Abstract
Microbial engineering often requires fine control over protein expression; for example, to connect genetic circuits 1-7 or control flux through a metabolic pathway 8-13. We have developed a predictive design method for synthetic ribosome binding sites that enables the rational control of a protein's production rate on a proportional scale. Experimental validation of over 100 predictions in Escherichia coli shows that the method is accurate to within a factor of 2.3 over a range of 100,000-fold. The design method also correctly predicts that reusing a ribosome binding site sequence in different genetic contexts can result in different protein expression levels. We demonstrate the method's utility by rationally optimizing a protein's expression level to connect a genetic sensor to a synthetic circuit. The proposed forward engineering approach will accelerate the construction and systematic optimization of large genetic systems.
Keywords: synthetic biology, translation, optimization, metabolic engineering, genetic circuit, RNA secondary structure
Introduction
Microbial engineering is a time-consuming procedure that often requires multiple rounds of trial-and-error genetic mutation. As it becomes possible to construct larger pieces of synthetic DNA 14, including whole genomes 15, automated methods for genetic circuit assembly and metabolic pathway optimization will be critically important. As genetic systems grow in size and complexity, the application of a trial-and-error approach to optimizing these systems is more difficult.
A genetic system's function is optimized by varying the sequences of its regulatory elements to control the expression levels of its protein coding sequences. Each rate-limiting step in gene expression offers the opportunity for rationally modulating the protein expression level. In bacteria, ribosome binding sites (RBSs) and other regulatory RNA sequences are effective control elements for translation initiation 16-19. As a consequence, they are commonly mutated to optimize genetic circuits, metabolic pathways, and the expression of recombinant proteins.
Previous studies have generated libraries of RBS sequences with the goal of optimizing the function of a genetic system 1, 7, 18. Generation and selection of a sequence library can become impractical as the number of participating proteins increases, especially if measuring the function requires a low-throughput assay or screen 6. For example, randomly mutating 4 nucleotides of an RBS generates a library of 256 sequences. The library size increases combinatorially with the number of proteins in the engineered system (16.7 million sequences for 3 proteins, 2.8×1014 sequences for 6 proteins).
A biophysical model of translation initiation would aid the optimization process by enabling the design of an RBS sequence to obtain a desired translation initiation rate. Using thermodynamics, the free energies of key molecular interactions involved in translation initiation have been characterized 20, 21. Thermodynamic models are made possible by measuring the sequence-dependent energetic changes during RNA folding and hybridization 22-26. These methods have enumerated and characterized the attributes of a RBS sequence that affect its translation initiation rate, but a predictive model that combines all of the interactions together has not been created and tested.
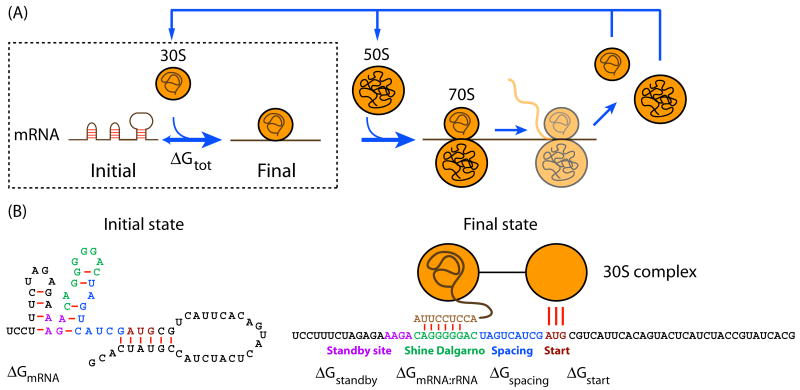
Bacterial translation consists of four phases: initiation, elongation, termination, and ribosome turnover (Figure 1A) 27. In most cases, translation initiation is the rate-limiting step. The translation initiation rate is determined by the summary effect of multiple molecular interactions, including the hybridization of the 16S rRNA to the RBS sequence, the binding of tRNAfMET to the start codon, the distance between the 16S rRNA binding site and the start codon, and the presence of RNA secondary structures that occlude either the 16S rRNA binding site or the standby site 20, 21, 28-31.
Figure 1.
A thermodynamic model of bacterial translation initiation. (A) The ribosome translates an mRNA transcript and produces a protein in a four step process: the rate-limiting assembly of the 30S pre-initiation complex, translation initiation, translation elongation, translation termination, and the turnover of ribosomal subunits and other factors. (B) The thermodynamic free energy change during the translation initiation step is determined by five molecular interactions that participate in the initial and final states of the system. See text for a description of each free energy term. The Watson-Crick base pairs and G:U wobbles (red lines) are shown.
We have developed an equilibrium statistical thermodynamic model to quantify the strengths of the molecular interactions between the 30S complex and an mRNA transcript and to predict the resulting translation initiation rate. The thermodynamic model describes the system as having two states separated by a reversible transition (Figure 1B). The initial state is the folded mRNA transcript and the free 30S complex. The final state is the assembled 30S pre-initiation complex on an mRNA transcript. The difference in Gibbs free energy between these two states is quantified by the Gibbs free energy change ΔGtot. The ΔGtot depends on the mRNA sequence surrounding a specified start codon and will become more negative when attractive interactions are present and more positive when mutually exclusive secondary structures are present.
The translation initiation rate r is related to the ΔGtot according to
| (1) |
where β is the Boltzmann factor for the system. The derivation of Equation 1 is presented in the Supplementary Methods. Importantly, Equation 1 describes the differences in translation initiation rate that result from differences in mRNA sequence. The amount of expressed protein E is proportional to the translation initiation rate where the proportionality factor K accounts for any ribosome-mRNA molecular interactions that are independent of mRNA sequence and any translation-independent parameters, such as the DNA copy number, the promoter's transcription rate, the mRNA stability, and the protein dilution rate (Supplementary Figure 1).
Given a specific mRNA sequence surrounding a start codon, called the subsequence, the ΔGtot is predicted according to the energy model:
| (2) |
where the reference state is a fully unfolded subsequence with ΔGref = 0.
The ΔGmRNA:rRNA term is the energy released when the last 9 nucleotides (nt) of the E. coli 16S rRNA – 3′-AUUCCUCCA-5′ – hybridizes and co-folds to the mRNA subsequence (ΔGmRNA:rRNA < 0). Intra-molecular folding within the mRNA is allowed. All possible hybridizations between the mRNA and 16S rRNA are considered to find the highest affinity 16S rRNA binding site. The binding site minimizes the sum of the hybridization free energy ΔGmRNA:rRNA and the penalty for non-optimal spacing ΔGspacing. Thus, the algorithm can identify the 16S rRNA binding site regardless of its similarity to the consensus Shine-Dalgarno sequence.
The ΔGstart term is the energy released when the start codon and the initiating tRNA anti-codon loop – 3′-UAC-5′ – hybridize together. The ΔGspacing is the free energy penalty caused by a non-optimal physical distance between the 16S rRNA binding site and the start codon (ΔGspacing > 0). When this distance is increased or decreased from an optimum of 5 nt (or ∼17 Å) 29, the 30S complex becomes distorted, resulting in a decreased translation initiation rate.
The ΔGmRNA is the work required to unfold the mRNA subsequence when it folds to its most stable secondary structure, called the minimum free energy structure (ΔGmRNA < 0). The ΔGstandby is the work required to unfold any secondary structures sequestering the standby site (ΔGstandby < 0) after 30S complex assembly. We define the standby site as the 4 nucleotides upstream of the 16S rRNA-binding site, which is its location in a previously studied mRNA 28.
To calculate the ΔGmRNA:rRNA, ΔGstart, ΔGmRNA, and ΔGstandby free energies, we use the NUPACK suite of algorithms, developed by Pierce and coworkers 32, with the Mfold 3.0 RNA energy parameters 22, 23. These free energy calculations do not have any additional fitting or training parameters and explicitly depend on the mRNA sequence. In addition, the free energy terms are not orthogonal; changing a single nucleotide can potentially affect multiple energy terms.
We designed a series of experiments to quantify the relationship between the aligned spacing s and the free energy penalty ΔGspacing. Thirteen synthetic RBSs are created where the aligned spacing is varied from 0 to 15 nucleotides while verifying that the ΔGmRNA:rRNA, ΔGmRNA, ΔGstart, and ΔGstandby free energies remain constant (Supplementary Table I). The translation initiation rates of RBS sequences are measured using a fluorescent protein measurement system (Methods). Steady-state fluorescence measurements are performed on E. coli cultures over a 24 hour period. Under these conditions, the average fluorescence measurement is expected to be proportional to the translation initiation rate r.
The quantitative relationship between the aligned spacing and ΔGspacing is obtained from the fluorescence measurements (Methods). According to the data, it is conceptually useful to treat the 30S complex as a model barbell connected by a rigid spring, where either stretching or compressive forces cause a reduction in entropy and an increase in the ΔGspacing penalty. We empirically fit these measured ΔGspacing values to either a quadratic (s > 5 nt) or a sigmoidal function (s < 5 nt). Following this parameterization, we tested the accuracy of these equations on an additional set of synthetic RBS sequences (Supplementary Figure 2).
For an arbitrary mRNA transcript, the thermodynamic model (Equation 2) is evaluated for each AUG or GUG start codon. The algorithm considers only a subsequence of the mRNA transcript, consisting of 35 nucleotides before and after the start codon. This subsequence includes the RBS and part of the protein coding sequence. The model predictions do not improve when longer subsequences are considered (Supplementary Figure 3).
The development of the thermodynamic model makes certain assumptions. Contributions related to the ribosomal S1 protein's potential preference for pyrimidine-rich sequences are omitted from the free energy model33. The model also assumes that the reversible transition between the initial and final state of 30S complex assembly reaches chemical equilibrium on a physiologically relevant timescale and without any long-lived intermediate states. The presence of overlapping or neighboring start codons, overlapping RBS and protein coding sequences, regulatory RNA binding sites, or RNAse binding sites also pose a challenge to the predictive accuracy of the thermodynamic model. The presence of multiple in-frame start codons, each with significant translation initiation, may distort its predictive accuracy. A genetic system can be designed to avoid many of these complications.
The thermodynamic model can be used in two ways. First, it can predict the relative translation initiation rate of an existing RBS sequence for a particular protein coding sequence on an mRNA transcript. We refer to this as “reverse engineering” because the RBS sequence already exists. Second, it can be used in conjunction with an optimization algorithm to identify a synthetic RBS sequence that is predicted to translate a given protein coding sequence at a user-selected rate. We refer to this mode as “forward engineering” because it generates a de novo sequence according to a user's specifications.
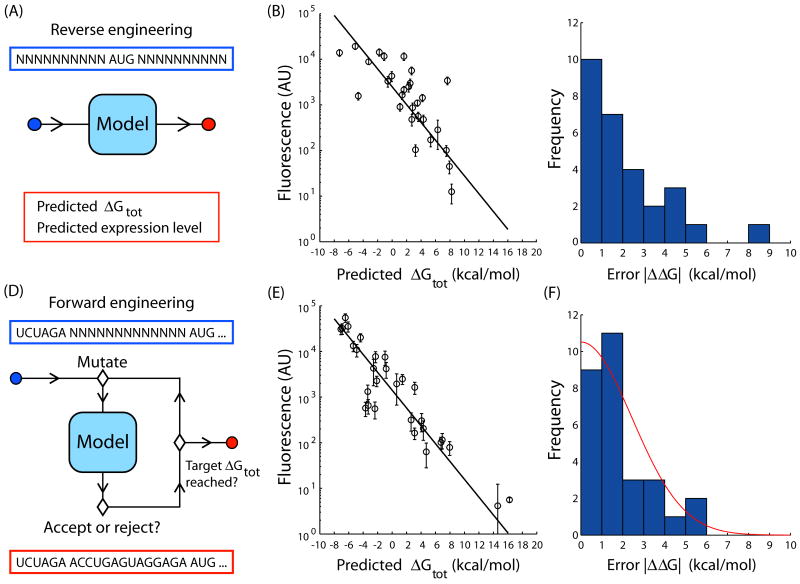
We use the thermodynamic model to predict the translation initiation rates of 28 existing RBS sequences (Figure 2A) that were obtained from a natural genome or taken from a list of commonly used sequences (Supplementary Table I). The lengths of these sequences, as measured by the distance from the transcriptional start site to the fluorescent protein's start codon, vary from 24 to 42 nucleotides. The steady-state protein fluorescences from the sequences are then assayed in the measurement system (Methods). The growth rates of the cell cultures did not correlate with protein fluorescence (Supplementary Figure 4). According to the theory (Equation 1), we expect a linear relationship between the predicted ΔGtot and the log protein fluorescence. Using linear regression, the squared correlation coefficient R2 is 0.54 with Boltzmann factor β = 0.45 ± 0.05 mol/kcal (Figure 2B). The average error is 〈|ΔΔG|〉 = 2.1 kcal/mol (Figure 2C).
Figure 2.
The design method has two modes of operation: (A) The method can predict the relative translation initiation rate of an existing RBS when placed in front of a protein coding sequence. The method calculates the ΔGtot from the input sequence. According to Equation 1, a linear relationship between the log protein fluorescence and the predicted ΔGtot is expected. (B) The fluorescence levels from 28 natural or existing RBSs in front of the RFP fluorescent protein are measured (circles) and compared to the predicted ΔGtot calculations. The error bars are calculated as the standard deviation of 6 measurements performed on two different days. The expected relationship is obtained (line, R2 = 0.54) with a slope β = 0.45 ± 0.05. (C) A histogram shows the distribution of error in the predicted ΔGtot, denoted by |ΔΔG|, of the sequences in B. The average of this distribution is 2.11 kcal/mol. (D) An optimization algorithm with Metropolis criteria, the sequence constraints, and simulated annealing uses iterations of mutation and selection to identify an RNA sequence that is predicted to have the target ΔGtot. (E) The fluorescence levels from 29 synthetic RBSs in front of RFP are measured (circles) and compared to the predicted ΔGtot calculations. The error bars are calculated as the standard deviation of at least 5 measurements performed on 2 different days. The expected linear relationship between log protein expression level and predicted ΔGtot is shown (line, R2 = 0.84) with slope β = 0.45 ± 0.01. (F) A histogram shows the distribution of the error, |ΔΔG|. The average of the distribution is 1.82 kcal/mol and fits well to a one-sided Gaussian distribution (red line) with standard deviation σ = 2.44 kcal/mol.
While these commonly used RBS sequences vary the protein expression by 1500 fold, the thermodynamic model predicts that both stronger and weaker RBSs are possible. For example, one of these RBS sequences contains a strong 16S rRNA binding site (ΔGmRNA:rRNA = −15.2 kcal/mol), but did not yield a high protein expression level due to a strong mRNA secondary structure and non-optimal spacing (ΔGmRNA = −11.4, ΔGspacing = 1.73 kcal/mol). By optimizing the RBS sequence towards a selected ΔGtot, we gain the ability to rationally control the translation initiation rate over a wide range with a proportional effect on the protein expression level.
Using the thermodynamic model, we developed an optimization algorithm that automatically designs an RBS sequence to obtain a desired relative protein expression level. The user inputs a specific protein coding sequence and a desired translation initiation rate. The rate can be varied over five orders of magnitude on a proportional scale. Equation 1 and the experimentally measured β = 0.45 mol/kcal is used to convert the user-selected translation initiation rate into the target ΔGtot. The method then generates a synthetic RBS sequence according to the desired specifications.
The design method combines the thermodynamic model of translation initiation with a simulated annealing optimization algorithm to design an RBS sequence that is predicted to have a target ΔGtot (Figure 2D). The RBS sequence is initialized as a random mRNA sequence upstream of the protein coding sequence. The method then creates new mRNA sequences by inserting, deleting, or replacing random nucleotides. For each new sequence, the ΔGtot is calculated and compared to the target ΔGtot. The sequences are then accepted or rejected according to the Metropolis criteria and three additional sequence constraints that are based on the model's assumptions (Methods). The procedure continues until the synthetic sequence has a predicted ΔGtot to within 0.25 kcal/mol of the target. For a given target ΔGtot, multiple solutions are possible, creating an ensemble of degenerate RBS sequences. The characterization of these ensembles is described in the Supplementary Discussion.
The forward design method is tested by generating 29 synthetic RBS sequences (Supplementary Table I) and comparing their predicted ΔGtot values to the measured protein fluorescences. The coding sequence for a red fluorescent protein is specified and the ΔGtot target is varied from −7.1 to 16.0 kcal/mol. The design method then generates a synthetic RBS sequence for each target ΔGtot. These RBS sequences vary in length from 16 to 35 nucleotides and were highly dissimilar. The steady-state protein fluorescence for each sequence is measured (Methods). The growth rates of the cell cultures did not significantly vary across sequences (Supplementary Figure 4). As expected from the theory (Equation 1), we obtain a linear relationship between the log protein fluorescence and the predicted ΔGtot with β = 0.45 ± 0.01 (R2 = 0.84) (Figure 2E). The average error is 〈|ΔΔG|〉 = 1.82 kcal/mol, corresponding to a 2.3-fold error in the protein expression level. The probability distribution of the ΔΔG for a synthetic RBS is well fit by a Gaussian distribution (Figure 2F).
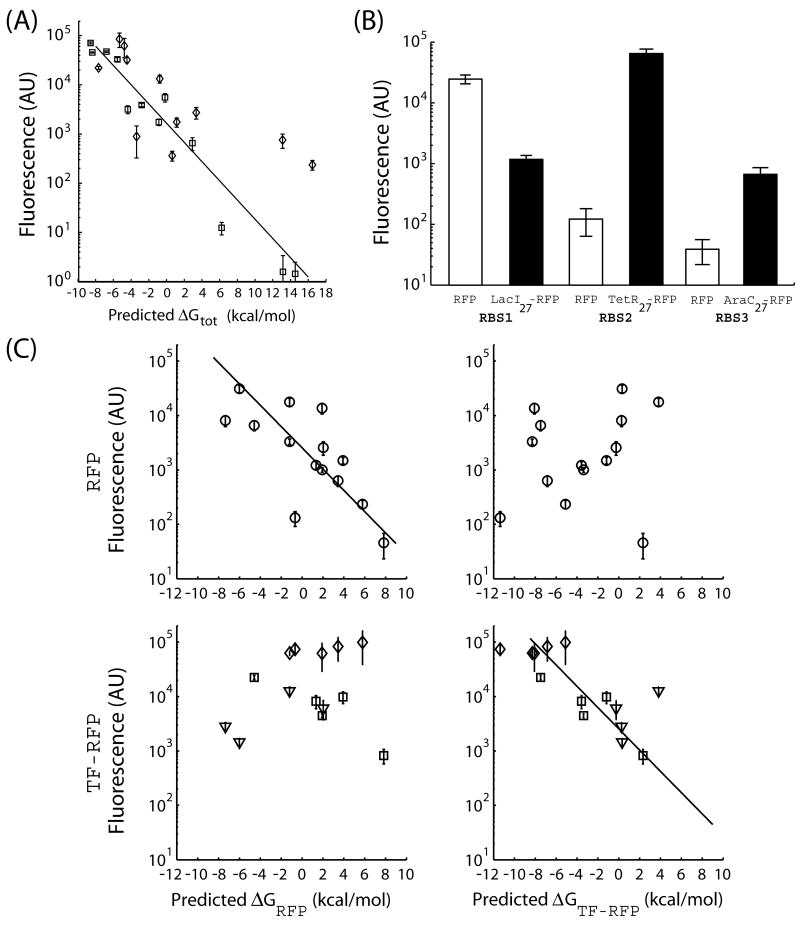
We next tested the ability of the design method to control the translation initiation rates of different proteins. Two chimeric proteins are constructed that fused the first 27 nucleotides from commonly used transcription factors to a red fluorescent protein (TetR27-RFP and AraC27-RFP). The design method is then used to generate 23 synthetic RBSs with ΔGtot targets ranging from −8.5 to 10.5 kcal/mol (Supplementary Table I). The thermodynamic model correctly predicts the translation initiation rates of the TetR27-RFP (R2 = 0.54) and AraC27-RFP (R2 = 0.95) chimeric protein coding sequences (Figure 3A). Notably, the linear relationship between the predicted ΔGtot and the log protein fluorescence yields a similar slope β = 0.45 ± 0.05 mol/kcal.
Figure 3.
The design method can control the expression level of different proteins by predicting the impact of changing the protein coding sequence. (A) The fluorescence levels from 23 synthetic RBSs in front of two different protein coding sequences are measured and compared to the predicted ΔGtot calculations. The two proteins are TetR27-RFP (diamonds) and AraC27-RFP (squares). The expected relationship between the log protein fluorescence and the predicted ΔGtot is obtained for each protein coding sequence (TetR27-RFP, R2=0.54; AraC27-RFP, R2 = 0.95). (B) Reusing the same RBS sequence with two different protein coding sequences can alter the translation initiation. Fluorescence levels from identical RBS sequences in front of either RFP (white bars) or a chimeric fluorescent protein (either LacI27-RFP, TetR27-RFP, or AraC27-RFP; black bars) are shown. (C) The design method must use the correct protein coding sequence to accurately predict the ΔGtot. The fluorescence levels from 14 pairs of RBS sequences in front of either RFP (black circles) or a chimeric fluorescent protein (LacI27-RFP, triangles; TetR27-RFP, diamonds; AraC27-RFP, squares) are measured. When the correct protein coding sequence is used to calculate the ΔGtot, the expected relationship between log protein fluorescence and ΔGtot is obtained (lines, R2 = 0.62 and R2 = 0.51). Otherwise, the thermodynamic model does not correctly predict the expression level (R2 = 0.04 and 0.02). The error bars calculated as the standard deviation of at least 6 measurements performed on 2 different days.
A common practice is to reuse the same well-characterized RBS sequence for the expression of different proteins. Interestingly, the thermodynamic model predicts that this can yield dramatically different translation initiation rates. This absence of modularity will occur when the RNA sequence, containing the RBS, forms strong secondary structures with one protein coding sequence, but not another 30.
We designed experiments to test the model's ability to predict the impact of changing the protein coding sequence on the translation initiation rate. We use the design method to generate 14 synthetic RBS sequences; these sequences are then placed upstream of two different protein coding sequences: the fluorescent protein (RFP) and a chimeric fluorescent protein (TF-RFP: LacI27-RFP, TetR27-RFP, or AraC27-RFP). The optimization procedure for these synthetic RBSs was modified to maximize the objective function |ΔGRFP − ΔGTF-RFP|, where ΔGRFP and ΔGTF-RFP are the predicted ΔGtot's when the RBS sequence is placed upstream of either the RFP or TF-RFP protein coding sequences, respectively. As predicted by the model, the translation initiation rates of these synthetic RBS sequences greatly change when they are reused with different protein coding sequences (Figure 3B); for example, replacing the fluorescent protein with the TetR27-RFP chimera resulted in a 530-fold increase in expression level.
The thermodynamic model can accurately predict these differences in translation initiation rate when the correct protein coding sequence is specified (R2 = 0.62 and 0.51, Figure 3C). When the incorrect protein coding sequence is used, the translation initiation rate is not accurately predicted (R2 = 0.04, 0.02). Consequently, when designing a RBS sequence, the beginning of the protein coding sequence must be included in the thermodynamic calculations.
Altogether, 119 predictions of the design method were tested, revealing that the translation initiation rate can be controlled over at least a 100,000-fold range. The thermodynamic model is most accurate when all free energy terms are included in the ΔGtot calculation (Supplementary Figure 5). By themselves, each free energy term is a poor predictor of the translation initiation rate (Supplementary Figure 6) and excluding one free energy term from the ΔGtot calculation results in a poorer prediction (Supplementary Figure 7). According to the distribution of the method's error (Figure 2F), an optimized RBS sequence has a 47% probability of expressing a protein to within 2-fold of the target. The probability increases to 72%, 85%, or 92% by generating two, three, or four optimized RBS sequences with identical target translation initiation rates (Supplementary Discussion).
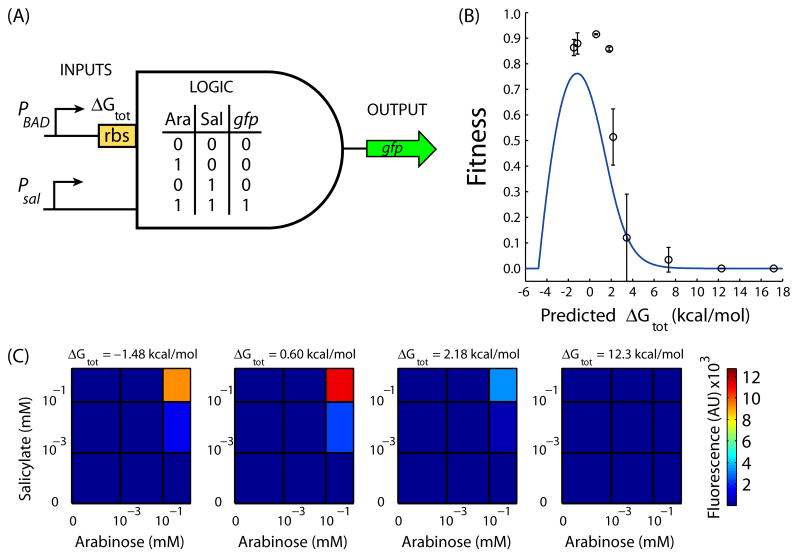
We now demonstrate how combining the design method with a quantitative model of a genetic system enables the efficient optimization of its RBS sequences towards a targeted system behavior. Here, our objective is to optimize the connection between the arabinose-sensing PBAD promoter and an AND gate genetic circuit7. The AND gate genetic circuit is regulated by the expression levels of two input promoters (PBAD and Psal) and controls the expression level of an output gene, which is selected to be a gfp reporter (Figure 4A). The desired AND logic requires that the output gene is only expressed when both input promoters are active. The digital accuracy of the AND logic is highest when the maximum expression level from the PBAD promoter is an optimal value between underexpression and overexpression. When the promoter is underexpressed, the gfp expression is never turned on; when overexpressed, transcriptional leakiness causes gfp expression to turn on even in the input's absence.
Figure 4.
Optimal connection of a sensor input to an AND gate genetic circuit. (A) A functional AND gate genetic circuit will only turn on the gfp reporter output when both the PBAD and Psal promoter inputs are sufficiently induced by arabinose and salicylate, respectively. (B) The quantitative model and design method predict a fitness curve F(ΔGtot) (blue line), relating the predicted ΔGtot of the PBAD promoter's RBS sequence to the quality of the genetic circuit's AND logic. The accuracy of this curve is tested by assaying the fitness of nine genetic circuit variants, each containing a synthetic RBS that was designed to possess a selected ΔGtot (black circles). (C) The amount of gfp fluorescence is shown in response to combinations of arabinose (0.0, 1.3×10-3, 8.3×10-2, and 1.3 mM) and salicylate (0.0, 6.1×10-4, 3.9×10-2, and 0.62 mM) for selected AND gate genetic circuits. These genetic circuits contain RBS sequences with predicted ΔGtot's of 12.3, 2.18, 0.60, and −1.48 kcal/mol. The error bars calculated as the standard deviation of 2 measurements of fitness performed on 2 different days.
The quantitative model relates the RBS sequence downstream of the PBAD promoter to the accuracy of the AND gate genetic circuit's function (Figure 4B). We use previously characterized transfer functions7 to relate the arabinose and salicylate concentrations to the expression levels of the PBAD and Psal promoters (I1 and I2) (Supplementary Figure 8). The PBAD promoter has a maximum protein expression level of gref = 590 au at full induction (x = 1.3 mM arabinose) and when using an RBS sequence with a predicted ΔGtot value of ΔGref = −1.05 kcal/mol. We then substitute I1 and I2 into the AND gate genetic circuit's transfer function to determine the output gene's expression level, which is in turn substituted into the fitness function F that quantifies the ability of the genetic system to carry out AND logic (Supplementary Methods).
We can interconvert between the maximum protein expression level of the PBAD promoter and the predicted ΔGtot of its RBS sequence according to the equation,
| (3) |
where g is called the gain. The experimentally measured β = 0.45 mol/kcal is utilized. Consequently, we create a quantitative curve F(ΔGtot) that relates the predicted ΔGtot of the PBAD promoter's RBS sequence to the fitness of the genetic system. The fitness curve identifies an optimal region at ΔGtot = −1.17 ± 2 kcal/mol where the genetic system will exhibit the best AND logic with respect to the PBAD promoter's RBS sequence (Figure 4B).
Using the forward engineering mode of the design method, we then generate 2 synthetic RBS sequences targeted to the optimum region of the genetic system's function (predicted ΔGtot = −1.48 and −1.15 kcal/mol). We also design 7 additional synthetic RBSs to test the accuracy of the F(ΔGtot) fitness curve, where the ΔGtot ranged from 0.60 to 17.2 kcal/mol. Each RBS sequence (32 to 35 nt) is inserted downstream of the PBAD promoter and the resulting genetic circuit's response to varying inducer concentrations is assayed (Figure 4C and Methods). The fitness values of these rationally mutagenized genetic systems are then compared to the predictions of the model and design method (Figure 4B). The insertion of two stronger RBS sequences (ΔGtot = −2.5 and −3.0 kcal/mol) cause the genetic system to exhibit a fatal growth defect.
Both optimally designed synthetic RBS sequences result in a successful connection between the arabinose-sensing PBAD promoter and the AND gate genetic circuit (mean fitness > 0.85, Figure 4B). The experimentally determined optimum in the F(ΔGtot) curve is nearby ΔGtot = 0.60 kcal/mol, which is only a 1.8 kcal/mol deviation from the model's prediction. The quantitative model and design method also correctly predict how the fitness of the genetic system deteriorates with an increasing ΔGtot. Thus, our approach enabled us to rationally connect two synthetic genetic circuits together to obtain a target behavior while performing only a few mutations and assays (additional design calculations are located in the Supplementary Discussion).
A central goal of synthetic biology is to program cells to carry out valuable functions. As we construct larger and more complicated genetic systems, models and optimization techniques will be required to efficiently combine genetic parts to achieve a target behavior. To accomplish this, biophysical models that link the DNA sequence of a part to its function will be necessary. As engineered genetic systems scale to the size of genomes, the integration of multiple design methods will enable the design of synthetic genomes on a computer to control cellular behavior.
Materials and Methods
Software Implementation
A software implementation of the design method has been named the RBS Calculator and is available at http://voigtlab.ucsf.edu/software. Visitors may use the RBS Calculator in two ways: first, to predict the translation initiation rate of each start codon on an mRNA sequence (reverse engineering); second, to optimize the sequence of a ribosome binding site to rationally control the translation initiation rate with a proportional effect on the protein expression level (forward engineering). The translation initiation rate is gauged on a proportional scale with a suggested range of 0.1 to 100000, although a larger range is potentially feasible. In reverse engineering mode, the software will warn visitors when ribosome binding sites fail to satisfy the sequence constraints or contain additional sequence complications.
A thermodynamic model of translation initiation
The mRNA subsequence S1 consists of the max(1, nstart − 35) to nstart nucleotides and the subsequence S2 consists of the max(1, nstart − 35) to nstart + 35 nucleotides, where nstart is the position of a start codon. The ΔGstart is −1.19 and −0.075 kcal/mol for AUG and GUG start codons, respectively 22.
Using the NuPACK ‘subopt’ algorithm 32 with Mfold 3.0 parameters at 37°C 22, 23, base pair configurations of the folded 16S rRNA and sequence S1 are enumerated, starting with the minimum free energy (mfe) configuration and continuing with suboptimal configurations, each with a corresponding ΔGmRNA:rRNA. For each configuration, the aligned spacing between the 16S rRNA binding site and start codon is calculated according to s = nstart − n1 − n2, where n1 and n2 are the rRNA and mRNA nucleotide positions in the farthest 3′ base pair in the 16S rRNA binding site. When the 30S complex is stretched (s > 5 nt), the ΔGspacing is calculated according to the quadratic equation,
| (4) |
where sopt = 5 nt, c1 = 0.048 kcal/mol/nt2, and c2 = 0.24 kcal/mol/nt. When the 30S complex is compressed (s < 5 nt), the ΔGspacing is calculated according to the sigmoidal function,
| (5) |
where c1 = 12.2 kcal/mol and c2 = 2.5 nt −1. The above parameter values are determined by minimizing the difference between the ΔGspacing values calculated from the experimental measurements (Supplementary Figure 2) and the evaluation of Equation 4 or 5. For each configuration, the ΔGspacing is added to the ΔGmRNA:rRNA. The configuration in the list with the lowest free energy is then identified as containing the predicted 16S rRNA binding site with a corresponding ΔGmRNA:rRNA. The protein coding sequence is excluded from S1 because ribosome binding excludes the formation of downstream secondary structures.
Using the NuPACK ‘mfe’ algorithm and Mfold parameters, the mfe configuration of sequence S2 is calculated and its free energy is designated ΔGmRNA. The standby site is the 4 nt region upstream of the 16S rRNA binding site. The energy required to unfold the standby site is determining by calculating the mfe of sequence S2 with and without preventing the standby site from forming base pairs. The difference between these mfe's is designated ΔGstandby. To calculate the mfe of sequence S2 with a standby site that is constrained to be single-stranded, the sequence is first split into two subsequences, their mfes are each calculated, and then summed together. The two subsequences are the nucleotides nstart − 35 to n3 − 4 and n3 to nstart + 35, where n3 is the most 5′ base pair in the 16S rRNA binding site and 4 is the standby site length.
The five energy terms are summed together to calculate the ΔGtot. Notably, selecting an alternate reference energy state simply adds a sequence-independent constant to the predicted ΔGtot, which becomes indistinguishable from the proportionality factor K.
The simulated annealing optimization algorithm
An initial RBS sequence is randomly generated and inserted in between a pre-sequence and protein coding sequence to create a sequence S. The ΔGtot of the sequence S is calculated and the objective function Oold = |ΔGtot − ΔGtarget| is evaluated. In an iterative procedure, the simulated annealing optimization algorithm randomly deletes, inserts, or replaces a nucleotide in the RBS sequence. The ΔGtot and objective function Onew are then recalculated. If the ΔGtot calculation of S invalidates the sequence constraints, then the mutation is immediately rejected. Otherwise, the mutation is accepted with probability max(1, exp([Oold − Onew]/TSA)), where TSA is the simulated annealing temperature. The TSA is continually adjusted to maintain a 5% to 20% acceptance rate.
There are three sequence constraints that prevent the optimization algorithm from generating a synthetic RBS sequence that may invalidate one of the thermodynamic model's assumptions. The first constraint calculates the energy required to unfold the 16S rRNA binding site on the mRNA sequence and rejects the ones that require more than 6 kcal/mol to unfold. The second constraint quantifies the presence of long-range nucleotide interactions. According to a growth model for random RNA sequences 34, the equilibrium probability of nucleotides i and j forming a base pair in solution is proportional to p = |i − j|−1.44. For each base pair in sequence S, we calculate p. If the minimum p is less than 6×10-3 then the sequence is rejected. Finally, the creation of new AUG or GUG start codons within the RBS sequence is disallowed.
Strains, media, and plasmid construction
The Luria-Bertani (LB) media (10 g/L tryptone, 5 g/L yeast extract, 10 g/L NaCl) is obtained from Fisher Scientific (Pittsburgh, PA). The supplemented minimal media contains M9 minimal salts (6.8 g/L Na2PO4, 3 g/L KH2PO4, 0.5 g/L NaCl, 1 g/L NH4Cl) from Sigma, 2 mM MgSO4 (Fischer Scientific), 100 μM CaCl2 (Fischer Scientific), 0.4% glucose (Sigma), 0.05 g/L leucine (Acros Organics, Belgium), 5 μg/mL chloramphenicol (Acros Organics), and an adjusted pH of 7.4. The expression system is a ColE1 vector with chloramphenicol resistance (derived from pProTet, Clontech). The expression cassette contains a σ70 constitutive promoter (BioBrick J23100), the RBS sequence, followed by the mRFP1 fluorescent protein reporter. XbaI and SacI restriction sites are located before the RBS and after the start codon. An RBS with a desired sequence is inserted into the expression vector using standard cloning techniques. Pairs of complementary oligonucleotides are designed with XbaI and SacI overhangs and the vector is digested with XbaI and SacI restriction enzymes (NEB, Ipswich, MA). Ligation of the annealed oligonucleotides with cut vector results in a nicked plasmid, which is transformed into E. coli DH10B cells. Sequencing is used to verify a correct clone.
The AND gate genetic circuit is composed of three plasmids: pBACr-AraT7940, pBR939b, and pAC-SalSer914 with kanamycin, ampicillin, and chloramphenicol resistance markers, respectively. The PBAD promoter maximum expression level was modified by inserting designed synthetic RBSs on plasmid pBACr-AraT7940. Plasmid pBACr-AraT7940 was digested with BamHI and ApaLI enzymes and pairs of oligonucleotides were designed to contain the desired RBS sequence and corresponding overhangs. Ligation, transformation, selection, and sequencing proceeded as described above.
Growth and fluorescence measurements
The fluorescent protein measurement system is composed of a constitutive promoter, a sequence containing a RBS, and the mRFP1 fluorescent protein reporter (Supplementary Figure 9). An annotated DNA sequence of the system (Genbank format) is available in the Supplementary Data.
Growth and fluorescence measurements are performed in 96-well high throughput format. A 96-well plate containing 200 μl LB and 50 μg/ml chloramphenicol is inoculated, from single colonies, with up to 30 different DH10B E. coli cultures in an alternating, staggered pattern that excludes the outer wells. Cultures are incubated overnight at 37°C with 250 RPM orbital shaking. A fresh 96-well plate containing 200μl supplemented minimal media is inoculated by overnight cultures using a 1:100 dilution. This plate is then incubated at 37°C in a Safire2 plate spectrophotometer (Tecan) with high orbital shaking. OD600 measurements are recorded every 3 minutes. Once a culture reaches an OD600 of 0.15 to 0.20 (4 to 6 hours), a sample of each culture is transferred to a new plate containing 200 μl PBS and 2 mg/ml kanamycin (Acros Organics) for flow cytometry measurements. This media replacement strategy is repeated twice more using fresh, pre-warmed plates containing supplementary minimal media (the first with a 1:10 dilution requiring 8 to 10 hours of growth and the second with a 1:7 dilution requiring 13 to 15 hours of growth). At least three samples are taken for each culture. The fluorescence distribution of each sample is measured with a LSRII flow cytometer (BD Biosciences). We use an ellipse in forward and side scatter space to gate at least 30 000 flow cytometer events. All distributions are unimodal. The autofluorescence distribution of DH10B cells is also measured. The arithmetic mean of each distribution is taken and the mean autofluorescence is substracted.
From single colonies, RBS variants of each AND gate genetic circuit are grown overnight in LB and antibiotics (50 μg/ml ampicillin, 25 μg/ml chloramphenicol, and 25 μg/ml kanamycin). A 96-well plate containing 200 μl LB, antibiotics, and sixteen different inducer concentrations (combinations of 0.0, 1.3×10-3, 8.3×10-2, and 1.3 mM arabinose with 0.0, 6.1×10-4, 3.9×10-2, and 0.62 mM sodium salicylate) are inoculated by overnight cultures using a 1:100 dilution. Plates are grown in a Safire2 plate spectrophotometer (Tecan) with high orbital shaking. OD600 and gfp fluorescence measurements are recorded every 10 minutes for 14 hours. Background autofluorescence is subtracted from each fluorescence measurement. This procedure is repeated twice for each variant. For each variant, the average and standard deviation of the fluorescence per OD600 for each inducer concentration at the final time point are then calculated.
Data analysis
The ΔGspacing is inferred from the fluorescent protein expression data E in the following way. The RNA sequences used to parameterize the model of ΔGspacing are predicted to have identical ΔGmRNA, ΔGmRNA:rRNA, ΔGstandby, and ΔGstart free energies. According to Equation 1, dividing the expression of a sequence with spacing s1 over another with spacing s2 and rearranging then yields the relation: ΔGspacing(s1) − ΔGspacing(s2) = −β−1log(E1/E2). The fluorescent protein expression at s = 5 nt was considered maximal and ΔGspacing(s = 5) is accordingly set to zero. Using an experimentally measured value of β = 0.45 mol/kcal, the model of ΔGspacing for each s is then determined.
Linear regression is used to determine the accuracy of the theory, which hypothesizes a linear relationship between the log average protein fluorescence E and the predicted ΔGtot data. The squared correlation coefficient R2 and slope −β are calculated according to −β = (NΣ(xiyi) − ΣxiΣyi) / (NΣ(xi2) − (Σxi)2) and R2 = (NΣ(xiyi) − ΣxiΣyi)2 / [(NΣ(xi2) − (Σxi)2)(NΣ(yi2) − (Σyi)2)], where N is the number of average expression levels recorded, y is log E, and x is ΔGtot. The standard deviation of β is calculated by substituting the log E data with the log(E+δE) and log(E−δE) data (δE : standard deviation of E) and calculating the average difference.
Supplementary Material
Acknowledgments
We are grateful to all members of the Voigt lab for technical advice and continued support. This work is supported by the Pew and Packard Foundations, Office of Naval Research, NIH EY016546, NIH AI067699, NSF BES-0547637, NSF TeraGrid TG-MCB080126T, and a Sandler Family Opportunity Award. C.A.V, H.S, and E.M. are part of the NSF SynBERC ERC (www.synberc.org). E.M is supported by an NSF Graduate Research Fellowship and an ASEE National Defense Science and Engineering Graduate Fellowship.
Footnotes
Author Contributions: HMS and CAV designed the study and wrote the manuscript. HMS developed the method. HMS and EAM performed the experiments.
References
- 1.Basu S, Gerchman Y, Collins CH, Arnold FH, Weiss R. A synthetic multicellular system for programmed pattern formation. Nature. 2005;434:1130–1134. doi: 10.1038/nature03461. [DOI] [PubMed] [Google Scholar]
- 2.Stricker J, et al. A fast, robust and tunable synthetic gene oscillator. Nature. 2008;456:516–519. doi: 10.1038/nature07389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Friedland AE, et al. Synthetic gene networks that count. Science. 2009;324:1199–1202. doi: 10.1126/science.1172005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ellis T, Wang X, Collins JJ. Diversity-based, model-guided construction of synthetic gene networks with predicted functions. Nat Biotechnol. 2009;27:465–471. doi: 10.1038/nbt.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yokobayashi Y, Weiss R, Arnold FH. Directed evolution of a genetic circuit. Proc Natl Acad Sci U S A. 2002;99:16587–16591. doi: 10.1073/pnas.252535999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tabor JJ, et al. A synthetic genetic edge detection program. Cell. 2009;137:1272–1281. doi: 10.1016/j.cell.2009.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anderson JC, Voigt CA, Arkin AP. Environmental signal integration by a modular AND gate. Mol Syst Biol. 2007;3:133. doi: 10.1038/msb4100173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dueber JE, et al. Synthetic protein scaffolds provide modular control over metabolic flux. Nat Biotechnol. 2009;27:753–759. doi: 10.1038/nbt.1557. [DOI] [PubMed] [Google Scholar]
- 9.Anthony JR, et al. Optimization of the mevalonate-based isoprenoid biosynthetic pathway in Escherichia coli for production of the anti-malarial drug precursor amorpha-4,11-diene. Metab Eng. 2008 doi: 10.1016/j.ymben.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 10.Atsumi S, Hanai T, Liao JC. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature. 2008;451:86–89. doi: 10.1038/nature06450. [DOI] [PubMed] [Google Scholar]
- 11.Hawkins KM, Smolke CD. Production of benzylisoquinoline alkaloids in Saccharomyces cerevisiae. Nat Chem Biol. 2008;4:564–573. doi: 10.1038/nchembio.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee KH, Park JH, Kim TY, Kim HU, Lee SY. Systems metabolic engineering of Escherichia coli for L-threonine production. Mol Syst Biol. 2007;3:149. doi: 10.1038/msb4100196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lutke-Eversloh T, Stephanopoulos G. Combinatorial pathway analysis for improved L-tyrosine production in Escherichia coli: identification of enzymatic bottlenecks by systematic gene overexpression. Metab Eng. 2008;10:69–77. doi: 10.1016/j.ymben.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 14.Czar MJ, Anderson JC, Bader JS, Peccoud J. Gene synthesis demystified. Trends Biotechnol. 2009;27:63–72. doi: 10.1016/j.tibtech.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 15.Gibson DG, et al. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science. 2008;319:1215–1220. doi: 10.1126/science.1151721. [DOI] [PubMed] [Google Scholar]
- 16.Isaacs FJ, et al. Engineered riboregulators enable post-transcriptional control of gene expression. Nat Biotechnol. 2004;22:841–847. doi: 10.1038/nbt986. [DOI] [PubMed] [Google Scholar]
- 17.Carrier TA, Keasling JD. Library of synthetic 5′ secondary structures to manipulate mRNA stability in Escherichia coli. Biotechnol Prog. 1999;15:58–64. doi: 10.1021/bp9801143. [DOI] [PubMed] [Google Scholar]
- 18.Pfleger BF, Pitera DJ, Smolke CD, Keasling JD. Combinatorial engineering of intergenic regions in operons tunes expression of multiple genes. Nat Biotechnol. 2006;24:1027–1032. doi: 10.1038/nbt1226. [DOI] [PubMed] [Google Scholar]
- 19.Chubiz LM, Rao CV. Computational design of orthogonal ribosomes. Nucleic Acids Res. 2008;36:4038–4046. doi: 10.1093/nar/gkn354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Smit MH, van Duin J. Secondary structure of the ribosome binding site determines translational efficiency: a quantitative analysis. Proc Natl Acad Sci U S A. 1990;87:7668–7672. doi: 10.1073/pnas.87.19.7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vellanoweth RL, Rabinowitz JC. The influence of ribosome-binding-site elements on translational efficiency in Bacillus subtilis and Escherichia coli in vivo. Mol Microbiol. 1992;6:1105–1114. doi: 10.1111/j.1365-2958.1992.tb01548.x. [DOI] [PubMed] [Google Scholar]
- 22.Xia T, et al. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs. Biochemistry. 1998;37:14719–14735. doi: 10.1021/bi9809425. [DOI] [PubMed] [Google Scholar]
- 23.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 24.Kierzek R, Burkard ME, Turner DH. Thermodynamics of single mismatches in RNA duplexes. Biochemistry. 1999;38:14214–14223. doi: 10.1021/bi991186l. [DOI] [PubMed] [Google Scholar]
- 25.Miller S, Jones LE, Giovannitti K, Piper D, Serra MJ. Thermodynamic analysis of 5′ and 3′ single- and 3′ double-nucleotide overhangs neighboring wobble terminal base pairs. Nucleic Acids Res. 2008;36:5652–5659. doi: 10.1093/nar/gkn525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Christiansen ME, Znosko BM. Thermodynamic characterization of the complete set of sequence symmetric tandem mismatches in RNA and an improved model for predicting the free energy contribution of sequence asymmetric tandem mismatches. Biochemistry. 2008;47:4329–4336. doi: 10.1021/bi7020876. [DOI] [PubMed] [Google Scholar]
- 27.Laursen BS, Sorensen HP, Mortensen KK, Sperling-Petersen HU. Initiation of protein synthesis in bacteria. Microbiol Mol Biol Rev. 2005;69:101–123. doi: 10.1128/MMBR.69.1.101-123.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Studer SM, Joseph S. Unfolding of mRNA secondary structure by the bacterial translation initiation complex. Mol Cell. 2006;22:105–115. doi: 10.1016/j.molcel.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 29.Chen H, Bjerknes M, Kumar R, Jay E. Determination of the optimal aligned spacing between the Shine-Dalgarno sequence and the translation initiation codon of Escherichia coli mRNAs. Nucleic Acids Res. 1994;22:4953–4957. doi: 10.1093/nar/22.23.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kudla G, Murray AW, Tollervey D, Plotkin JB. Coding-sequence determinants of gene expression in Escherichia coli. Science. 2009;324:255–258. doi: 10.1126/science.1170160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Smit MH, van Duin J. Translational standby sites: how ribosomes may deal with the rapid folding kinetics of mRNA. J Mol Biol. 2003;331:737–743. doi: 10.1016/s0022-2836(03)00809-x. [DOI] [PubMed] [Google Scholar]
- 32.Dirks RM, Bois JS, Schaeffer JM, Winfree E, Pierce NA. Thermodynamic Analysis of Interacting Nucleic Acid Strands. SIAM Review. 2007;49:65–88. [Google Scholar]
- 33.Sengupta J, Agrawal RK, Frank J. Visualization of protein S1 within the 30S ribosomal subunit and its interaction with messenger RNA. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:11991–11996. doi: 10.1073/pnas.211266898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.David F, Hagendorf C, Wiese KJ. A growth model for RNA secondary structures. Journal of Statistical Mechanics: Theory and Experiment. 2008:P04008. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.