgm plants were not chlorotic, but were retarded in growth to the same extent as pgm single mutants. In contrast, the all-Columbia dpe1/mex1 plants were extremely small and chlorotic, as was the case for the Columbia-Wassilewskija isolate (Niittylä et al., 2004). The phenotype of the dpe1/mex1 double mutant was so severe that the plants failed to grow to maturity in a 12-h photoperiod. However, in continuous low light, growth was improved such that the double mutant did eventually flower and produce approximately 100 seeds per plant (Supplemental Figure 1C). Growth was also improved by the provision of exogenous sugars, though both mex1 and dpe1/mex1 were still chlorotic and grew much more slowly than the wild-type (Supplemental Figure 2). This severe phenotype was rescued by introducing the pgm mutation to create the dpe1/mex1/pgm triple mutant. These plants were similar in appearance to the pgm single mutants (Figure 2A).
Figure 2.
Modulation of the Chlorotic and Malto-Oligosaccharide-Accumulating Phenotypes of mex1 by Introducing dpe1 and pgm Mutations.
(A) Photographs of plants (upper panel) grown in a 12-h photoperiod. All plants were 5 weeks old. Note the extreme chlorotic dwarf phenotype of dpe1/mex1 double mutants and that blocking starch synthesis through introduction of the pgm mutation rescues chlorosis of both mex1 and dpe1/mex1. Harvesting plants at the end of the night and staining them for the presence of starch with iodine (lower panel) reveals that the starch-excess phenotype visible in dpe1 and mex1 is lost in the dpe1/mex1 double mutant. Inset shows an enlargement of a stained dpe1/mex1 plant. Note the dark-staining youngest leaves (see also Supplemental Figure 1D).
(B) Chlorophyll content of mature leaves of the wild-type and the mutants shown in (A), measured using a SPAD chlorophyll meter and expressed on a leaf area basis. Each value is the mean ± SE of nine replicate measurements on different leaves from at least three individual plants. n.d., not determined (leaves were too small for accurate analysis). The chlorophyll content of mex1 is significantly lower than the wild-type but that of mex1/pgm is not significantly different from pgm (student's t-test, p ≤ 0.5).
(C) Maltose (dark bars) and maltotriose contents (light bars) at the end of the night. Values are the mean ± SE of three to five replicate samples, each comprising a single rosette extracted using ethanol (see Methods). Note the 10-fold difference in scale of the two y-axes.
We analyzed the starch and malto-oligosaccharide contents of mex1/pgm, dpe1/mex1, dpe1/mex1/pgm, and the respective single mutants. Both mex1 and dpe1 are sex mutants (Figure 2A; Critchley et al., 2001; Niittylä et al., 2004), whereas pgm is essentially starchless (Caspar et al, 1985). Unexpectedly, iodine staining revealed that, unlike the mex1 and dpe1 parental lines, the mature leaves of the dpe1/mex1 double mutant were virtually free of starch. Compared with the wild-type, mex1 and dpe1 mutants accumulated high levels of maltose and maltotriose, respectively (Figure 2C; Critchley et al., 2001; Niittylä et al., 2004). The dpe1/mex1 double mutant accumulated both maltose and maltotriose, although the levels were decreased relative to the single mutant parental lines. The pgm mutant contained very low levels of malto-oligosaccharides, as did the mex1/pgm and dpe1/mex1/pgm mutant combinations (Figure 2C), showing that starch synthesis is a prerequisite for malto-oligosaccharide accumulation.
The low-starch phenotype of mature dpe1/mex1 leaves was surprising. Therefore, we analyzed these plants in more detail. Light microscopy showed that the very youngest leaves did contain starch (Figure 2A and Supplemental Figure 1D–1F). Quantitative measurements on seedlings revealed that, unlike the wild-type and single mutants, the starch content of dpe1/mex1 decreased over time, as the proportion of mature tissue increased (Supplemental Table 1). These data, and the presence of malto-oligosaccharides in dpe1/mex1, suggest that the low-starch phenotype is not due to an inability to make starch per se, but is more likely related to the development of the extreme chlorotic phenotype.
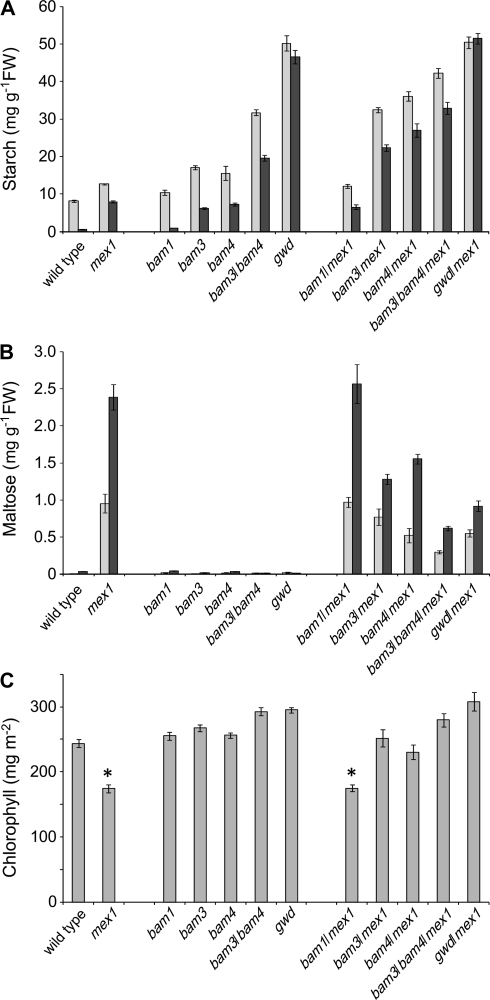
To test further whether the accumulation of starch breakdown intermediates triggers chlorosis, we limited malto-oligosaccharide production in mex1 using genetics. At least three β-amylase (BAM) proteins are involved in the production of maltose during starch breakdown: single mutations in either BAM3 or BAM4 reduce both maltose levels at night and starch breakdown, resulting in a sex phenotype (Fulton et al., 2008). BAM3 is a key active enzyme, whereas BAM4 is thought to regulate starch degradation. BAM1 is also an active enzyme but the bam1 mutant does not have a sex phenotype due to functional overlap between BAMs 1 and 3 (Fulton et al., 2008). We introduced mutations in BAM1, BAM3, or BAM4 into the mex1 background and analyzed the mutant combinations (Figure 3). Introduction of the bam1 mutation had no effect on the phenotype: starch, maltose, and chlorophyll content were the same as in mex1. In contrast, introduction of the bam3 or bam4 mutations increased starch, decreased maltose, and increased chlorophyll content compared with mex1. In the bam3/bam4/mex1 triple mutant, these effects were greater than in the bam3/mex1 and bam4/mex1 double mutants. We also introduced the gwd (sex1–3) mutation into mex1. GWD phosphorylates amylopectin at the starch granule surface, thereby enabling BAM action (Edner et al., 2007). The gwd/mex1 double mutants also had increased starch, decreased maltose, and increased chlorophyll content compared with mex1 (Figure 3).
Figure 3.
Modulation of Starch and Maltose Accumulation and of the Chlorotic Phenotype of mex1 by Introducing bam and gwd Mutations.
(A) Starch contents of the wild-type, and selected double or triple mutants in which enzymes of starch breakdown are missing in addition to MEX1. Plants were of the same developmental stage (20–43 d old, depending on the genotype, approximately 10-leaf stage) and were harvested at the end of the 12-h photoperiod (light bars) and at the end of the night (dark bars). Samples were extracted using perchloric acid (see Methods). Values are the mean ± SE of eight replicate samples, each comprising a single rosette.
(B) Maltose content of plants (as in (A)) harvested at the end of the 12-h photoperiod (light bars) and 4 h into the night (dark bars). Values are the mean ± SE of eight replicate samples, each comprising a single rosette.
(C) The chlorophyll content of mature leaves (leaf numbers 5–7 of 27–50-day-old plants, depending on the genotype) of the genotypes shown in (A). Chlorophyll was measured in acetone extracts and expressed on a leaf area basis. Each value is the mean ± SE of six replicate measurements on leaves from individual plants. Asterisks indicate lines carrying the mex1 mutation that differ significantly (student's t-test, p ≤ 0.5) in their chlorophyll content from the respective parental line (e.g. bam1/mex1 compared to bam1).
The mex1 Mutant Has Low Chlorophyll, Fewer Chloroplasts, and Decreased Cell Size
To understand the basis of the mex1 phenotype, we analyzed leaf structure and chloroplast numbers in mature leaves. First, we measured leaf thickness using an indicating caliper. This revealed that mex1 leaves were thinner than wild-type leaves (203 ± 10 and 263 ± 3 μm thick, respectively; mean ± SE of measurements on three replicate plants). Consistent with this, chlorophyll content of mature mex1 leaves was comparable to that of the wild-type when expressed on a fresh weight basis (1.13 ± 0.12 and 1.18 ± 0.05 mg g−1, respectively).
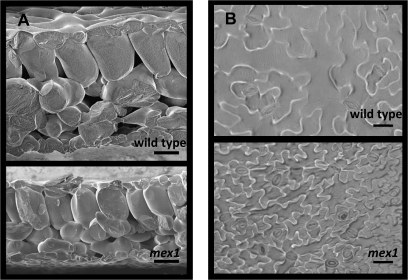
Scanning electron microscopy of freeze-fractured leaves confirmed the reduction of leaf thickness in mex1. No change in leaf structure was observed, but cell size in mex1 was reduced compared with the wild-type (Figure 4A). The reduced cell size in mex1 was also observed by analyzing epidermal imprints by light microscopy (Figure 4B). A reduced cell size should result in an increased cell density, even in thinner leaves. Thus, if the chlorophyll content of mex1 cells was the same as the wild-type, darker leaves rather than lighter leaves would be expected both on a leaf area and on a fresh weight basis. Therefore, we measured the number of chloroplasts in leaf mesophyll cells (Table 1). The average number of chloroplasts in wild-type mesophyll cells was 73, whereas mex1 cells had less than half that number. In contrast, young wild-type and mex1 leaves had similar numbers of chloroplasts per cell (approximately 10).
Figure 4.
Leaf Thickness and Cell Size in Wild-Type and mex1 Plants.
(A) Scanning electron micrographs of a mature leaf cross-section of wild-type and mex1 (4 and 5 weeks old, respectively; leaf numbers 6–8). Note the smaller cell size and reduced leaf thickness in mex1. Bars = 50 μm.
(B) Imprints of epidermal cells from the abaxial side of mature wild-type and mex1 leaves obtained using nail polish and viewed by light microscopy. Plants were 5 weeks old. Leaf numbers were 7–9 and 6–8 for the wild-type and mex1, respectively. Bars = 50 μm.
We determined chlorophyll fluorescence parameters to investigate whether photosynthetic performance was altered in mex1 (Supplemental Table 2). Ground state fluorescence (F0) was increased by 53% in mex1 relative to the wild-type. Subsequent determination of maximal fluorescence (Fm) in the dark-adapted state revealed that the maximum quantum efficiency of photosystem II (Fv/Fm) was decreased by 10% compared with the wild-type. The determination of the maximum fluorescence in the light-adapted state (Fm’) and calculation of the Genty parameter (φPSII) showed that the overall efficiency of PSII reaction centers in the light is decreased in mex1.
Chloroplast Degradation Is Triggered in mex1
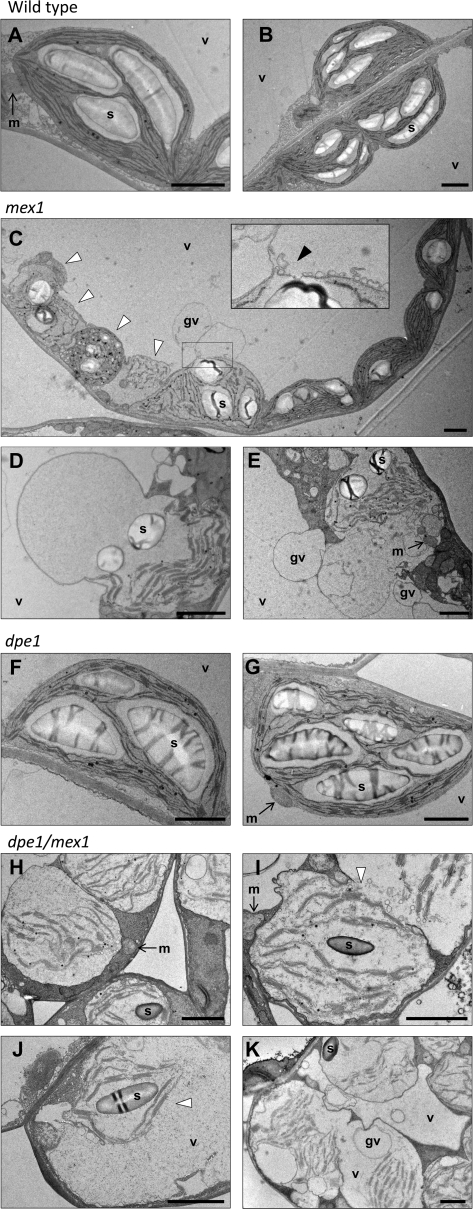
We analyzed the cellular ultrastructure of mesophyll cells in mex1 leaves using transmission electron microscopy. Compared with the chloroplasts in the leaves of the wild-type, mex1 chloroplasts were more variable in their appearance (Figure 5A–5E). Some resembled the wild-type chloroplasts, but often contained fewer granal stacks, which were frequently disorganized in appearance. In other cases, the chloroplasts appeared as swollen or spherical, with a disrupted thylakoid membrane system and many plastoglobules (Figure 5C). Frequently, cells were observed in which chloroplasts appeared to be fusing with other cellular compartments (vesicles or globular vacuoles; Figure 5C–5E). In young mex1 leaves, fewer differences to the wild-type were observed, but misshapen chloroplasts could still be observed (not shown).
Figure 5.
Chloroplast Ultrastructure in the Wild-Type, mex1, dpe1, and the dpe1/mex1 Double Mutant.
In each transmission electron micrograph, the bar = 2 μm. s, starch; v, vacuole; gv, globular vacuole; m, mitochondrion.
(A, B) Wild-type mesophyll cell chloroplasts.
(C) Multiple chloroplasts in a mesophyll cell from a mex1 mutant. Note the normal-looking chloroplasts on the right and the aberrant chloroplasts on the left (white arrowheads), which are misshapen, contain disorganized thylakoid membranes, and many plastoglobules. The swollen chloroplast in the center is associating with the adjacent globular vacuole. The inset shows an enlargement of the region of interaction (black arrowhead).
(D, E) Chloroplasts from mesophyll cells of the mex1 mutant that are either greatly swollen or that have fused with globular vacuoles. Many mesophyll cells of mex1 contained similar structures but none was seen in the wild-type.
(F, G) Chloroplasts from mesophyll cells of the dpe1 mutant. Note the enlarged starch granules.
(H) Swollen chloroplasts in three adjacent mesophyll cells of dpe1/mex1.
(I) Two fusing compartments, both containing chloroplast remnants, in a mesophyll cell of dpe1/mex1. Note the vesicles at the fusion site (white arrowhead).
(J) Remnants of a single chloroplast within a large vacuole in a mesophyll cell of dpe1/mex1. Note the absence of the chloroplast envelope (white arrowhead).
(K) A single cell of dpe1/mex1 containing multiple swollen chloroplasts and multiple vacuoles. One vacuole contains the remnants of three chloroplasts. Note also the numerous globular vacuoles. Cells with this appearance were common.
We also examined the chloroplasts in dpe1/mex1 double mutants (Figure 5H–5K). In this case, a much more extreme phenotype was observed than in mex1. There were almost no chloroplasts that resembled those of the wild-type. All were swollen and misshapen, and had altered thylakoid membrane systems (Figure 5H). Many cells also appeared to contain chloroplast components (thylakoid membranes, starch granules, plastoglobules) within the vacuolar compartment, which were electron-dense compared with wild-type vacuoles. In some cases, the chloroplast components were still associated with each other, but were not bordered by any visible membranes (Figure 5J). In other cases, the chloroplast components were separated and spread throughout the vacuoles, which were smaller and more numerous than in the wild-type. Even in the youngest dpe1/mex1 leaves that we could prepare for microscopy, severely aberrant chloroplasts and chloroplast remnants in vacuoles could be observed (not shown). Cell sections that did not contain such visible remnants of chloroplast structures often had electron dense vacuoles.
These phenotypes suggest that in mex1, chloroplasts are being degraded via an autophagy-like process and that in dpe1/mex1 double mutants, this process is initiated earlier in the development of the leaf. We did not observe structures similar to those in mex1 and dpe1/mex1 in either the wild-type (Figure 5A and 5B) or the dpe1 single mutant (Figure 5F and 5G), though the latter had large starch granules.
Microarray Analysis Reveals Reprogramming of Metabolic and Cellular Processes in mex1
To investigate the possible underlying cause(s) of the chlorotic phenotype of mex1, we performed microarray analyses on young leaves, which were as green as wild-type leaves, and mature leaves that contained less chlorophyll (Figure 1 and Table 1). Three experimental replicates were performed for each genotype. A gene list was compiled that contained all genes that were designated as ‘present’ on one or more of the 12 arrays (for details on sampling and microarray analysis, see Methods). Two complementary approaches were used for the comparisons of the gene expression levels (Supplemental Figure 3). First, the lists were subject to statistical analyses (Welch t-test; p ≤ 0.1) and two-fold expression-level cut-offs (i.e. genes whose expression in mex1 is either more than twice or less than half that of the wild-type) to determine subsets of genes that were significantly altered in expression in mex1 relative to the wild-type. These subsets were then functionally annotated. Second, changes in expression were visualized using MAPMAN (Thimm et al., 2004). In this case, only the statistical cut-off was applied so that more subtle, widespread changes could also be visualized. A fourth experimental replicate was used to conduct quantitative RT–PCR for 18 genes. The results were largely consistent with those obtained from the microarrays (Supplemental Table 3).
In mex1, 1750 genes from young leaves and 4747 genes from mature leaves were significantly differently regulated compared to the equivalent tissues in the wild-type. Subsequent application of the two-fold cut-off resulted in lists of 145 (77 up- and 68 down-regulated) and 894 (474 up- and 420 down-regulated) genes from young and mature leaves, respectively (Supplemental Dataset 1). Only a small proportion of these genes were altered in expression in the same way in both tissues. Functional annotation enrichment analysis of the gene lists in Supplemental Dataset 1 was performed with BiNGO and DAVID software (Dennis et al., 2003; Maere et al., 2005; Huang et al., 2007) using the major Gene Ontology (GO) categories (‘biological process’, ‘molecular function’, and ‘cellular component’) and protein domains defined by public databases.
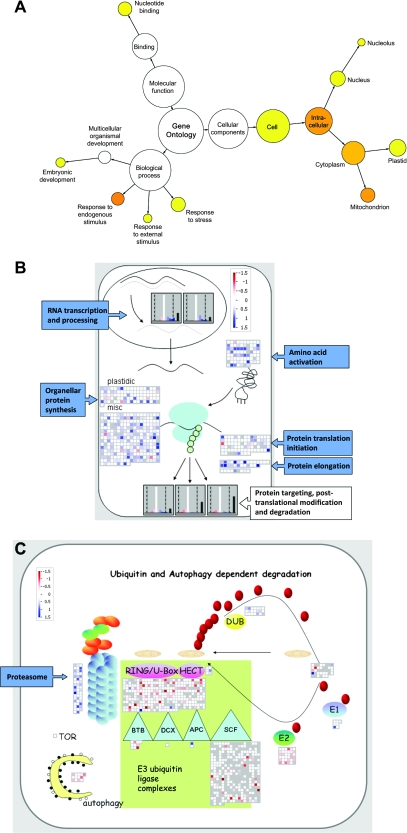
Of the altered transcripts in young mex1 leaves, genes involved in flavonoid metabolism were significantly over-represented, due to the induction of a number of genes (Supplemental Figure 4A). Increased flavonoid production (e.g. anthocyanins) is commonly associated with stress responses and with elevated sugar levels (Winkel-Shirley, 2002; Solfanelli et al., 2006), although there was no increase in anthocyanin content in the young leaves of mex1 (not shown). Of the altered transcripts in mature mex1 leaves, there were many significantly over-represented gene categories (Figure 6A). The biological processes included ‘responses to internal stimuli’ (primarily down-regulated), ‘responses to external stimuli’, and ‘responses to stress’ (a mixture of both up- and down-regulated). Genes annotated with the molecular function ‘nucleotide-binding’ (e.g. aspects of RNA processing) and associated with cellular components ‘nucleus’, ‘mitochondrion’, and ‘plastid’ were also over-represented, due to widespread gene induction. In particular, a large number of proteins containing PPR domains were up-regulated. Most PPR proteins characterized thus far are involved in organellar RNA metabolism (splicing, stability, and editing; e.g. Fisk et al., 1999; Meierhoff et al., 2003; Kotera et al., 2005). Similar conclusions could be drawn from MAPMAN displays (Figure 6B), which revealed an induction of genes associated with transcription and RNA processing, amino acid activation, and protein biosynthesis. Several transcripts encoding proteasome components were increased, and there were decreases in a number of transcripts encoding components of E3 ubiquitin ligase complexes (Figure 6C). Genes involved in autophagy were unchanged or slightly down-regulated. Collectively, these data suggest that there may be changes in protein turnover in mature mex1 leaves compared to the wild-type.
Figure 6.
Alterations in the Transcriptome of mex1 Revealed by Microarray Analysis.
(A) Gene Ontology (GO) analysis of transcripts significantly altered in abundance in mature leaves of mex1, relative to the wild-type. Over-represented categories relative to the genomic average are visualized using Cytoscape (Shannon et al., 2003) and colored yellow (p ≤ 5 × 10−2) to orange (p ≤ 5 × 10−7).
(B) Transcript levels of genes involved in RNA metabolism and translation visualized using MAPMAN. Transcripts are shown either individually as squares, or collectively as frequency histograms. Changes in mature mex1 leaves relative to the wild-type are shown in color (blue, up-regulated; red, down-regulated; white, no change) on a log2 scale as indicated. Gray squares in the displays and black bars on the histograms indicate genes called as ‘not present’ on all of the microarrays. In the gene categories shown, there were almost no changes in young mex1 leaves relative to the wild-type (not shown).
(C) Transcript levels of genes involved in protein degradation visualized using MAPMAN, as in (B). In the gene categories shown, there were almost no changes in young mex1 leaves relative to the wild-type (not shown).
MAPMAN analysis of the mature leaf microarray data also revealed changes in transcripts of genes involved in metabolism and cellular response pathways. Genes involved in the light reaction of photosynthesis (notably light-harvesting complex proteins), cell wall biosynthesis and modification (notably expansins and arabinogalactan proteins), raffinose metabolism, and trehalose metabolism were decreased (Supplemental Figure 4B). Transcripts of genes of nucleotide biosynthesis were increased. There were significant changes in the transcripts of genes involved in stress responses. The trend was down-regulation of genes involved in biotic stress responses and up-regulation of those involved in abiotic stress responses (notably heat-shock proteins). Genes involved in the cell cycle and development showed a mixed response (Supplemental Figure 4D). Several thioredoxin and glutaredoxin transcripts were decreased. There were far fewer changes in the transcriptome of young mex1 leaves, and the patterns were not consistent with those seen in the mature leaves (Supplemental Figure 4A and 4C).
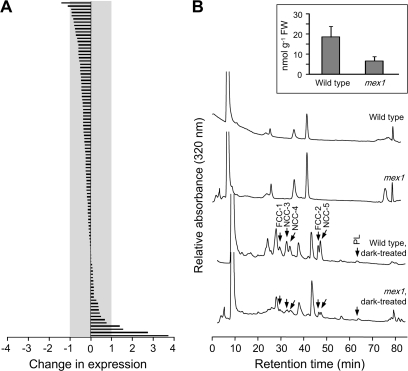
Chloroplast autophagy has been reported to accompany leaf senescence (Wittenbach et al., 1982; Ono et al., 1995; Minamikawa et al., 2001; Wada et al., 2009). Therefore, we analyzed our microarray data for changes in 105 senescence-induced genes identified previously by suppression subtraction hybridization and confirmed using Northern blot analysis (Gepstein et al., 2003; Supplemental Dataset 1). In mex1, three-quarters of these genes had lower expression values compared with the wild-type, and most of the changes were less than two-fold (Figure 7A). We also investigated the response of 827 genes whose transcription was found to be at least three-fold up-regulated in mature rosette leaves at a mid-senescence stage, compared with non-senescent leaves, using microarray analysis (Buchanan-Wollaston et al., 2005). In our microarrays studies, only 24 of these 827 genes were more than three-fold up-regulated in mex1 compared with the wild-type. Only 67 were more than two-fold up-regulated (Supplemental Dataset 1). Widely used senescence marker genes such as SAG12 and SAG13 were not up-regulated. These data suggest that the large-scale transcriptional changes associated with senescence are not occurring in mex1.
Figure 7.
Senescence-Related Gene Expression and Chlorophyll Degradation Are Not Detected in mex1.
(A) Changes in expression in mex1 relative to the wild-type of 105 genes previously identified as senescence-induced (Gepstein et al., 2003). Expression levels obtained from the ATH1 GeneChip analyses of mature leaves are given on a log2 scale. Note that most expression levels differ less than two-fold from the wild-type (indicated by the gray area).
(B) HPLC analysis of extracts of mature leaves of the wild-type and mex1. Chlorophyll catabolites were not detectable in either line. After induction of senescence by incubating the leaves in the dark for 5 d, fluorescent chlorophyll catabolites (FCCs), non-fluorescent chlorophyll catabolites (NCCs), and pheophorbide-like catabolites (PLs), identified by their characteristic spectra, were readily detectable in both lines. The quantification of total NCCs from analyses of four replicate, dark-treated samples, each comprising eight 7-mm leaf discs, is inset.
We also analyzed leaf extracts of the wild-type and mex1 for the presence of chlorophyll catabolites, which accumulate during senescence, but none was detectable. However, after detaching the leaves and incubating them in the dark for 5 d to induce senescence, chlorophyll catabolites were detected, albeit at lower levels than in the wild-type, illustrating that senescence could be triggered in mex1 (Figure 7B).
DISCUSSION
We previously speculated about the existence of a link between malto-oligosaccharide accumulation in the chloroplast stroma and the development of leaf chlorosis in mex1 (Niittylä et al., 2004). This work supports and extends this hypothesis. We provide a body of genetic and biochemical evidence that the degree of chlorosis is influenced by the extent of malto-oligosaccharide accumulation. Furthermore, our data reveal the cellular basis for the chlorosis and allow us to generate testable hypotheses about what triggers it.
Malto-Oligosaccharide Accumulation Is Correlated with Chlorosis
Starch breakdown is the major pathway leading to malto-oligosaccharide production in the leaves of Arabidopsis. Thus, starch synthesis is a prerequisite for maltose accumulation in the mex1 mutant. Consequently, it is not surprising that mex1/pgm double mutants do not accumulate maltose. The fact that these plants are also not chlorotic was the first indication that the maltose accumulation and chlorosis are linked (Niittylä et al., 2004). Here, we show that mex1 chlorosis is also reduced by blocking starch breakdown upstream of maltose either by mutation of β-amylase, which produces maltose (i.e. the bam3/mex1 double mutant), or by mutating GWD, which acts at the start of the starch breakdown pathway (Zeeman et al., 2007b). In contrast, the removal of DPE1 in the mex1 background greatly increases the severity of chlorosis. This is consistent with the idea that malto-oligosaccharides somehow trigger chlorosis as the loss of DPE1 results in maltotriose accumulation in addition to maltose. However, as both dpe1 and mex1 are sex mutants, it would have been reasonable to hypothesize that the dpe1/mex1 double mutant would also contain high levels of starch. This is not the case, and the double mutants are almost free of starch. Furthermore, in mature tissues, maltose and maltotriose accumulate to lower levels in comparison to the single mutant parental lines. Our results show that the double mutant is capable of synthesizing starch, which was visible in young leaves. We suggest that degradation of this starch leads rapidly to high malto-oligosaccharide levels in the very young leaves, provoking an early onset of chlorosis. This, in turn, affects the photosynthetic capacity, resulting in a decrease of carbon available for starch synthesis. Evidence that malto-oligosaccharide accumulation is the underlying cause of the severe chlorotic phenotype of dpe1/mex1 comes again from the fact that the introduction of the pgm mutation rescues the phenotype.
Other mutations also result in the accumulation of maltose in Arabidopsis leaves. Loss of the cytosolic glucosyltransferase DPE2, which metabolizes maltose exported from the chloroplast via MEX1, results in the accumulation of 100 times as much maltose as the wild-type (about twice as much as in mex1; Lu and Sharkey, 2004; Chia et al., 2004). However, unlike mex1, maltose accumulates both inside and outside the chloroplast in dpe2 (Lu et al., 2006). Similarly, maltose accumulation in both the cytosol and chloroplast has been reported in Arabidopsis plants deficient in two isoforms of starch branching enzyme (be2/be3 double mutants; Dumez et al., 2006). In the latter case, plants were also reported to be chlorotic in appearance. Although this was not reported in the descriptions of the dpe2 mutants (Lu and Sharkey, 2004; Chia et al., 2004), we have observed a small decrease in the chlorophyll content per unit area of mature dpe2 leaves (Stettler and Zeeman, unpublished data). Our investigations into the chlorotic phenotypes of these plants are ongoing.
Induction of Chloroplast Degradation Causes mex1 Chlorosis
The visibly chlorotic phenotype of mature mex1 leaves is influenced by several different factors. Leaves of mex1 are thinner than wild-type leaves, due to a reduction in cell size, but in itself, this would not cause leaves to appear chlorotic. The reduced chlorophyll content is primarily due to a decrease of over 50% in the number of chloroplasts per cell in mature mex1, relative to the wild-type. This phenotype develops as mex1 leaves mature. The young mex1 leaves emerge with almost the same chlorophyll content and chloroplast numbers as the wild-type, but as the leaves mature, chloroplast numbers increase over seven-fold in the wild-type, but only 3.5-fold in mex1. A decrease in chloroplast biogenesis or an induction (or increase) in the rate of chloroplast degradation could account for this difference.
The changes in gene expression revealed by our microarray analysis are not consistent with a decrease in chloroplast biogenesis as an explanation for the low chloroplast numbers. The most prominent transcriptional changes in mex1 relative to the wild-type included increases in the expression of genes encoding proteins involved in RNA metabolism, amino acid activation, protein folding, and proteosome function. Collectively, these changes indicate a possible increase in protein turnover. However, the change in the pattern of genes encoding components of ubiquitin ligase complexes suggests that the protein targets of the proteasome may differ in mex1 compared to the wild-type. Furthermore, there was increased expression of genes encoding proteins targeted to the chloroplast and mitochondrion, notably PPR proteins, which are involved in organellar gene expression. Thus, chloroplast biogenesis may actually be increased in mex1, rather than decreased.
The most likely cause of the reduced chloroplast number in mex1 was revealed by the microscopic analysis of cellular ultrastructure. The formation of vacuole-like structures containing partially degraded chloroplast components could be seen in most mesophyll cells of mature mex1 leaves, but not in the wild-type. Not all mex1 chloroplasts were simultaneously affected; chloroplasts with a normal appearance could be seen alongside aberrant chloroplasts. If chloroplast biogenesis is increased, as suggested by the microarray data, this may partly compensate for the induction of chloroplast degradation. The resultant chloroplast turnover would explain the diversity in chloroplast appearance that we observed. In young mex1 leaves, which were as green and contained as many chloroplasts as the wild-type, structures indicative of chloroplast degradation were rare (not shown).
The chloroplast degradation phenotype of the dpe1/mex1 double mutant was much more severe and appeared to be triggered earlier in leaf development than in the mex1 single mutant. Even the youngest leaves imaged contained highly aberrant chloroplasts and none had a wild-type appearance. Vacuole-like structures containing chloroplast remnants were widespread in mature leaves. This can also help to explain the surprisingly low starch and malto-oligosaccharide levels in this line. As chloroplasts are degraded, so will be their contents, including the malto-oligosaccharides and starch. It is likely that enzymes capable of glucan breakdown, distinct from those in the chloroplast, occur in lytic vacuoles.
The Trigger for Chloroplast Degradation in mex1
Despite the link between malto-oligosaccharide accumulation and chlorosis, the nature of the signal and intermediate steps that trigger chloroplast degradation remain unclear. Our transmission electron micrographs suggest that chloroplasts are subject to autophagy. Attempts to stain autophagosomes with the molecular marker monodansylcadaverine (Contento et al., 2005) to confirm this were inconclusive (not shown). Genes known to be associated with autophagy were unchanged in expression or slightly down-regulated in mex1. This does not necessarily exclude them from being involved in the mex1 phenotype, as gene up-regulation does not always accompany the induction of autophagy (Klionsky et al., 2008). For example, upon nutrient starvation in Arabidopsis cell cultures, induction of several autophagy genes occurs only transiently (Rose et al., 2006). Furthermore, it is also possible that additional, as yet unidentified genes are involved in the process that we report.
The degradation of whole chloroplasts during senescence has recently been shown to occur via autophagy (Wada et al., 2009). However, our data are not consistent with the idea that senescence per se has been induced. First, mature, chlorotic mex1 leaves do not die rapidly, but persist on the plant. Second, expression of senescence-associated genes is not induced in mex1 leaves relative to the wild-type. Third, senescence-associated chlorophyll catabolites are not present in chlorotic mex1 leaves. However, conditions that induce senescence cause the accumulation of chlorophyll catabolites in both the wild-type and mex1. While these data suggest that mex1 leaves are not senescing prematurely, it remains possible that the same mechanism that mediates chloroplast degradation during senescence has been induced separately in mex1.
Carbon starvation has been shown to initiate autophagy in heterotrophic rice, sycamore, and Arabidopsis cell cultures (Chen et al., 1994; Aubert et al., 1996; Rose et al., 2006). The inability to export starch breakdown products might result in carbon starvation in mex1 leaves at night. This would be exacerbated in dpe1/mex1, as DPE1 produces glucose that can be exported from the chloroplast on a distinct transporter to maltose (Rost et al., 1996). However, nighttime starvation also occurs in pgm mutants (Thimm et al., 2004) and in mutants that are blocked in starch breakdown upstream of maltose production (e.g. the gwd mutant; Messerli et al., 2007), but these mutants are not chlorotic. Furthermore, provision of exogenous sugars (in the form of sucrose, glucose, or maltose) did not rescue the chlorotic phenotype of mex1 or dpe1/mex1, even if it did improve their growth. Therefore, carbon starvation per se is unlikely to be the trigger of chloroplast degradation in mex1.
Niwa et al. (2004) proposed that chloroplasts could be selectively degraded via autophagy, serving a ‘quality control’ function. It remains possible that the excessive accumulation of maltose could somehow signal chloroplast dysfunction. First, maltose or other malto-oligosaccharides could, at high levels, be sensed inside the chloroplast and mark them for autophagy. However, the components of such a signaling system are not known, and it is not clear what its physiological function would be. Second, the accumulation of high levels of malto-oligosaccharides inside the chloroplast might cause an intracellular osmotic stress. The effect of this on the plant cell is not clear, as most studies of osmotic stress focus on differences between the cell and the external environment. Many of the aberrant chloroplasts observed in mex1 and the dpe1/mex1 appeared swollen, which might indicate an osmotic imbalance between the chloroplast and the cytosol. However, if mutants that accumulate maltose in both cytosol and chloroplast also display autophagy, it would undermine this explanation. Third, maltose is a reducing sugar and, at high concentrations, may interfere with important chloroplast functions. Our analysis of photosynthetic parameters suggested that several aspects of photosynthesis have changed in mex1. The increase in F0 could indicate the dissociation of light harvesting complexes from the photosystems. This, in turn, could result from an over-reduction of the plastoquinone pool, potentially brought about by the elevated maltose levels. The plastid redox status has been associated with retrograde signaling from the chloroplast to the nucleus (Pfannschmidt et al., 1999), although the signaling process is not well defined and has not previously been associated with chloroplast degradation. Furthermore, the measured increase in F0 could be a consequence of the autophagic process rather than associated with its cause. Thus, the speculative link needs further experimental testing.
Our ongoing analysis of this autophagy-like process includes the isolation of re-mutagenized dpe1/mex1 lines in which the chlorotic phenotype is suppressed. Interestingly, some of the suppressor lines contain elevated maltose, showing that the two aspects of the double mutant phenotype—the malto-oligosaccharide accumulation and chloroplast degradation—can be dissociated. Such suppressor lines may lack components underlying either the mechanism of chloroplast degradation or the putative signaling pathway involved in triggering it.
METHODS
Plant Growth and Harvesting
Arabidopsis thaliana L., ecotype Columbia (Col-0), was grown in a nutrient-rich, medium-grade, peat-based compost in a Percival AR95 or AR66 growth chamber (CLF Plant Climatics, Emersacker, Germany). The conditions were a constant temperature of 20°C, 60% relative humidity, with a 12-h photoperiod (150 μmol quanta m−2 s−1), a 16-h photoperiod (150 μmol quanta m−2 s−1), or continuous light (100 μmol quanta m−2 s−1), as specified. Seeds were sown by hand and stratified for 2 d at 4°C. After seedling establishment, individuals were pricked out into 200-ml pots. To provide supplementary sugars, plants were grown on agar plates containing half-strength Murashige and Skoog salts supplemented with 0.5% (w/v) sucrose, glucose, or maltose. Leaf thickness was measured using an indicating calliper (Etalon, Switzerland).
Mutants
Details of the mutants used in this study are given in Supplemental Table 4. The mutations in mex1-1, pgm, and bam3-1 are point mutations, for which we developed cleaved amplified polymorphic sequence (CAPS) markers. Plants homozygous for the mutation in gwd (sex1-3; Yu et al., 2001) were identified by immunoblotting with a GWD-specific antibody. The mutation in bam4-1 is caused by a T-DNA insertion. All mutants are in the Columbia background. Lines carrying multiple mutations were obtained by crossing and selecting homozygous plants of the required genotypes from the segregating F2 populations using PCR- and CAPS-based genotyping. Primer sequences are given in Supplemental Table 4.
Starch and Malto-Oligosaccharide Measurements
For the measurement of starch, the perchloric acid extraction method described by Delatte et al. (2005) was used, with slight modifications. Two sampling strategies were used. Samples comprising either entire individual rosettes (20–43 d old, approximately 100 mg fresh weight), or the aerial parts of 5–14-day-old seedlings (pooled plants resulting in 10–30 mg tissue per genotype), were frozen in 96-format collection tubes and pulverized while still frozen using a Mixer Mill (Retsch, Haan, Germany). The frozen powder was extracted in 800 μl ice-cold 1 M perchloric acid for 5 min with intermittent mixing. All the subsequent steps were carried out between 0 and 4°C. After centrifugation (10 min, 2000 g, 4°C), the insoluble material in the pellet was washed once with water and three times with 80% (v/v) ethanol (2 ml wash volume per ml of original extract volume). Starch in the insoluble material was measured by determining the glucose released by treatment with α-amylase and amyloglucosidase as described previously (Smith and Zeeman, 2006). The supernatant (soluble fraction) was adjusted to pH 5 by adding 2 M KOH, 0.4 M MES, 0.4 M KCl (approximately 0.4 ml per ml of supernatant). Precipitated potassium perchlorate was removed by centrifugation (2000 g, 15 min, 4°C) and malto-oligosaccharides in the supernatant were measured as described in Fulton et al. (2008).
An additional method for the determination of malto-oligosaccharide contents was used. Rosettes were harvested in 1–4 ml 80% (v/v) ethanol (depending on sample size) and incubated for 10 min at 80°C. An aliquot of 800 μl was transferred to a new tube and the ethanol was evaporated using a vacuum concentrator. The samples were dissolved in 100 μl water and malto-oligosaccharides analyzed as above.
Chlorophyll, Chlorophyll Catabolite, and Anthocyanin Measurements
Chlorophyll was measured either using a SPAD 502 chlorophyll meter (Konica Minolta, Dietikon, Switzerland) or by homogenizing four 7-mm diameter leaf discs from mature leaves of individual plants in 1 ml 98% (v/v) acetone. Extracts were kept in the dark and cell debris was removed by centrifugation (1 min, 1600 g, at 4°C). The absorption of the extracts was determined at 645 and 663 nm and chlorophyll content calculated according to Arnon (1949).
Chlorophyll catabolites were extracted from and analyzed as described previously (Pružinská et al., 2007). Briefly, samples comprising eight leaf disks (7-mm diameter) were frozen in liquid N2 and pulverized in a pestle and mortar. Tetrapyrrolic chlorophyll catabolites (RCCs, FCCs, and NCCs) were extracted in 300 μl medium containing 20 mM Tris pH 8 and 80% (v/v) methanol. After centrifugation (5 min, 16 000 g, 4°C), the supernatant was analyzed by HPLC. Pigments were identified by their absorption spectra and quantified by comparison with standard curves established using authentic standards. To induce senescence in control samples, eight leaf disks were incubated on a wet filter paper in a closed Petri dish in the dark for 5 d at 20°C. Pigments were extracted and analyzed as above.
Anthocyanins were extracted by homogenization of samples (10 mg) in 600 μl ice-cold acidified methanol (containing 1% (v/v) HCl). Water (400 μl) was added and mixed. Chloroform (500 μl) was added to remove chlorophyll through mixing and centrifugation (1 min, 14 000 g, 4°C). Anthocyanin content in the aqueous phase was determined spectrophotometrically at 538 nm.
Chlorophyll Fluorescence Measurements
Chlorophyll fluorescence parameters were determined using a FluorCam system (Photon Systems Instruments, Brno, Czech Republic) according to the manufacturer's instructions. Prior to the measurement, plants were dark-adapted for 15 min to set all PSII centers to the open state.
Chloroplast Counting
Chloroplast numbers per cell were determined using the method described by Pyke and Leech (1991). Leaf pieces were vacuum-infiltrated and incubated in phosphate-buffered saline solution (PBS) containing 3.5% glutaraldehyde for 1 h at 25°C. The leaf tissue was once washed and then incubated with 0.1 M EDTA for 3.5 h at 60°C to soften the tissue. The leaf pieces were macerated on a microscope slide and analyzed by light microscopy.
Electron Microscopy
Procedures for transmission electron microscopy were as described in Delatte et al. (2005) with the following modifications: leaf samples were fixed in 2% glutaraldehyde in 0.1 M sodium cacodylate buffer, pH 7.4 for 4 h at 4°C. Tissue was washed six times in cold 0.1 M sodium cacodylate buffer, pH 7.4, post-fixed overnight in 1% (w/v) aqueous osmium tetroxide, 0.1 M sodium cacodylate buffer, pH 7.4 at 4°C, then washed a further six times in cold 0.1 M sodium cacodylate buffer, pH 7.4 and once with water. Then, sections were dehydrated in an ethanol series and infiltrated and embedded in epoxy resin (Spurr's, Agar Scientific, Stansted, UK). Ultra-thin sections were cut with a diamond knife and stained sequentially with uranyl acetate and Reynold's lead citrate. Stained sections were examined using a Phillips BioTwin CM100 or a FEI Morgagni 268 electron microscope.
For freeze-fracture and cryo-scanning electron microscopy, a piece (approximately 5 × 8 mm) was cut out of the middle of the leaf (omitting the main vein) using micro-scissors. The pieces were inserted upright into a cryo-holder for scanning electron microscopy and frozen in liquid N2. After transfer into the freeze-drying and coating unit (BAF 060, Balzers, Liechtenstein) at –120°C, the samples were fractured perpendicular to the cryo-holder surface. Contaminations of ice crystals on the surface were removed by freeze-etching for 5 min at –90°C. The samples were then coated with a 5-nm layer of tungsten at –120°C, and subsequently transferred and imaged at –120°C in a Zeiss Gemini 1530 FEG scanning electron microscope (Zeiss Oberkochen, Germany), using 2 kV acceleration voltage.
Microarray and Quantitative RT–PCR Analyses
For the microarray analysis, three independent batches of plants were grown in a 12-h photoperiod. Plants were harvested after 4 weeks (wild-type) or 5 weeks (mex1). At these ages, the plants had reached the same developmental stage (rosettes with 15–16 leaves). Mature and young leaves (leaf numbers 6–8 and 13–15, respectively) from each genotype were harvested from each batch 4 h before the end of the day, resulting in a total of 12 samples. Within each batch, material was pooled from 12–15 plants per genotype. A fourth, independent batch was grown for quantitative RT–PCR analysis from which six leaves (numbers 6–8) of at least three plants were harvested. Samples were frozen in liquid N2 and pulverized using a mortar and pestle. Total RNA was extracted using Trizol (Invitrogen, Leek, Netherlands) and purified using the RNeasy Kit from Qiagen (Hombrechtikon, Switzerland) according to the supplier's instructions.
For microarray analysis, the quality and purity of the RNA were confirmed using an Agilent 2100 Bioanalysier (Agilent, Waldbronn, Germany). Total RNA samples (2 μg) were reverse-transcribed, yielding double-stranded cDNA, which was transcribed in vitro in the presence of biotin-labeled nucleotides using an IVT Labelling Kit (Affimetrix Inc., Santa Clara, CA), and purified and quantified using BioRobot Gene Exp-cDNA Target Prep (Qiagen AG, Switzerland). Labeled cRNA was fragmented and hybridized to Affymetrix ATH1 GeneChip arrays for 16 h at 45°C according to Affymetrix protocols. Arrays were washed on an Affymetrix Fluidics Station 450 using the EukGE-WS2v4_450 protocol. An Affimetrix GeneChip Scanner 3000 was used to measure the fluorescence intensity of the arrays.
Raw data processing was performed using Affymetrix Gene Chip Operating Software (GCOS1.4), which determined ‘present’ and ‘absent’ calls based on the signal intensity ratio between perfect-match and miss-match oligos on the array. Cell intensities were calculated and summarized for the respective probe sets by means of the MAS5 logarithm (Hubbell et al., 2002). Labeling efficiency and hybridization performance were controlled through monitoring of the housekeeping genes (GAPDH and ACO7), and of the poly A spike and the prokaryotic controls (BIOB, BIOC, CREX, BIODN). To compare values between arrays, global scaling was used to normalize the trimmed mean of each chip to a target intensity (TGT value) of 500 as recommended by Affymetrix. Quality-control measures including adequate scaling factors (between 1 and 3 for all samples) and appropriate number of present calls (calculated by application of a signed-rank call algorithm; Liu et al., 2002) were considered before subsequent analysis with GeneSpring GX software (Agilent Technologies). The results were normalized to the 50th percentile per chip and normalized to median per gene.
Functional annotation enrichment analysis was performed using BiNGO (www.psb.ugent.be/cbd/papers/BiNGO/index.htm; Maere et al., 2005), DAVID (http://david.abcc.ncifcrf.gov; Dennis et al., 2003; Huang et al., 2007), and MAPMAN software (Thimm et al., 2004).
For quantitative RT–PCR analysis, total RNA was treated with DNase I (Roche). The synthesis of cDNA was performed using the SuperScript III First Strand Kit (Invitrogen) with oligo-dT primers. Quantitative RT–PCR was carried out using the Fast SYBR Green Master Mix and a 7500 Fast Real Time PCR Instrument (Applied Biosystems, Lincoln, CA) according to the supplier's recommendations. The mean value of three technical replicates was normalized using the PP2A transcript used as a control. The primer sequences (5’ to 3’) of the PP2A gene were TTACGTGGCCAAAATGATGC and GTTCTCCACAACCGCTTGGT. Gene-specific primer sequences are given in Supplemental Table 3.
SUPPLEMENTARY DATA
Supplementary Data are available at Molecular Plant Online.
FUNDING
This work was funded by the ETH Zurich, the Swiss National Science Foundation (National Centre of Competence in Research—Plant Survival), and SystemsX.ch (Plant Growth in a Changing Environment).
Supplementary Material
Acknowledgments
We thank Sabine Klarer for help growing plants, Martine Trevisan for technical assistance, Phillip Zimmermann, Lars Hennig, Hubert Rehrauer, and Marzanna Kuenzli for advice and assistance with the microarray analysis, Teresa Koller for assistance with quantitative RT–PCR, Miriam Lucas, Stephan Handschin, Heinz Gross, and Roger A. Wepf for advice and assistance with electron microscopy, and Jean-David Rochaix for advice on chlorophyll fluorescence measurements. The GWD antibody was a gift from Dr G. Ritte and Prof. M. Steup, University of Potsdam, Germany. The DPE1 antibody was a gift from Prof. S. Smith, University of Western Australia, Australia. No conflict of interest declared.
References
- Arnon DI. Copper enzymes in isolated chloroplasts: polyphenoloxidase in Beta vulgaris. Plant Physiol. 1949;24:1–15. doi: 10.1104/pp.24.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert S, Gout E, Bligny R, Marty-Mazars D, Barrieu F, Alabouvette J, Marty F, Douce R. Ultrastructural and biochemical characterization of autophagy in higher plant cells subjected to carbon deprivation: control by the supply of mitochondria with respiratory substrates. J. Cell Biol. 1996;133:1251–1263. doi: 10.1083/jcb.133.6.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan-Wollaston V, et al. Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J. 2005;42:567–585. doi: 10.1111/j.1365-313X.2005.02399.x. [DOI] [PubMed] [Google Scholar]
- Buléon A, Colonna P, Planchot V, Ball S. Starch granules: structure and biosynthesis. Int. J. Biol. Macromol. 1998;23:85–112. doi: 10.1016/s0141-8130(98)00040-3. [DOI] [PubMed] [Google Scholar]
- Caspar T, Huber SC, Somerville C. Alterations in growth, photosynthesis, and respiration in a starchless mutant of Arabidopsis thaliana (L.) deficient in chloroplast phosphoglucomutase activity. Plant Physiol. 1985;79:11–17. doi: 10.1104/pp.79.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MH, Liu LF, Chen YR, Wu HK, Yu SM. Expression of α-amylases, carbohydrate metabolism, and autophagy in cultured rice cells is coordinately regulated by sugar nutrient. Plant J. 1994;6:625–636. doi: 10.1046/j.1365-313x.1994.6050625.x. [DOI] [PubMed] [Google Scholar]
- Chia T, Thorneycroft D, Chapple A, Messerli G, Chen J, Zeeman SC, Smith SM, Smith AM. A cytosolic glucosyltransferase is required for conversion of starch to sucrose in Arabidopsis leaves at night. Plant J. 2004;37:853–863. doi: 10.1111/j.1365-313x.2003.02012.x. [DOI] [PubMed] [Google Scholar]
- Contento AL, Xiong Y, Bassham DC. Visualisation of autophagy in Arabidopsis using the fluorescent dye monodansylcadaverine and a GFP–AtATG8e fusion protein. Plant J. 2005;42:598–608. doi: 10.1111/j.1365-313X.2005.02396.x. [DOI] [PubMed] [Google Scholar]
- Critchley JH, Zeeman SC, Takaha T, Smith AM, Smith SM. A critical role for disproportionating enzyme in starch breakdown is revealed by a knock-out mutation in Arabidopsis. Plant J. 2001;26:89–100. doi: 10.1046/j.1365-313x.2001.01012.x. [DOI] [PubMed] [Google Scholar]
- Delatte T, Trevisan M, Parker ML, Zeeman SC. Arabidopsis mutants Atisa1 and Atisa2 have identical phenotypes and lack the same multimeric isoamylase, which influences the branch point distribution of amylopectin during starch synthesis. Plant J. 2005;41:815–830. doi: 10.1111/j.1365-313X.2005.02348.x. [DOI] [PubMed] [Google Scholar]
- Delatte T, Umhang M, Trevisan M, Eicke S, Thorneycroft D, Smith SM, Zeeman SC. Evidence for distinct mechanisms of starch granule breakdown in plants. J. Biol. Chem. 2006;281:12050–12059. doi: 10.1074/jbc.M513661200. [DOI] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- Dumez S, Wattebled F, Dauvillee D, Delvalle D, Planchot V, Ball SG, D'Hulst C. Mutants of Arabidopsis lacking starch branching enzyme II substitute plastidial starch synthesis by cytoplasmic maltose accumulation. Plant Cell. 2006;18:2694–2709. doi: 10.1105/tpc.105.037671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edner C, et al. Glucan, water dikinase activity stimulates breakdown of starch granules by plastidial β-amylases. Plant Physiol. 2007;145:17–28. doi: 10.1104/pp.107.104224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisk DG, Walker MB, Barkan A. Molecular cloning of the maize gene CRP1 reveals similarity between regulators of mitochondrial and chloroplast gene expression. EMBO J. 1999;18:2621–2630. doi: 10.1093/emboj/18.9.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton DC, et al. BETA-AMYLASE4, a noncatalytic protein required for starch breakdown, acts upstream of three active β-amylases in Arabidopsis chloroplasts. Plant Cell. 2008;20:1040–1058. doi: 10.1105/tpc.107.056507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gepstein S, Sabehi G, Carp MJ, Hajouj T, Nesher MFO, Yariv I, Dor C, Bassani M. Large-scale identification of leaf senescence-associated genes. Plant J. 2003;36:629–642. doi: 10.1046/j.1365-313x.2003.01908.x. [DOI] [PubMed] [Google Scholar]
- Hejazi M, Fettke J, Haebel S, Edner C, Paris O, Frohberg C, Steup M, Ritte G. Glucan, water dikinase phosphorylates crystalline maltodextrins and thereby initiates solubilization. Plant J. 2008;55:323–334. doi: 10.1111/j.1365-313x.2008.03513.x. [DOI] [PubMed] [Google Scholar]
- Hörtensteiner S. Chlorophyll degradation during senescence. Annu. Rev. Plant Biol. 2006;57:55–77. doi: 10.1146/annurev.arplant.57.032905.105212. [DOI] [PubMed] [Google Scholar]
- Huang DW, et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35:W169–W175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbell E, Liu WM, Mei R. Robust estimators for expression analysis. Bioinformatics. 2002;18:1585–1592. doi: 10.1093/bioinformatics/18.12.1585. [DOI] [PubMed] [Google Scholar]
- Kaplan F, Guy CL. RNA interference of Arabidopsis β-amylase8 prevents maltose accumulation upon cold shock and increases sensitivity of PSII photochemical efficiency to freezing stress. Plant J. 2005;44:730–743. doi: 10.1111/j.1365-313X.2005.02565.x. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotera E, Tasaka M, Shikanai T. A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts. Nature. 2005;433:326–330. doi: 10.1038/nature03229. [DOI] [PubMed] [Google Scholar]
- Kötting O, et al. SEX4, a glucan phosphatase, dephosphorylates amylopectin at the granule surface during starch breakdown in Arabidopsis leaves. Plant Cell. 2009;21:334–346. [Google Scholar]
- Lin TP, Preiss J. Characterization of D-enzyme (4-α-glucanotransferase) in Arabidopsis leaf. Plant Physiol. 1988;86:260–265. doi: 10.1104/pp.86.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WM, et al. Analysis of high density expression microarrays with signed-rank call algorithms. Bioinformatics. 2002;18:1593–1599. doi: 10.1093/bioinformatics/18.12.1593. [DOI] [PubMed] [Google Scholar]
- Lorberth R, Ritte G, Willmitzer L, Kossmann J. Inhibition of a starch-granule-bound protein leads to modified starch and repression of cold sweetening. Nature Biotech. 1998;16:473–477. doi: 10.1038/nbt0598-473. [DOI] [PubMed] [Google Scholar]
- Lu Y, Sharkey TD. The role of amylomaltase in maltose metabolism in the cytosol of photosynthetic cells. Planta. 2004;218:466–473. doi: 10.1007/s00425-003-1127-z. [DOI] [PubMed] [Google Scholar]
- Lu Y, Steichen JM, Weise SE, Sharkey TD. Cellular and organ level localization of maltose in maltose-excess Arabidopsis mutants. Planta. 2006;224:935–943. doi: 10.1007/s00425-006-0263-7. [DOI] [PubMed] [Google Scholar]
- Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of Gene Ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- Meierhoff K, Felder S, Nakamura T, Bechtold N, Schuster G. HCF152, an Arabidopsis RNA binding pentatricopeptide repeat protein involved in the processing of chloroplast psbB–psbT–psbH–petB–petD RNAs. Plant Cell. 2003;15:1480–1495. doi: 10.1105/tpc.010397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messerli G, Partovi Nia V, Trevisan M, Kolbe A, Schauer N, Geigenberger P, Chen J, Davison AC, Fernie AR, Zeeman SC. Rapid classification of phenotypic mutants of Arabidopsis via metabolite fingerprinting. Plant Physiol. 2007;143:1484–1492. doi: 10.1104/pp.106.090795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamikawa T, Toyooka K, Okamoto T, Hara-Nishimura I, Nishimura M. Degradation of ribulose-bisphosphate carboxylase by vacuolar enzymes of senescing French bean leaves: immunocytochemical and ultrastructural observations. Protoplasma. 2001;218:144–153. doi: 10.1007/BF01306604. [DOI] [PubMed] [Google Scholar]
- Niittylä T, Messerli G, Trevisan M, Chen J, Smith AM, Zeeman SC. A previously unknown maltose transporter essential for starch degradation in leaves. Science. 2004;303:87–89. doi: 10.1126/science.1091811. [DOI] [PubMed] [Google Scholar]
- Niwa Y, Kato T, Tabata S, Seki M, Kobayashi M, Shinozaki K, Moriyasu Y. Disposal of chloroplasts with abnormal function into the vacuole in Arabidopsis thaliana cotyledon cells. Protoplasma. 2004;223:229–232. doi: 10.1007/s00709-004-0037-7. [DOI] [PubMed] [Google Scholar]
- Ono K, Hashimoto H, Katoh S. Changes in the number and size of chloroplasts during senescence of primary leaves of wheat grown under different conditions. Plant Cell Physiol. 1995;36:9–17. [Google Scholar]
- Pfannschmidt T, Nilsson A, Allen JF. Photosynthetic control of chloroplast gene expression. Nature. 1999;397:625–628. [Google Scholar]
- Pružinská A, Anders I, Aubry S, Schenk N, Tapernoux-Luthi E, Muller T, Krautler B, Hortensteiner S. In vivo participation of red chlorophyll catabolite reductase in chlorophyll breakdown. Plant Cell. 2007;19:369–387. doi: 10.1105/tpc.106.044404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke KA, Leech RM. Rapid image analysis screening procedure for identifying chloroplast number mutants in mesophyll cells of Arabidopsis thaliana (L.) Heynh. Plant Physiol. 1991;96:1193–1195. doi: 10.1104/pp.96.4.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose TL, Bonneau L, Der C, Marty-Mazars D, Marty F. Starvation-induced expression of autophagy-related genes in Arabidopsis. Biol. Cell. 2006;98:53–67. doi: 10.1042/BC20040516. [DOI] [PubMed] [Google Scholar]
- Rost S, Frank C, Beck E. The chloroplast envelope is permeable for maltose but not for maltodextrins. Biochim. Biophys. Acta. 1996;1291:221–227. doi: 10.1016/s0304-4165(96)00068-2. [DOI] [PubMed] [Google Scholar]
- Schäfer G, Heber U, Heldt HW. Glucose-transport into spinach-chloroplasts. Plant Physiol. 1977;60:286–289. doi: 10.1104/pp.60.2.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheidig A, Fröhlich A, Schulze S, Lloyd JR, Kossmann J. Downregulation of a chloroplast-targeted β-amylase leads to a starch-excess phenotype in leaves. Plant J. 2002;30:581–591. doi: 10.1046/j.1365-313x.2002.01317.x. [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AM, Zeeman SC. Quantification of starch in plant tissues. Nature Protocols. 2006;1:1342–1345. doi: 10.1038/nprot.2006.232. [DOI] [PubMed] [Google Scholar]
- Solfanelli C, Poggi A, Loreti E, Alpi A, Perata P. Sucrose-specific induction of the anthocyanin biosynthetic pathway in Arabidopsis. Plant Physiol. 2006;140:637–646. doi: 10.1104/pp.105.072579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, Blasing O, Gibon Y, Nagel A, Meyer S, Kruger P, Selbig J, Muller LA, Rhee SY, Stitt M. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004;37:914–939. doi: 10.1111/j.1365-313x.2004.02016.x. [DOI] [PubMed] [Google Scholar]
- Wada S, Ishida H, Izumi M, Yoshimoto K, Ohsumi Y, Mae T, Makino A. Autophagy plays a role in chloroplast degradation during senescence in individually darkened leaves. Plant Physiol. 2009;149:885–893. doi: 10.1104/pp.108.130013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber APM, Servaites JC, Geiger DR, Kofler H, Hille D, Groner F, Hebbeker U, Flügge UI. Identification, purification, and molecular cloning of a putative plastidic glucose translocator. Plant Cell. 2000;12:787–801. doi: 10.1105/tpc.12.5.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weise SE, Weber AP, Sharkey TD. Maltose is the major form of carbon exported from the chloroplast at night. Planta. 2004;218:474–482. doi: 10.1007/s00425-003-1128-y. [DOI] [PubMed] [Google Scholar]
- Winkel-Shirley B. Biosynthesis of flavonoids and effects of stress. Curr. Opin. Plant Biol. 2002;5:218–223. doi: 10.1016/s1369-5266(02)00256-x. [DOI] [PubMed] [Google Scholar]
- Wittenbach VA, Lin W, Hebert RR. Vacuolar localization of proteases and degradation of chloroplasts in mesophyll protoplasts from senescing primary wheat leaves. Plant Physiol. 1982;69:98–102. doi: 10.1104/pp.69.1.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu TS, et al. The Arabidopsis sex1 mutant is defective in the R1 protein, a general regulator of starch degradation in plants, and not in the chloroplast hexose transporter. Plant Cell. 2001;13:1907–1918. doi: 10.1105/TPC.010091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeman SC, ap Rees T. Changes in carbohydrate metabolism and assimilate partitioning in starch-excess mutants of Arabidopsis. Plant Cell Environ. 1999;22:1445–1453. [Google Scholar]
- Zeeman SC, Delatte T, Messerli G, Umhang M, Stettler M, Mettler T, Streb S, Reinhold H, Kötting O. Starch breakdown: recent discoveries suggest distinct pathways and novel mechanisms. Functional Plant Biol. 2007a;34:465–473. doi: 10.1071/FP06313. [DOI] [PubMed] [Google Scholar]
- Zeeman SC, Northrop F, Smith AM, ap Rees T. A starch-accumulating mutant of Arabidopsis thaliana deficient in a chloroplastic starch-hydrolyzing enzyme. Plant J. 1998;15:357–365. doi: 10.1046/j.1365-313x.1998.00213.x. [DOI] [PubMed] [Google Scholar]
- Zeeman SC, Smith SM, Smith AM. The diurnal metabolism of leaf starch. Biochem. J. 2007b;401:13–28. doi: 10.1042/BJ20061393. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.