Abstract
Class III myosins are important for the function and survival of photoreceptors and ciliary hair cells. Although vertebrates possess two class III myosin genes, myo3A and myo3B, recent studies have focused on Myo3A because mutations in the human gene are implicated in progressive hearing loss. Myo3B may compensate for defects in Myo3A, yet little is known about its distribution and function. This study focuses on Myo3B expression in the mouse retina. We cloned two variants of myo3B from mouse retina and determined that they are expressed early in retinal development. In this study we show for the first time in a mammal that both Myo3B and Myo3A proteins are present in inner segments of all photoreceptors. Myo3B is also present in outer segments of S opsin-immunoreactive cones but not M opsin dominant cones. Myo3B is also detected in rare cells of the inner nuclear layer and some ganglion cells. Myo3B may have diverse roles in retinal neurons. In photoreceptor inner segments Myo3B is positioned appropriately to prevent photoreceptor loss of function caused by Myo3A defects.
Keywords: Photoreceptors, blue cones, ganglion cells, immunocytochemistry, Myosin3A
INTRODUCTION
Among unconventional myosins, class III myosins are particularly unusual in that they probably exhibit both signaling and motor functions. A protein kinase domain is located at the N-terminus of these myosins, and in all of the class III myosins studied to date, from both invertebrates and vertebrates, this kinase has been shown to phosphorylate its own kinase and/or myosin domain as well as other substrates (Ng et al., 1996; Komaba et al., 2003; Dosé et al., 2007; Kempler et al., 2007). While no motor activity has been demonstrated for the two invertebrate class III myosins that have been studied (Hicks et al., 1996; Kempler et al., 2007), vertebrate class III myosins are molecular motors (Erickson et al., 2003; Komaba et al., 2003; Kambara et al., 2006; Dosé et al., 2007).
Class III myosin transcripts have been detected in a variety of vertebrate tissues including retina, cochlea, brain, kidney, testes, intestine and pancreas (Dosé and Burnside, 2000, 2002; Walsh et al., 2002; Dosé et al., 2003). Although their specific functions are largely unknown and may differ in different cell types, much evidence suggests class III myosins are important for the normal function and maintenance of sensory cells. Class III myosins were first discovered in Drosophila and then in Limulus; in both of these animals myosin III expression is specific to rhabdomeral photoreceptors (Edwards and Battelle, 1987; Montell and Rubin, 1988; Battelle et al., 1998). Drosophila myosin III is the ninaC gene product required for normal photoreceptor function and survival (Montell and Rubin, 1988; Porter and Montell, 1993; Porter et al., 1993, 1995; Hofstee et al., 1996; Li et al., 1998; Chyb et al., 1999). Limulus myosin III undergoes circadian changes in phosphorylation in photoreceptors (Edwards and Battelle, 1987; Edwards et al., 1990; Battelle et al., 1998; Cardasis et al., 2007; Kempler et al., 2007) and may be involved in some of the dramatic circadian changes in structure and function that occur in these photoreceptors.
Class III myosins are also present in the photoreceptors of vertebrates. Vertebrate genomes contain two distinct class III myosin genes, myo3A and myo3B (Dosé et al., 2003). Transcripts for both were cloned from retinal cDNA of fish (Dosé et al., 2003) and humans (Dosé and Burnside, 2000; 2002), and in both of these species myosin IIIA protein (Myo3A) is present in photoreceptors (Dosé et al., 2003; 2004). An additional finding that emphasizes the importance of class III myosins in sensory cells is that mutations in human myosin IIIA (hMYO3A) are linked to progressive hearing loss DFNB 30 (Walsh et al., 2002); furthermore, mMYO3A was recently localized to a region of cochlear and vestibular hair cells that defines a previously unidentified compartment at the tips of the stereocilia (Schneider et al., 2006). mMyo3A cDNA was originally cloned from whole eye cDNA but the protein was not localized to retina (Walsh et al., 2002).
Because of the association between mutations in hMYO3A and hearing loss, most studies to date have focused on this protein. The results of two recent studies that examined the motor activity of hMYO3A differ in detail, but both suggest the protein spends considerable time bound to actin, and it may be a processive motor (Kambara et al., 2006; Dosé et al., 2007). The precise functions of the kinase activity of class III myosins are not yet known, but studies of both human and fish Myo3As demonstrate that deleting the kinase domain dramatically influences acto-Myo3A interactions (Erickson et al., 2003; Lin-Jones et al., 2004; Schneider et al., 2006; Dosé et al., 2008).
MYO3A is present in human photoreceptors and vestibular hair cells (Walsh et al., 2002; Dosé et al., 2004; Schneider et al., 2006) in addition to the cochlear hair cells, yet patients with mutations in MYO3A exhibit no apparent defects in vision or vestibular function. A possible explanation for this puzzling observation is that hMYO3B may be co-expressed with hMYO3A in some cells and that there may be functional redundancy between these two proteins. These speculations cannot be evaluated without additional knowledge of the distribution and biochemistry of Myo3B.
Myo3B is the focus of this study. We describe here the cloning of two variants of myo3B from mouse retina and compare these with myo3B transcripts from humans and myo3A transcripts from mouse. We also describe the tissue distribution of mouse Myo3B (mMyo3B) transcripts and protein, the developmental expression pattern of mMyo3B transcripts and protein in retina, and the cellular and subcellular distributions of mMyo3B in retina. Additionally, we describe the distribution of the Myo3A protein in retina and provide the first evidence of co-localization of Myo3A and Myo3B in mammalian photoreceptor inner segments (IS).
MATERIALS AND METHODS
Animals
C57BL/6J mice maintained on a 12 hr light, 12 hr dark cycle were used for all experiments, and all animal procedures were approved by the University of Florida’s Institutional Animal Care and Use Committee. Eyes were enucleated from animals sacrificed by either decapitation (animals younger than postnatal day 14) or by CO2 asphyxiation followed by cervical dislocation.
Reagents
Unless otherwise specified all reagents were purchased from Fisher Scientific (Pittsburg, PA) or Sigma Aldrich (St. Louis, MO).
Cloning mmyo3B from mouse retina
We initiated the cloning of myo3s from mouse retina before myo3B was annotated in the mouse genome; therefore we followed the strategy for cloning class III myosins described by Dosé and Burnside (2000). Total RNA was isolated during the day from adult C57BL/6J mouse retinas with TRIZOL Reagent (Invitrogen, Life Technologies, CA), then reverse transcribed with Superscript II (Invitrogen) and the oligo dT primer PA 142 (GACTTCAGGCTAGCATCGATGCATGGGTCGT15). This pool of cDNA was further enriched for kinases by PCR with a degenerate forward primer corresponding to the amino acid sequence TPFWMAPE, which is highly conserved in all kinases, and a reverse primer (5′-CATCGATGCATGGGTCGT-3′), a nested primer for PA142. The kinase enriched cDNA was subsequently used as a template in PCR with degenerate primers designed against a conserved motif in kinases (GITAIE, 5′-GGNATHACNGCNATHGA-3′), and a conserved motif in myosins (NPPHIFAV, 5′-CNACNGCRAANAYRTGNGGNGGRTT-3′), to amplify a fragment that would span the kinase/myosin junction of class III myosins. PCR products from this amplification were cloned into pGEM-T (Promega) and sequenced using the BigDye termination method.
Nucleotide sequences obtained from 20 clones were compared with sequences in the GenBank, EMBL, DDBJ and PDB databases using the BLASTX program of the NCBI. Fourteen of the clones contained an identical 579 bp fragment that spanned a kinase/myosin junction and showed 66% identity to human myo3A. The sequence of this fragment was used to design gene-specific forward and reverse nested primers to amplify by RACE (rapid amplification of cDNA ends) the 5′ region of the presumptive myo3 cDNA from total mouse retinal RNA. The 5′ RACE reactions produced a single 840 bp product that extended 5′ of a predicted start codon. Nearly full length myo3 cDNAs were amplified from RACE Ready retinal cDNA (RACE Ready cDNA protocol, Ambion, Austin TX). We used two forward gene-specific primers (5′ CAGCCTCGGAGGTGAGCGGTG 3′ and 5′ CTATAACCCCATGATGCTTGGCCT 3′) designed against sequences located 5′ of the predicted start codon and including the start codon, respectively, a reverse gene-specific primer designed against myo3B sequences amplified from mouse testes cDNA that were predicted to be located near the 3′ end of the coding region (5′ CTTGGCTTCCTCTGGCTCCCT 3′), and a nested reverse gene specific primer (5′ GGTTATAGTACATGGTGTCCT 3′) (Dosé, unpublished). Three clones of the amplified product were sequenced.
The remaining 3′ end of myo3B was cloned using a forward gene-specific primer designed against sequences located near the 3′ end of the coding region (5′ CTGGTTCCGAAGAAGGTCTC 3′) and the 3′ outer primer of the 3′ RLM RACE protocol (Ambion).
Additional analyses of sequences encoding the loop2 actin binding region of the motor domain were performed. Total RNA from adult mouse retinas was isolated as described above, reverse transcribed (RLM RACE protocol, Ambion), then amplified using gene-specific primers that flank the region encoding loop2 (forward: 5′ TTCTCAATTCCTTTGACCAAAACAGGT 3′; reverse:, 5′ GTCATCATTAGGCTTAATGCAACG 3′). The PCR products were cloned into pCR 4-TOPO and sequenced.
All PCR and RACE reactions were performed using LATaq polymerase (Takara, Madison, WI) and Eppendorf Mastercycler.
Real time quantitative PCR (qPCR)
Total RNA was isolated mid afternoon as described above from the retinas of at least three light-adapted animals at each postnatal (P) age and from different tissues of adult animals between three and four months of age. The RNA was reverse transcribed with random primers using Superscript II (Invitrogen, Carlsbad, CA); reverse transcription was followed by qPCR with gene-specific primers in the presence of SYBR Green. The relative standard curve method was applied in which the myo3B transcripts detected were normalized to 18s rRNA measured in separate aliquots of the same cDNA sample. Adult retina served as the reference group. Standard curves were generated with 18s rRNA primers (forward: 5′ AGTCCCTGCCCTTTGTACACA 3′; reverse: 5′ CCGAGGGCCTCACTAAACC 3′) and each of two sets of myo3B specific primers. One set was designed to span the transition between the kinase and the myosin domains (forward: 5′ CAAAAACAGCTGGCCAAGGT 3′; reverse: 5′ CTTTCATGCCTGGTTTTAGCAA 3′); a second set was designed to amplify the loop2 region of the myosin domain (forward: 5′ AACTGTTCTCAATTCCTTTGACCAA 3′; reverse: 5′ TCGAGGATGCTGTGATCTTAG 3′). The regions amplified by both sets of primers are present in transcripts encoding both mmyo3B variants cloned from mouse retina, and both also span an intron-exon junction thus eliminating the possibility of amplifying genomic DNA. Primers were designed using the Primer Express 2.0 software (Applied Biosystems, Foster City, CA). All qPCR reactions were performed in triplicate using an ABI PRISM 7000 sequence detection system and the default one-step PCR protocol (50°C for 2 min, 95°C for 10 min, and 40 cycles of 95°C for 15 sec, 60°C for 1 min).
Antibody production
Anti-mMyo3B
cDNA encoding the tail domain of mMyo3B (amino acids 1104–1132 was amplified from reverse transcribed total mouse retinal RNA using primers that added NdeI and HindIII restriction sites to the 5′ and 3′ ends, respectively, and then subcloned into pET28-a using the NdeI and HindIII restriction enzymes. The plasmid was transformed into Rosetta 2(DE3) E. coli cells (Novagen, Madison, WI) and the antigen was expressed as an insoluble protein with an N-terminal His6 tag. The antigen was solubilized in 6 M urea, enriched by standard Ni+ chelation chromatography, purified further by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and injected into rabbits as described previously (Battelle et al., 2001). The antiserum from one of the rabbits (4J10 mMyo3B) was used for all mMyo3B experiments in this study.
In preliminary studies, the antiserum stained Western blots of the antigen and a truncated form of the antigen consisting of the C-terminal end of the tail domain (amino acids 1226–1322 in Figure 1). Thus the antiserum should detect the gene products of both mmyo3B isoforms cloned from retina because its immunoreactivity does not require the N-terminal half of the tail domain or the region deleted in variant 2.
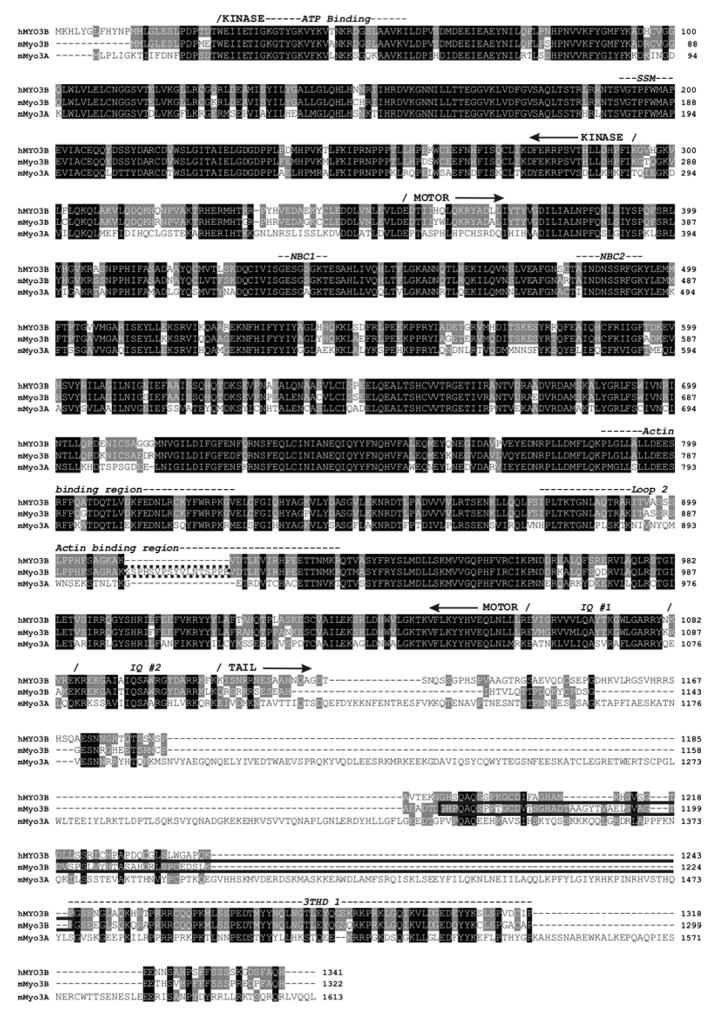
Figure 1.
Amino acid sequence alignment of full length mouse Myo3B (mMyo3B) with human MYO3B (hMYO3B) and mouse Myo3A (mMyo3A) (Clustal X, Young, 1976). Black boxes indicate amino acids that are identical or similar in all three sequences, and gray boxes indicate amino acids that are identical or conserved in two of the three sequences. The amino acid numbering for each sequence is indicated at the right. The amino acids inserted in loop2 and deleted from the tail of variant 2 cloned from mouse retina are shown outlined with dotted and solid lines respectively. The kinase, myosin, and IQ domains are labeled and were predicted using the InterProScan program from EMBL-EBI. The ATP-binding region (Prosite) and the ste20 kinase family signature motif (SSM, Dan et al., 2001) within the kinase domain are also labeled. The nucleotide binding cleft (NBC1 or P-loop, and NBC2), the actin binding region, and the loop2 actin binding region located within the myosin domain were predicted from an alignment with chicken skeletal muscle myosin (Accession # P13538, Sellers, 1999). Tail homology domain I (THDI) is a region within the tail domain that shows high amino acid identity among the three sequences.
Anti-mMyo3A
cDNA encoding exon 30 of mMyo3A (amino acids 1140–1424 in Figure 1) was amplified from genomic DNA, and was subcloned into pET28-a. The mMyo3A fragment was expressed as a His-tagged fusion protein in Rosetta 2(DE3) E. coli cells and enriched by Ni+ chelation chromatography as described above. The resulting antigen was dialyzed against PBS, mixed with an equal volume of RIBI’s adjuvant and injected subcutaneously into two rabbits. The antiserum from one of the animals was used for all mMyo3A experiments.
Myo3B antiserum absorption controls
Attempts were made to affinity purify Myo3B-specific antibodies with procedures modified from Bretscher (1983). Urea solubilized mMyo3B antigen that had been enriched by Ni+ affinity chromatography as described above was dialyzed against H2O and dried under vacuum in a SpeedVac concentrator. The antigen (5–10 mg) was dissolved in 1 ml of borate buffer (0.1 M H3BO3, 0.025 M NaB4O7, 0.075 M NaCl, pH 8.4) with 0.1% SDS. After 1 g of CNBr-activated sepharose (3–4 ml volume when swollen) had been washed and activated as directed by the manufacturer, dissolved antigen was incubated with shaking with the resin overnight at room temperature. The next morning the resin was washed sequentially with borate buffer containing 0.1% SDS, borate buffer, and 0.1 M NaCl; then it was incubated for 2 hr with 1 M ethanolamine-HCl to block unbound sites, and washed sequentially with 0.1 M NaCl, 0.01 M HCl, and borate buffer. The mMyo3B antiserum (100 μl, diluted 1:10 in TBS) was incubated with the resin overnight with shaking at 4°C then the resin was washed with at least 50 column volumes of the borate buffer described above adjusted to pH 7.4 and then with 5–10 volumes of 0.25 M glycine pH 2.7 to elute the specific antibodies. The resulting elutes exhibited very low OD280 readings (≤0.05), nevertheless they were concentrated to the initial antiserum volume and tested on Western blots of retinal homogenates. No immunostaining was detected indicating that no specific, non-denatured immunoglobulins were recovered from the column. Attempts to elute specific antibodies using glycine pH 9.5, triethylamine pH 11.5, lithium chloride, magnesium chloride, and water also failed. Therefore the affinity column was used to produce serum from which the specific antibodies had been removed. Total antiserum was incubated with resin linked to the mMyo3B antigen as described above. The flow through from the column and the borate buffer washes were combined, concentrated to the initial volume of antiserum applied to the column and filter dialyzed into PBS.
As additional controls, separate aliquots of the mMyo3B antiserum were processed in parallel as described above using resin with no protein attached or resin coupled to an irrelevant antigen: a heterologously expressed His-tagged polypeptide with the sequence of the C-terminus of Limulus opsin1 (Battelle et al., 2001). Western blots and tissue sections immunostained with total antiserum or antiserum that had been processed with each of the three different resins were compared to determine specific staining.
Affinity purification of mMyo3A antiserum
mMyo3A antiserum was processed as described above through an affinity column to which the mMyo3A antigen had been bound. In this case, the low pH glycine eluate contained active antibodies. This eluate was neutralized, concentrated to 1 ml and used on Western blots and tissue sections. To evaluate further the specificity of the affinity purified antibody, it was incubated with mMyo3A antigen that had been blotted on to nitrocellulose (Battelle et al., 2001).
Western blot analyses
Tissues were dissected and homogenized with a glass-glass homogenizer in ice cold homogenizing buffer (HB: 50 mM MOPS pH 7.3, 1 mM EDTA, 1 mM EGTA, 160 mM KCl, 1 mM DTT, 10 mM ATP, 10 mM MgCl2, 0.25 U/ml aprotinin, 100 μM leupeptin, 1 μM pepstatin, 1 mM calpeptin, 5 μM calpain inhibitor III, 200 μM PMSF, 1 mM benzamidine, 2 mM phenanthroline, 10 μg/μl TAME, 5 μl/ml P8340), and the homogenate was centrifuged for 20 min at 100,000 × g in an airfuge to obtain the soluble fraction. An aliquot of each soluble fraction was removed for protein determinations using either a Lowry (Lowry et al., 1951) or a Coomassie Plus assay (Thermo-Scientific, Rockford, IL). Proteins in the remaining supernatant were denatured by sonication in a bath sonicator (W-225, Heat Systems-Ultrasonics, Farmingdale, NY) in the presence of 0.25 volumes of 4 × SDS sample buffer, and the SDS-denatured proteins were separated by SDS-PAGE on 7.5% acrylamide gels. In each experiment, 40 μg of protein were loaded onto each lane of the gel. After the proteins were separated they were transferred to Immobilon-P™ PVDF using standard protocols and fixed with fast green. The membranes were then rinsed, blocked and incubated with primary antibodies and then an alkaline phosphatase conjugated secondary antibody (Jackson ImmunoResearch, West Grove, PA) as described previously (Battelle et al., 2000). The secondary antibody was visualized using the Alkaline Phosphate Conjugate Substrate Kit from BioRad (Hercules, CA). Total mMyo3B antiserum and mMyo3B antisera that had been processed through the three affinity columns as described above were diluted 1:500. Affinity purified and absorbed mMyo3A antibodies were diluted 1:200 and the secondary antibody was diluted 1:5000.
To quantify mMyo3B in soluble fractions of tissue homogenates, Western blots immunostained with mMyo3B antiserum were scanned (hp200c) and the intensities of the mMyo3B immunoreactive bands were quantified using ImageQuant™ software (GE Healthcare, Piscataway, NJ). The intensity of the mMyo3B immunoreactive band in each extract was normalized to the mMyo3B immunoreactive band detected in an extract of adult retinas on the same blot.
Tissue fixation
Right and left eyes from adult animals (P35-P60) were processed separately, and the superior half of each eye was marked to maintain orientation. The scleras of enucleated light-adapted eyes were punctured, and the eyes were fixed overnight at 4°C in 4% paraformaldehyde in phosphate-buffered saline (PBS, pH 7.4). The next morning the cornea from the inferior half of the eye and the lens were removed, and after continued incubation in the fixative for 1 hr, the eyes were rinsed in PBS and incubated for at least 16 hr at 4°C in 30% sucrose in PBS containing 0.02% NaN3. Eyecups were embedded and frozen in OCT (Tissue Tek) and 18 μm frozen sections were cut using a Leica CM3050S cryostat. Sections were collected on gelatin-coated slides and stored at −20°C until further processing.
Immunocytochemistry: heat-induced antigen retrieval, immunostaining, and image collection
Heat-induced antigen retrieval was routinely used to detect mMyo3B-like immunoreactivity (ir) on frozen sections of fixed tissue and to detect mMyo3A-ir in selected experiments as indicated in the legends to figures. For antigen retrieval, retinal sections were incubated for 10 min in 10 mM sodium citrate, pH6 and heated to 85–95°C in a water bath (Kawai et al., 1994). After sections cooled to room temperature, they were washed in H2O and immunostained as described in Calman et al. (1991).
The dilutions of the primary antibodies used were determined empirically. The mMyo3B antiserum was diluted between 1:100 and 1:500 as indicated in the legends to the figures. The concentrated column eluate containing affinity purified mMyo3A antibody was used undiluted without antigen retrieval or at a 1:5 dilution following antigen retrieval. Antibodies directed against blue (sc-14363, Lot # L1906) and red/green (sc-22117, Lot # I1206) opsins were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA) and used at 1:100, dilution. The rhodopsin antibody (Adamus et al., 1991) was used at 1:200 dilution. The appropriate secondary antibodies conjugated to either Alexa Fluor 488 or 546 (Invitrogen, Carlsbad, CA) were used at a dilution of 1:200 or 1:400. Peanut agglutinin and phalloidin, both labeled with Alexa-546, were used at a 1:200 dilution. To visualize nuclei, sections were incubated for 10 min in DRAQ-5 (Biostatus Limited, Shepshed, UK) diluted to 1:1000 in TBS (20 mM Tris-HCl, pH 7.5, 500 mM NaCl).
Fluorescent images were collected using a Leica laser scanning confocal microscope (LSCM SP2) (Leica Microsystems, Bannockburn, IL). Each confocal image shown is either a 1 μm optical section or the maximum projection of an optical stack of 3–5 1 μm thick optical sections as indicated in the figure legends. Fluorescent images of tissue sections incubated with antisera that had been preincubated with and without antigen were compared using identical gain settings and included the same number of 1 μm optical sections. Double-labeled images were collected using a sequential scanning protocol to avoid bleed through from one channel to the other. Digital images produced by the confocal microscope were assembled using CorelDRAW Graphics Suite X3 (Corel; Ottawa, ON, Canada).
RESULTS
Cloning mMyo3B (Accession numbers AY830392 and AY830393)
We sequenced three clones of mmyo3B that begin 5′ of the start codon and end with nucleotides encoding amino acid 1274 (Fig. 1). The 3′ end of the open reading frame and the 3′ untranslated region were sequenced from clones of subsequent 3′ RACE products that extended from nucleotides encoding amino acid 1226 (Fig. 1) to the poly A tail. Figure 1 shows the predicted full length protein encoded by these sequences aligned with human MYO3B and mouse Myo3A.
Two of the three nearly full length clones were identical to one another (variant 1); the third clone (variant 2) had a 51 nt insert in a region of the transcript encoding loop2 of the motor domain (outlined with a dotted line in Fig. 1) and a 183 nt deletion in a region encoding the tail domain (outlined with a solid line in Fig. 1). We anticipated finding splice variants within the tail of mmyo3B because multiple tail domain splice variants were identified in human myo3B (Dosé and Burnside, 2002). We did not expect a splice variant in loop2 although loop2 splice variants have been described in nonmuscle class II myosins (Takahashi et al., 1992; Li et al., 2008). To confirm that both motor domain splice variants of myo3B are represented in retinal RNA, we examined 58 clones of a PCR product that was amplified from reverse transcribed retinal RNA with primers that flanked the region encoding loop2. All of the clones had inserts of either approximately 300 nt or about 250 nt, except for one which was considerably smaller (about 200 nt). The sequences of 16 representative clones with the longest insert matched the loop2 sequence of variant 2; the sequences of 10 representative clones with the shorter insert matched the loop2 sequence of variant 1. The sequence of the one very small insert was unrelated to mmyo3. Thus, transcripts encoding two variants of loop2 in the myosin motor domain are present in retinal RNA. If other splice variants of this region are present, they are rare. Other tail domain splice variants of mmyo3B may be present in the retina, but we did not search for these in this study.
A comparison of our sequences with sequences in the GenBank, EMBL and DDBJ databases using the BLASTn program of the NCBI revealed that they are most similar to hmyo3B. Variant 1, with the shorter loop2 and the longer tail, corresponds to annotated myo3B sequences on chromosome 2 of the mouse genome (http://vega.sanger.ac.uk/Mus_musculus/geneview?gene=OTTMUSG00000013112;db=core). The transcript consists of 34 exons, and the predicted start codon is within exon 2. In variant 2, the 51 nt insert in the region encoding loop2 occurs between predicted exons 23 and 24, and the inserted sequence is located within the intron between exons 23 and 24 close to exon 24. The deletion in the tail domain is a deletion of predicted exons 30 and 31.
During our cloning attempts we did not detect transcripts for myo3A although a myo3A is clearly expressed in retina (Fig. 5).
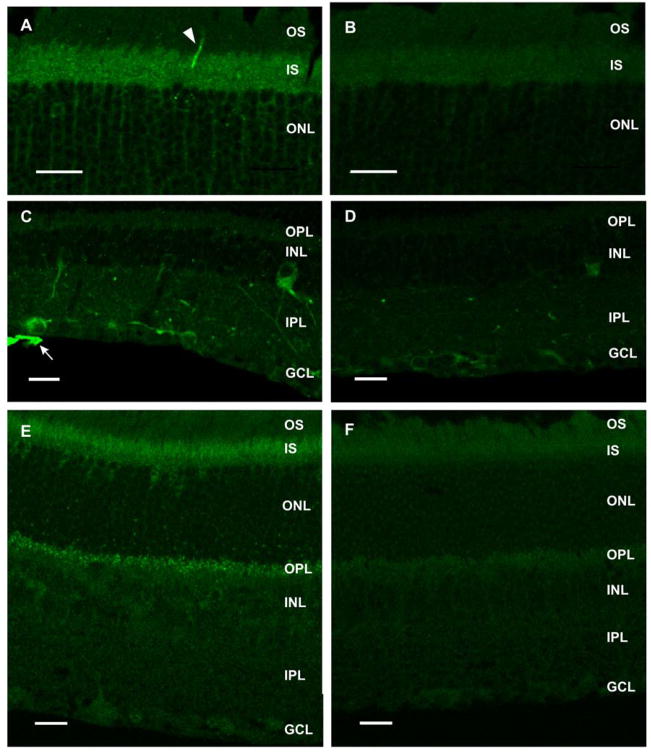
Figure 5.
mMyo3B and mMyo3A-ir in adult mouse retina. Confocal images of fixed frozen sections processed for antigen recovery. Images A–D are 1 μm thick optical sections whereas E and F are the maximum projections of optical stacks of 4 images. A–D: mMyo3B-ir in retina. Sections were either incubated with the mMyo3B antiserum that had been preincubated with the His-tagged C terminus of Limulus opsin1 (A, C) or with the Myo3B antigen (B, D). Antisera dilutions used were 1:250 for A and B, and 1:500 for C and D. The values of the gain were the same between A and B and C and D. A: Intense mMyo3B-ir is observed in the outer segment (OS) of a cone (arrowhead). mMyo3B is also present but less concentrated in the inner segments (IS) of all photoreceptors and in cytoplasm surrounding photoreceptor nuclei in the outer nuclear layer (ONL). B: A different section from the same eye incubated with antiserum that had been preincubated with mMyo3B antigen showing only background staining. C: mMyo3B is present in cells in the inner nuclear (INL) and ganglion cell (GC) layers and labels GC axons intensely (arrow). D: Section from the same eye incubated with antiserum that had been preincubated with mMyo3B antigen showing much reduced staining. E–F: mMyo3A-ir in retina. Sections were incubated either with the mMyo3A affinity purified antibody or affinity purified antibody that had been preincubated with mMyo3A antigen. E: Overview of the staining observed in the retina using affinity purified mMyo3A antibody. mMyo3A is present in the outer portion of the IS as well as in putative photoreceptor terminals shown as punctate in the outer plexiform layer (OPL). F: Section from the same eye incubated with mMyo3A antibody that had been preincubated with mMyo3A antigen. Only background staining is observed. GCL, ganglion cell layer; IPL, inner plexiform layer. Bars=20 μm.
Figure 1 shows a ClustalX alignment of full length mMyo3B as determined from this study (variant 1 plus the loop2 insert) with mMyo3A (Accession # AAM34501) and variant 2 of hMYO3B (NM138995). Dosé and Burnside (2002) identified 6 hMYO3B variants which are identical in their kinase and myosin domains and vary in their neck and tail domains due to alternative splicing. Of these, variant 2 is most similar to the mouse sequences we cloned. Pairwise alignments (EMBOSS) of the predicted amino acid sequences of mMyo3B with variant 2 of hMYO3B, and mMyo3B with mMyo3A show that the Myo3Bs from mouse and humans are more similar to one another (83% identical and 88% similar) than are Myo3B and 3A both from mouse (47% identical and 60% similar). Table 1 summarizes the percent identity and similarity of amino acids in different regions of mMyo3B compared to hMYO3B and mMyo3A.
Table 1.
Amino acid homology of mMyo3B and hMYO3B and of mMyo3B and mMyo3A
| Domaina | mMyo3B | hMyo3Bc | mMyo3Ad | ||
|---|---|---|---|---|---|
| Amino acidsb | Identical | Similar | Identical | Similar | |
| Full-length | 1–1322 | 83 | 88 | 47 | 60 |
| Kinase | 15–281 | 95 | 98 | 75 | 87 |
| Myosin | 325–1063 | 90 | 94 | 63 | 80 |
| Myosin Loop2 Le | 868–933 | 67 | 72 | 29 | 45 |
| Myosin Loop2 Sf | 82 | 88 | 35 | 51 | |
| IQ1 | 1064–1086 | 86 | 95 | 40 | 72 |
| IQ2 | 1090–1103 | 91 | 91 | 47 | 69 |
| Entire Tail | 1104–1322 | 51 | 60 | 13 | 18 |
| C-terminal Tail | 1226–1322 | 80 | 85 | 25 | 34 |
| 3 THD1 | 1227–1300 | 86 | 90 | 38 | 53 |
Domains listed are indicated in Figure 1
Amino acid numbers below are based on those for mMyo3B shown in Figure 1 and indicate the amino acids included in each domain.
Numbers are the percent of amino acids that are identical and similar in a pairwise comparison of mMyo3B and mMyo3A (EMBOSS pairwise alignment).
Numbers are the percent of amino acids that are identical and similar in a pairwise comparison of mMyo3B and mMyo3A (EMBOSS pairwise alignment).
Longer variant of loop 2 in mMyo3B variant 2
Shorter variant of loop 2:868–933 minus the 17 amino acid insert highlighted in Figure 1 found in mMyo3B variant 1
The kinase and myosin head domains of the three Myo3 sequences are the most conserved, with the Myo3B sequences from mouse and humans showing a strikingly high level of identity in these regions: kinase (95%) and myosin head (90%). Even within loop2 of the myosin head domain, a region which is one of the least conserved among myosins (Sellers, 1999), the loop2 sequence in mMyo3B variant 1 is 82% identical to human Myo3Bs. The tail domains of the three sequences are considerably less conserved, but the C-terminal portions of their tail domains and, in particular, a small region within this C-terminal region corresponding to the tail homology domain I (3THDI) identified by Dosé et al. (2003), show higher levels of identity than the tail as a whole. The identity between mouse and human 3B in this region is again strikingly high. Like class III B myosins of other species, mMyo3B lacks a second THD (3THDII) located at the C-terminus of Myo3As (Dosé et al., 2003). These sequence comparisons suggest that the biochemical properties and functions of mouse and human Myo3B will be similar to one another and may be different from those of Myo3As.
Quantification of mmyo3B transcript levels in different adult tissues and in postnatally developing retina
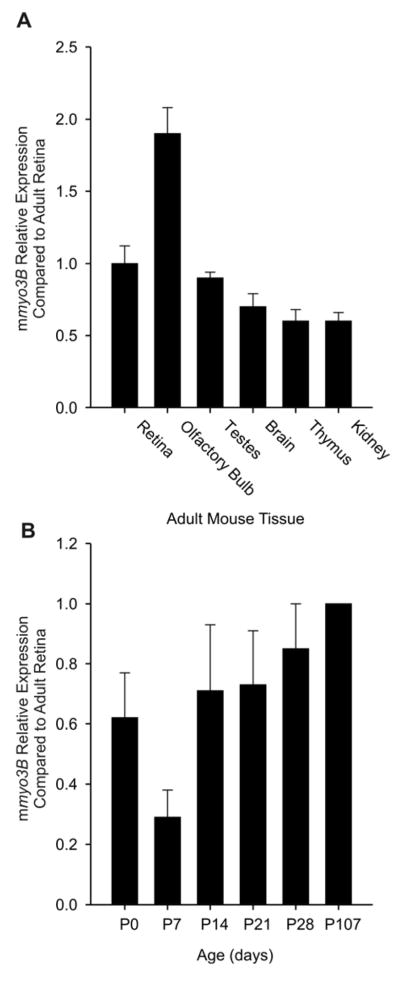
Figure 2A shows the results of a single qPCR assay of the relative levels of myo3B transcripts in various adult mouse tissues expressed relative to the level measured in adult retina. Similar results were obtained from a replicate assay of tissues from a second group of animals. mmyo3B transcripts were detected in a number of different tissues but not in all tissues examined. In the olfactory bulb, transcript levels were nearly twice those measured in retina, in testes they were similar to those in the retina, but in brain tissue from which the olfactory bulb had been removed, thymus and kidney, transcript levels were somewhat lower than those in retina. Olfactory epithelium, cornea, lens, eye cup tissues that remain after removing the retina, intestine, ovary, and liver did not exhibit detectable levels of mmyo3B transcripts.
Figure 2.
qPCR assays of A: Relative levels of mmyo3B transcripts in RNA extracted from various tissues and B: Relative mmyo3B transcripts in RNA extracted from retinas at different times during postnatal development. All tissues were dissected in the afternoon from light-adapted animals. RNA was isolated and reverse transcribed as described in Methods. Measurements of mmyo3B transcript levels were first normalized to the level of 18s rRNA measured separately on the same plate from the same RNA extract using the primers described in Methods. The normalized values obtained from different tissues or retinas of different ages were plotted relative to the normalized values obtained from the adult (P107) retina. A: All tissues were from adult animals. The primers amplified a region of the mmyo3B transcript encoding the transition between the kinase and myosin domains, and should detect all mmyo3B isoforms. The results of a single qPCR assay are shown. The means ± the standard errors of the means plotted are for three determinations of the same RNA sample (technical replicates). Similar relative levels of mmyo3B transcripts were observed in a separate qPCR assay using RNA extracted from tissues of a second group of animals. mmyo3B transcripts were detected in retina, olfactory bulb, testes, brain tissue from which the olfactory bulbs had been removed, thymus, and kidney. B: Retinas were dissected from animals at the different postnatal (P) ages indicated. The primers amplified a region of the mmyo3B transcript encoding the 5′ end of loop2 in the motor domain and that spanned the junction between exons 22 and 23. They should amplify all myo3Bs expressed in retina. Shown are the means ± the standard errors of the means of qPCR assays performed on three different groups of animals. Samples from each group were assayed in triplicate. A one-way ANOVA revealed no statistically significant change in relative mmyo3B transcript levels during development.
We are particularly interested in the function of mMyo3B in the retina; therefore we also examined the relative levels of its transcripts during postnatal retinal development (Fig. 2B). The relative mmyo3B transcript levels do not change significantly during postnatal retinal development (one-way ANOVA); although at P7, they were consistently lower than at any other time. The physiological relevance of this dip is not yet clear. Most importantly these assays revealed that mmyo3B is expressed at birth which is before most rod photoreceptors differentiate and before opsin mRNA is first detected (Liou et al., 1994).
mMyo3B and mMyo3A antibody characterization
mMyo3B antiserum
Affinity purification of Myo3B antibodies failed because Myo3B antibodies bound irreversibly to the Myo3B affinity column used in the purification protocol (see Materials and Methods for more details). Therefore, we used Myo3B antiserum in all of our studies. Myo3B antiserum stained several bands on Western blots of retinal soluble extracts (Fig. 3A, lane S). One of these was a major band with an apparent molecular mass of approximately 130 kDa which is close to the predicted sizes of the two variants of mMyo3B cloned (150 and 146 kDa). This immunostained band was depleted on replicate blots incubated at the same time with antiserum that had been preincubated with mMyo3B antigen (Fig. 3A, lane AM) but not on blots immunostained with antiserum preincubated with Limulus opsin1 antigen (Fig. 3A, lane AO) or no antigen (data not shown). This demonstrates that the 130 kDa immunoreactive band is mMyo3B, and that the staining of the other bands is non-specific.
Figure 3.
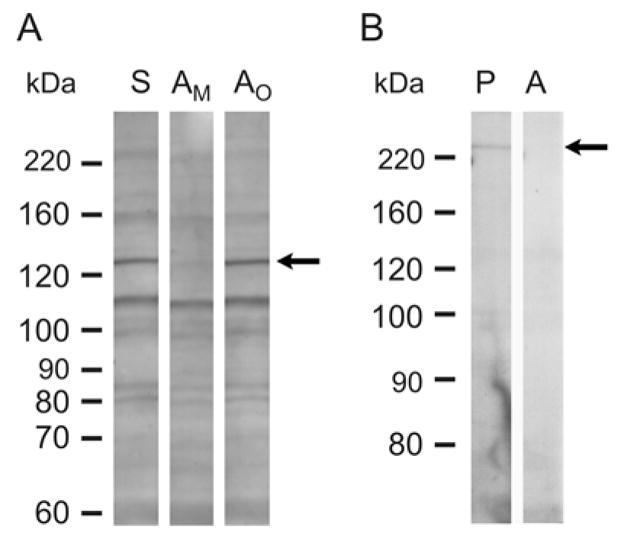
Western blots of soluble proteins from adult retinas immunostained with mMyo3B or mMyo3A. A. mMyo3B staining with total antiserum (S) and mMyo3B antiserum preincubated with Myo3B antigen (AM) or a His-tagged polypeptide with the sequence of the C-terminus of Limulus opsin1 antigen (AO). Antisera were used at a 1:500 dilution. The mMyo3B antiserum (S) and the antiserum preincubated with Limulus opsin1 (AO) immunostained the same multible protein bands in the retinal extract. On a replicate Western blot incubated in parallel with antiserum preincubated with Myo3B (AM) antigen a 130 kDa immunoreactive band (arrow) was selectively eliminated. B. mMyo3A staining with affinity purified antiserum (P) and affinity purified antiserum that had been preincubated with Myo3A antigen (A). Antisera were used at a 1:200 dilution. The immunopurified antiserum (P) stained a single band at 220 kDa which was not detected on blots incubated with antiserum that had been preincubated with Myo3A antigen. Molecular weight standards are shown on the left of each blot.
mMyo3A antiserum
Affinity purified mMyo3A antibody stained a single band of approximately 220 kDa on a Western blot (Fig. 3B, lane P), close to the predicted size of mMyo3A (185 kDa). The band was depleted on a replicate blot of affinity purified antibody that had been preincubated with mMyo3A antigen (Fig. 3B, lane A).
Quantification of mMyo3B protein in different tissues and the developing retina using Western blots
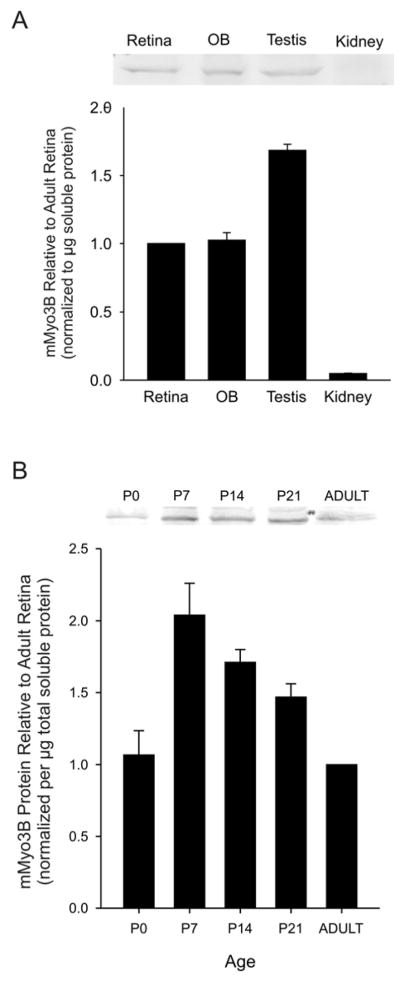
Assays for mMyo3B-like immunoreactivity (mMyo3B-ir) on Western blots of soluble extracts of retina, olfactory bulb, testis and kidney, four tissues with relatively high levels of mmyo3B transcripts (Fig. 2A), showed that the concentration of mMyo3B was significantly higher in testis (one-way ANOVA followed by a two-tailed T-test) compared to the other tissues examined (Fig. 4A) and that mMyo3B concentrations in the retina and olfactory bulb were similar. Surprisingly, we did not detect mMyo3B-ir in kidney extracts. The concentration of mMyo3B may be below our level of detection; alternatively, mMyo3B isoforms expressed in kidney may not be detected with our antiserum.
Figure 4.
A: (Upper Portion). Representative Western blot showing mMyo3B-immunostained protein in soluble extracts of various tissues from adult mice (P35-P60). Each lane was loaded with 40 μg of protein. Tissues from three different sets of animals were assayed and each biological replicate was assayed at least in triplicate. The mMyo3B immunoreactive bands were quantified using densitometry and normalized to the mMyo3B immunoreactive band in the retinal extract on the same blot. (Lower Portion). Average results ± the standard errors of the means of the biological replicates. The concentration of mMyo3B was highest in soluble extracts of testis and somewhat lower in extracts of retina and olfactory bulb. mMyo3B-ir was not detected in the kidney. B: (Upper Portion). Representative Western blot showing mMyo3B-immunostained protein in soluble extracts of retinas dissected in the afternoon from light adapted animals at different postnatal ages (P0-Adult). Three different groups of animals were assayed and each biological replicate was assayed at least in triplicate. Each lane contained 40 μg of protein. The mMyo3B immunoreactive bands were quantified using densitometry and normalizing to the mMyo3B band detected in the extract of the adult retinas analyzed on the same blot. (Lower Portion). Average results ± the standard errors of the means of the biological replicates. The concentration of mMyo3B in soluble retinal extracts peaked at P7-P14 and gradually decreased after P21 and until adulthood.
In developing retina, mMyo3B was clearly detected at P0 (Fig. 4B), which is consistent with our detection of the transcript in the retina at this age (Fig. 2B). The concentration of mMyo3B among soluble proteins increased significantly from P0 to P7 (one-way ANOVA followed by a two-tailed T-test), remained high through P21, and then fell to the adult (P35-P60) concentration which was the same as that measured at P0.
Cellular distribution of mMyo3B and mMyo3A in the adult retina
Validation of immunostaining on retinal sections
mMyo3B was detected on retinal sections by comparing the immunostaining obtained with mMyo3B antiserum to that obtained with mMyo3B antiserum that had been preincubated with mMyo3B antigen, an irrelevant antigen (Limulus opsin1) or no antigen. Retinal sections incubated with mMyo3B antiserum or with mMyo3B antiserum that had been preincubated with either Limulus opsin 1 or no antigen showed the same distribution of mMyo3B-ir. Figures 5A and 5C show the distribution of mMyo3B-ir obtained with antiserum that had been preincubated with Limulus opsin1. Figures 5B and 5D show images of sections incubated in parallel with mMyo3B antiserum that had been preincubated with the mMyo3B antigen.
In the outer retina, mMyo3B-ir was detected in photoreceptor IS and in the outer nuclear layer (ONL) in cytoplasm surrounding photoreceptor cell nuclei. However, mMyo3B-ir was most intense in some photoreceptor outer segments (OS) (Fig. 5A) with the distribution and appearance of cone OS. In the inner retina, mMyo3B-ir was detected in a few cells in the inner nuclear layer (INL) and in some cells in the ganglion cell layer (GCL) (Fig. 5C). The most intense staining was seen in GC axons (Fig. 5C). The immunoreactivity observed in each retinal layer was either eliminated or greatly reduced on sections incubated with the same concentration of antiserum that had been preincubated with mMyo3B antigen (Fig. 5B and 5D). Thus, the immunostaining observed on tissue sections is specific for mMyo3B.
mMyo3A-ir was detected in the outer portion of the IS and in the cytoplasm surrounding some photoreceptors nuclei positioned adjacent to the outer limiting membrane where most cone cell bodies are typically located (Carter-Dawson and LaVail, 1979). Punctate staining was also observed in a region of the OPL that contains photoreceptor terminals (Fig. 5E). No mMyo3A-ir was observed on sections incubated with the same concentration of affinity purified antibody that had been preincubated with antigen (Fig. 5F) indicating that the mMyo3A-ir observed on sections is specific.
Myo3B in photoreceptors
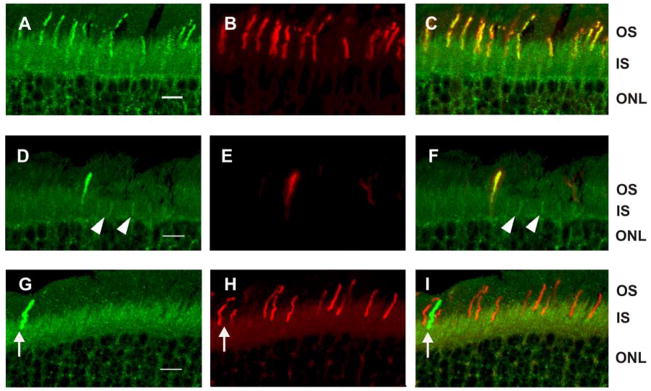
When the distribution of mMyo3B in photoreceptors was examined separately in ventral (Fig. 6, A–C) and dorsal (Fig. 6, D–I) retina, many mMyo3B immunoreactive OS were seen in the ventral retina and fewer in the dorsal retina. Cones expressing short wavelength sensitive opsin (S opsin) are also abundant in the ventral retina and decline in number along the ventral-dorsal axis (Szél et al., 1992; Applebury et al., 2000; Haverkamp et al., 2005); therefore we tested whether mMyo3B might be uniquely present in the OS of S opsin immunoreactive cones by double-labeling retinal sections for mMyo3B and either S opsin or midwavelength sensitive opsin (M opsin). mMyo3B-ir and S opsin-ir consistently co-localized in the OS of cones in both the ventral (Fig. 6, A–C) and dorsal retina (Fig. 6, D–F), but mMyo3B was not detected in M opsin immunoreactive OS in the dorsal retina (Fig. 6, G–I). Furthermore, no M opsin-ir was detected in the few mMyo3B immunoreactive OS in the dorsal retina (Fig. 6, G–I).
Figure 6.
mMyo3B is concentrated in S opsin immunoreactive OS. Fixed frozen sections were incubated with mMyo3B antiserum (1:100 dilution) and either blue- or red/green-sensitive opsin antibody (1:100 dilution). Each image is the maximum projection of an optical stack of 3–5 images (1 μm thick optical sections). Additionally, scanning from each channel was done in sequence to avoid bleed through from one channel to the other. A–C: mMyo3B (A, green) and S opsin-ir (B, red) in the ventral retina. C is a merged image of A and B. mMyo3B is concentrated in cone OS that are also S opsin immunoreactive. mMyo3B-ir is also present in all photoreceptor IS and appears more concentrated in cone IS. D–F: mMyo3B (D, green) and S opsin-ir (E, red) in the dorsal retina. F is the merged image of D and E. mMyo3B is highly concentrated in one cone OS, which is also labeled with S opsin. mMyo3B is also present in other putative cone IS (arrowheads). G–I: mMyo3B (G, green) and M opsin-ir (H, red) in the dorsal retina. I is the merged image of G and H. mMyo3B is present in photoreceptor IS and is concentrated in one cone OS (arrows) in which M opsin is not detected. mMyo3B-ir is not detected in OS that are M opsin immunoreactive. Abbreviations as in Fig. 5. Bars=10 μm.
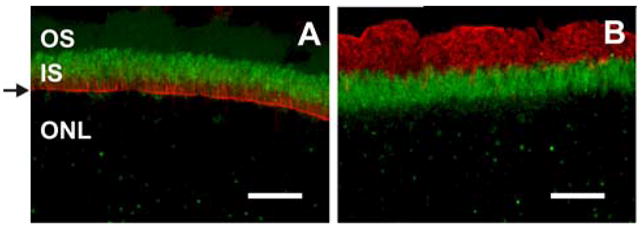
Although the mMyo3B staining pattern of photoreceptor OS was clearly different between dorsal and ventral retina, the staining pattern of IS was similar (see for example Figures 6A, 6D, and 7A). IS consistently showed less intense mMyo3B-ir than the S opsin immunoreactive OS, but more strikingly, in both the ventral and dorsal retina, some IS with the predicted distribution of cones stained more brightly for mMyo3B than the more abundant rod IS. This suggests that compared to rods, mMyo3B is more concentrated in the IS of all cones. To test this, we double-labeled photoreceptors in the dorsal retina for mMyo3B and fluorescently labeled peanut agglutinin which binds to galactose-galactosamine disaccharide moieties in the interphotoreceptor matrix surrounding the outer and IS of all cones (Blanks and Johnson, 1983). As is seen in Figure 7, each of the more brightly mMyo3B labeled IS in the dorsal retina is also labeled with peanut agglutinin. Thus, mMyo3B is present in the IS of all rods and cones, is more concentrated in the IS of cones compared to rods and is detected in S opsin but not M opsin immunoreactive OS. To test whether Myo3B-ir in rods extended to the OS we immunostained retinas with Myo3B and rhodopsin, a marker for rod OS. mMyo3B and rhodopsin did not colocalize (supplemental figure 1); therefore we conclude that Myo3B is not present in rod OS.
Figure 7.
mMyo3B labels all cone IS more intensely than rod IS. Fixed, frozen sections of the dorsal retina were incubated with the mMyo3B antiserum (1:100 dilution) and Alexa-546 labeled peanut agglutinin (PNA) (1:200 dilution). Images were obtained as described in Fig. 6. mMyo3B-ir (A, green) is observed in one brightly labeled cone OS (arrowhead). All of the photoreceptor IS exhibit mMyo3B-ir but some IS are more brightly mMyo3B immunoreactive (arrows). The mMyo3B positive OS and the more brightly Myo3B immunoreactive IS stain with PNA (B, red). C is the merged image of A and B. Abbreviations as in Fig. 5. Bar=10 μm.
Myo3A in photoreceptors
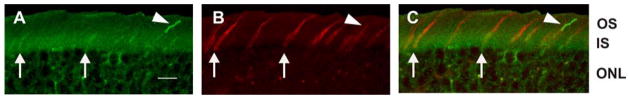
Like Myo3B, Myo3A was detected in the IS of all photoreceptors (Fig. 5), however the distributions of Myo3A and Myo3B within IS appeared somewhat different. While Myo3B-ir was detected throughout the IS (Fig. 5), Myo3A-ir was most intense in the distal portion of the IS. Myo3A-ir also was not associated uniformly with the actin cytoskeleton of photoreceptors because filamentous actin (F-actin), as visualized with fluorescently labeled phalloidin, is more abundant in the proximal portion of the IS and in the outer limiting membrane (Fig. 8A). A similar comparison between the distribution of mMyo3B and F-actin was not possible because the antigen retrieval protocol used to detect mMyo3B immunoreactivity destroyed phalloidin staining. To further verify the differential localization of mMyo3B and mMyo3A in the IS we immunostained sections from the same eye with mMyo3A or mMyo3B and a nuclear marker (DRAQ5). We observed consistently that mMyo3B-ir was detected throughout the IS (Fig. 9A) and was present adjacent to the ONL. On the other hand, mMyo3A-ir was more concentrated in the distal portion of the IS (Fig. 9B).
Figure 8.
mMyo3A concentrates in the distal portion of the photoreceptor IS, does not distribute uniformly with F-actin and does not extend into OS. Fixed frozen sections were incubated without antigen recovery with affinity purified mMyo3A antibodies (undiluted) and A. fluorescently labeled phalloidin (1:200 dilution) or B. a monoclonal antibody directed against rhodopsin (1:200 dilution). Images were obtained as described in Fig. 6. A: mMyo3A-ir (green) is concentrated in the distal portion of the IS whereas F-actin, as visualized with fluorescently labeled phalloidin (red) is most abundant in the proximal portion of the IS and in the outer limiting membrane (arrow). B: mMyo3A (green) does not co-localize with rhodopsin (red) indicating that mMyo3A does not extend to the OS. Abbreviations as in Fig. 5. Bars=20 μm.
Figure 9.
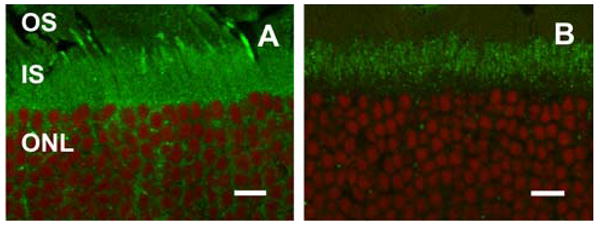
mMyo3B and mMyo3A exhibit differential localization in the IS of photoreceptors. Confocal images of retinal sections labeled with either mMyo3B (A, green) or Myo3A (B, green) and the nuclear marker DRAQ5 (red). Images were obtained as described in Fig. 6. A. Myo3B-ir is present throughout the IS and in the OS of some cones. B. mMyo3A is more concentrated in the distal portion of the IS indicated by the gap between mMyo3A-ir and the nuclei of the ONL. Abbreviations as in Fig. 5. Bars=10 μm.
To test whether mMyo3A-ir extends into the OS, sections were double-labeled with mMyo3A and rhodopsin (Fig. 8B). mMyo3A and rhodopsin-ir did not co-localize, therefore Myo3A is excluded from OS. However, mMyo3A-ir was clearly detected in the OPL in structures with the location and distribution of photoreceptor terminals. No Myo3A-ir was detected in the inner layers of the retina.
mMyo3B in the inner retina
mMyo3B-ir was detected in cells of the inner retina; a few cells in the INL and in cells in the GCL (Fig. 10). Inner retinal mMyo3B immunoreactive cells from 4 different animals were counted to determine their number and distribution. At least 29 sections (18 μm thick) were counted per animal; these included sections from the mid-nasal and mid-temporal retina, as well as sections that spanned the optic nerve head. Each section included the dorsal and ventral region.
Figure 10.
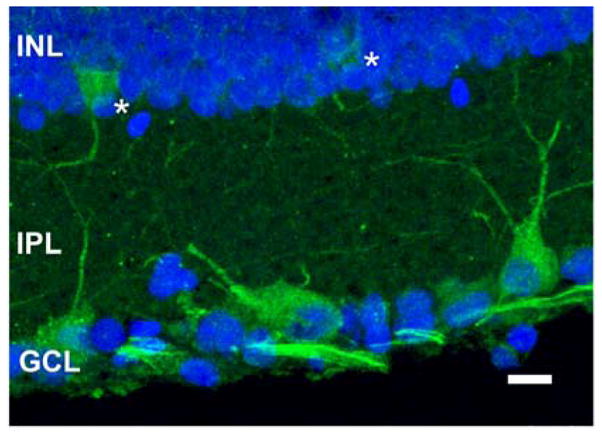
mMyo3B labels cells in the INL and GCL. Confocal image of a retinal section stained with the mMyo3B antiserum (green, 1:500 dilution) and DRAQ5 (blue, 1:1000 dilution). The image was obtained as described in Fig. 6. mMyo3B-ir is seen in the cytoplasm of two cells in the INL (asterisks), in a number of cells in the GCL and in dendrites in the IPL probably from cells in the GCL and INL. Ganglion cell axons are brightly labeled for mMyo3B. Abbreviations as in Fig. 5. Bar=10 μm
Only a few mMyo3B immunoreactive cells were detected in the INL (Fig. 10). The number of cells stained per retinal section ranged from 0 to 6 with an average of 1.4 cells. The median number of labeled cells was 1 and the most frequent number was 0. Most mMyo3B immunoreactive cells were located in the peripheral half of the dorsal and ventral retina with more cells detected in the ventral retina (114/203 cells). No consistent gradient was observed across the nasal-temporal axis.
In the GCL, some mMyo3B immunoreactive cells are clearly GCs because GC axons label brightly for Myo3B. An average of 23.3 cells were mMyo3B immunoreactive per retinal section, and in most sections, about half were large and brightly labeled (Fig. 10). In both ventral and dorsal retina brightly labeled GCs were mostly distributed between the periphery and mid-periphery of the section; the GCL of the central retina was relatively free of mMyo3B immunoreactive cells. We did not observe a gradient across the nasal-temporal axis.
DISCUSSION
We show here that mouse retina expresses at least two variants of Myo3B which differ from one another in both the motor and tail domains of the protein, and that Myo3B is expressed in several other mouse tissues. In retina, Myo3B is present in photoreceptor IS, within the cytoplasm surrounding photoreceptor nuclei in the ONL, a small number of cells in the INL and in some but not all ganglion cells. Surprisingly, Myo3B is highly concentrated in the OS of mouse cones expressing S opsin, but not in the OS of other cones. We also show that Myo3B co-localizes with Myo3A in portions of photoreceptor IS, but unlike Myo3B, Myo3A does not extend into OS. Myo3A is detected in the OPL in putative photoreceptor synaptic terminals where we did not detect Myo3B.
Myo3B variants expressed in mouse retina
The insert and deletion in mmyo3B variant 2 occur in regions encoding loop2 of the motor domain and in the tail domain, respectively, two functionally important regions of the protein. Loop2 is part of the actin binding interface of the motor domain and important for determining actin binding affinity and the rate of ATP hydrolylsis (Geeves et al., 2005). The tail domains of class III myosins are critical for determining their subcellular localization and cargo (Porter et al., 1992; Erickson et al., 2003). Thus, the two mMyo3B variants identified may differ functionally. Other loop2 variants are probably rare in retina, based on the results of our screen for such variants in retinal cDNA, but evidence discussed below suggests that at least in tissues other than the retina, additional tail variants are likely.
Tail splice variants of hMYO3B have also been identified (Dosé and Burnside, 2002) but no loop2 variants have been reported so far. A 19 kb intron spans the exon 23–24 junction of hMYO3B, site of the loop2 insertion in mMyo3B; therefore it is possible that loop2 variants also exist in the human protein.
Myo3B and Myo3A co-expression in photoreceptors
Co-expression of Myo3B with Myo3A in photoreceptors and functional redundancy between these two proteins has been proposed to explain why patients with mutations in Myo3A who experience hearing loss have no defects in vision (Walsh et al., 2002). The presence of both Myo3A and Myo3B in the IS of mammalian photoreceptors indicates that Myo3B is positioned appropriately to compensate for Myo3A defects. Our studies extend the previous studies of Burnside and collaborators who showed that both Myo3A and Myo3B are present in the IS of fish photoreceptors (Dosé et al., 2003; 2004). The functions of mMyo3A and mMyo3B may overlap; however, differences in their primary sequences (Fig. 1), particularly in their tail domains, and their distinct distributions in the retina, suggest their functions are not identical.
Myo3B in photoreceptor outer segments
We anticipated finding mMyo3B in photoreceptor IS because they contain an extensive actin cytoskeleton (Fig. 8 and Woodford and Blanks, 1989), and all class III myosins that have been studied bind F-actin (Hicks et al., 1996; Dosé et al., 2003; Komaba et al., 2003; Kambara et al., 2006; Dosé et al., 2007; Kempler et al., 2007). Other F-actin-containing regions of the mammalian photoreceptor where we anticipated finding class III myosins were the connecting cilium (Wolfrum and Schmitt, 2000) and the distal end of the connecting cilium (Chaitin et al., 1984). Finding mMyo3B in cone OS was unexpected. To the best of our knowledge, there is no actin cytoskeleton in OS, therefore the binding partners and functions of mMyo3B in OS remain to be discovered.
Also unexpected was finding mMyo3B in the OS of only those cones that express detectable levels of S opsin (Fig. 6). It has been proposed that mouse retina contains only one type of cone (Applebury et al., 2000). S opsin immunoreactive cones are clearly more abundant in ventral compared to dorsal retina (Applebury et al., 2000; Haverkamp et al., 2005; Nikonov et al., 2006; this paper, Fig. 6), but most cones in the mouse retina express both S and M opsin (Applebury et al., 2000; Nikonov et al., 2006). What changes between ventral and dorsal retina is the relative level of the two visual pigments expressed in each cone. S opsin expression is similar in cones across the retina, while M opsin expression is lower in cones of ventral compared to dorsal retina (Applebury et al., 2000; Nikonov et al., 2006). Nevertheless, some cones in the dorsal mouse retina, called M opsin dominant cones, express very little, if any, S opsin (Applebury et al., 2000; Haverkamp et al., 2005; Nikonov et al., 2006). In addition, rare cones have been identified in both the ventral and dorsal mouse retina that express S opsin only (Haverkamp et al., 2005).
In ventral retina, mMyo3B and S opsin consistently co-localize in OS (Fig. 6, A–C). We did not detect M opsin-ir in OS of ventral retina probably because of the low level of M opsin expressed in these cones (Applebury et al., 2000) and the sensitivity limits of the M opsin antibody we used. But since S and M opsin are co-expressed in most cones of ventral retina, mMyo3B must be present in OS of dual-expressing cones. In dorsal retina, the few mMyo3B immunoreactive OS detected also consistently show S opsin-ir (Fig. 6, D–F), but mMyo3B-ir is not detected in M opsin-immunoreactive cones (Fig. 6, G–H). In addition, mMyo3B immunoreactive OS show no detectable M opsin-ir. The mMyo3B positive/M opsin negative OS are typically surrounded by other M opsin positive OS, and Applebury et al. (2000) has shown that neighboring M opsin dominant cones in dorsal retina express similar levels of M opsin. It is therefore highly likely we would have detected M opsin-ir in these mMyo3B immunoreactive OS of the dorsal retina had it been present.
Finding mMyo3B positive OS in dorsal retina that are consistently S opsin positive and M opsin negative supports the idea that some cones in the mouse retina express S opsin only (Haverkamp et al., 2005). Our results also show that M opsin dominant cones differ biochemically from other cones in the mouse retina in that while mMyo3B is present in their IS (Fig. 7), it is not present in their OS (Fig. 6). Since vertebrate class III myosins are actin-based molecular motors, there also may be undiscovered structural differences between the M opsin dominant cones and other cones in the mouse retina. The functional consequences of the presence of mMyo3B in OS will require a more complete understanding of the functions of the protein. Subtle electrophysiological differences have been observed between M opsin dominant and dual opsin expressing cones (Nikonov et al., 2006), but the biochemistry underlying these differences is not known.
Myo3B in the inner retina
mMyo3B-ir in the inner retina is restricted to a few cell types. GC somata, their dendrites and axons are the most brightly mMyo3B immunoreactive structures in the retina. But not all cells in the GCL are labeled (Fig. 10). In the INL a few cells are consistently labeled and in many instances we detected mMyo3B-ir in their projections into IPL (Fig. 10). The identity of the mMyo3B immunoreactive cells in the GCL and INL is clearly of interest and will be pursued in future studies.
Does our antiserum detect all mMyo3B isoforms?
Mouse kidney contains mmyo3B transcripts as measured by qPCR (Fig. 2A), but no mMyo3B-ir was detected on Western blots of soluble proteins from kidney (Fig. 4A). Thus, our antiserum probably does not detect all Myo3B variants expressed in mouse. Results of preliminary Western blots indicated our antiserum would detect proteins produced from the two mmyo3B variants we cloned from retina (See Methods). However, six myo3B variants were cloned from human retina. One of these lacks the C-terminal half of the tail domain and in another the sequence of the C-terminal half of the tail domain is almost entirely different from that of the other variants (Dosé and Burnside, 2002). The expression in mouse of myo3B variants that differ significantly in their tail domains could explain the discrepancy between our qPCR and Western blot results with the kidney. Such variants could be present in retina. If true, our antiserum may be showing the distribution of a specific subset of mMyo3B proteins with similar tail domains.
Conclusions
Since Myo3A and Myo3B are both expressed in photoreceptor IS, Myo3B could provide functional redundancy for Myo3A in photoreceptors and explain the absence of a retinal phenotype in humans with progressive hearing loss produced by defective MYO3A. Our finding that Myo3B is also highly concentrated in the OS of mouse cones expressing S opsin but not in the OS of M-dominant cones has revealed a previously unrecognized difference in the biochemistry of the OS of S opsin and M opsin dominant cones. But since OS are not known to contain F-actin, and Myo3B is a predicted actin-based motor, the role Myo3B might play in S-cone OS remains mysterious.
Supplementary Material
Acknowledgments
Author contributions: Cloning, J.S.D. with A.C.D.; qPCR, J.S.D.; Immunochemistry and immunocytochemistry, C.K.; Manuscript, C.K. and B-A.B. with contributions from each of the authors. We thank Lynn Milstead for preparing figures and Dr. W. Clay Smith for the rhodopsin antibody. This work was supported by the following: NSF-IOS 0517273 to B-A.B, NIH-EY01750 to BB, NIH- EY08571 to the Department of Ophthalmology, University of Florida School of Medicine; NIH- S10RR14638 to the Whitney Laboratory, and Whitney Laboratory internal funds.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adamus G, Zam ZS, Arendt A, Palczewski K, McDowell JH, Hargrave PA. Anti-rhodopsin monoclonal antibodies of defined specificity: Characterization and application. Vision Res. 1991;31:17–31. doi: 10.1016/0042-6989(91)90069-h. [DOI] [PubMed] [Google Scholar]
- Applebury ML, Antoch MP, Baxter LC, Chun LLY, Falk JD, Farhangfar F, Kage K, Krzystolik MG, Lyass LA, Robbins JT. The murine cone photoreceptor: A single cone type expresses both S and M opsins with retinal spatial patterning. Neuron. 2000;27:513–523. doi: 10.1016/s0896-6273(00)00062-3. [DOI] [PubMed] [Google Scholar]
- Battelle BA, Andrews AW, Calman BG, Sellers JR, Greenberg RM, Smith WC. A myosin III from Limulus eyes is a clock-regulated phosphoprotein. J Neurosci. 1998;18:4548–4559. doi: 10.1523/JNEUROSCI.18-12-04548.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battelle BA, Andrews AW, Kempler KE, Edwards SC, Smith WC. Visual arrestin in Limulus is phosphorylated at multiple sites in the light and in the dark. Vis Neurosci. 2000;17:813–822. doi: 10.1017/s0952523800175145. [DOI] [PubMed] [Google Scholar]
- Battelle BA, Dabdoub A, Malone MA, Andrews AW, Cacciatore C, Calman BG, Smith WC, Payne R. Immunocytochemical localization of opsin, visual arrestin, myosin III, and calmodulin in Limulus lateral eye retinular cells and ventral photoreceptors. J Comp Neurol. 2001;435:211–225. doi: 10.1002/cne.1203. [DOI] [PubMed] [Google Scholar]
- Blanks JC, Johnson LV. Selective Lectin Binding of the Developing Mouse Retina. J Comp Neurol. 1983;221:31–41. doi: 10.1002/cne.902210103. [DOI] [PubMed] [Google Scholar]
- Bretscher A. Purification of an 80,000-Dalton Protein That Is a Component of the Isolated Microvillus Cytoskeleton, and Its Localization in Non-Muscle Cells. J Cell Biol. 1983;97:425–432. doi: 10.1083/jcb.97.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calman BG, Lauerman MA, Andrews AW, Schmidt M, Battelle BA. Central Projections of Limulus Photoreceptor Cells Revealed by a Photoreceptor-Specific Monoclonal-Antibody. J Comp Neurol. 1991;313:553–562. doi: 10.1002/cne.903130402. [DOI] [PubMed] [Google Scholar]
- Cardasis HL, Stevens SM, McClung S, Kempler KE, Powell DH, Eyler JR, Battelle BA. The actin-binding interface of a myosin III is phosphorylated in vivo in response to signals from a circadian clock. Biochemistry. 2007;46:13907–13919. doi: 10.1021/bi701409f. [DOI] [PubMed] [Google Scholar]
- Carter-Dawson LD, LaVail MM. Rods and cones in the mouse retina. 1 Structural analysis using light and electron microscopy. J Comp Neurol. 1979;188:245–262. doi: 10.1002/cne.901880204. [DOI] [PubMed] [Google Scholar]
- Chaitin MH, Schneider BG, Hall MO, Papermaster DS. Actin in the photoreceptor connecting cilium - Immunocytochemical localization to the site of outer segment disk formation. J Cell Biol. 1984;99:239–247. doi: 10.1083/jcb.99.1.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chyb S, Hevers W, Forte M, Wolfgang WJ, Selinger Z, Hardie RC. Modulation of the light response by cAMP in Drosophila photoreceptors. J Neurosci. 1999;19:8799–8807. doi: 10.1523/JNEUROSCI.19-20-08799.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan I, Watanabe NM, Kusumi A. The Ste20 group kinases as regulators of MAP kinase cascades. Trends Cell Biol. 2001;11:220–230. doi: 10.1016/s0962-8924(01)01980-8. [DOI] [PubMed] [Google Scholar]
- Dosé AC, Burnside B. Cloning and chromosomal localization of a human class III myosin. Genomics. 2000;67:333–342. doi: 10.1006/geno.2000.6256. [DOI] [PubMed] [Google Scholar]
- Dosé AC, Burnside B. A class III myosin expressed in the retina is a potential candidate for Bardet-Biedl syndrome. Genomics. 2002;79:621–624. doi: 10.1006/geno.2002.6749. [DOI] [PubMed] [Google Scholar]
- Dosé AC, Hillman DW, Wong C, Sohlberg L, Lin-Jones J, Burnside B. Myo3A, one of two class III myosin genes expressed in vertebrate retina, is localized to the calycal processes of rod and cone photoreceptors and is expressed in the sacculus. Mol Biol Cell. 2003;14:1058–1073. doi: 10.1091/mbc.E02-06-0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosé AC, Lin-Jones J, Burnside B. Myosin III in photoreceptors: What does it do? In: Williams DS, editor. Cell Biology and Disease of the Outer Retina. World Scientific Press; New Jersey: 2004. pp. 133–162. [Google Scholar]
- Dosé AC, Ananthanarayanan S, Moore JE, Burnside B, Yengo CM. Kinetic mechanism of human myosin IIIA. J Biol Chem. 2007;282:216–231. doi: 10.1074/jbc.M605964200. [DOI] [PubMed] [Google Scholar]
- Dosé AC, Ananthanarayanan S, Moore JE, Corsa AC, Burnside B, Yengo CM. The kinase domain alters the kinetic properties of the myosin IIIA motor. Biochemistry. 2008;47:2485–2496. doi: 10.1021/bi7021574. [DOI] [PubMed] [Google Scholar]
- Edwards SC, Battelle BA. Octopamine-Stimulated and Cyclic AMP-Stimulated Phosphorylation of a Protein in Limulus Ventral and Lateral Eyes. J Neurosci. 1987;7:2811–2820. doi: 10.1523/JNEUROSCI.07-09-02811.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards SC, Andrews AW, Renninger GH, Wiebe EM, Battelle BA. Efferent Innervation to Limulus Eyes Invivo Phosphorylates a 122 Kd Protein. Biol Bull. 1990;178:267–278. doi: 10.2307/1541828. [DOI] [PubMed] [Google Scholar]
- Erickson FL, Corsa AC, Dosé AC, Burnside B. Localization of a class III myosin to filopodia tips in transfected HeLa cells requires an actin-binding site in its tail domain. Mol Biol Cell. 2003;14:4173–4180. doi: 10.1091/mbc.E02-10-0656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geeves MA, Fedorov R, Manstein DJ. Molecular mechanism of actomyosin-based motility. Cell Mol Life Sci. 2005;62:1462–1477. doi: 10.1007/s00018-005-5015-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haverkamp S, Wassle H, Duebel J, Kuner T, Augustine GJ, Feng GP, Euler T. The primordial, blue-cone color system of the mouse retina. J Neurosci. 2005;25:5438–5445. doi: 10.1523/JNEUROSCI.1117-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks JL, Liu XR, Williams DS. Role of the NinaC proteins in photoreceptor cell structure: Ultrastructure of NinaC deletion mutants and binding to actin filaments. Cell Motil Cytoskeleton. 1996;35:367–379. doi: 10.1002/(SICI)1097-0169(1996)35:4<367::AID-CM8>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Hofstee CA, Henderson S, Hardie RC, Stavenga DG. Differential effects of ninaC proteins (p132 and p174) on light-activated currents and pupil mechanism in Drosophila photoreceptors. Vis Neurosci. 1996;13:897–906. doi: 10.1017/s0952523800009147. [DOI] [PubMed] [Google Scholar]
- Kambara T, Komaba S, Ikebe M. Human myosin III is a motor having an extremely high affinity for actin. J Biol Chem. 2006;281:37291–37301. doi: 10.1074/jbc.M603823200. [DOI] [PubMed] [Google Scholar]
- Kawai K, Serizawa A, Hamana T, Tsutsumi Y. Heat-Induced Antigen Retrieval of Proliferating Cell Nuclear Antigen and P53 Protein in Formalin-Fixed, Paraffin-Embedded Sections. Pathol Int. 1994;44:759–764. doi: 10.1111/j.1440-1827.1994.tb02923.x. [DOI] [PubMed] [Google Scholar]
- Kempler K, Toth J, Yamashita R, Mapel G, Robinson K, Cardasis H, Stevens S, Sellers JR, Battelle BA. Loop 2 of Limulus myosin III is phosphorylated by protein kinase A and autophosphorylation. Biochemistry. 2007;46:4280–4293. doi: 10.1021/bi062112u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komaba S, Inoue A, Maruta S, Hosoya H, Ikebe M. Determination of human myosin III as a motor protein having a protein kinase activity. J Biol Chem. 2003;278:21352–21360. doi: 10.1074/jbc.M300757200. [DOI] [PubMed] [Google Scholar]
- Li HS, Porter JA, Montell C. Requirement for the NINAC kinase/myosin for stable termination of the visual cascade. J Neurosci. 1998;18:9601–9606. doi: 10.1523/JNEUROSCI.18-23-09601.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Lalwani AK, Mhatre AN. Alternative splice variants of MYH9. DNA Cell Biol. 2008;27:117–125. doi: 10.1089/dna.2007.0661. [DOI] [PubMed] [Google Scholar]
- Lin-Jones J, Parker E, Wu M, Dosé A, Burnside B. Myosin 3A transgene expression produces abnormal actin filament bundles in transgenic Xenopus laevis rod photoreceptors. J Cell Sci. 2004;117:5825–5834. doi: 10.1242/jcs.01512. [DOI] [PubMed] [Google Scholar]
- Liou GI, Matragoon S, Overbeek PA, Fei YJ. Identification of a Retina-Specific Footprint within the Retina-Specific Regulatory Region of the Human Interphotoreceptor Retinoid-Binding Protein Gene. Biochem Biophys Res Commun. 1994;203:1875–1881. doi: 10.1006/bbrc.1994.2406. [DOI] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein Measurement with the Folin Phenol Reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Montell C, Rubin GM. The Drosophila Ninac Locus Encodes 2 Photoreceptor Cell Specific Proteins with Domains Homologous to Protein-Kinases and the Myosin Heavy-Chain Head. Cell. 1988;52:757–772. doi: 10.1016/0092-8674(88)90413-8. [DOI] [PubMed] [Google Scholar]
- Ng KP, Kambara T, Matsuura M, Burke M, Ikebe M. Identification of myosin III as a protein kinase. Biochemistry. 1996;35:9392–9399. doi: 10.1021/bi960181a. [DOI] [PubMed] [Google Scholar]
- Nikonov SS, Kholodenko R, Lem J, Pugh EN. Physiological features of the S- and M-cone photoreceptors of wild-type mice from single-cell recordings. J Gen Physiol. 2006;127:359–374. doi: 10.1085/jgp.200609490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter JA, Hicks JL, Williams DS, Montell C. Differential localizations of and requirements for the 2 Drosophila NinaC kinase myosins in photoreceptor cells. J Cell Biol. 1992;116:683–693. doi: 10.1083/jcb.116.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter JA, Montell C. Distinct Roles of the Drosophila-Ninac Kinase and Myosin Domains Revealed by Systematic Mutagenesis. J Cell Biol. 1993;122:601–612. doi: 10.1083/jcb.122.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter JA, Yu MJ, Doberstein SK, Pollard TD, Montell C. Dependence of Calmodulin Localization in the Retina on the Ninac Unconventional Myosin. Science. 1993;262:1038–1042. doi: 10.1126/science.8235618. [DOI] [PubMed] [Google Scholar]
- Porter JA, Minke B, Montell C. Calmodulin-Binding to Drosophila Ninac Required for Termination of Phototransduction. EMBO J. 1995;14:4450–4459. doi: 10.1002/j.1460-2075.1995.tb00124.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider ME, Dosé AC, Salles FT, Chang WS, Erickson FL, Burnside B, Kachar B. A new compartment at stereocilia tips defined by spatial and temporal patterns of myosin IIIa expression. J Neurosci. 2006;26:10243–10252. doi: 10.1523/JNEUROSCI.2812-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellers JR. Myosins. Oxford University Press; New York: 1999. [Google Scholar]
- Szel A, Rohlich P, Caffe AR, Juliusson B, Aguirre G, Vanveen T. Unique Topographic Separation of 2 Spectral Classes of Cones in the Mouse Retina. J Comp Neurol. 1992;325:327–342. doi: 10.1002/cne.903250302. [DOI] [PubMed] [Google Scholar]
- Takahashi M, Kawamoto S, Adelstein RS. Evidence for inserted sequences in the head region of nonmuscle myosin specific to the nervous system. Cloning of the cDNA encoding the myosin heavy chain-B isoform of vertebrate nonmuscle myosin. J Biol Chem. 1992;267:17864–17871. [PubMed] [Google Scholar]
- Walsh T, Walsh V, Vreugde S, Hertzano R, Shahin H, Haika S, Lee MK, Kanaan M, King MC, Avraham KB. From flies’ eyes to our ears: Mutations in a human class III myosin cause progressive nonsyndromic hearing loss DFNB30. Proc Natl Acad Sci USA. 2002;99:7518–7523. doi: 10.1073/pnas.102091699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfrum U, Schmitt A. Rhodopsin transport in the membrane of the connecting cilium of mammalian photoreceptor cells. Cell Motil Cytoskeleton. 2000;46:95–107. doi: 10.1002/1097-0169(200006)46:2<95::AID-CM2>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Woodford BJ, Blanks JC. Localization of actin and tubulin in developing and adult mammalian photoreceptors. Cell Tissue Res. 1989;256:495–505. doi: 10.1007/BF00225597. [DOI] [PubMed] [Google Scholar]
- Young RW. Visual cells and concept of renewal. Invest Ophthalmol Vis Sci. 1976;15:700–725. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.