Many natural products with antibiotic, anticancer and antifungal properties are synthesized by nonribosomal peptide synthetases (NRPSs) and polyketide synthases (PKSs)1. Genome sequencing has revealed great NRPS/PKS biosynthetic capacity, yet the analytical challenge is in accessing the natural products and their biosynthetic machinery2. By virtue of their huge size (often >2000 amino acids) and unique marker ions deriving from their common cofactor, we have adapted mass spectrometry-based proteomics for selective detection of expressed NRPS/PKS gene clusters in microbial proteomes, without requiring genome sequence information. Here we show unambiguous detection of known NRPS/PKS systems in members of the genera Bacillus and Streptomyces, and when used for screening 22 environmental isolates uncovered production of undetected natural products from the zwittermicin A biosynthetic gene cluster3. Additionally, we discovered a NRPS cluster encoding a new 7-residue lipopeptide. Overall, this “protein-first” strategy provides an entirely new entrée to quickly find expressed gene clusters generating new natural products at a level comparable and complementary to bioassay- and sequence-based discovery platforms.
With over half of the nearly one thousand new chemical entities introduced as antibacterial and or anticancer drugs over the past few decades being natural products or their derivatives there has been a resurgence of interest in natural products4. The natural product discovery process has diverse implementations, and strategies continue to evolve as more microbial genomes become available. Traditional discovery platforms employ a bioassay-guided strategy, where an iterative cycle of metabolite fractionation and bioassay panels attempt to isolate the chemical compound responsible for the observed bioactivity5. This process rediscovers known compounds most of the time, highlighting the inefficiency of this “dereplication” bottleneck. In development of the complementary platform presented here, we set out to circumvent some of the limitations of bioassay-based screening, including the bias that arises when screening against only one drug target or indicator cell line. As shown here, we have developed a process (Fig. 1) that allows targeted detection of peptide- and polyketide-type natural products in a molecular screening approach.
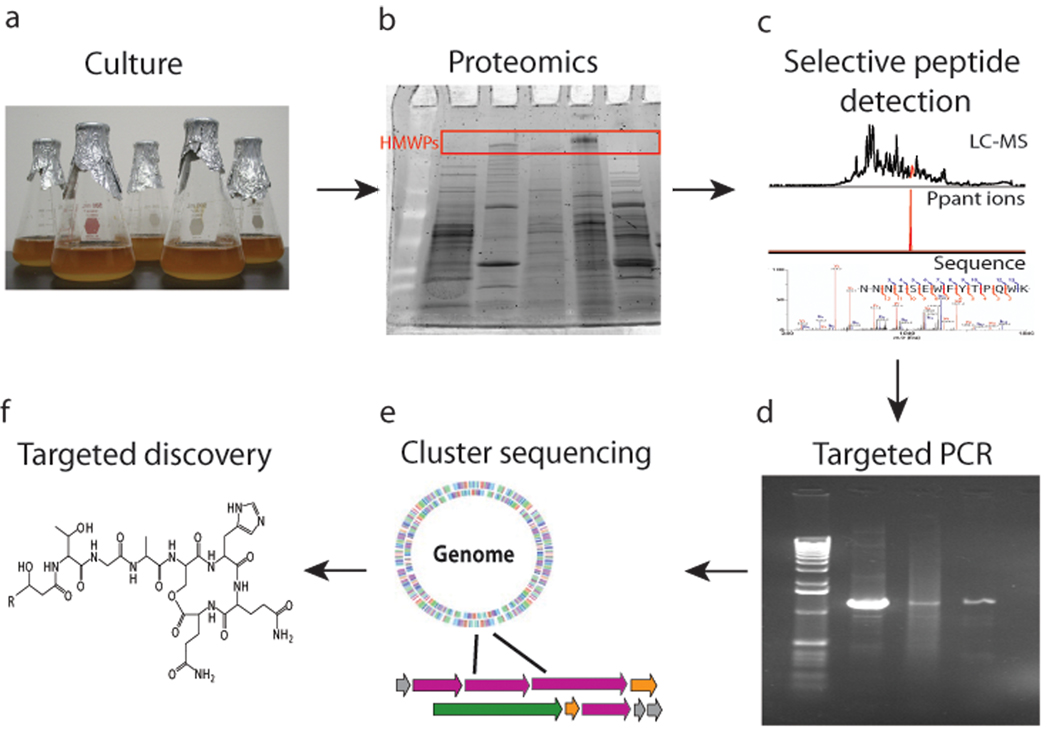
Figure 1.
The workflow for PrISM. a, Microbial strains are grown in liquid culture. b, The proteome of the strain is subjected to proteomics or in-gel digestion of high molecular weight bands. c, LC-FTMSn is conducted on the resulting peptide mixture, with expressed T domain active site peptides identified by the Ppant ejection assay. d, Peptide sequences are used to generate primers to amplify DNA sequence portions of the expressed gene cluster. e, The gene cluster is identified and sequenced, which informs targeted detection of the natural product produced as depicted in panel f.
Systems-biology approaches, such as genomics, transcriptomics, and metabolomics, are now being adapted to update natural product discovery platforms and bypass the dereplication bottleneck of bioassay-directed discovery. With ever more sequenced genomes in hand, one can use bioinformatic analysis to predict the biosynthetic potential of an organism. There have been a few successful attempts where this sequence-based approach has successfully guided the search for new natural products6, 7. There is, however, a great disparity between the genetic potential for natural product production and the actual expression of biosynthetic gene clusters under laboratory culture conditions. While several reports demonstrate diverse methods to force expression of “cryptic” gene clusters, accessing novel compounds and the enzymes that make them is still a low-throughput affair8–10. We therefore use proteomics to screen for expressed biosynthetic gene clusters producing new natural products, without the requirement for DNA sequence information a priori. Discovery of a gene cluster and its associated metabolite in tandem can expedite the downstream goal of pathway engineering to improve yield, bioactivity or bioavailability.
Two highly valued families of natural products are polyketides (PKs) and peptides produced nonribosomally (NRPs) by large enzymes called polyketide synthases (PKSs) and nonribosomal peptide synthetases (NRPSs), respectively. The therapeutic value of NRPs and PKs as antibiotics, antiproliferatives and immunosuppressants, combined with the lack of methods to detect expressed NRPS and PKS gene clusters in discovery mode, prompted us to develop the method called PrISM (short for the Proteomic Investigation of Secondary Metabolism), based on microbial proteomics. Many NRPS and PKS enzymes are enormous (often >> 200 kDa) and have many domains that act as a molecular “assembly-line”1 to create complex natural product scaffolds. The various domains are responsible for substrate activation, condensation and tailoring, while the growing natural product is covalently tethered to “carrier” regions, also called thiolation (T) domains, that uniformly harbor a phosphopantetheinyl (Ppant) cofactor bound to a serine residue in their active sites11.
Even the earliest implementations of mass spectrometry (MS) to detect covalent intermediates on the “thiotemplate” family of NRPS/PKS enzymes noted the facile release of the Ppant cofactor; its phosphodiester linkage is labile during tandem MS (MS/MS) (Supplementary Figs. 1–2)12, conceptually similar to the ion chemistry used in modern phospho-proteomics. More recently, modern mass spectrometry has revealed a great diversity of covalent chemistry occurring on NRPS and PKS enzymes in vitro, with the Ppant ejection assay now established for single enzymes using liquid chromatography-mass spectrometry (LC-MS) (Supplementary Fig. 1)13, 14. Benchtop15 and high-performance Fourier Transform mass spectrometers (FTMS)16 have both been used for investigation of NRPS and PKS intermediates in reconstituted systems. FTMS provides mass accuracy of <2 part-per-million (ppm) for ions diagnostic of Ppant (i.e. m/z 261.1267 and m/z 359.1036), which translates here into high selectivity for detection of Ppant-containing peptides in complex proteomes (Fig. 1c, middle) and forms one aspect of the integrated approach described here.
For development of the overall PrISM workflow (Fig. 1), three systems with increasing complexity were interrogated and the detailed discussion is provided in the Supplementary Information. First, a di-domain enzyme from the gramicidin S system (PheAT from GrsA, 70 kDa)17, was analyzed using shotgun proteomics in conjunction with the Ppant ejection assay. The single tryptic peptide (Asp564-Lys575; 1,638.70 Da) from PheAT harboring the Ppant arm was detected in the proteomic background of Escherichia coli (Supplementary Fig. 3). Next, the native producer of gramicidin S, Bacillus brevis ATCC 9999, was analyzed as it entered early stationary phase17 when production of this 10-mer NRPS product was verified by LC-FTMS of the crude extract. In this system, four of the five carrier peptides from the GrsA (127 kDa) and GrsB (510 kDa) proteins were detected in a shotgun proteomics experiment (Supplementary Fig. 4). High molecular weight bands from SDS-PAGE gels of B. brevis were also analyzed by in-gel digestion and nanocapillary LC-MS/MS (nanoLC-MS) to identify NRPSs encoded by grsA and grsB (Supplementary Table 1). A similar overall result was obtained for the phosphinothricin tripeptide system in the significantly more complicated proteomic background of the native producer, Streptomyces viridochromogenes DSM 4073618 (Supplementary Fig. 5).
With proof-of-concept experiments in hand for the first half of the Figure 1 workflow (Fig. 1a–c) in both Gram-positive (e.g. Bacillus brevis and Streptomyces viridochromogenes) and Gram-negative bacteria (e.g. Escherichia coli), we set out to apply this method to strains isolated from the environment without the benefit of DNA sequence or other information a priori. Heat-treated soil samples were used to isolate Bacillus spores19, and 22 isolates were stored for analysis after 16S rDNA sequencing for taxonomic dereplication. SDS-PAGE was used as an initial screen for these 22 strains, each grown for 2 days at 30°C in nutrient broth with sampling at 8, 16, 24, 36 and 48 h. Five of the 22 strains showed expression of high molecular weight proteins (HMWPs) (consistent with NRPS/PKS expression) for at least one time point, including the NK2018 strain which was subjected to full proteome analysis and nanoLC-MS of in-gel digests of HMWPs.
Using shotgun proteomics combined with the Ppant ejection assay, twenty cation exchange fractions (from separation of a tryptic digest of the NK2018 proteome by strong cation exchange chromatography) were analyzed by reverse phase (RP) LC-MS/MS using a linear ion trap-FTMS (ThermoFisher LTQ-FT) operating at 12 Tesla. Such analysis of NK2018 detected Ppant-containing peptides, the most prominent being a 2+ peptide at m/z 1,038.98 (2,075.94 Da) that showed all expected Ppant elimination marker ions during MS/MS analysis14. These MSn experiments on the Ppant-producing peptide provided sufficient de novo sequence information for its identification as the ACP active site peptide from fatty acid biosynthesis (Supplementary Fig. 6). The ten amino acid sequence generated, [GADSPpant(I/L)DVVE(I/L)], was sufficient for differentiation of this peptide as a fatty acyl ACP (AcpP), because the sequence motif flanking the active site Ser is distinct from that found in either NRPS or PKS20. The identification of this peptide provides a positive control for identification of phosphopantetheinylated peptides and de novo generation of long stretches of peptide sequence information, a critical step for design of good primers for PCR.
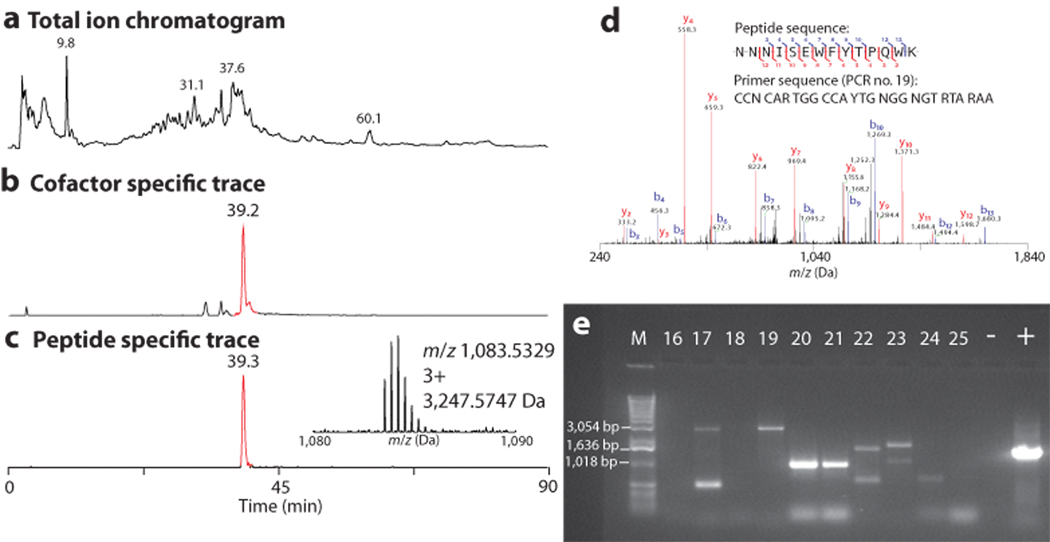
For the targeted analysis of proteins >200 kDa, SDS-PAGE gel bands harboring HMWPs of interest from NK2018 were subjected to in-gel trypsin digestion and nanoLC-MS data were collected using a 12 Tesla LTQ-FT for high resolution detection of intact peptides and the phosphopantetheinyl ejection ions. Unit-resolution MS/MS data were collected in a data-dependent fashion on the six most abundant precursors for a total loop time of 3 s that was executed throughout an entire 90 min LC-MS experiment (Fig. 2a–c). Fragmentation data were processed by manual de novo sequencing and batch searching with the open mass spectrometry search algorithm (OMSSA)21 against the NCBI nonredundant protein database (nr). In the analysis of the HMWP band (Supplementary Fig. 7), four phosphopantetheinylated peptides were observed. Figure 2a–c compares the total ion chromatogram (TIC, Fig. 2a) to the selected ion chromatogram (SIC, Fig. 2b) for Ppant ejection and a SIC (Fig. 2c) for a 3+ peptide at m/z 1,083.5329 (3,247.57 Da) (Fig. 2c inset) which co-eluted with the Ppant product from Figure 2b. This 3+ species was verified as a bona fide peptide from the active site of a NRPS carrier domain (Supplementary Fig. 8).
Figure 2.
Identification of an expressed T domain active site peptide in the NK2018 proteome by the on-line Ppant ejection assay using nanoLC-MS. a, Total ion chromatogram (TIC) from a nanoLC-FTMS analysis of a single SDS-PAGE gel slice containing NK2018 HMWPs. b, Selected ion chromatogram (SIC) for the elution of the Ppant ejection ion of interest (Pant-CAM, m/z 318.1482). c, SIC for the phosphopantetheinylated peptide eluting at 39.3 min and a phosphopantetheinylated peptide observed in nanoLC-MS analysis (inset). See the Supplementary Discussion for detailed information on the identification of this peptide. d, Representative results from proteomic analysis of NK2018. Shown is the MS/MS spectrum for a peptide from C1S2. Inset: The de novo peptide sequence and the degenerate primer designed from the given peptide sequence. e, Representative results of PCR amplification using degenerate primers designed from peptide sequences determined de novo. See Supplementary Table 4 for a complete list of all PCR amplified products and the PCR numbers corresponding to the lanes in Figure 2c.
When MS/MS data collected from all peptides were searched (not just those harboring the Ppant modification), the top three predicted protein identifications were NRPS-PKS proteins from Bacillus cereus AH1134. The genome of this strain was recently sequenced by the J. Craig Venter Institute, and the annotations of 37 contigs were uploaded to NCBI in late 2008. The peptides identified arose from expression of two separate gene clusters, labeled here as cluster #1 (C1) and cluster #2 (C2), producing at least three different NRPS/PKS synthases (C1S2 (ZmaA), C1S6 (ZmaK) and C2S2) (Fig. 3a and Supplementary Figs. 9–10) on two separate contigs (C1 from contig GenBank accession number ABDA02000035 and C2 from contig Gen Bank accession number ABDA02000007). All peptides with homology to B. cereus AH1134 predicted to derive from NRPS/PKS gene products are listed in Supplementary Table 2. The best database search results were manually validated (e.g. Fig. 2d) and provided enough sequence information to design degenerate PCR primers (Supplementary Tables 3–4) that ultimately obtained stretches of DNA sequence that were >94% identical to B. cereus AH1134 (Supplementary Table 5). Figure 2e shows the results of 11 representative PCRs, showing ample microsequence to convert peptide MS/MS data into DNA sequence of expressed NRPS/PKS gene clusters (see Supplementary Fig. 11 for complete gel image).
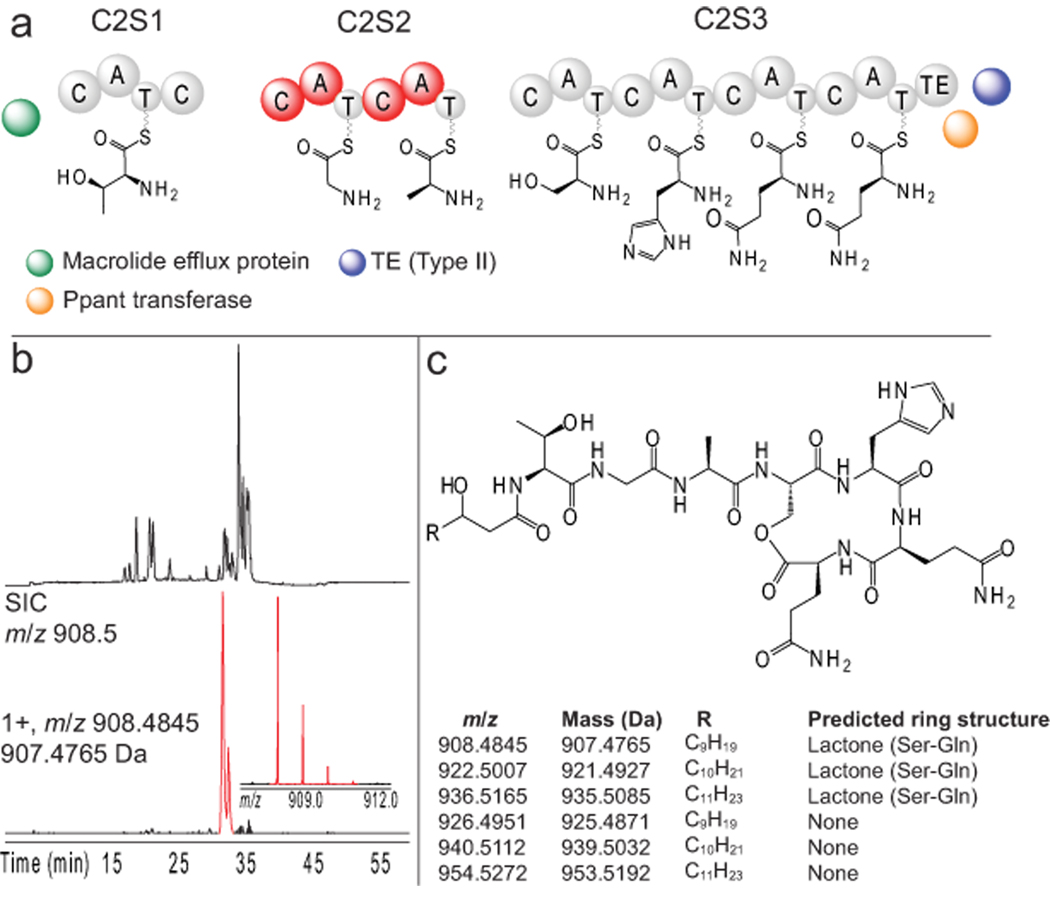
Figure 3.
Identification of new lipoheptapeptides in NK2018. a, Domain organization of cluster #2 based upon the gene sequence in B. cereus AH1134. Amino acid substrates were selected based upon bioinformatic analysis and the structure of the detected peptides. Peptides from the domains in red were identified by nanoLC-MS. b, Base peak chromatogram (top) of a NK2018 culture supernatant sample and a SIC (red, bottom) for the species at m/z 908.4845. The mass spectrum of the 1+ charge state of the ion is shown (inset). c, Putative structure for the lipoheptapeptides detected, including the species detected in Figure 3b. The structures are predicted to differ in the length of the fatty acid tail and in the formation of a lactone ring (table inset). Abbreviations: C – condensation domain, A – adenylation domain, T – thiolation domain, TE – thioesterase.
The integrated data from PrISM along with targeted PCR (Fig. 2e) show direct evidence for expression of two gene clusters from Bacillus strain NK2018 with the basic architecture of those observed in the newly-sequenced B. cereus AH1134; the high sequence identity between the two strains was sufficient for the assumption that the two clusters in this strain are orthologous. The large synthetases identified by LC-MS/MS are highlighted in red in Figure 3a and Supplementary Figure 9, and the individual domains within the synthetases represented in the MS/MS data are highlighted in red in Figure 3a and Supplementary Figure 9. A large number of the genes in cluster #1 are orthologous to those that produce the aminopolyol antibiotic zwittermicin A (ZmA). Zwittermicin A has a broad spectrum of activity against both Gram-positive and Gram-negative bacteria as well as certain eukaryotes22. A total synthesis was recently reported23 and the sequence of individual biosynthetic proteins has been reported over the past four years3. A 2009 report based on the analogous B. cereus AH1134 sequence we found expressed in NK2018 revealed that the ZmA biosynthetic gene cluster is much larger than expected and predicted to produce three ZmA-related small molecules3. Targeted searching for such molecules allowed detection of zwittermicin A and a previously undetected methionine-containing NRPS product (Supplementary Fig. 9), thus completing the PrISM approach (cf. Fig. 1f). Additionally, two of the phosphopantheinylated peptides observed during nanoLC-MS of the HMWP in-gel digestion match within 3 ppm to thiolation domain active site peptides from ZmaB and ZmaK (Supplementary Fig. 8).
Evidence for expression of a second cluster from Bacillus strain NK2018 was uncovered by detection of peptides from the two-module NRPS protein depicted as C2S2 in Figure 3a. Annotation of the flanking ~50 kb of sequence from B. cereus AH1134 around the gene for the protein identified from this cluster is shown in Figure 3a and Supplementary Figure 10. Three genes predicted to encode NRPSs are present in this gene cluster, along with the nearby efflux protein, a phosphopantetheinyl transferase and a type II thioesterase. There are homologs of this gene cluster known in other B. cereus strains such as B4264 and G9842, and multiple B. weihenstephanensis strains, but it is clearly an orphan with no corresponding natural product known. As discussed below, the PrISM platform has enabled identification of this new gene cluster via detection of the expressed gene products and linked this expression with secondary metabolite production.
NRPS rules24, 25 were used to predict a seven-residue, NRPS-type natural product with amino acids including serine (Ser), alanine (Ala) and threonine (Thr), in addition to two glutamine (Gln) or glutamic acid (Glu) residues at the C-terminus. Targeted analysis of NK2018 extracts for peptides of this type uncovered a set of six related species at m/z 908.4845, 922.5007, 926.4951, 936.5165, 940.5112, and 954.5272 (Supplementary Fig. 12) that were analyzed and sequenced using MS/MS (Supplementary Figs. 13–16). A sequence of six amino acids common to all was generated by de novo sequencing: Gly-Ala-Ser-His-Gln-Gln, a reasonable match to the adenylation domain substrates predicted by tools of the field24, 25. One empirical formula predicted for the species at m/z 908.4845 (within 1 ppm error) was C40H65N11O13; the lipoheptapeptide shown in Figure 3c is a putative structure for this chemical formula that is strongly supported by the MS data and a previous report of lipopeptides of similar structure26 (Supplementary Fig. 17).
Tandem mass spectrometry clearly shows these six species (Supplementary Figs. 12–16), which differ by exactly 18.0103 Da and 14.0162 Da, have highly related fragmentation patterns (Supplementary Figs. 14–16). These mass differences are unambiguously due to differences of CH2 and H2O, which are best explained by incorporation of longer fatty acid chains and lactone ring formation, respectively. The intact and fragment masses support the assignments of ring open form (+18 Da) and the lactone ring as drawn in Figure 3c and reported for the homologous kurstakins26 (Supplementary Fig. 17). Further, amide fragmentation adjacent to the Thr residue localizes the 14 Da variations and the hydroxyl group to the fatty acid tail, with the OH group assigned to position 3 based on the precedents from other Bacillus lipopeptides such as surfactin. However, the hydroxyl group is not involved with lactone ring formation, a conclusion supported by detailed interpretation of MS/MS data (see Supplementary Figs. 14–16). The structure of these reported natural products and associated bioinformatic analysis strongly support the assignment of cluster #2 as the previously unreported biosynthetic gene cluster for these compounds. The unique domain organization of this cluster involves two extra condensation (C) domains, one which we hypothesize to facilitate NRPS initiation by loading the fatty acyl chain in conjunction with the type II thioesterase, analogous to SrfD in surfactin biosynthesis27.
This report extends prior work on isolated NRPS/PKS enzymes13 and initial reports of microbial proteomics28, 29 or selective labeling of phosphopantetheinylated proteins30 into a general method for targeted proteome analysis; NRPS, PKS and fatty acid biosynthetic gene products from diverse organisms are detected with antibody-like specificity for Ppant-containing proteins common to all “thiotemplate” systems. Detection of a biosynthetic enzyme in a microbial proteome provides a high value entrée into an unsequenced genome at the protein level. The integrated PrISM approach augments genomic methods and directs efforts toward expressed genes, a strong indication that the corresponding natural product is also being produced.
Given the potential of systems biology approaches to direct natural products discovery in ways different from classical bioassay-based discovery, let us compare aspects of PrISM with DNA-, RNA- and small molecule-based methods. The rapid increase in genome sequence information opens the door for development of sequence-based discovery and characterization methods (e.g. genome mining, RNA-based analysis and heterologous expression of full gene clusters). Genome mining provides information on the biosynthetic potential of an organism, but does not reveal which secondary metabolic pathways will actually be expressed; proteomics achieves direct observation of gene expression, and detects whether enzymes are correctly post-translationally modified. Induction of “cryptic” gene clusters and heterologous expression of full pathways have been achieved, but are quite challenging2. There are well-developed RNA-based methods for monitoring gene expression, such as reverse-transcriptase PCR (RT-PCR) or transcriptomics, but these are not generally used in the context of an unsequenced genome. A structure-based approach using direct chemical screening can reveal small molecules produced by strains under a variety of growth conditions. It does not, however, provide information on the biosynthetic machinery for those metabolites. As technology improves for all “-omic” analyses, PrISM will fill an important gap and accelerate development of complementary approaches in the systems biology of natural products research.
Challenges in our implementation of PrISM included the translation of sequence information from MS into PCR products and a self-imposed bias toward >100 kDa NRPS/PKS systems. Future improvements in sample processing and mass spectrometry will provide increased data quality for eased design of degenerate primers. We also note that a global proteomics approach can, in principle, provide a more complete picture of an organism’s biosynthetic capacity. All challenges familiar to natural product structure elucidation still apply to PrISM, including elucidation of complex polyketide structures.
In the future, it will be interesting to see how molecular screening methods contribute to natural product discovery as they ramp up for application to hundreds or even thousands of strains. We project that PrISM will be most valuable for screening conditions and strains where novel NRP/PK scaffolds are produced. PrISM achieves dereplication at the biopolymer level where relatives of well-characterized systems are quickly flagged (because of high sequence identity to known domains and modules). For such cases, one will elect to simply not continue down the PrISM work flow (cf. Fig. 1) or choose to spot check the putatively homologous cluster with a small set of easy PCRs.
PrISM positions proteomics as an initial survey in the natural product discovery pipeline. Subsequent small molecule detection modalities (e.g. bioassays, MS, or NMR) can then be used to assess structure and activity. Therefore, we have achieved a net reversal of the traditional “small molecule first” discovery process. Streamlined versions of PrISM will realize efficiencies of scale and detect enzyme fingerprints from strains cultured on plates, fungi, and even environmental samples for meta-proteomics, such as complex marine microorganism-invertebrate assemblages31, with extension to all types of secondary metabolism also possible.
Methods
Materials
Trypsin (TRL3) for digests of bacterial proteomes in shotgun proteomics was purchased from Worthington Biochemicals. Sequencing grade trypsin (Promega) was used for all in-gel digestions. Escherichia coli BL21(DE3) cells were purchased from EMD Biosciences. All other chemicals used were purchased from either ThermoFisher Scientific or Sigma-Aldrich unless otherwise noted.
Cloning of GrsA PheAT
All cloning was performed in E. coli strain DH5α. All PCR used Phusion Hot Start Polymerase (Finnzymes) and PCR grade dNTPs (Invitrogen). Restriction enzymes were obtained from Invitrogen and T4 DNA ligase was from New England Biolabs. PCR products and restriction digested DNA were purified with Qiaquick gel extraction and PCR cleanup kits (Qiagen). Sfp was amplified with primers F 5 ’-CCATATGATGAAGATTTACGGAATTTAT ATGGAC-3 ’ and R 5 ’-CCTGGTACCTTATAAAAGCTCTTCGTACGAGACC-3’ containing the NdeI and KpnI restriction sites respectively (underlined) using the plasmid pUC-8 Sfp as the template. The PCR product was cleaned up prior to digestion with NdeI and KpnI. The linear fragment was purified from a 1% agarose gel prior to ligation to similarly cut and purified pET-Duet-1 previously modified to contain the BamHI to HindIII fragment of pQE-60 (Qiagen) to yield pET-Duet-1-Sfp. PheAT was amplified from plasmid pQE-60 PheATE using primers F 5'-ATATCCATGGTAAACAGTTCTAAAAG-3' and R 5'-ATCGGATCCATTTGGTCTATACAAC-3’ containing the NcoI and BamHI restriction sites respectively (underlined). The PCR product was cleaned up prior to digestion with NcoI and BamHI and gel purified prior to ligation to similarly cut pET-Duet-1 Sfp to yield pET-Duet-1 PheAT-His6 Sfp. Sequence was confirmed by sequencing at the UIUC Core DNA Sequencing Facility, 334 Edward R. Madigan Laboratory, 1201 W. Gregory Drive, Urbana, IL 61801.
Preparation of samples for proteomic investigations of PheAT
100 mL of Luria-Burtani (LB) broth supplemented with ampicillin (final concentration 100 µg/mL) were inoculated with one colony of E. coli BL21(DE3) transformed with pET-Duet-1 PheAT-His6 Sfp and grown overnight at 37°C with shaking at 225 rpm. Ten mL of the starter culture were added to 1 L of LB supplemented with ampicillin (final concentration 100 µg/mL) and placed at 37°C with shaking at 225 rpm until an OD600 of approximately 0.6. At this time, the incubation temperature was dropped to 20°C and IPTG was added to a final concentration of 1 mM. The culture was incubated with shaking at 18°C for an additional 20 h, at which time 500 mL cell culture were harvested by centrifugation (10 min, 4°C, 4,400 x g). Cell pellets were resuspended in lysis buffer (25 mM Tris-HCl, pH 7.5–7.8) and lysed by sonication (4 cycles of 30 s sonication on ice followed by 30 s incubation on ice). The soluble lysate was collected by centrifugation (15 min, 4°C, 47,810 x g). Protein concentration of the soluble lysate was determined by the Bradford assay. Five mg of protein were digested in a reaction mixture of 0.05 M NH4HCO3 (pH 7.8), 3 M urea, and trypsin (ratio of 1:10 total trypsin:substrate) by incubating at 30°C for 20 min, and the reaction was quenched by freezing at −80 °C or the addition of SCX solvent A.
Preparation of B. brevis samples
Bacillus brevis ATCC 9999, purchased from the American Type Culture Collection, was grown on nutrient agar plates overnight at 37°C. One colony from growth was selected and added to 50 mL YP + NaCl growth media (9 g peptone, 5 g yeast extract, 5 g NaCl in 1 L, pH 7.2–7.25) and incubated with shaking at 250 rpm at 37°C overnight. Ten mL of the starter culture were added to 2 L YP+NaCl and placed at 37°C with shaking at 250 rpm. Previous reports have shown that maximal production of the natural product gramicidin S produced by B. brevis occurs during the entry to stationary phase17. The OD600 of the culture was monitored to determine when cells were to be harvested, and after approximately 15 min of identical OD600 measurements (indicating stationary phase, approximately 8 h after culture inoculation) cells were harvested by centrifugation (10 min, 17,600 x g, 4°C). Cell pellets were resuspended in lysis buffer (25 mM Tris-HCl, pH 7.5–7.8) and lysed by sonication (4 cycles of 30 s sonication on ice followed by 30 s incubation on ice). The soluble lysate was collected by centrifugation (15 min, 4°C, 47,810 x g). Protein concentration was determined by the Bradford assay. Five mg of protein were digested in a reaction mixture of 0.05 M NH4HCO3 (pH 7.8), 3 M urea, and trypsin (ratio of 1:10 trypsin:substrate) by incubating at 30°C for 20 min, and the reaction was quenched by freezing at −80°C or addition of SCX solvent A.
Preparation of S. viridochromogenes samples
Streptomyces viridochromogenes DSM 40736 was grown on solid ISP2 medium (Difco) for 4–5 days at 30°C. One colony was selected and added to 15 mL of MYG media (1 L contains 10 g malt extract, 4 g yeast extract, 4 g glucose, pH 7.3) in a baffled flask for 4–5 days at 30°C with shaking at 225 rpm. Seven mL of the starter culture were fully homogenized using a sterile glass homogenizer and added to 1 L MYG. The culture was incubated at 30°C with shaking at 225 rpm until significant phosphinothricin tripeptide (PTT) production was observed by bioassay. Protocols for performing the bioassay for PTT production have been described previously18. In brief, B. subtilis ATCC 6633 was grown in minimal media (1 L contains 3 g KH2PO4, 7 g K2HPO4, 0.5 g sodium citrate-dihydrate, 0.1 g MgSO4-7H2O, 1 g (NH4)2SO4, and 2 g glucose) at 37°C until an OD600 of approximately 0.4. 200 µL of the culture was plated on minimal media (same recipe as above, with addition of 12 g agar / L). Six mm paper disks were placed on top of the plated lawn, and 9 µL of supernatant from S. viridochromogenes growth was placed on the disk (9 µL of MYG media was used as a control). The plates were placed at 37°C for overnight growth and monitoring of PTT production. The bioassays were performed daily after the first overnight growth of the 1 L cultures. After PTT production was observed, cells were harvested by centrifugation (20 min, 4°C, 17,600 x g) and resuspended in lysis buffer (25 mM Tris-HCl, pH 7.5–7.8). Cells were lysed by two passages through a French press operating at high pressure, and the soluble lysate was collected by centrifugation (45 min, 4°C, 47,810×g). Trypsin digests were performed as previously described.
Isolation of strains of Bacillus and preparation of proteome samples
Soil was collected from Haughton, LA, USA and Bacillus strains were isolated by heat treatment and dilution plating on nutrient agar. Strain NK2018 was chosen for further analysis based on presence of high molecular weight bands on an SDS-PAGE gel (Supplementary Fig. 7). A starter culture of nutrient broth (50 mL) was inoculated with a single colony of NK2018 and grown overnight. An aliquot (5 mL) was used to inoculate 1 L nutrient broth for 24 h or 48 h growth at 30°C. Cells were isolated via centrifugation (10,000×g, 10 min, 4°C) and resuspended in a minimal amount of 100 mM NH4HCO3, pH 7.8. Cells were lysed by sonication (5 cycles of 30 s sonication on ice followed by 30 s incubation on ice) and debris was cleared by centrifugation (20 min, 4°C, 47,810 x g). Protein concentration was estimated by the BCA assay (ThermoPierce).
Proteomic analysis of PheAT, B. brevis and S. viridochromogenes
SCX chromatography was carried out using a Shimadzu Prominence HPLC. The column used for SCX analysis was a Polysulfoethyl A column (PolyLC) with 4.6 mm inner diameter and length of 200 mm. An entire tryptic digest was loaded onto the SCX column and eluted with a step gradient (Supplementary Table 6) using solvents A (20 mM citric acid (pH 2.65), 25% MeCN) and B (20 mM citric acid (pH 2.65), 1 M NH4Cl, 25% MeCN) flowing at 0.5 mL/min. Fractions were collected every 2 min in 96 well plates and analyzed by FTMS immediately or stored at −20°C until further analysis. SCX fractions were subjected to RPLC-MS/MS according to the following method. Online RPLC-MS/MS data were collected using a ThermoFisher 12 T LTQ-FT Ultra coupled to an Agilent autosampler and Agilent HP1100 binary pump HPLC system. The column used for all RPLC analysis was a Jupiter C18 or C4 1 mm × 150 mm column (Phenomenex). The gradient used for all RPLC analysis is provided in Supplementary Table 7, with solvent A being water + 0.1% formic acid and solvent B being acetonitrile + 0.1% formic acid flowing at 100 µL/min. 150–300 µL of each SCX fraction was injected onto the RPLC column for MS analysis.
Parameters for MS analysis
All MS methods included the following events: (1) FT scan, m/z 500–2000, (2) FT scan, source induced dissociation (SID) = 75, detect m/z 200–600, (3) data-dependent MS/MS on the top X (X=3 for high-resolution MS/MS data collection and X=6 or 10 for unit resolution MS/MS data collection) peaks in a given spectrum using collision induced dissociation (CID) or infrared multi-photon dissociation (IRMPD).
Data analysis and peptide identification for full proteome analysis
All data were analyzed using QualBrowser, part of the Xcalibur software packaged with the ThermoFisher LTQ-FT and custom in-house software. Selected ion chromatograms (SICs) were generated for the Ppant ejection ions of interest. Based upon the elution of the Ppant ejection ion, the time of elution was analyzed for the presence of predicted active site peptides of the proteins in question (masses calculated based upon published sequences). Tandem MS data generated by CID or IRMPD was analyzed manually.
Gel-based proteomic analysis
An aliquot of the soluble proteome (350 µL) was added to 100 µL 2X SDS-PAGE loading buffer and incubated at 95°C for 5 min before loading onto a BioRad SDS-PAGE gel (Tris-HCl gradient gel, 4 – 20%, 10 × 30 µL wells). The gel was stained with colloidal Coomassie G-250 and the band at ~225 kDa was excised with a razor blade and chopped into pieces smaller than 2 mm3 (Supplementary Figs. 7 and 18). These gel samples were then destained, reduced with DTT, alkylated with iodoacetamide and digested with trypsin. Peptides were extracted, lyophilized, rehydrated with 0.1% acetic acid and bomb-loaded onto a self-packed C4 nano-LC guard column (75µ × 10 cm, 10–20 µm particle size). This guard column was then placed upstream of a ProteoPepII C18 column (75 µ × 10 cm, New Objective) and peptides were eluted over an 90 min linear gradient of water and acetonitrile with 0.1 % formic acid at a flow rate of 300 nL/min (produced by an Eksigent 1D nano-LC) into a 12 T ThermoFisher LTQ-FT Ultra. Samples were analyzed using the online Ppant ejection assay as well as data-dependent low resolution CID on the top six precursors. LC-MS/MS data from each in-gel digestion run were processed into DTA files with BioWorks 3.2 (ThermoFisher, San Jose, CA) and concatenated into encapsulated XML. These data were automatically searched (using a custom Perl script) against the nr protein database with OMSSA as described in the main text, using standard settings for the detection of intact peptides at high resolution (FTMS) and MS/MS fragment ions with unit resolution (ITMS) (0.01 Da intact peptide tolerance, 0.4 Da fragment ion tolerance). Individual files were then combined into one master file with a custom Perl script for viewing with the OMSSA browser. Phosphopantetheinylated peptides were observed as the carboxyamidomethylated-pantetheinyl (Ppant-Cam) ejection products (318.1482 Da), generated from alkylation of the free Ppant thiol with iodoacetamide during the standard in-gel digestion procedure.
Genomic analysis of NK2018
Genomic DNA was isolated from 5 mL cells (overnight culture at 30°C in nutrient broth) using a Qiagen DNeasy Blood and Tissue DNA Kit. PCR was performed on BioRad DNA Engine thermal cycler. For degenerate primer design based on MS/MS data alone, peptides with regions of low degeneracy were chosen and both a degenerate primer set as well as primers with the most likely codon usage (predicted from sequenced B. cereus ATCC 10987) were synthesized (Supplementary Table 3). Reactions (per 25 µL) were composed of 5 µL 5x GoTaq Flexi reaction buffer (Promega), 2 µL 2.5 mM dNTPs, 1.3 µL 50 mM MgCl2, 0.5 µL template DNA, 0.25 µL Taq polymerase, 0.25 µL each primer (100 µM each), and 15.45 µL H2O. Based on the annealing temperature of the primers to the template DNA and the length of the PCR product, one of two PCR cycles was used. PCR method 1 (~60°C Tm or shorter products) was comprised of these steps: 3 min at 94°C to denature DNA, followed by 35 cycles consisting of 30 s at 94°C, 30 s at 55°C for annealing and 2 min at 68°C for elongation. PCR method 2 (~50°C Tm or longer products) differs from the first method in the annealing temperature (50°C) and the elongation time (3.5 min). Both methods are concluded with 10 min of elongation at 68°C. PCR products were separated on a 1% agarose gel (in 1× TAE, ethidium bromide: 0.5 µg/mL) and visualized with a UV transilluminator. Product sizes were compared to a 1 kb standard (Invitrogen). Several PCR products were sequenced after gel purification by the W. M. Keck Center for Comparative and Functional Genomics at the University of Illinois. Supplementary Table 2 summarizes the peptides identified by an OMSSA search of the MS/MS data generated from nanoLC-MS of the excised gel band. See Figure 3 and Supplementary Figure 9 for the location of the peptide within a NRPS/PKS in cluster #1 (C1) or cluster #2 (C2). Supplementary Table 3 summarizes the primer sequences designed from two sources, the first being the available C1 and C2 sequences from B. cereus AA1134 (GenBank accession numbers ABDA02000035 and ABDA02000007, http://msc.jcvi.org/bacillus_cereus/bacillus_cereus_ah1134/index.shtml) and the second being the peptide sequences reported in Supplementary Table 2. Supplementary Table 4 summarizes the 26 PCRs completed in this study, while Supplementary Figure 11 is an agarose gel separation of the products of the PCRs. In Supplementary Tables 3–4, those rows in white correspond to primers and PCRs completed using B. cereus AH1134 sequence and those rows in gray correspond to primers and PCRs completed using sequence information generated from nanoLC-MS analysis. See Supplementary Table 5 the results of sequencing of selected PCR products.
Identification of NK2018 natural products
A 100 mL starter culture of NK2018 was grown in supplemented M9 minimal medium (per 1 L: 800 mL H2O, 200 mL M9 salts (a 1 L 10× solution contains 64 g Na2HPO4·7H2O, 15 g KH2PO4, 2.5 g NaCl, and 2.5 g NH4Cl), 20 mL 20% glucose, 1 mL 1 M MgSO4, 100 µL 1 M CaCl2, and 10 mL Difco nutrient broth)) at 30°C for 2 days. An aliquot of this starter culture was used to inoculate (at a 1:100 dilution) either a 100 mL or 500 mL volume of M9 minimal media (minus nutrient broth) and was grown for 3–10 days at 30°C. Culture purity was assessed by examination by light microscope and by conducting PCRs #3, #6, #9, #14 and/or #26 (from Supplementary Table 4, data not shown) on genomic DNA purified from each culture as described in the Supplementary Information. Cultures were harvested by centrifugation (10,000 x g, 10 min) and the culture supernatant filter sterilized. The cell pellets were used to prepare protein samples as described previously for PrISM analysis. These samples were used to confirm the presence of HMWPs by SDS-PAGE (Supplementary Figure 18) and for PrISM proteome analysis (using the gel-based proteomic analysis as described above) to confirm the species as NK2018 and to assay production of NRPS/PKS-related proteins (data not shown). Culture supernatant was concentrated 15–20 fold through rotary evaporation and stored at −80°C until further analysis. LC-MS analysis was conducted on all culture supernatants using a Phenomenex Gemini-NX C18 column (5 um particle size, 110 Ǻ pore size) with either a 2 mm or 4.6 mm inner diameter. All LC-MS analysis was conducted on a ThermoFisher LTQ-FT operating at 7 T and connected in-line with Surveyor MS pump and autosampler. An external fraction collector was added if HPLC fractions were to be collected simultaneously with MS analysis. For LC-MS analysis, a shallow gradient was used as shown in Supplementary Table 8, where solvent A was H2O + 0.1% HCOOH and solvent B was MeCN + 0.1% HCOOH. Each FTMS analysis included a full scan (m/z 300–2000) with data-dependent MS/MS on the top 3 ions in each full scan. The MS/MS data were analyzed using an in-house software package and ThermoFisher Xcalibur Qualbrowser. Additional LC-MS separation of these samples was conducted, collecting HPLC fractions at 1 min intervals and drying the fractions under vacuum. Fractions were resuspended in electrospray solution (49% H2O, 49% MeOH, 2% formic acid) and analyzed by direct infusion into a ThermoFisher LTQ-FT Ultra operating at 12 T using a TriVersa robot system for sample delivery. Extensive MSn analysis was conducted on the species of interest in order to gain as much structural information as possible; this information is summarized in Supplementary Figures 13–16 and Supplementary Table 9.
Accessing mass spectrometry and protein sequence data
The data for this manuscript is open access according to the Science Commons CC0 license and can be downloaded from the Tranche network (proteomecommons.org/tranche) using the hashes in Supplementary Table 10. These hashes may be used to prove exactly what files were published as part of this manuscript's dataset, and the hashes may also be used to check that the data has not changed since publication.
Supplementary Material
Acknowledgements
We thank William Metcalf for providing the S. viridochromogenes DSM 40736 and B. subtilis ATCC 6633 strains and Taq polymerase, and him along with Wilfred van der Donk and Peter Yau for technical assistance. We would also like to thank Dana Dlott and the following members of the Kelleher Research Group for their assistance in this work: Leonid Zamdborg, Jeff Osuji, Josh Norris, Jordon Anderson, and Heidi Hannon. This work was supported in part by National Institutes of Health Grants (N.L.K; R01 GM 067725-07, P01 GM 077596-03), NIH Chemistry Biology Interface Training Grant (P.M.T), NIH Molecular Biophysics Training Grant (B.S.E), and NIH Cell & Molecular Biology Training Grant NIH Grant (S.B.B). P.M.T was also supported by an ACS-Division of Analytical Chemistry Fellowship sponsored by Eli Lilly and Company. S.B.B. is currently supported by an ACS-Division of Analytical Chemistry Fellowship sponsored by the Society for Analytical Chemists of Pittsburgh.
Footnotes
Reprints and permissions information are available at www.nature.com/reprints.
Supplementary Information (Supplementary Discussion, Figures, & Tables) is linked to the online version of the paper at www.nature.com/naturebiotechnology. The data collected for this manuscript is open access according to the Science Commons CC0 license and can be downloaded from the Tranche network (proteomecommons.org/tranche) using the hashes provided in Supplementary Table 10.
Author Contributions S.B.B designed and performed proteomic analyses, performed gel-based analyses, identified the natural products discussed herein, conducted LC-MS analyses and wrote the paper. B.S.E. isolated and characterized strains, performed gel-based analyses, performed LC-MS analyses, designed and executed genomic analyses of NK2018 and wrote the paper. P.M.T. assisted in experimental design and performed gel-based analyses and wrote the paper. I.N. assisted in experimental design and conducted LC-MS analyses. NK designed experiments and wrote the paper. S.B.B and B.S.E. contributed equally to this study. All authors discussed the results and commented on the manuscript.
References
- 1.Fischbach MA, Walsh CT. Assembly-line enzymology for polyketide and nonribosomal Peptide antibiotics: logic, machinery, and mechanisms. Chem Rev. 2006;106:3468–3496. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- 2.Zerikly M, Challis GL. Strategies for the discovery of new natural products by genome mining. Chembiochem. 2009;10:625–633. doi: 10.1002/cbic.200800389. [DOI] [PubMed] [Google Scholar]
- 3.Kevany BM, Rasko DA, Thomas MG. Characterization of the complete zwittermicin A biosynthesis gene cluster from Bacillus cereus. Appl Environ Microbiol. 2009;75:1144–1155. doi: 10.1128/AEM.02518-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Newman DJ, Cragg GM. Natural products as sources of new drugs over the last 25 years. J Nat Prod. 2007;70:461–477. doi: 10.1021/np068054v. [DOI] [PubMed] [Google Scholar]
- 5.Weinstein MJ, Wagman GH. Antibiotics: isolation, separation, and purification. New York: Elsevier Scientific Publishing Company, New York; 1978. [Google Scholar]
- 6.Lautru S, Deeth RJ, Bailey LM, Challis GL. Discovery of a new peptide natural product by Streptomyces coelicolor genome mining. Nat Chem Biol. 2005;1:265–269. doi: 10.1038/nchembio731. [DOI] [PubMed] [Google Scholar]
- 7.Knappe TA, et al. Isolation and structural characterization of capistruin, a lasso peptide predicted from the genome sequence of Burkholderia thailandensis E264. J Am Chem Soc. 2008;130:11446–11454. doi: 10.1021/ja802966g. [DOI] [PubMed] [Google Scholar]
- 8.Zazopoulos E, et al. A genomics-guided approach for discovering and expressing cryptic metabolic pathways. Nat Biotechnol. 2003;21:187–190. doi: 10.1038/nbt784. [DOI] [PubMed] [Google Scholar]
- 9.Bode HB, Bethe B, Hofs R, Zeeck A. Big effects from small changes: possible ways to explore nature's chemical diversity. Chembiochem. 2002;3:619–627. doi: 10.1002/1439-7633(20020703)3:7<619::AID-CBIC619>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 10.Brakhage AA, et al. Activation of fungal silent gene clusters: a new avenue to drug discovery. Prog Drug Res. 2008;66(1):3–12. doi: 10.1007/978-3-7643-8595-8_1. [DOI] [PubMed] [Google Scholar]
- 11.Lambalot RH, et al. A new enzyme superfamily - the phosphopantetheinyl transferases. Chem Biol. 1996;3:923–936. doi: 10.1016/s1074-5521(96)90181-7. [DOI] [PubMed] [Google Scholar]
- 12.Stein T, et al. Detection of 4'-phosphopantetheine at the thioester binding site for L-valine of gramicidinS synthetase 2. FEBS Lett. 1994;340:39–44. doi: 10.1016/0014-5793(94)80169-x. [DOI] [PubMed] [Google Scholar]
- 13.Dorrestein PC, Kelleher NL. Dissecting non-ribosomal and polyketide biosynthetic machineries using electrospray ionization Fourier-Transform mass spectrometry. Nat Prod Rep. 2006;23:893–918. doi: 10.1039/b511400b. [DOI] [PubMed] [Google Scholar]
- 14.Dorrestein PC, et al. Facile detection of acyl and peptidyl intermediates on thiotemplate carrier domains via phosphopantetheinyl elimination reactions during tandem mass spectrometry. Biochemistry. 2006;45:12756–12766. doi: 10.1021/bi061169d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meluzzi D, Zheng WH, Hensler M, Nizet V, Dorrestein PC. Top-down mass spectrometry on low-resolution instruments: characterization of phosphopantetheinylated carrier domains in polyketide and non-ribosomal biosynthetic pathways. Bioorg Med Chem Lett. 2008;18:3107–3111. doi: 10.1016/j.bmcl.2007.10.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crawford JM, et al. Deconstruction of iterative multidomain polyketide synthase function. Science. 2008;320:243–246. doi: 10.1126/science.1154711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matteo CC, Glade M, Tanaka A, Piret J, Demain AL. Microbiological Studies on the Formation of Gramicidin S Synthetases. Biotechnology and Bioengineering. 1975;17:129–142. [Google Scholar]
- 18.Blodgett JA, Zhang JK, Metcalf WW. Molecular cloning, sequence analysis, and heterologous expression of the phosphinothricin tripeptide biosynthetic gene cluster from Streptomyces viridochromogenes DSM 40736. Antimicrob Agents Chemother. 2005;49:230–240. doi: 10.1128/AAC.49.1.230-240.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Travers RS, Martin PA, Reichelderfer CF. Selective Process for Efficient Isolation of Soil Bacillus spp. Appl Environ Microbiol. 1987;53:1263–1266. doi: 10.1128/aem.53.6.1263-1266.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mercer AC, Burkart MD. The ubiquitous carrier protein--a window to metabolite biosynthesis. Nat Prod Rep. 2007;24:750–773. doi: 10.1039/b603921a. [DOI] [PubMed] [Google Scholar]
- 21.Geer LY, et al. Open mass spectrometry search algorithm. J Proteome Res. 2004;3:958–964. doi: 10.1021/pr0499491. [DOI] [PubMed] [Google Scholar]
- 22.Silo-Suh LA, Stabb EV, Raffel SJ, Handelsman J. Target range of zwittermicin A, an aminopolyol antibiotic from Bacillus cereus. Curr Microbiol. 1998;37:6–11. doi: 10.1007/s002849900328. [DOI] [PubMed] [Google Scholar]
- 23.Rogers EW, Dalisay DS, Molinski TF. (+)-Zwittermicin A: assignment of its complete configuration by total synthesis of the enantiomer and implication of D-serine in its biosynthesis. Angew Chem Int Ed Engl. 2008;47:8086–8089. doi: 10.1002/anie.200801561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Challis GL, Ravel J, Townsend CA. Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains. Chem Biol. 2000;7:211–224. doi: 10.1016/s1074-5521(00)00091-0. [DOI] [PubMed] [Google Scholar]
- 25.Rausch C, Weber T, Kohlbacher O, Wohlleben W, Huson DH. Specificity prediction of adenylation domains in nonribosomal peptide synthetases (NRPS) using transductive support vector machines (TSVMs) Nucleic Acids Res. 2005;33:5799–5808. doi: 10.1093/nar/gki885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hathout Y, Ho YP, Ryzhov V, Demirev P, Fenselau C. Kurstakins: a new class of lipopeptides isolated from Bacillus thuringiensis. J Nat Prod. 2000;63:1492–1496. doi: 10.1021/np000169q. [DOI] [PubMed] [Google Scholar]
- 27.Steller S, et al. Initiation of surfactin biosynthesis and the role of the SrfD-thioesterase protein. Biochemistry. 2004;43:11331–11343. doi: 10.1021/bi0493416. [DOI] [PubMed] [Google Scholar]
- 28.Schley C, Altmeyer MO, Swart R, Muller R, Huber CG. Proteome analysis of Myxococcus xanthus by off-line two-dimensional chromatographic separation using monolithic poly-(styrene-divinylbenzene) columns combined with ion-trap tandem mass spectrometry. J Proteome Res. 2006;5:2760–2768. doi: 10.1021/pr0602489. [DOI] [PubMed] [Google Scholar]
- 29.Rodriguez-Garcia A, Barreiro C, Santos-Beneit F, Sola-Landa A, Martin JF. Genome-wide transcriptomic and proteomic analysis of the primary response to phosphate limitation in Streptomyces coelicolor M145 and in a DeltaphoP mutant. Proteomics. 2007;7:2410–2429. doi: 10.1002/pmic.200600883. [DOI] [PubMed] [Google Scholar]
- 30.Meier JL, Mercer AC, Burkart MD. Fluorescent profiling of modular biosynthetic enzymes by complementary metabolic and activity based probes. J Am Chem Soc. 2008;130:5443–5445. doi: 10.1021/ja711263w. [DOI] [PubMed] [Google Scholar]
- 31.Simmons TL, et al. Biosynthetic origin of natural products isolated from marine microorganism-invertebrate assemblages. Proc Natl Acad Sci U S A. 2008;105:4587–4594. doi: 10.1073/pnas.0709851105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.