Abstract
While most animal–bacterial symbioses are reestablished each successive generation, the mechanisms by which the host and its potential microbial partners ensure tissue colonization remain largely undescribed. We used the model association between the squid Euprymna scolopes and Vibrio fischeri to examine this process. This light organ symbiosis is initiated when V. fischeri cells present in the surrounding seawater enter pores on the surface of the nascent organ and colonize deep epithelia-lined crypts. We discovered that when newly hatched squid were experimentally exposed to natural seawater, the animals responded by secreting a viscous material from the pores of the organ. Animals maintained in filtered seawater produced no secretions unless Gram-negative bacteria, either living or dead, were reintroduced. The viscous material bound only lectins that are specific for either N-acetylneuraminic acid or N-acetylgalactosamine, suggesting that it was composed of a mucus-containing matrix. Complex ciliated fields on the surface of the organ produced water currents that focused the matrix into a mass that was tethered to, and suspended above, the light organ pores. When V. fischeri cells were introduced into the seawater surrounding the squid, the bacteria were drawn into its fluid-filled body cavity during ventilation and were captured in the matrix. After residing as an aggregate for several hours, the symbionts migrated into the pores and colonized the crypt epithelia. This mode of infection may be an example of a widespread strategy by which aquatic hosts increase the likelihood of successful colonization by rarely encountered symbionts.
Most animals and plants obtain their essential microbial symbionts by horizontal transmission, the process by which a host becomes colonized by specific microorganisms acquired from the surrounding environment after embryogenesis. Because these potential symbionts usually represent only a small fraction of the ambient microbial assemblage, host species must develop mechanisms by which they increase the probability of being colonized by appropriate microbes, while discouraging colonization by nonspecific ones. One of the best-understood examples of this process is the formation of root-nodule associations between leguminous plants and N2-fixing bacteria. In the bacteria-rich structured environment of soil, the plant host creates a gradient of root exudates that serves as a chemoattractant (1). These exudates are specifically detected and metabolized by symbiotically competent bacteria in the family Rhizobiaceae. Flavonoid compounds present in these exudates induce symbiosis-specific bacterial nod genes and, through a subsequent “molecular conversation” between the host and microbe, the bacteria are able to enter the plant via a specialized, host-derived structure known as the infection thread (2).
In contrast to soil, aquatic environments typically have low to undetectable concentrations of the specific symbiont cells that inoculate and colonize the tissues of animal hosts (3–6). In addition, the fluid nature of aquatic environments, which are commonly dominated by turbulent flow and high shear stress, restricts the long-term stability of chemoattractant gradients (7). These features suggest that establishing a symbiosis in an aquatic habitat presents a challenge and, thus far, mechanisms evolved by aquatic animals to overcome these constraints and ensure colonization by symbionts have remained largely a mystery. In this study, we used the model animal–bacterial symbiosis between the Hawaiian sepiolid squid Euprymna scolopes and the marine luminous bacterium Vibrio fischeri to examine this process.
On hatching, juvenile E. scolopes have a nascent light-emitting organ in the center of their mantle cavity (Fig. 1a). This organ is initially uncolonized and has a complex array of ciliated epithelial cells on each lateral surface (8). Each array consists of a pair of appendages whose tips appose, forming a ring that extends laterally away from the center of the organ. Successful colonization of the host by V. fischeri cells induces the loss of the ciliated surface epithelium, a finding that provided the first indication that this tissue is important only for inoculation (9). The ciliated field surrounds a set of three pores at the base of each appendage (Fig. 1b). During initiation of the symbiosis, bacteria must enter these pores, travel down ducts, and colonize the internal crypt spaces of the organ (10). When they are present, V. fischeri cells typically colonize the light organ of the juvenile squid within 12 h after it hatches; however, in the absence of V. fischeri, the organ remains uncolonized by any of the other bacterial species present in seawater (8).
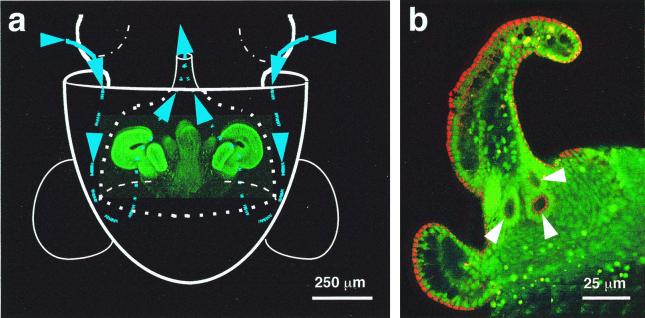
Figure 1.
The path of V. fischeri cells to the site of inoculation of the E. scolopes light organ. (a) Diagram illustrating an outline of the host's body (solid white lines), superimposed over a laser-scanning confocal micrograph (LSM) of the nascent light organ, indicating the relative size and position of the organ within the host's mantle cavity. The organ is circumscribed by the posterior portion of the excurrent funnel (dotted white lines). Ventilatory movements of the host draw ambient seawater (blue arrows and lines) containing V. fischeri cells into the mantle cavity. The water travels into the funnel where, before being vented back into the environment, it encounters complex ciliated fields (bright green) on the lateral surfaces of the organ. The fields entrain water into the vicinity of pores on the light organ surface. (b) Higher-magnification LSM of one side of a hatchling light organ, showing the location of the three pores (arrows) that lie at the base of the appendages of each ciliated field.
A theoretical consideration of the scale over which the initiation of colonization occurs indicates that, for efficient inoculation of the organ, V. fischeri cells must be captured or enriched for in some way. Every half-second the juvenile squid ventilates about 1.3 μl of ambient seawater through its mantle cavity. Constituting less than 0.1% of the total ambient bacteria, V. fischeri occurs at fewer than 500 cells per ml in nature (6). Thus, on average no more than a single V. fischeri cell, occupying about one-millionth the volume of the mantle cavity, will be present during each ventilation. Without mechanisms to harvest them, the symbionts would have to find one of the six 10-μm pores on the light organ surface in less than 1 sec before being expelled.
Using V. fischeri cells labeled with a green fluorescent protein (GFP), we characterized the colonization of the E. scolopes light organ by confocal microscopy. The results provide evidence that reciprocal signaling between the host and environmental microbes results in a bacteria-induced, host-derived structure composed of mucus-like material. This structure, together with a complex ciliated field, creates a dynamic ciliary-mucus current that mediates the aggregation of potential symbionts from the ambient environment and promotes their colonization of specific host tissues.
Materials and Methods
Construction of a GFP-Encoding Plasmid.
The gene encoding a high-efficiency, red-shifted GFP derivative in pQBI63 (Quantum Biotechnologies, Montreal) was cloned into the shuttle vector pLS6 (11), generating pKV111, which was mobilized by triparental mating and maintained by chloramphenicol selection (5 μg/ml).
Bacterial Inoculations.
Five bacterial strains were used as inocula. Cells of V. fischeri ES114 or Vibrio parahaemolyticus KNH1, both carrying pKV111, or Bacillus cereus 43-25 (12) or Listeria monocytogenes NF-L512 (donated by S. Kathariou, Univ. of Hawaii), carrying a chromosomally inserted GFP gene, were grown to mid-logarithmic phase before inoculation. Strain KNH1, isolated from Kaneohe Bay, Hawaii, was identified as V. parahaemolyticus on the basis of the sequence of its 16S rRNA gene (99.2% identity over 1,269 bp). Red-fluorescent, heat-killed Escherichia coli (Molecular Probes) were rehydrated in filter-sterilized seawater (FSSW) before use. Hatchling squid were rinsed and placed in 1 ml of either FSSW or natural seawater, containing approximately 106 nonsymbiotic bacteria per ml, and an inoculum of between 103 and 106 cells of one of the test strains was added. Within this range the potential for aggregate formation was independent of the total bacterial concentration used.
Microscopy.
Hatchling squid tissue was either unstained or stained for 30 min in seawater containing either 0.001% acridine orange or 0.005% CellTracker Orange (Molecular Probes). Animals were then anesthetized in a 1:1 solution of 7.5% MgCl2 and FSSW. After dissection unstained animals were viewed by differential interference contrast (DIC), and stained animals were viewed by fluorescence, on a Zeiss LSM 510 laser-scanning confocal microscope.
Lectin Labeling of Secreted Matrix Material.
Newly hatched squid were either exposed to V. fischeri cells or maintained in FSSW for between 6 and 10 h. Animals were then treated for 30 min with one of several fluorescently labeled lectins (see Table 1) at a final concentration of 50 μg/ml, and then viewed by laser-scanning confocal microscopy.
Table 1.
Lectin labeling of host-secreted material
| Lectin | Glycan specificity | Labeling of host mucus-like secretion |
|---|---|---|
| Wheat germ agglutinin | N-acetylneuraminic acid/ N-acetylglucosamine | + |
| WGA succinylated | N-acetylglucosamine | − |
| Sophora japonica agglutinin | N-acetylgalactosamine | + |
| Concanavalin A | α-d-mannose/α-d-glucose | − |
| Ulex europaeus agglutinin | Fucose | − |
Results and Discussion
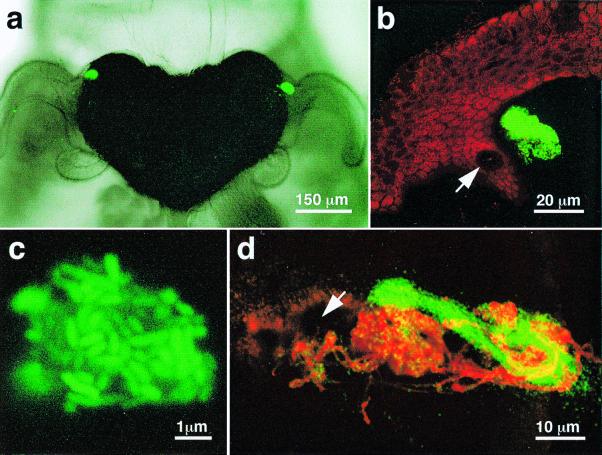
We found that within a few hours after V. fischeri cells were introduced into seawater containing newly hatched E. scolopes, currents created by the ciliated appendages amassed the bacteria into aggregates that became embedded in a mucus-like material suspended just above the light organ pores (Fig. 2a–c; Table 1). Wheat germ agglutinin and Sophora japonica agglutinin, which bind to two sugars commonly found in mucin, N-acetylneuraminic acid and N-acetylgalactosamine, respectively, were the only two lectins tested that labeled the host secretions (Fig. 2d; Table 1). Ciliary-mucus currents are a common means by which invertebrate tissues interact with food particles in the environment (13), but their involvement in the initiation of a symbiotic association has not been previously reported. Because transiently occurring fields of cilia have been noted in other juvenile animals near sites where symbionts colonize, and these fields disappear once colonization takes place (14, 15), this type of inoculation strategy may be used in a variety of other aquatic associations.
Figure 2.
Bacterial aggregations that form during inoculation of the E. scolopes light organ. (a) Differential interference contrast image superimposed over a fluorescent image of the organ of a newly hatched host squid that had been exposed to GFP-labeled V. fischeri. Within 3 h after inoculation, the labeled bacteria have formed two dense aggregations on either side of the organ near the base of the anterior appendage of the ciliated field. (b) In a higher-magnification LSM, an aggregation of GFP-labeled bacteria could be seen suspended just above a light-organ pore (arrow). (c) A very highly magnified LSM of one of these aggregates confirmed that it was a dense assemblage of the GFP-labeled V. fischeri. (d) An LSM of an aggregate containing GFP-labeled V. fischeri cells 8 h after inoculation. The mucus-like matrix originating from the host's pore (arrow) was stained with fluorescently labeled wheat germ agglutinin (red).
In inoculation experiments, three species of Gram-negative bacteria were capable of inducing juvenile E. scolopes to form aggregates (Table 2). In contrast, squid that were maintained in bacteria-free seawater did not produce these mucus-like secretions, even in the presence of bacteria-sized latex beads. However, when the latex beads and V. fischeri cells were coincubated, both the beads and the bacteria became aggregated (Fig. 3a). Thus, although the presence of bacteria was required to induce the formation of the secretions, the ability to adhere was not specific to bacterial cells. The two species of Gram-positive bacteria tested were incapable of inducing aggregate formation. The ability of the Gram-negative species to induce aggregate formation and the inability of the Gram-positive species to do so suggests that some component specific to the outer membrane or cell wall of Gram-negative bacteria may be responsible for initiating this phenomenon. Lipopolysaccharide (LPS), a characteristic constituent found only on the surfaces of Gram-negative bacteria, has been reported to induce the secretion of mucus by host cells during a pathogenic bacterial infection (16). However, the addition of purified LPS to seawater in concentrations that are sufficient to induce other biological responses in cell culture (17) did not lead to either mucus production or the aggregation of latex beads (Table 2). Thus, some other (or additional), and as yet unknown, signal must be required. Regardless of its identity, an inductive trigger may ensure that the mucus-like secretions are not produced prematurely, but only after the squid has exited the sterile confines of the egg, and entered bacteria-containing seawater from which it can obtain its inoculum of V. fischeri cells.
Table 2.
Inducers of aggregate formation and their subsequent activities
| Potential inducer | Formation of aggregates | Migration to the pores | Colonization of the light organ |
|---|---|---|---|
| Gram-negative bacteria | |||
| V. fischeri | + | + | + |
| V. fischeri (nonmotile) | + | − | − |
| V. parahaemolyticus | + | + | − |
| Escherichia coli | |||
| (heat-killed) | + | − | − |
| Gram-positive bacteria | |||
| Bacillus cereus | − | − | − |
| Listeria monocytogenes | − | − | − |
| Polystyrene microbeads | |||
| Alone | − | − | − |
| With V. fischeri cells | + | − | − |
| With purified | |||
| lipopolysaccharide | − | − | − |
Figure 3.
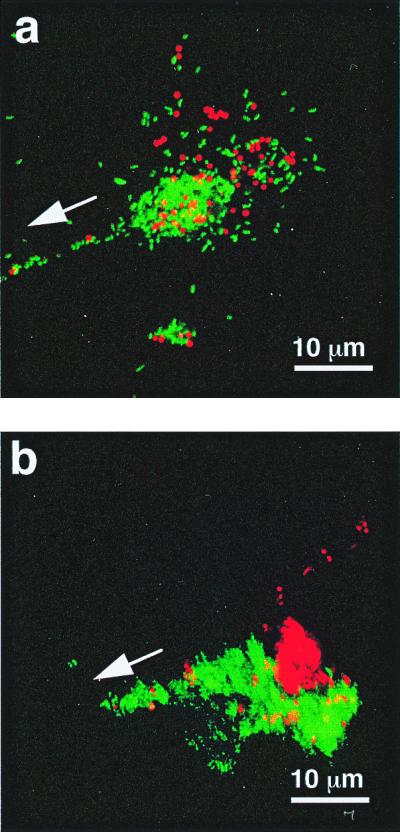
The aggregation and segregation of polystyrene beads within the mucus-like secretions induced by the presence of V. fischeri cells. (a) Red-fluorescent polystyrene beads (Molecular Probes), 1 μm in diameter, were incubated with GFP-labeled V. fischeri and visualized by laser-scanning microscopy. After a 2- to 4-h incubation, bacteria and beads were found randomly distributed in aggregations. (b) Six hours after inoculation, the bacteria and beads had become segregated as the V. fischeri cells migrated in the direction of the pores (arrows).
Analysis of the behavior of V. fischeri cells in the aggregates demonstrated that they play a dynamic and responsive role within these structures. Cells migrated down the strands of the mucus-like material that bridge between the aggregation centers and the light organ pores (Fig. 3). In contrast, latex beads did not move from the site of their initial aggregation, and became segregated from the migrating cells (Fig. 3b); similarly, nonmotile mutants of V. fischeri did not migrate (Table 2), an observation that would explain the inability of these mutants to colonize the light organ (18). The inability of nonmotile V. fischeri cells to move toward the pores suggests that the directional migration of wild-type V. fischeri in the aggregates was a result of bacterial locomotion and chemotaxis, and not the result of an active process controlled by E. scolopes, such as the currents created by the host's ciliated fields. Although V. parahaemolyticus cells also migrated to the pores, they rarely reached the crypts, and they were unable to colonize successfully (Table 2), revealing that the establishment of the association requires the completion of several distinct steps.
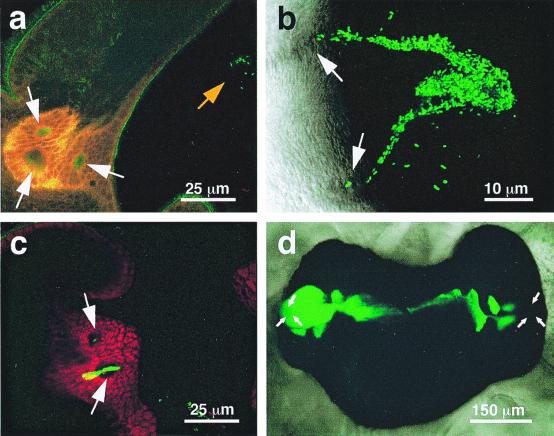
During the initial stages of the inoculation process, V. fischeri cells require a minimum residence time within the matrix of the mucus-like secretion before initiating colonization. Aggregation of the bacteria began within the first hour after exposure of juvenile squids to the inoculum (Fig. 4a). The cells continued to accumulate in the secreted matrix over the next few hours, and by 4–6 h after inoculation V. fischeri cells were observed migrating down the mucus-like secretions and into the pores (Fig. 4 b and c). Continued movement of the bacteria into the internal crypt spaces, and their subsequent proliferation there, resulted in normal colonization (Fig. 4d).
Figure 4.
Stages in the process of infection and colonization of the squid light organ. (a) After a 1-h exposure to GFP-labeled V. fischeri, an LSM revealed a small aggregate (orange arrow) forming above a pore of the light organ. (b) Between 2 and 4 h after inoculation, bacteria were seen as streams migrating from the aggregate to the pores. (c) Between 4 and 6 h after inoculation, a mass of GFP-labeled V. fischeri cells appeared to be migrating through a pore and into a duct of the light organ. Cells within the duct appear yellow. (d) Differential interference contrast image of the fully colonized light organ of E. scolopes, showing the population of GFP-labeled symbionts (green). The location of the pores is indicated by white arrows in all panels.
The molecular interactions between the host's secretions and the bacteria in the aggregates have yet to be determined. For example, yet to be resolved is how V. fischeri cells are able to migrate after their association with the mucus-like material. One possible mechanism is that V. fischeri adheres to the secretion but is able to detach. V. fischeri has a homologue to the Vibrio cholerae hap gene (P. Fidopiastis and E.G.R., unpublished data), which encodes a hemagglutinin/protease and “detachase” that allows V. cholerae to degrade mucin and detach from the mucosal surfaces of epithelial cells (19). Alternatively, instead of binding tightly to the mucus-like material, V. fischeri cells may be loosely enough associated with the matrix that they are able to migrate through it and into the pores.
The nature of the several-hour delay is at present unknown, but this period may be a time during which the bacteria exchange signals with each other and/or their host. V. fischeri is one of a group of bacterial species that exhibits density-dependent quorum sensing, wherein certain genes are induced only under conditions where the bacteria are in high concentration, such as when they are in association with plants and/or animals (20). Because the host appears to either collect and/or enrich for V. fischeri cells in the aggregates, these structures, which function as a host-derived biofilm (21), may provide sites where density-dependent quorum-sensing genes are induced and diffusible signals are generated (22).
We have demonstrated that juvenile E. scolopes harvest their bacterial partner by secreting a mucus-like matrix in which potential symbionts aggregate before developing the capacity to initiate colonization. Ciliary currents created by the host cause bacteria to aggregate within this material near sites of colonization. After residing for several hours within these aggregates, the symbionts migrate to and infect specific host tissues. With the exception of the involvement of a ciliated epithelium, many aspects of this process are strikingly similar to the infection of leguminous plant tissues by nitrogen-fixing rhizobia. In response to environmental bacteria, the plant host secretes flavonoids (1) and a “mucigel” in which dense microcolonies of rhizobial cells become aggregated near the vicinity of root hairs that are susceptible to colonization (23). The molecular signaling between the plant and microbe that mediates these events is well characterized (1, 2), and perhaps the broad similarities between this symbiosis and the squid–vibrio association reflect the presence of shared, ancient molecular mechanisms. Interestingly, both colonization events involve the use of host exudates that are generally associated with antimicrobial responses—i.e., mucus secretion in animals (24) and flavonoid production in plants (25). Thus, processes that serve as host “defense responses” appear to play additional roles in the more common cross-taxa interactions that occur between eukaryotes and their commensal and mutualistic bacterial partners.
The discovery that the squid–vibrio association uses ciliary-mucus currents to harvest symbionts provides not only an example of what may be a generally important phenomenon during symbiont colonization of host tissues in aquatic environments, but also a model system for discovering the mechanisms underlying this process. This phenomenon presents several interesting questions for future investigation: how do bacteria induce the formation of this matrix, and what are the signal transduction pathways through which the host responds; are V. fischeri cells enriched within the aggregates; what is the basis of the nascent symbionts' required residence time within the aggregates; and, how do the symbionts direct their migration from these aggregates to their target tissue? The answers to these questions are relevant not only to understanding the interactions between V. fischeri and its host, but also to discovering how other associations have evolved the use of ciliary mucus currents and specific symbiont responses to achieve intimate long-term relationships.
Acknowledgments
We thank J. Handelsman, S. Kathariou, and K. Visick for supplying strains and plasmids and C. Unabia and M. Hadfield for supplying fluorescent labeled lectins. Support was provided by grants from the National Science Foundation (IBN-9904601) and the National Institutes of Health (RO1-RR12294).
Abbreviations
- LSM
laser-scanning confocal micrograph
- GFP
green fluorescent protein
References
- 1.VanRhijn P, Vanderleyden J. Microbiol Rev. 1995;59:124–142. doi: 10.1128/mr.59.1.124-142.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fisher R F, Long S R. Nature (London) 1992;357:655–660. doi: 10.1038/357655a0. [DOI] [PubMed] [Google Scholar]
- 3.Haygood M G. Crit Rev Microbiol. 1993;19:191–216. doi: 10.3109/10408419309113529. [DOI] [PubMed] [Google Scholar]
- 4.Gros O, Darrasse A, Durand P, Frenkiel L, Moueza M. Appl Environ Microbiol. 1996;62:2324–2330. doi: 10.1128/aem.62.7.2324-2330.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muller-Parker G, D'Elia C F. In: Life and Death of Coral Reefs. Birkeland C, editor. New York: Chapman and Hall; 1997. pp. 96–113. [Google Scholar]
- 6.Ruby E G, Lee K-H. Appl Environ Microbiol. 1998;64:805–812. doi: 10.1128/aem.64.3.805-812.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blackburn N, Fenchel T, Mitchell J. Science. 1998;282:2254–2256. doi: 10.1126/science.282.5397.2254. [DOI] [PubMed] [Google Scholar]
- 8.McFall-Ngai M J, Ruby E G. Science. 1991;254:1491–1494. doi: 10.1126/science.1962208. [DOI] [PubMed] [Google Scholar]
- 9.Montgomery M K, McFall-Ngai M J. Development (Cambridge, UK) 1994;120:1719–1729. doi: 10.1242/dev.120.7.1719. [DOI] [PubMed] [Google Scholar]
- 10.McFall-Ngai M J, Ruby E G. BioScience. 1998;48:257–265. [Google Scholar]
- 11.Visick K L, Ruby E G. In: Bioluminescence and Chemiluminescence. Hastings J W, Kricka L J, Stanley P E, editors. New York: Wiley; 1997. pp. 119–122. [Google Scholar]
- 12.Dunn A K, Handelsman J. Gene. 1999;226:297–305. doi: 10.1016/s0378-1119(98)00544-7. [DOI] [PubMed] [Google Scholar]
- 13.Brusca R C, Brusca G J. Invertebrates. Sunderland, MA: Sinauer; 1990. [Google Scholar]
- 14.Kaufman M R, Ikeda Y, Patton C, Van Dykhuizen G, Epel D. Biol Bull. 1998;194:36–43. doi: 10.2307/1542511. [DOI] [PubMed] [Google Scholar]
- 15.Southward E C. J Mar Biol Assoc UK. 1988;68:465–487. [Google Scholar]
- 16.Jeffery P K, Li D. Eur Respir J. 1997;10:1655–1662. doi: 10.1183/09031936.97.10071655. [DOI] [PubMed] [Google Scholar]
- 17.Dean D F, Bochsler P N, Carroll R C, Olchowy T W J, Neilsen N R, Slauson D O. Am J Vet Res. 1998;59:445–451. [PubMed] [Google Scholar]
- 18.Graf J, Dunlap P V, Ruby E G. J Bacteriol. 1994;176:6986–6991. doi: 10.1128/jb.176.22.6986-6991.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Finkelstein R A, Boesman-Finkelstein M, Chang Y, Hase C C. Infect Immun. 1992;60:472–478. doi: 10.1128/iai.60.2.472-478.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gray K M. Trends Microbiol. 1997;5:184–188. doi: 10.1016/S0966-842X(97)01002-0. [DOI] [PubMed] [Google Scholar]
- 21.Costerton J W, Stewart P S, Greenberg E P. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 22.Visick K L, Ruby E G. In: Cell-Cell Signaling in Bacteria. Dunny G M, Winans S C, editors. Washington, DC: Am. Soc. Microbiol.; 1999. pp. 333–352. [Google Scholar]
- 23.Gage D J, Bobo T, Long S R. J Bacteriol. 1996;178:7159–7166. doi: 10.1128/jb.178.24.7159-7166.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kato T, Owen R L. In: Handbook of Mucosal Immunology. Ogra P L, Mestecky J, Lamm M E, Strober W, McGhee J R, Bienenstock J, editors. San Diego: Academic; 1994. pp. 11–26. [Google Scholar]
- 25.Kondorosi A. In: Plant-Microbe Interactions: Molecular and Genetic Perspectives. Kosuge T, Nester E W, editors. Vol. 3. New York: McGraw-Hill; 1989. pp. 383–420. [Google Scholar]