Summary
The absence of cytotoxic T lymphocyte activity and the failure of MHC class I–restricted T cell receptor (TCR) transgenic thymocytes to mature in CD8α-deficient mice suggest that CD8 may be essential for CD8 lineage commitment. We report that variants of the antigenic peptide that delete TCR transgenic thymocytes from CD8 wild-type but not CD8α-deficient mice can restore positive selection of CD8 lineage cells in the absence of CD8. The positively selected cells down-regulate CD4, up-regulate TCR, respond to the antigenic peptide, and express CD8β mRNA. Interestingly, there was no enhanced selection of CD4+ T cells, implying that the TCR–MHC interaction, even in the absence of CD8, provided instructive signaling for commitment to the CD8 lineage. Our results are discussed in terms of recent models of T cell lineage commitment.
Introduction
The CD8 molecule on the surface of T cells serves as a coreceptor for T cell receptor (TCR) recognition of major histocompatibility (MHC) class I–associated peptides. CD8 may be involved in positive selection either by stabilizing TCR–MHC interactions or by enhancing the activation of essential intracellular signaling components (Zamoyska, 1994). CD8 exists on the cell surface as a disulfide-linked transmembrane heterodimer of α and β chains (Parnes, 1989). In the absence of CD8β expression, CD8α homodimers reach the surface, whereas in the absence of CD8α, CD8β is retained and destroyed within the cell. The extracellular immunoglobulin domains of CD8 homodimers and heterodimers bind the membrane-proximal region of MHC class I molecules (Potter et al., 1987; Salter et al., 1990; Sun et al., 1995). Soluble CD8α homodimers bind MHC class I with approximately 3-fold lower affinity than do soluble CD8αβ heterodimers (Garcia et al., 1996). Studies with both whole cells and purified components have shown that CD8 stabilizes the interaction between the TCR and MHC-associated peptide ligands (Lueshcher et al., 1995; Garcia et al., 1996; Renard et al., 1996).
The CD8α cytoplasmic tail associates with the Src family tyrosine kinase, Lck, a molecule critically involved in TCR-mediated signaling (Veillette et al., 1988; Rudd et al., 1989; Miceli and Parnes, 1993). In contrast, the CD8β tail lacks a motif for interaction with Lck or any other known signaling molecules. Despite this and the rather small effect of CD8β on the binding affinity for class I, eliminating CD8β expression dramatically influences CD8 lineage development. Thymocytes from CD8β-deficient mice produce very few CD8 single positive cells (Crooks and Littman, 1994; Fung-Leung et al., 1994), and thymocytes overexpressing a tailless CD8β molecule are also poorly selected into this lineage (Itano et al., 1994). These findings raise the possibility that CD8β may contribute in some unknown but critical way to the selection of class I–restricted T cells.
In CD8α-deficient (CD8°) mice, MHC class I–restricted cytotoxic T lymphocytes (CTL) appear to be completely absent (Fung-Leung et al., 1991a; Fung-Leung et al., 1991b; Fung-Leung et al. 1993a; Bachmann et al., 1995). No significant CD4−TCRhi T cell population is observed in the CD8° mice in excess of the numbers normally observed in CD8 wild-type mice. More remarkable than this phenotypic characteristic is the paucity of cytolytic function after in vivo introduction of potent immunogens such as lymphocytic choriomeningitis virus (Fung-Leung et al., 1991b), vesicular stomatitis virus (Bachmann et al., 1995), or immunization with allogeneic cells (Fung-Leung et al., 1991a). A number of lines of mice transgenic for MHC class I–restricted TCRs also fail to give rise to CD8 lineage cells when crossed onto the CD8° background (Fung-Leung et al., 1993). In summary, detection of CD8 “wannabes” in CD8α-deficient mice is difficult. This is in contrast with CD4° mice, which readily generate MHC class II–restricted responses to foreign antigens mediated by CD4−CD8−TCRhi CD4 “wannabes” (Locksley et al., 1993; Rahemtulla et al., 1994).
Further indications of the different requirements for commitment to the CD4 versus CD8 lineage come from studies using anti-TCR antibodies to drive positive selection. In MHC-deficient (class II°, β2m°) mice, mature CD4 single positive cells, but not CD8 single positive T cells, can be selected by intraperitoneal injection of anti-TCR antibodies (Muller and Kyewski, 1993, 1995). Similarly, in fetal thymic organ culture (FTOC) of MHC-deficient lobes, deaggregated anti-TCR antibodies preferentially promote the selection of CD4 lineage cells (Takahama et al., 1994). Other studies analyzing the co-receptor reexpression patterns of immature thymocyte subsets have suggested that CD8 lineage commitment is dependent on TCR interactions with MHC class I and therefore may involve a necessary CD8-dependent signal. In contrast, CD4 lineage–committed cells are apparent in the thymus of MHC class II–deficient mice using this assay (Suzuki et al., 1995).
We wanted to address whether expression of CD8 on the thymocyte surface is essential for positive selection of the CD8+ CTL lineage. To do this, we used a system in which we can control the affinity of the interaction between a transgenic TCR and the MHC-associated peptide ligands that drive selection (Alam et al., 1996). OT-1 TCR transgenic mice express a TCR that recognizes the octameric peptide OVA257–263 in the context of H-2Kb, and thymocytes expressing this receptor are positively selected into CD8 single positive cells by recognition of natural peptide ligands presented by H-2Kb (Hogquist et al., 1994). Variants of the antigenic ovalbumin peptide (OVAp) that act as antagonists or weak agonists for CTL bearing the OT-1 TCR have been defined (Jameson et al., 1993; Barnden et al., 1994; Hogquist et al., 1994; Jameson et al., 1994). We have shown previously that such variants can restore positive selection when added to FTOC of OT-1 transgenic, β2m° mice (Hogquist et al., 1994). In the absence of the CD8 coreceptor, OT-1 thymocytes are not positively selected by normal levels of H-2Kb. We asked whether selection could be restored, in this case, by the addition to FTOC of OVAp variant peptides that serve as strong agonists for the OT-1 transgenic T cells.
Our results show that positive selection of thymocytes bearing an MHC class I–restricted TCR can be restored in the absence of CD8 expression by increasing the affinity of the TCR–MHC ligand interaction. Curiously, this TCR–MHC class I interaction, even in the absence of CD8, appears to direct commitment to the CD8 lineage.
Results
Identification of Peptide Variants for Selection of CD8°OT-1 Thymocytes
We reasoned that peptide–MHC complexes that had higher affinity for the TCR than those previously used to mediate positive selection of CD8WTOT-1 thymocytes, but that did not delete CD8°OT-1 thymocytes, would provide candidate ligands for positive selection of CD8°OT-1 thymocytes. Variants of OVAp (SIINFEKL) that have been described as agonists for T cells bearing the OT-1 TCR, either in CTL targeting or thymocyte deletion assays (Jameson et al., 1993; Barnden et al., 1994; Hogquist et al., 1994; Jameson et al., 1994), were tested for their ability to induce deletion and CD69 up-regulation of CD8°OT-1 thymocytes. In an in vitro deletion assay, OVAp deleted CD8WTOT-1 thymocytes approximately 100-fold more effectively on a dose response curve than it did CD8°OT-1 thymocytes (Figures 1 a and 1c). The dose response curve for deletion correlates well with the induction of CD69 expression on the thymocytes (Figures 1b and 1d). Thus, CD8°OT-1 thymocytes are approximately 100-fold less sensitive to deletion and CD69 induction by OVAp than their counterpart CD8WT thymocytes.
Figure 1. In Vitro Deletion and CD69 Expression of CD8°OT-1 and CD8WTβ2m°OT-1 Thymocytes by OVAp Variants.
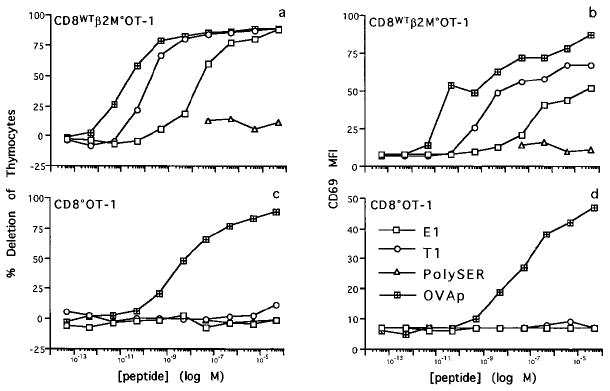
Thymocytes from CD8°OT-1 and CD8WTβ2m°OT-1 were cultured with irradiated EL4 cells and titrations of OVAp (crossed squares), T1 (circles), E1 (open squares), or polySER (triangles) peptides for 18 hr. Cells were harvested and analyzed for expression of CD4, CD8, and CD69 by flow cytometry. The percentage deletion of viable thymocytes (a and c) and CD69 mean fluorescence intensity (MFI) for total thymocytes (b and d) are shown.
E1 (EIINFEKL), an OVAp variant previously shown to be a weak agonist of the OT-1 TCR (Jameson et al., 1993; Hogquist et al., 1994; Jameson et al., 1994), induced deletion and CD69 expression in CD8WTOT-1 thymocytes approximately 1000-fold less efficiently than did OVAp (Figures 1a and 1b) but had no effect on CD8°OT-1 thymocytes at any concentration tested (Figures 1c and 1d). The T1 (TIINFEKL) variant is a strong agonist (Barnden et al., 1994) and induced deletion and CD69 expression in CD8WTOT-1 thymocytes only about 10-fold less efficiently than OVAp (Figures 1a and 1b). Surprisingly, given its similarity to OVAp in the dose response curve, T1 also had no effect on CD8°OT-1 thymocytes at any concentration tested (Figures 1c and 1d). A control peptide that binds to H-2Kb, polySER (SSYSYSSL) (Hogquist et al., 1994), did not induce deletion of either CD8WTOT-1 or CD8°OT-1 thymocytes (Figure 1). Several other OVAp variants that have been described as strong agonists for the OT-1 TCR, A2, H7, and R7 (Barnden et al., 1994) were found to delete both CD8°OT-1 and CD8WTOT-1 thymocytes. Furthermore, nontransgenic C57BL/6 thymocytes were not deleted by any of the peptides tested (data not shown).
The E1 and T1 variants were also tested for their ability to target killing by a CD8WTOT-1 CTL line in a short-term 51Cr-release assay (Table 1). CD8WTOT-1 CTL killed targets coated with OVAp and T1. However, when a saturating amount of an anti-CD8 monoclonal antibody was added to the assay to block CD8–MHC interactions, lysis of OVAp targets required approximately 100-fold more peptide to achieve the level of killing observed in wells in the absence of blocking antibody, whereas T1 targets were not lysed at any peptide concentration. Similarly, E1 targeted for low levels of lysis, which was completely blocked by the addition of an anti-CD8 antibody (data not shown).
Table 1.
CD8 Dependence of CD8WTOT-1 CTL–Mediated Cytolysisa
| % Specific Lysis in the Presence of MAb | ||
|---|---|---|
| Peptide | No MAb | αCD8 MAb |
| OVAp 10−8 M | 51 | 35 |
| OVAp10−10 M | 38 | 27 |
| T1 10−8 M | 33 | 5 |
| T1 10−10 M | 5 | 5 |
CTL line from OT-1 TCR transgenic mice used at 1:1 effector:target ratio with 51Cr-labeled EL4 target cells precoated with the indicated peptides. The assay was performed in the presence or absence of saturating amounts of blocking anti-CD8 monoclonal antibody (MAb).
We find that for immature thymocyte deletion, thymocyte CD69 up-regulation (Figure 1), and CTL-mediated lysis (Table 1), the loss of the CD8 coreceptor can be compensated for by an approximately 100-fold increase in OVAp concentration. These findings are consistent with data that show that presentation by cells bearing an α3-mutant H-2Kb molecule that interferes with CD8–MHC binding requires 100-fold more OVAp to stimulate OVA-specific CTL clones than cells expressing wild-type H-2Kb (Knall et al., 1994).
E1 and T1 Peptides Delete CD8WTOT-1 Thymocytes in FTOC
FTOC of CD8WTOT-1 thymic lobes allows the efficient positive selection of CD8 single positive cells without the addition of peptide (Figure 2). When the agonist peptides E1, T1, or OVAp were added to cultures at 50 μM, the cells recovered from FTOC were predominantly CD4−CD8− and Vα2hi, as shown previously for OVAp (Figure 2) (Hogquist et al., 1994; Hogquist et al., 1995). Also, cell recovery from cultures treated with agonist peptides was decreased 2- to 4-fold compared to control cultures without added peptide (Table 2). Thus, peptides that induce deletion of adult CD8WTOT-1 thymocytes in the suspension culture assay also induce deletion in FTOC.
Figure 2. Agonist Peptides E1, T1, and OVAp Cause Deletion of CD8WTOT-1 Thymocytes in FTOC.
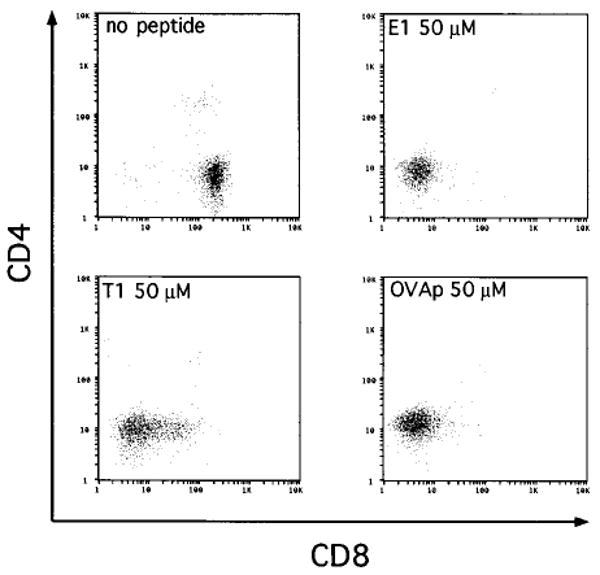
FTOC was performed with CD8WTOT-1 transgenic thymic lobes in the presence of media or 50 μM E1, T1, or OVA peptides. Thymocytes from cultured lobes were harvested after 7 days, counted, and analyzed for expression of CD4 and CD8 by flow cytometry.
Table 2.
Total Cell Yield and Recovery of CD4−Vα2hi Cells from FTOC
| Thymus Donor | No Peptide | E1a | T1 | OVAp |
|---|---|---|---|---|
| CD8°OT-1 | 5.5 (2.5)b | 4.8 (2.1) | 6.6 (2.0) | 1.7 (0.8) |
| 0.7c | 3.3 | 5.4 | 1.1 | |
| CD8WTOT-1 | 4.2 (0.8) | 1.5 (0.4) | 2 (0.1) | 1 (0.1) |
All peptides were present at 50 μM.
Cell yield per lobe × 10−5 (standard deviation).
CD4−Vα2hi cell yield per lobe × 10−5.
E1 and T1 Peptides Positively Select CD8°OT-1 Thymocytes in FTOC
Culture of CD8°OT-1 thymic lobes in the absence of added peptide resulted in the generation of a cell population that was largely CD4+ and that expressed low to intermediate levels of Vα2 (Figures 3a and 3b). These cells appeared to be nonselected and probably correspond to the CD4+CD8+ subset in wild-type mice. Only 13% of the recovered cells had up-regulated Vα2 expression and down-regulated CD4 (Figure 3b).
Figure 3. Agonist Peptides E1, T1, and OVAp Select CD4−Vα2hi Cells in CD8°OT-1 FTOC.
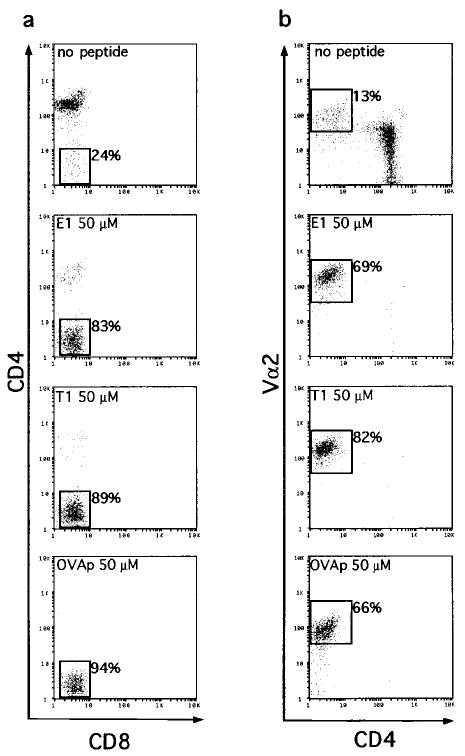
CD8°OT-1 transgenic thymic lobes were cultured in the presence of media or 50 μM E1, T1, or OVA peptides. Thymocytes from cultured lobes were harvested on day 7, counted, and analyzed for expression of CD4, CD8, and Vα2 by flow cytometry. (a) CD8 versus CD4 expression for each culture condition. (b) CD4 versus Vα2 expression for each culture condition.
Inclusion of OVAp, E1, or T1 peptides in the FTOC led to a dramatic change in the recovered populations (Figure 3). The recovery of CD4+ thymocytes was decreased, with a corresponding increase in the recovery of Vα2hi cells that were CD4−. Another peptide variant, VOVA, which has been shown to be an even weaker agonist for the OT-1 TCR than E1 (Hogquist et al., 1994), induced deletion and CD69 expression of CD8WTOT-1 thymocytes only at very high concentrations and not at any concentration for CD8°OT-1 thymocytes. VOVA had no effect on CD8°OT-1 thymocytes in FTOC (data not shown). Although the superficial outcomes with OVAp, E1, and T1 peptides were similar, further analysis showed that while OVAp cultures represented the production of unresponsive CD4−CD8− cells, E1 and T1 cultures represented positive selection of functional CD8 lineage cells. Selection of thymocytes with wild-type coreceptor expression was studied by observing the transition of CD4+CD8+TCRlo cells to either CD8+TCRhi cells (positive selection) or to CD4−CD8−TCRhi cells (deletion). In the absence of CD8 expression, it becomes more difficult to distinguish the outcome of FTOC by surface phenotype alone. Even though we had previously shown that E1 and T1 do not cause deletion of CD8°OT-1 thymocytes in suspension culture deletion assays (Figure 1), it was conceivable that they could delete in FTOC. Therefore, we further characterized the CD4−Vα2hi cells generated in FTOC by these peptides.
First, the absolute cell recoveries from FTOC differed dramatically depending on the peptide added. Notably, similar numbers of cells were recovered from cultures to which E1 or T1 was added and cultures to which no peptide was added, suggesting that E1 and T1 were not mediating deletion (Table 2). Cultures to which OVAp was added, on the other hand, yielded approximately 3.5-fold fewer cells than cultures to which E1, T1, or no peptide was added. This is similar to the deletion observed in FTOC of CD8WTOT-1 thymocytes cultured with the agonist peptides E1, T1, and OVAp, which showed a 2- to 4-fold decrease in cell recovery compared to FTOC in which cells were positively selected in the absence of exogenous peptide (Table 2). In addition, in terms of the yield of cells with a selected phenotype (i.e., CD4−Vα2hi), both E1 and T1 promoted a 3- to 7-fold higher yield above that of the control, no-peptide cultures, whereas OVAp cultures did not (Table 2). The variant peptide, VOVA, which appeared to have no phenotypic effect in FTOC of CD8°OT-1 thymic lobes, yielded numbers of cells similar to those of cultures that received no peptide (data not shown).
To characterize further the maturation state of the peptide-selected cells, TCR, HSA, and H-2Kb expression levels were analyzed (Figure 4). Following FTOC, Vα2 was markedly up-regulated in cultures receiving E1 and T1 peptides, as expected for cells that have undergone positive selection (Figures 3b and 4a). The OVAp-selected cells showed only a modest up-regulation of Vα2 expression (Figures 3b and 4a). Although the CD4− cells generated in FTOC to which OVAp, E1, or T1 peptide had been added were all HSAlo (Figure 4b) and had up-regulated H-2Kb, the OVAp-selected CD4− cells differed from the E1- and T1-selected cells in that they expressed slightly higher levels of H-2Kb (Figure 4c). In contrast to the cells recovered from FTOC receiving peptide, cells from the control FTOC receiving no peptide were Vα2lo, and the few CD4− cells present in these cultures did not significantly down-regulate HSA or up-regulate H-2Kb expression. Thus, these cells do not represent mature cells that have escaped the requirement for CD8.
Figure 4. Phenotypic Analysis of Peptide Selected CD8°OT-1 Cells Recovered from FTOC.
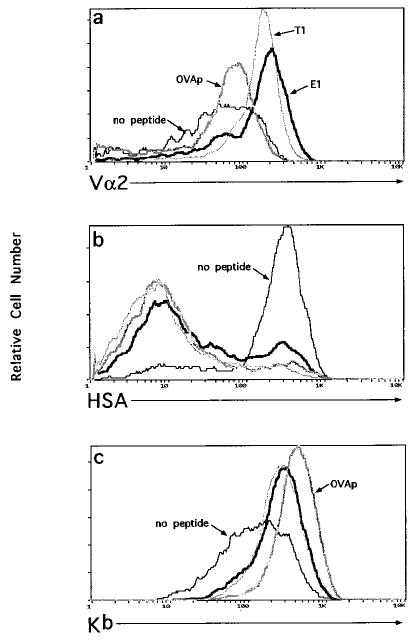
Thymocytes from CD8°OT-1 FTOC were stained for CD4 and CD8 with either Vα2 (total thymocytes) (a), HSA (CD4− gated thymocytes) (b), or H-2Kb (CD4− gated thymocytes) (c). Histograms of Vα2, HSA, and H-2Kb expression are shown for cells recovered from the following FTOC conditions: no peptide (thin solid line), 50 μM OVAp (thick gray line), T1 (thin gray line), or E1 (thick solid line).
E1- and T1-Selected Thymocytes Respond to OVAp
CD8°OT-1 thymocytes recovered from FTOC were tested for their ability to respond to cognate antigen in a proliferation assay. Cells recovered from FTOC were cultured with irradiated antigen-presenting cells in the presence or absence of OVAp. Minimal proliferation in response to stimulation with OVAp was observed for cells from FTOC to which no peptide or OVAp had been added (Figure 5). In contrast, a vigorous response to OVAp was observed for E1- and T1-selected cells. This result demonstrated that functional cells were selected in FTOC by E1 and T1, whereas OVAp induced deletion and functional tolerance of CD8°OT-1 thymocytes.
Figure 5. CD8°OT-1 Thymocytes Selected by OVAp Variants Proliferate in Response to Antigen.
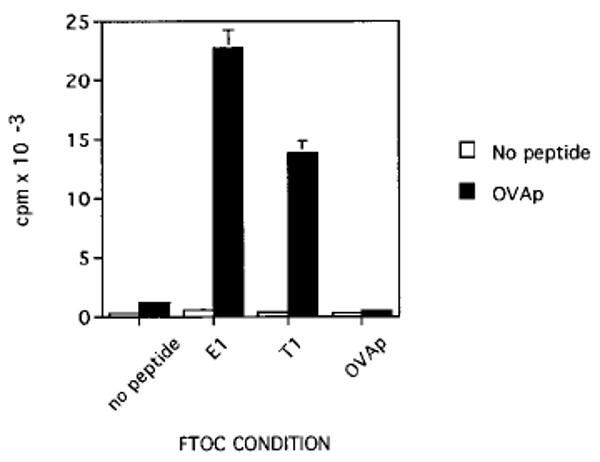
FTOC was performed with CD8°OT-1 transgenic fetuses as described in Figure 3. Thymic lobes were cultured for 7 days in the presence of media or 50 μM E1, T1, or OVA peptides. At the end of FTOC, cells were harvested and added to cultures at one quarter-lobe equivalent per well with irradiated TAP° splenocytes as antigen-presenting cells with or without 100 nM OVAp. Cultures were pulsed for 8 hr with [3H]thymidine after 36 hr of culture. Data are shown as the mean and standard deviation and are representative of three experiments.
To confirm that the observed antigen-specific proliferation was due to the selected CD4−Vα2hi cells and not to proliferation of CD4+ cells expressing the OT-1 TCR, identical proliferation assays were pulsed with bromodeoxyuridine (BrdU), and CD4 and Vα2 expression was analyzed to allow phenotypic analysis of the proliferating cells. As expected, BrdU incorporation in response to antigen stimulation was observed only in cells from E1- and T1-selected FTOC (Figure 6a). The proliferating BrdU+ cells from E1-selected FTOC were nearly all CD4− and Vα2hi (Figure 6b). Similar results were obtained from antigen-specific proliferation of T1-selected cells (data not shown). Therefore, the CD4−Vα2hi cells that were selected by E1 and T1 in FTOC were the cells that proliferated in response to OVAp.
Figure 6. E1 and T1 Selected CD8°OT-1 Thymocytes Responding to OVAp Are Vα2hiCD4−.
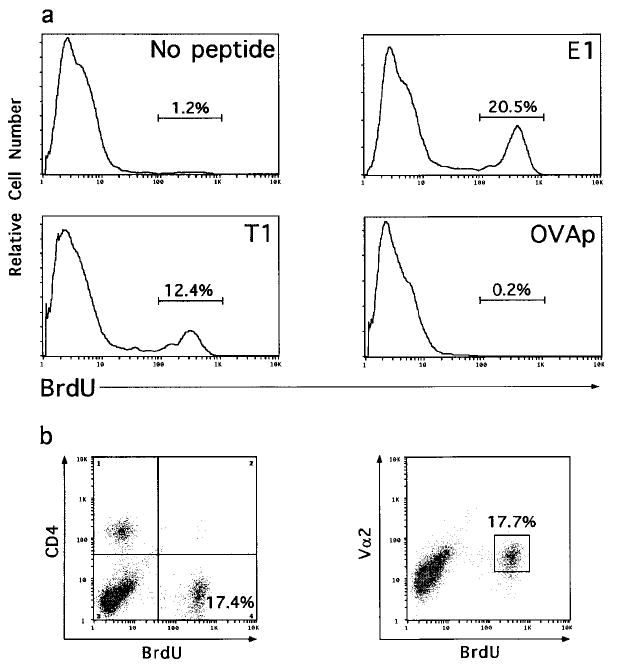
FTOC and proliferation assays were performed as described in Figure 5, except that after 24 hr in culture, wells were pulsed with BrdU. Cells were then harvested and surface stained for expression of CD4 and Vα2 prior to fixation, permeabilization, and intracellular staining for BrdU. BrdU staining is shown for cells cultured with OVAp (100 nM) following selection in FTOC with no peptides or 50 μM E1, T1, or OVA peptides (a). BrdU versus CD4 expression and BrdU versus Vα2 expression are shown for thymocytes derived from the E1-selected FTOC (b). BrdU was incorporated into less than 0.6% of the cells cultured with media alone for all FTOC conditions.
E1- and T1-Selected Thymocytes Express CD8β mRNA
Although the CD4−Vα2hi thymocytes selected by E1 and T1 in FTOC from CD8°OT-1 mice cannot express surface CD8 molecules, we were interested in ascertaining whether they expressed CD8β mRNA as a marker for their commitment to the CD8 lineage. By reverse transcription polymerase chain reaction (RT-PCR), both CD8β and CD4 messages were detected in total adult CD8° and CD8°OT-1 thymocytes as well as in CD8°OT-1 cells recovered from FTOC cultured in the absence of added peptide, showing that it is possible to detect CD8β expression in CD8α-deficient thymocytes (data not shown). Cells harvested from E1 and T1 FTOC were sorted into CD4−Vα2hi (selected) and CD4+Vα2lo (nonselected) populations for RT-PCR analysis. Levels of CD8β, CD4, and HPRT mRNA were compared to the CD4−Vα2hi sorted cells obtained from OVAp FTOC (Figure 7). CD8β message was detected in the CD4−Vα2hi population derived from E1- and T1-selected cultures. CD8β message was also detected in the CD4+Vα2lo populations derived from E1- and T1-selected cultures, as would be expected of double positive thymocytes. However, no detectable CD8β message was observed in the CD4−Vα2hi population recovered from FTOC to which OVAp was added. CD4 message was detected only in the CD4+Vα2lo populations. Because there was no detectable CD4 mRNA in the CD4−Vα2hi population, the CD8β message observed is unlikely to be contributed by a low level of contaminating CD4+Vα2lo cells. Importantly, cells undergoing deletion induced by OVAp in FTOC, of which the majority are CD4−Vα2hi, did not express any detectable CD4 or CD8β mRNA; they appeared to be true CD4−CD8− cells. This indicates that CD4−Vα2hi cells that were selected by E1 and T1 in FTOC were of the CD8 lineage and confirms that this selective process was clearly distinct from thymocyte deletion induced in the presence of OVAp.
Figure 7. E1 and T1 Selected CD8°OT-1 Thymocytes Express CD8β mRNA.
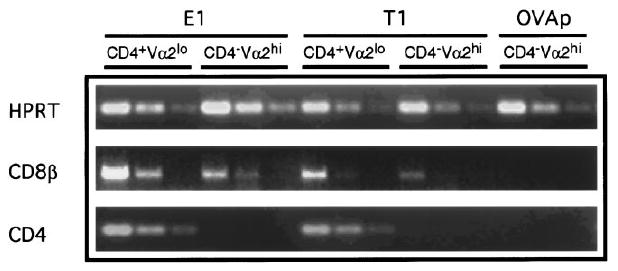
FTOC was performed with CD8°OT-1 transgenic fetuses as described in Figure 3. Thymocytes were harvested and stained for expression of CD4, CD8, and Vα2. Thymocytes from E1 and T1 cultures were sorted for CD4+Vα2lo and CD4−Vα2hi populations. Thymocytes from lobes cultured with OVAp were sorted for CD4−Vα2hi populations. Following sorting, each population contained less than 0.3% contamination with the other sorted population. Total RNA was isolated and cDNA was synthesized. HPRT normalized template was used for PCR amplification of CD4 and CD8β mRNA titrated in 3-fold serial dilutions.
Discussion
To investigate further the role of the CD8 molecule in thymocyte lineage commitment, we have used variants of the cognate peptide to manipulate the affinity of the selecting ligand for the OT-1 TCR in the absence of CD8 expression. CD8°OT-1 thymocytes are deleted in FTOC and in suspension cultures by the cognate antigen, OVAp, although less efficiently than are OT-1 thymocytes expressing CD8. However, even in the absence of CD8, weaker agonists can provide the necessary signal for the positive selection of mature cells that have up-regulated TCR and H-2Kb expression, have down-regulated CD4 and HSA (Figures 3 and 4), respond to antigen (Figures 5 and 6), and express CD8β mRNA (Figure 7). This we interpret as illustrating that an increase in the affinity of the TCR for the positively selecting ligand can overcome the need for CD8 coreceptor involvement in this maturation step. It is unlikely that the cells described here correspond to the CD4−CD8−TCRhi cells selected in TCRαβ transgenic mice that appear to be of the γδ T cell lineage (Bruno et al., 1996). The cells positively selected by agonists in FTOC from CD8°OT-1 thymic lobes proliferate to nominal antigen and express CD8β mRNA.
In this report we have shown that by providing strong agonist ligands for the TCR, we can restore positive selection into the CD8 lineage in the absence of the CD8 molecule. This finding suggests that there may be no essential requirement for CD8 lineage commitment that can be provided only by the CD8 molecule. This possibility then raises the question as to why CD8° mice themselves select undetectable numbers of CD8 lineage cells, particularly in comparison to CD4° mice, which develop easily detectable CD4 lineage cells. To explain this puzzle, we present the following model (diagrammed in Figure 8). In this scheme, TCR affinity for the selecting MHC ligand and the degree of coreceptor involvement in the signaling event play distinguishable roles in the process of thymocyte selection. We propose that under normal conditions in which mice express a diverse TCR repertoire as well as both coreceptors, the selection of CD4 and CD8 lineages occurs at different windows of TCR–MHC affinity. According to this model, CD8 lineage cells are normally positively selected at higher TCR–MHC affinities for the thymic ligand than are CD4 lineage cells. Thus, the ability of thymic ligands to positively select class I–restricted cells may depend precisely on the peptide presented, and for this reason the density of the appropriate ligand on the stromal cell may be low. In contrast, positive selection of CD4 lineage cells on MHC class II ligands may be less peptide specific, involving many low-affinity interactions with a diversity of MHC class II ligands on the stromal cell.
Figure 8. A Model of CD4/CD8 Lineage Commitment in Which the Coreceptor Involvement and TCR–MHC Affinity Contribute Separately to Lineage Choice.
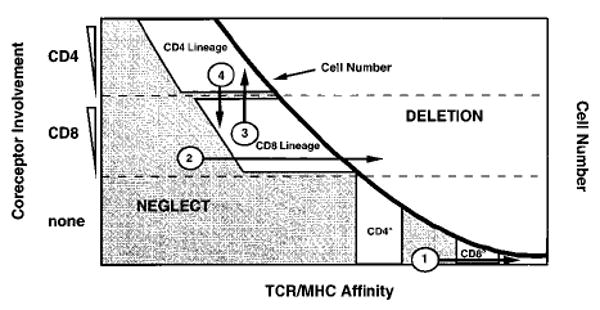
The number of cells with randomly generated TCRs at each TCR–MHC affinity explains the observation that CD4 “wannabes” are present in CD4° mice, but no CD8 “wannabes” are observed in CD8° mice. We propose that the window for positive selection of CD8 lineage cells is set at a higher TCR–MHC affinity than for CD4 lineage cells. Very few naturally generated TCRs are generated that will react with adequate affinity with MHC to compensate for the loss of CD8; however, many more cells with TCRs are generated that will interact with sufficient affinity to mediate selection into the CD4 lineage when CD4 is absent. This model also shows that no alteration in the strength of TCR–MHC interaction will redirect lineage choice from CD8 to CD4 or vice versa. Instead, increasing or decreasing TCR–MHC affinity allows only the transit from neglect, through selection of a single lineage, to deletion (arrows 1 and 2; see Discussion). Alternatively, the alteration of coreceptor contribution by signaling can redirect lineage choice, as is proposed by the quantitative, instructive model (arrows 3 and 4; see Discussion).
There is no firm evidence for this contention that CD4 cells are positively selected at lower TCR–MHC affinities than are cells of the CD8 lineage. However, in addition to explaining observations in the coreceptor-deficient mice, this hypothesis may provide an explanation for why mice engineered to express a single covalent MHC class II–peptide complex (Ignatowicz et al., 1996) or mice expressing a single, MHC class II–associated peptide due to deletion of H2-M appear to select a broad CD4 TCR repertoire (Fung-Leung et al., 1996; Martin et al., 1996; Miyazaki et al., 1996). Similar experiments have not been reported for mice expressing a single selecting peptide bound to MHC class I.
We have shown here that it is possible to compensate for the lack of CD8 expression by increasing the affinity of the TCR for the positively selecting ligand. Similarly, although it has not been demonstrated, it is assumed that the abundant CD4 “wannabes” that appear in the CD4° mice were positively selected at higher TCR–MHC affinity. The critical difference, for our argument, is that there are far fewer TCRs generated with sufficiently high affinity for MHC class I than for MHC class II to compensate for the loss of CD8 expression, explaining the dearth of CD8 “wannabes” in CD8° mice despite the abundance of CD4 “wannabes” in the CD4° mice (Figure 8).
We can also use the scheme in Figure 8 to discuss other recent work and additional implications that come from the present report. Two recent reports have provided strong support for a quantitative, instructive model for the CD4/CD8 lineage decision (Itano et al., 1996; Matechak et al., 1996). Both show that the commitment to the CD4 or CD8 lineage can be switched by altering the coreceptor involvement. According to this model, signals from the TCR of immature double positive thymocytes that can be classified as of low intensity, or low Lck involvement, favor commitment to the CD8 lineage. High-intensity signaling, or strong Lck involvement, would favor commitment to the CD4 lineage. One study showed that the involvement of the CD4 cytoplasmic tail—which associates with Lck more efficiently than does the CD8 tail (Veillette et al., 1988, Wiest et al., 1993; Ravichandran and Burakoff, 1994)—favored CD4 over CD8 lineage commitment during thymocyte development (Itano et al., 1996). Thus, the F5 (H-2Db-restricted) TCR transgenic thymocytes, which normally differentiate into the CD8 lineage in the presence of wild-type CD8, commit to the CD4 lineage in the presence of a transgenic hybrid coreceptor in which the CD8α cytoplasmic domain is replaced by the CD4 cytoplasmic domain (Figure 8, arrow 3). The OT-1 transgenic thymocytes behave similarly when expressing this hybrid coreceptor (K. A. H., unpublished). The other report showed that TCR interactions with MHC class II by AND (H-2Ek-restricted) TCR transgenic thymocytes, which could cause CD4 lineage development (or deletion at higher ligand densities) in the presence of CD4, actually promoted CD8 lineage commitment in CD4° mice (Matechak et al., 1996). These class II–restricted CD8+ cells were dependent on class I in a non-allele-specific manner, suggesting that the CD8 coreceptor was participating in this lineage choice as well (Figure 8, arrow 4).
One prediction of the quantitative instructive model is that by increasing the strength of the signal that a thymocyte receives via TCR–MHC interactions, an MHC class I–restricted thymocyte could be selected into the CD4 lineage. Our present data together with work published previously using OT-1 transgenic mice make the point that switching from CD8 to CD4 lineage commitment cannot occur by increasing or decreasing TCR affinity for the positively selecting ligand (Figure 8, arrows 1 and 2). Previously, we have shown that extensive titration of agonist peptides into FTOC of the OT-1 TCR transgenic thymocytes on a β2m° background led from no selection, into the CD8 lineage, and then to deletion of CD8 lineage cells (Hogquist et al., 1994; Hogquist et al., 1995). Importantly, we cannot push cells into the CD4 lineage by altering the ligand affinity or density. This may not be surprising with OT-1 thymocytes that express CD4 and CD8, since commitment to the CD4 lineage would result in down-regulation of CD8 and concomitant loss of signaling from the class I ligand.
However, in the case of OT-1 mice bred to the CD8° background, it is not obvious why we cannot push cells into the CD4 lineage by changing the MHC ligand for positive selection. The three ligands we have used here, E1, T1, and OVAp, vary greatly in the strength of their agonist properties (Figures 1 and 2), yet none of them induces any commitment to the CD4 lineage. One way to explain the failure of T1, a stronger agonist than E1, to select CD4+ cells from CD8°OT-1 thymocytes may be to postulate that cells that commit to the CD4 lineage following the TCR–class I interaction retain CD4 expression, which then inhibits continued signaling through the TCR by sequestering Lck (Teh et al., 1991). However, given the efficient and roughly equal selection of CD8 lineage cells by both T1 and E1 ligands, we think it is more likely that the thymocytes are preferentially committing to the CD8 lineage as a consequence of the TCR–MHC class I interaction.
In conclusion, this model separates the sum of signals received by developing thymocytes into the contribution of the coreceptor and the strength of TCR–MHC interactions. As discussed, previous reports have shown that the alteration of coreceptor contribution can redirect lineage commitment, and we propose, based on our data, that even in the absence of coreceptor, the TCR–MHC interaction may deliver an instructive signal that directs the lineage choice of developing thymocytes.
Experimental Procedures
Animals
OT-1 TCR transgenic mice that express a Vα2, Vβ5 T cell receptor from a C57BL/6-derived CTL clone recognizing the naturally processed peptide of ovalbumin, OVA257–264, presented by H-2Kb (Hogquist et al., 1994) were bred to CD8α° (Fung-Leung et al., 1991a), β2-microglobulin° (Koller et al., 1990), or C57BL/6 (Jackson Laboratory, Bar Harbor, ME) mice. Timed matings were set up by placing one male in a cage with three females for 16 hr. The day that the male was removed was considered day 1 of gestation.
Peptides
Octameric, H-2Kb binding peptides (OVA257–264 [SIINFEKL]; position 1 variants E1 [EIINFEKL] and T1 [TIINFEKL]; VOVA [RGYNYEKL]; and polySER [SSYSYSSL]) were synthesized by Research Genetics (Huntsville, AL) or with an Applied Biosystems Synergy peptide synthesizer (Foster City, CA). Peptides were purified by high-performance liquid chromatography (Research Genetics or Shimadzu, Kyoto, Japan) and quantitated by a BCA assay (Pierce, Rockford, IL). All peptides used were determined to bind H-2Kb with similar affinity using the RMA-S stabilization assay (Schumacher et al., 1990).
In Vitro Thymocyte Deletion Assay
Thymocytes from CD8°OT-1 mice or nonselecting OT-1 (β2m°OT-1) mice were harvested, and 5 × 105 thymocytes and 104 irradiated (30,000 rad) EL4 cells were aliquotted into 96-well round-bottom microtiter plates in RP10 media (RPMI 1640 [GIBCO-BRL, Life Technologies, Grand Island, NY] with 10% fetal bovine serum, penicillin, streptomycin, l-glutamine, and 2-mercaptoethanol). Peptides were added to indicated final concentrations. Cultures were incubated at 37°C, 5% CO2 for 18 hr. Following culture, thymocytes were stained for three-color flow cytometric analysis with biotin-conjugated CD69 followed by fluorescein isothiocyanate (FITC)-conjugated anti-CD8, phycoerythrin (PE)-conjugated anti-CD4(Pharmingen, San Diego, CA), and Tri-color–conjugated streptavidin (Caltag Laboratories, South San Francisco, CA). Thirty thousand events were collected for each sample. To calculate percentage deletion, the number of viable cells falling into a CD4hi gate for CD8° thymocytes or CD4hiCD8hi gate for CD8WT thymocytes was used in the following formula: percentage deletion = 100 − (100 × [percentage viable CD4hi(CD8hi) cells with peptide/percentage viable CD4hi(CD8hi) cells without peptide]) (Barnden et al., 1994). Mean fluorescence intensity of CD69 staining for total ungated cells was used to compare CD69 induction.
CTL Lysis Assay
A CTL line was established from RAG°OT-1 mice. EL4 cells were labeled with [51Cr]sodium chromate for 1 hr at 37°C. Targets were washed three times, and 104 cells were placed in wells of a 96-well round-bottom plate. 104 CTL effectors were added to each well. Test peptides were added at various concentrations. Anti-CD8 (2.43) ascites was added at a 1:200 dilution for the duration of the assay where indicated. Plates were incubated at 37°C for 4 hr. Supernatant from each well was counted, and the percentage specific lysis was calculated.
Fetal Thymic Organ Culture
Thymic lobes were harvested from mice on day 16 of gestation as previously described (Hogquist et al., 1993). Lobes were placed on cellulose ester filters (Millipore, Bedford, MA) that were set on Gelfoam sponges (Upjohn, Kalamazoo, MI) in RP10 media without or with peptide at 50 μM. Lobes were incubated in a humidity chamber at 37°C, 5% CO2. Media without or with peptide were replenished each 24 hr. Thymocytes were released from lobes by pressing through a steel mesh on day 7 of culture.
Proliferation Assay
Thymocytes were harvested from FTOC on day 7 as described above. Cells were washed three times, and quarter-lobe equivalents were added to a 96-well flat-bottom plate with 5 × 105 irradiated TAP° splenocytes with or without OVAp at a final concentration of 100 nM. After 36 hr at 37°C, 5% CO2, wells were pulsed for 8 hr with 0.5 μCi of [3H]thymidine and harvested, and incorporated radioactivity was measured (Hogquist et al., 1995). Alternatively, after 24 hr, BrdU (Sigma Chemical, St. Louis, MO) was added to cultures to a final concentration of 5 μg/ml for 18 hr. BrdU-pulsed cultures were then prepared for flow cytometric analysis to determine the phenotype of proliferating cells.
Flow Cytometry and Cell Sorting
Thymocytes were stained for FACS analysis with biotinylated anti-Vα2 (B20.1.1), anti-HSA (Pharmingen, San Diego, CA), or anti-H-2Kb (Y3) (American Type Culture Collection, Rockville, MD) followed by FITC-conjugated anti-CD8, PE-conjugated anti-CD4 (Pharmingen, San Diego, CA), and Tri-color–conjugated streptavidin (Caltag Laboratories).
Cells from BrdU-pulsed cultures were prepared for flow cytometry as previously described (Tough and Sprent, 1994). In brief, cells were surface stained with anti-Vα2, followed by PE-conjugated anti-CD4 and Tri-color–conjugated streptavidin. Next, cells were resuspended in 0.5 ml of 0.15 M NaCl, and 1.2 ml of 95% ethanol was added dropwise while mixing followed by 30 min of incubation. Cells were washed with phosphate-buffered saline (PBS) and then incubated in 1% paraformaldehyde with 0.01% Tween 20 for 30 min at room temperature followed by 30 min on ice. Cells were washed again in PBS and then incubated for 10 min with 50 Kunitz units/ml of DNAse I (Sigma Chemical) in 0.15 M NaCl, 4.2 mM MgCl2, and 10 μM HCl. Cells were centrifuged and supernatants removed, and 10 μl of FITC-conjugated anti-BrdU (Becton Dickinson, Mountain View, CA) was added for 30 min and followed by a wash with PBS. Samples were analyzed on a FACScan (Becton Dickinson) and analyzed using Reproman Software (True Facts Software, Seattle, WA). For all samples, dead cells were excluded based on forward and side scatter.
For sorting of pure populations of thymocytes from FTOC, cells were harvested and stained for CD8, CD4, and Vα2 as described above. Samples were sorted using a FACStarPLUS (Becton Dickinson, Mt. View, CA). Sorted populations were contaminated with less than 0.3% of the other sorted population.
Detection of HPRT, CD4, and CD8β mRNA by RT-PCR
Total RNA was isolated from sorted populations of thymocytes using RNA-STAT-60 (TEL-TEST “B,” Friendswood, TX) with yeast tRNA as a carrier. RNA was reverse transcribed using oligo (dT)12–18 primers and SUPERSCRIPT II reverse transcriptase (GIBCO-BRL, Life Technologies). The levels of PCR product for HPRT (hypoxanthine phosphoribosyltransferase), a housekeeping gene, were used to normalize input amounts of template for all reactions. Three-fold dilutions of approximately equivalent amounts of cDNA were used as template in PCR with primers specific for HPRT, CD4, or CD8β. The number of cycles of amplification for each primer set was determined to yield product in the linear range. Primers used to detect HPRT were specific for exons 7/8 5′-GATACAGGCCAGACTTTG TTG-3′ and exon 9 5′-GGTAGGCTGGCCTATAGGCT-3′. Primers used to detect CD4 were specific for exon 4 5′-GATCGTTTCCTCTC ATCATC-3′ and exon 6 5′-CAGGGCTGGAAGAAAGAATC-3′. Primers used to detect CD8β were specific for exon 2 5′-CTCTCTGGA GCAGCTCTGCCC-3′ and exon 3 5′-GGTTGGGGCAGTTGTAGGA AGG-3′. Half of the product from each PCR reaction was electrophoresed on a 1.75% agarose gel, which was then stained with ethidium bromide.
Acknowledgments
Correspondence should be addressed to M. J. B. We wish to thank Dr. Stephen Hedrick for helpful discussions and for first separating coreceptor and affinity contributions in a phase diagram. CD8α-deficient mice were the gift of Dr. Tak Mak. Also, we acknowledge the excellent work of D. Wilson for care of the animals, and M. Zollman and B. Dere for technical assistance. This work was supported by the following grants: National Institutes of Health A1–29802, A1–39560, and P32CA09537, and the Howard Hughes Medical Institute.
References
- Alam SM, Travers PJ, Wung JL, Nasholds W, Redpath S, Jameson SC, Gascoigne NE. T-cell receptor affinity and thymocyte positive selection. Nature. 1996;381:558–559. doi: 10.1038/381616a0. [DOI] [PubMed] [Google Scholar]
- Bachmann MF, Oxenius A, Mak TW, Zinkernagel RM. T cell development in CD8-/- mice: thymic positive selection is biased toward the helper phenotype. J Immunol. 1995;155:3727–3733. [PubMed] [Google Scholar]
- Barnden MJ, Heath WR, Rodda S, Carbone FR. Peptide antagonists that promote positive selection are inefficient at T cell activation and thymocyte deletion. Eur J Immunol. 1994;24:2452–2456. doi: 10.1002/eji.1830241029. [DOI] [PubMed] [Google Scholar]
- Bruno L, Fehling HJ, von Boehmer H. The alpha beta T cell receptor can replace the gamma delta receptor in the development of gamma delta lineage cells. Immunity. 1996;5:343–352. doi: 10.1016/s1074-7613(00)80260-5. [DOI] [PubMed] [Google Scholar]
- Crooks ME, Littman DR. Disruption of T lymphocyte positive and negative selection in mice lacking the CD8 beta chain. Immunity. 1994;1:277–285. doi: 10.1016/1074-7613(94)90079-5. [DOI] [PubMed] [Google Scholar]
- Fung-Leung WP, Schilham MW, Rahemtulla A, Kundig TM, Vollenweider M, Potter J, van Ewijk W, Mak TW. CD8 is needed for development of cytotoxic T cells but not helper T cells. Cell. 1991a;65:443–449. doi: 10.1016/0092-8674(91)90462-8. [DOI] [PubMed] [Google Scholar]
- Fung-Leung WP, Kundig TM, Zinkernagel RM, Mak TW. Immune response against lymphocytic choriomeningitis virus infection in mice without CD8 expression. J Exp Med. 1991b;174:1425–1429. doi: 10.1084/jem.174.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung-Leung WP, Louie MC, Limmer A, Ohashi PS, Ngo K, Chen L, Kawai K, Lacy E, Loh DY, Mak TW. The lack of CD8 alpha cytoplasmic domain resulted in a dramatic decrease in efficiency in thymic maturation but only a moderate reduction in cytotoxic function of CD8+ T lymphocytes. Eur J Immunol. 1993a;23:2834–2840. doi: 10.1002/eji.1830231117. [DOI] [PubMed] [Google Scholar]
- Fung-Leung WP, Wallace VA, Gray D, Sha WC, Pircher H, Teh HS, Loh DY, Mak TW. CD8 is needed for positive selection but differently required for negative selection of T cells during thymic ontogeny. Eur J Immunol. 1993b;23:212–216. doi: 10.1002/eji.1830230133. [DOI] [PubMed] [Google Scholar]
- Fung-Leung WP, Kundig TM, Ngo K, Panakos J, De SHJ, Wang E, Ohashi PS, Mak TW, Lau CY. Reduced thymic maturation but normal effector function of CD8+ T cells in CD8 beta gene-targeted mice. J Exp Med. 1994;180:959–967. doi: 10.1084/jem.180.3.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung-Leung WP, Surh CC, Liljedahl M, Pang J, Leturcq D, Peterson PA, Webb SR, Karlsson L. Antigen presentation and T cell development in H2-M-deficient mice. Science. 1996;271:1278–1281. doi: 10.1126/science.271.5253.1278. [DOI] [PubMed] [Google Scholar]
- Garcia KC, Scott CA, Brunmark A, Carbone FR, Peterson PA, Wilson IA, Teyton L. CD8 enhances formation of stable T-cell receptor/MHC class I molecule complexes. Nature. 1996;384:577–581. doi: 10.1038/384577a0. [DOI] [PubMed] [Google Scholar]
- Hogquist KA, Gavin MA, Bevan MJ. Positive selection of CD8+ T cells induced by major histocompatibility complex binding peptides in fetal thymic organ culture. J Exp Med. 1993;177:1469–1473. doi: 10.1084/jem.177.5.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogquist KA, Jameson SC, Heath WR, Howard JL, Bevan MJ, Carbone FR. T cell receptor antagonist peptides induce positive selection. Cell. 1994;76:17–27. doi: 10.1016/0092-8674(94)90169-4. [DOI] [PubMed] [Google Scholar]
- Hogquist KA, Jameson SC, Bevan MJ. Strong agonist ligands for the T cell receptor do not mediate positive selection of functional CD8+ T cells. Immunity. 1995;3:79–86. doi: 10.1016/1074-7613(95)90160-4. [DOI] [PubMed] [Google Scholar]
- Ignatowicz L, Kappler J, Marrack P. The repertoire of T cells shaped by a single MHC/peptide ligand. Cell. 1996;84:521–529. doi: 10.1016/s0092-8674(00)81028-4. [DOI] [PubMed] [Google Scholar]
- Itano A, Cado D, Chan FK, Robey E. A role for the cytoplasmic tail of the beta chain of CD8 in thymic selection. Immunity. 1994;1:287–290. doi: 10.1016/1074-7613(94)90080-9. [DOI] [PubMed] [Google Scholar]
- Itano A, Salmon P, Kioussis D, Tolaini M, Corbella P, Robey E. The cytoplasmic domain of CD4 promotes the development of CD4 lineage T cells. J Exp Med. 1996;183:731–741. doi: 10.1084/jem.183.3.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson SC, Carbone FR, Bevan MJ. Clone-specific TCR receptor antagonists of major histocompatibility complex class I-restricted cytotoxic T cells. J Exp Med. 1993;177:1541–1550. doi: 10.1084/jem.177.6.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson SC, Hogquist KA, Bevan MJ. Specificity and flexibility in thymic selection. Nature. 1994;369:750–752. doi: 10.1038/369750a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knall C, Ingold A, Potter TA. Analysis of coreceptor versus accessory molecule function of CD8 as a correlate of exogenous peptide concentration. Mol Immunol. 1994;31:875–883. doi: 10.1016/0161-5890(94)90007-8. [DOI] [PubMed] [Google Scholar]
- Koller GH, Marrack P, Kappler JW, Smithies O. Normal development of mice deficient in beta-2M, MHC class I proteins and CD8+ T cells. Science. 1990;248:1227–1230. doi: 10.1126/science.2112266. [DOI] [PubMed] [Google Scholar]
- Locksley RM, Reiner SL, Hatam F, Littman DR, Killeen N. Helper T cells without CD4: control of leishmaniasis in CD4-deficient mice. Science. 1993;261:1448–1451. doi: 10.1126/science.8367726. [DOI] [PubMed] [Google Scholar]
- Lueshcher IF, Vivier E, Layer A, Mahiou J, Godeau F, Malissen B, Romer P. CD8 modulation of T-cell antigen receptor-ligand interactions on living cytotoxic T lymphocytes. Nature. 1995;373:353–356. doi: 10.1038/373353a0. [DOI] [PubMed] [Google Scholar]
- Martin WD, Hicks GG, Mendiratta SK, Leva HI, Ruley HE, Van Kaer L. H2-M mutant mice are defective in the peptide loading of class I molecules, antigen presentation, and T cell repertoire selection. Cell. 1996;84:543–550. doi: 10.1016/s0092-8674(00)81030-2. [DOI] [PubMed] [Google Scholar]
- Matechak EO, Killeen N, Hedrick SM, Fowlkes BJ. MHC class II–specific T cells can develop in the CD8 lineage when CD4 is absent. Immunity. 1996;4:337–347. doi: 10.1016/s1074-7613(00)80247-2. [DOI] [PubMed] [Google Scholar]
- Miceli MC, Parnes JR. Role of CD4 and CD8 in T cell activation and differentiation. Adv Immunol. 1993;5:59–122. doi: 10.1016/s0065-2776(08)60498-8. [DOI] [PubMed] [Google Scholar]
- Miyazaki T, Wolf P, Tourne S, Waltzinger C, Dierich A, Barois N, Ploegh H, Benoist C, Mathis D. Mice lacking H2-M complexes enigmatic elements of the MHC class II peptide loading pathway. Cell. 1996;84:531–541. doi: 10.1016/s0092-8674(00)81029-6. [DOI] [PubMed] [Google Scholar]
- Muller KP, Kyewski BA. T cell receptor targeting to thymic cortical epithelial cells in vivo induces survival, activation and differentiation of immature thymocytes. Eur J Immunol. 1993;23:1661–1670. doi: 10.1002/eji.1830230740. [DOI] [PubMed] [Google Scholar]
- Muller KP, Kyewski BA. Intrathymic T cell receptor (TcR) targeting in mice lacking CD4 or major histocompatibility complex (MHC) class II: rescue of CD4 T cell lineage without co-engagement of TcR/CD4 by MHC class II. Eur J Immunol. 1995;25:896–902. doi: 10.1002/eji.1830250406. [DOI] [PubMed] [Google Scholar]
- Parnes JR. Molecular biology and function of CD4 and CD8. Adv Immunol. 1989;44:265–311. doi: 10.1016/s0065-2776(08)60644-6. [DOI] [PubMed] [Google Scholar]
- Potter TA, Bluestone JA, Rajan TV. A single amino acid substitution in the alpha 3 domain of an H-2 class I molecule abrogates reactivity with CTL. J Exp Med. 1987;166:956–966. doi: 10.1084/jem.166.4.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahemtulla A, Kundig TM, Narendran A, Bachmann MF, Julius M, Paige CJ, Ohashi PS, Zinkernagel RM, Mak TW. Class II major histocompatibility complex-restricted T cell function in CD4-deficient mice. Eur J Immunol. 1994;24:2213–2218. doi: 10.1002/eji.1830240942. [DOI] [PubMed] [Google Scholar]
- Ravichandran KS, Burakoff SJ. Evidence for differential intracellular signalling via CD4 and CD8 molecules. J Exp Med. 1994;179:727–732. doi: 10.1084/jem.179.2.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renard V, Romero P, Vivier E, Malissen B, Luescher IF. CD8 beta increases CD8 coreceptor function and participation in TCR-Ligand binding. J Exp Med. 1996;184:2439–2444. doi: 10.1084/jem.184.6.2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd C, Helms S, Barber EK, Schlossman SF. The CD4/CD8:p56lck complex in T lymphocytes: a potential mechanism to regulate T-cell growth. Biochem Cell Biol. 1989;67:581–589. doi: 10.1139/o89-090. [DOI] [PubMed] [Google Scholar]
- Salter RD, Benjamin RJ, Wesley PK, Buxton SE, Garrett TP, Clayberger C, Krensky AM, Norment A, Littman DR, Parham P. A binding site for the T-cell co-receptor CD8 on the a3 domain of HLA-A2. Nature. 1990;345:41–43. doi: 10.1038/345041a0. [DOI] [PubMed] [Google Scholar]
- Schumacher TN, Heemels MT, Neefjes JJ, Kast WM, Melief CJ, Ploegh HL. Direct binding of peptide to empty MHC class I molecules on intact cells and in vitro. Cell. 1990;62:563–567. doi: 10.1016/0092-8674(90)90020-f. [DOI] [PubMed] [Google Scholar]
- Sun J, Leahy DJ, Kavathas PB. Interaction between CD8 and major histocompatibility complex (MHC) class I mediated by multiple contact surfaces that include the a2 and a3 domains of MHC class I. J Exp Med. 1995;182:1275–1280. doi: 10.1084/jem.182.5.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Punt JA, Granger LG, Singer A. Asymmetric signaling requirements for thymocytes commitment to the CD4+ versus CD8+ T cell lineages: a new perspective on thymic commitment and selection. Immunity. 1995;2:413–425. doi: 10.1016/1074-7613(95)90149-3. [DOI] [PubMed] [Google Scholar]
- Takahama Y, Suzuki H, Katz KS, Grusby MJ, Singer A. Positive selection of CD4+ T cells by ligation without aggregation even in the absence of MHC. Nature. 1994;371:67–70. doi: 10.1038/371067a0. [DOI] [PubMed] [Google Scholar]
- Teh HS, Garvin AM, Forbush KA, Carlow DA, Davis CB, Littman DR, Perlmutter RM. Participation of CD4 coreceptor molecules in T-cell repertoire selection. Nature. 1991;349:241–243. doi: 10.1038/349241a0. [DOI] [PubMed] [Google Scholar]
- Tough DF, Sprent J. Turnover of naive- and memory-phenotype T cells. J Exp Med. 1994;179:1127–1135. doi: 10.1084/jem.179.4.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veillette A, Bookman MA, Horak EM, Bolen JB. The CD4 and CD8 T cell surface antigens are associated with the internal membrane tyrosine-protein kinase p56lck. Cell. 1988;55:301–308. doi: 10.1016/0092-8674(88)90053-0. [DOI] [PubMed] [Google Scholar]
- Wiest DV, Yan L, Jefferson J, Benvenieste P, Tsokos M, Klausner RD, Glimcher LH, Samelson LE, Singer A. Regulation of T cell receptor expression in immature CD4+CD8+ thymocytes by p56lck tyrosine kinase: basis for differential signaling by CD4 and CD8 in immature thymocytes expressing both coreceptor molecules. J Exp Med. 1993;178:1701–1712. doi: 10.1084/jem.178.5.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamoyska R. The CD8 coreceptor revisted: one chain good, two chains better. Immunity. 1994;1:243–246. doi: 10.1016/1074-7613(94)90075-2. [DOI] [PubMed] [Google Scholar]


