Abstract
Aims:
To determine the effect of alcohol (EtOH) on endothelial angiogenic activity and to delineate the cell signaling mechanisms involved.
Methods and Results:
Treatment of human umbilical vein endothelial cells (HUVEC) with EtOH (1-100 mM, 24 h) dose-dependently increased their network formation on matrigel (an index of angiogenesis) with a maximum response (2.5 - 3 fold increase) at 25 mM. Ethanol also stimulated the proliferation (by cell counts and PCNA expression) and migration (by scratch wound assay) of HUVEC. In parallel cultures, EtOH stimulated Notch receptor (1 and 4) and Notch target gene (hrt-1, 2, 3) mRNA and protein expression and enhanced CBF-1/RBP-Jk promoter activity. EtOH also stimulated, at a mRNA and protein level, the expression of Angiopoietin 1 (Ang1) and its Tie2 receptor in these cells. Knockdown of Notch 1 or 4 by siRNA, or inhibition of Notch-mediated CBF-1/RBP-Jk regulated gene expression by RPMS-1, inhibited both ethanol-induced Ang1/Tie2 expression in HUVEC and their network formation on matrigel. Moreover, knockdown of Ang1 or Tie2 by siRNA inhibited ethanol-induced endothelial network formation.
Conclusions:
These data demonstrate that ethanol, at levels consistent with moderate consumption, enhances endothelial angiogenic activity in vitro by stimulating a novel Notch/CBF-1/RBP-JK - Ang1/Tie2- dependent pathway. These actions of ethanol may be relevant to the cardiovascular effects of alcohol consumption purported by epidemiological studies.
INTRODUCTION
Epidemiological studies have reported a biphasic effect of alcohol on cardiovascular disease. Moderate alcohol consumption (generally defined as up to 2 drinks per day) is a negative risk factor for atherosclerosis and its clinical sequelae; coronary heart disease, ischemic stroke and peripheral vascular disease 1-3. On the other hand, chronic alcohol abuse is associated with a higher incidence of cardiovascular disorders and increased morbidity and mortality 2-4, and has been identified as a significant risk factor for some cancers, including those of the upper alimentary tract and breast 5. The precise cell signaling mechanisms mediating these diverse effects of alcohol are not fully understood.
Angiogenesis, the formation of new capillaries from the preexisting vasculature by migration, proliferation and structural rearrangement of endothelial cells, plays a fundamental role in physiology and pathology 6. It is beneficial in some clinical circumstances, such as in tissue damage after reperfusion of ischemic tissue or cardiac failure and in wound healing, but maladaptive in other situations, such as cancer, arthritis and intraplaque formation 6. Recent studies have demonstrated a role for Notch signaling during angiogenesis 7-12. The Notch pathway is an evolutionarily conserved intercellular signaling mechanism that is important in vascular development, playing a key role in vascular cell fate decisions 13 14. Notch receptors and ligands are transmembrane proteins; four Notch receptors (Notch 1-4) and five ligands (Jagged-1 and -2, Delta-1, -3, -4) have been identified in mammals. Studies using constitutively activated Notch receptors missing their extracellular domains (i.e., Notch IC or NICD) have shown that Notch signaling determines proliferation, differentiation and more recently apoptosis in vascular smooth muscle cells 15-17. Notch IC is translocated to the nucleus where it interacts with the CSL family of transcription factors (CBF-1/RBP-Jk, Su (h) and LAG-1) to become a transcriptional activator that can then modulate the expression of Notch target genes that regulate cell fate decisions. These include the “Hairy Enhancer of Split” (hes) gene and HES related transcription factors (Hrt's) that are critically involved in mammalian cell differentiation 18 19. Like vascular smooth muscle cells, Notch receptors are also expressed on adult vascular endothelial cells 20 21 22 23, though relatively little is known about their regulation and function in this cell type.
In addition to the Notch pathway, the angiopoietin family of growth factors has been the focus of growing interest in angiogenesis research 24. Angiopoietin-1 (Ang1) is a ligand for the Tie2 (tyrosine kinase with immunoglobulin-like loop and EGF homology domains) receptor expressed exclusively on endothelial cells. Besides enhancing endothelial cell migration on fibronectin and collagen in a Tie2-dependent way 25, Ang1 can also induce endothelial cell adhesion, spreading, focal contact formation and migration in a Tie2-independent manner 26 as well as rescue endothelial cells from growth factor deprivation-induced apoptosis 27.
While ethanol has been reported to variably affect angiogenesis, particularly in the context of wound healing and tumorogenesis 28, 29, an interaction between ethanol and the Notch pathway has not been previously investigated. We report here that ethanol stimulates the expression of Notch receptors and downstream target genes in human endothelial cells and furthermore that ethanol-stimulated angiogenic activity (network formation on Matrigel) is Notch/CBF-1/RBP-Jk- Angiopoietin1 dependent.
MATERIALS AND METHODS
Endothelial cell isolation and culture
Endothelial cells (HUVEC) were prepared by established methods as previously described 30. Briefly, HUVEC were harvested from pooled, unidentified human umbilical cord veins by adding 0.1% collagenase (Gibco Laboratories, Grand Island, NY) for 30 min. The cells were grown to confluence in Medium 199 (Gibco) supplemented with 10% heat inactivated FCS (Gemini Bio-Products, West Sacramento, CA), penicillin-streptomycin (Gibco), fungizone (Gibco) and endothelial cell growth factor (BD Biosciences, San Jose, CA). Cells were assessed for endothelial cell phenotype by morphology, expression of von Willebrand Factor antigen and PECAM. HUVEC between passages 2-6 were used in all experiments.
Network formation on matrigel
The wells of 96-well tissue culture plates were coated with Matrigel basement membrane matrix (100 μL per well, Becton Dickinson, Franklin lakes, NJ), which was allowed to solidify at 37°C for 30 minutes, according to the manufacturers instructions, before plating the cells. HUVEC (3×103 cells), which had been treated with or without ethanol for 24 h, were then plated at 125 μL per well onto the surface of the Matrigel and incubated at 37°C. After 16 hours the cells were photographed with the use of a CCD digital camera (Spot RT, Diagnostics Instruments, Inc) at ×4 magnification. Network formation was quantified by measuring the length of the network of connected cells in each well by drawing a line over them and measuring the length of the line in pixels with the use of SpotSoftware Version 4.6 (Diagnostic Instruments, Inc) essentially as described by us previously 31.
Cell Counts
HUVEC were seeded at 5 × 103 cells/well onto 6-well plates. They were first incubated in 0.5% FCS containing media for 24, then treated with fresh growth media (containing 10% FCS) +/− ethanol and the cells counted 24 and 48h later. The average of 3 wells was quantified using a hemocytometer. In parallel experiments, protein lysates were extracted and proliferating cell nuclear antigen (PCNA) expression was determined by Western blot analysis.
Scratch Wound Assay
HUVEC were grown to confluence and treated for 24 h with 0.5% serum media. Thereafter, each dish was divided into a 2×3 grid. With the use of a 1-200 μl pipette tip, a linear wound was made in each hemisphere of the dish. The scratch resulted in a cell-free gap or ‘wound’ of approximately 1.0 mm between two adjoining areas of HUVEC. Immediately after wounding, growth media (containing 10% FCS) with or without ethanol was added. Under a 40X lens with an attached SPOT camera (Diagnostic Instruments, Inc., Sterling Heights, MI), images were taken of the intersections of the linear wound and each grid line. This resulted in eight fields per dish. Cells were allowed to migrate over 24 h at 37°C. Each field was measured at Time 0 and at 24 h. The area covered by cells that had migrated into the wound was determined using an area measurement program (SpotSoftware, Version 4.6, Diagnostic Instruments). Duplicate dishes were used for each condition (16 measurements total). Experiments were performed in at least seven separate dishes, and the results were averaged.
Notch-Expressing Vectors and Plasmid Preparation
Epstein Barr virus-encoded gene product that binds CBF-1 (RPMS-1) was a kind gift from Prof. Paul J. Farrell, Ludwig Institute for Cancer Research, Imperial College School of Medicine, London, UK. Plasmids were prepared for transfection according to manufacturer instructions using a Qiagen plasmid Midi Kit (Qiagen, Valencia, CA) as described previously 32.
siRNA transfection
For gene silencing studies, the Gene Pulser Xcell™ system (Biorad, Hercules, CA) was used for transient transfection of HUVEC with gene-specific siRNA. Briefly, 2×105 cells were transfected with 2 μg of siRNA targeting Notch 1 or Notch 4 or Ang1 or Tie2 (Ambion, Austin, TX) or a scrambled negative control siRNA (Ambion, cat #4611) in 75 μl of siRNA electroporation buffer. Following transfection, cells were treated with or without ethanol for 24 h.
Reporter Gene Analysis
Transient transfection of HUVEC was performed using the Gene Pulser Xcell™ system (Bio-Rad). Cells were transfected with 5 μg of the firefly luciferase reporter plasmid containing the human CBF-1 promoter sequence (pGa98-1-6 was a gift from B. Kempkes) 33. The vector pGa981-6 contains the hexamerized 50-bp Epstein-Barr virus nuclear antigen 2 (EBNA-2) response element of the TP-1 promoter (ERE-TP1) in front of the minimal β-globin promoter driving the luciferase gene. 0.5 μg of the Renilla luciferase control vector (pRL-SV40; Promega) was co-transfected with the CBF-1 Luc as an internal control to normalize for transfection efficiency. Following transfection, cells were allowed to recover for 24 h before being exposed to 0, 25 mM and 50 mM ethanol for 24 h. Dual luciferase assays were performed with the Dual-Luciferase Reporter assay system (Promega). Briefly, cells were washed with PBS and harvested in 1× passive lysis buffer. Firefly and Renilla luciferase activities were read in a microplate luminometer using luciferase assay reagent II and Stop and Glo reagent, respectively. The data are represented as the ratio of firefly to Renilla luciferase activity.
Preparation of Cell Lysates
Harvested HUVEC were pelleted by low-speed centrifugation. The cell pellet was placed in ice-cold lysis buffer and subjected to ultrasonication with a sonic dismembrator (Fischer Scientific, Pittsburg, PA). Samples were divided into aliquots and stored at −80°C before use for Western blot analysis. Protein concentration was measured by the method of Bradford, with BSA used as a standard.
Western Blotting
Cell lysates were analysed for Notch 1 IC, Notch 4 IC, Ang1 and Tie2 expression by Western blot. Proteins were separated by SDS-PAGE and electrophoretically transferred to nitrocellulose membrane (Hybond-C, Amersham Pharmacia Biotech, Piscataway, NJ) by using a Mini Trans-Blot Cell (Bio-Rad, Hercules, CA) at 80 V for 1 hour. Equal transfer and loading of proteins was was confirmed by ponceau staining. Anti-Notch 1 and 4 IC antibodies were obtained from Upstate (Lake Placid, NY). Anti-Ang1 and anti-Tie2 antibodies were obtained from Chemicon (Temecula, CA).
Quantitative real-time RT-PCR (QRTPCR)
Total RNA was isolated from cells using TRIzol™ (Invitrogen, Carlsbad, CA) according to the manufacturer's specifications. Total RNA (1-2 μg) was reverse-transcribed using iscript™ cDNA Synthesis kit from BIO-RAD (Carlsbad, CA). The gene-specific oligonucleotide sequences were; Notch 1, forward 5′ CAGGGTGTGCACTGTGAGAT 3′, reverse 5′ GACAGGCACTCGTTGACATC 3′, Notch 4, forward 5′ CTAGGGGCTCTTCTCGTCCT 3′, reverse 5′ CAACTTCTGCCTTTGGCTTC 3′, HRT1, forward 5′ CGAGGTGGAGAAGGAGAGTG 3′, reverse 5′ CTGGGTACCAGCCTTCTCAG 3′, HRT2, forward 5′ GTACCTGAGCTCCGTGGAAG 3′, reverse 5′ AGTTGTGGAGAGGCGACAAG 3′, HRT3, forward 5′ GGTGGGACAGGATTCTTTGA 3′, reverse 5′ AGCTGTTGAGGTGGGAGAGA 3′, Ang1, forward 5′ GAAGGGAACCGAGCCTATTC 3′, reverse 5′ GGGCACATTTGCACATACAG 3′, Tie2, forward 5′ TACACCTGCCTCATGCTCAG 3′, reverse 5′ TTCACAAGCCTTCTCACACG 3′, gapdh, forward 5′ CGAGATCCCTCCAAAATCAA 3′, reverse 5′ TTCACACCCATGGACGAACAT 3′. For quantitative measurement of mRNA, Real-Time RT-PCR was performed using the Stratagene MX3005 machine and the SYBER green jumpstart PCR kit (Sigma, St. Louis, MO) as described by the manufacturer.
Statistics
The data shown are the mean ± S.E.M. n=number of individual experiments, with a minimum of 3 independent experiments performed. Statistical significance was estimated using the following analysis: Unpaired Student's t-test for comparison of two groups; Wilcoxon signed rank test for the densitometric data. When >2 groups were present, ANOVA (factorial design) was used (GraphPad Prism). A value of P<0.05 was considered significant.
RESULTS
Ethanol stimulates endothelial cell pro-angiogenic activity
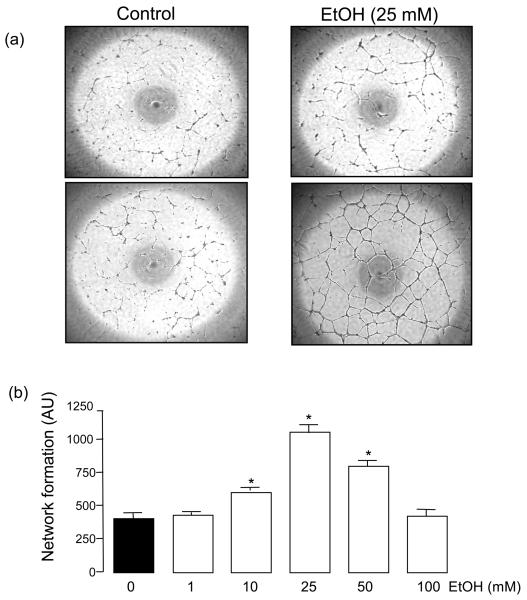
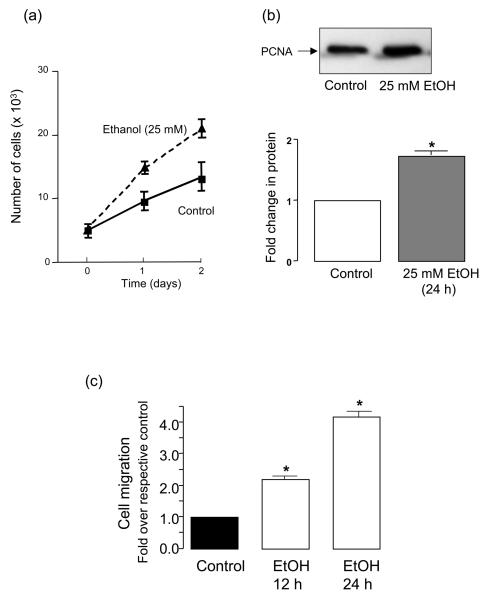
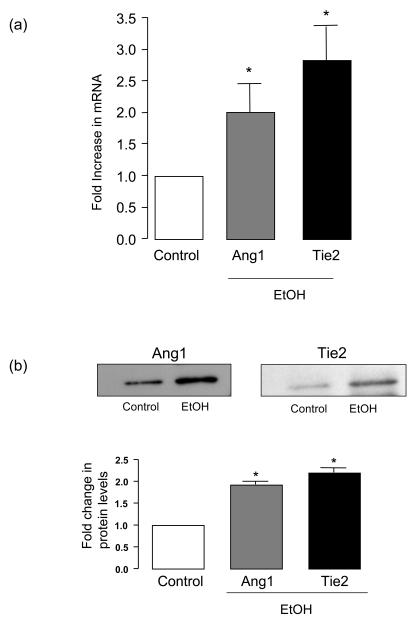
Exposure of human umbilical vein endothelial cells (HUVEC) to ethanol (EtOH 1-100 mM, 24 h) dose-dependently increased their network formation on Matrigel with a maximum response (2.5-3 fold) at 25 mM (Figure 1). Similar results were obtained in human coronary artery and bovine lung microvascular endothelial cells (results not shown). Ethanol treatment also stimulated HUVEC proliferation (assessed by cell counts and PCNA expression) (Figure 2a,b) and migration (assessed by scratch wound assay) (Figure 2c). The increase in HUVEC network formation, proliferation and migration following ethanol treatment was concomitant with increased angiopoietin-1 (Ang1) and tyrosine kinase receptor Tie2 mRNA (Figure 3a) and protein (Figure 3b) levels in these cells.
Figure 1. Ethanol stimulates endothelial cell angiogenic activity.
HUVEC were treated with or without ethanol (EtOH, 1-100mM) for 24 h. Angiogenic activity was then assessed by measuring network formation on Matrigel as described in Methods. (a) Representative images showing network formation on Matrigel of control and ethanol treated (EtOH, 25 mM) cells from 2 different experiments and (b) the dose-response cumulative data, mean ± SEM (n=3); *P<0.05 vs control.
Figure 2. Ethanol stimulates HUVEC growth and migration.
HUVEC were treated with growth media in the absence (control) or presence of ethanol (25 mM) as described in Methods. (a) Cell counts of parallel triplicate wells were made on a daily basis. (b) Proliferating cell nuclear antigen (PCNA) protein expression in control and ethanol treated (25 mM, 24 h) endothelial cells; representative Western blot (top), cumulative densitometric data, n=3 (bottom). (c) Bar graph showing increased migration (scratch wound assay) by ethanol treated HUVEC at 12 and 24 h compared to control cells. *p<0.05 vs respective control.
Figure 3. Ethanol stimulates endothelial cell Angiopoietin 1/Tie2 expression.
HUVEC were treated without (control) or with EtOH (25mM) for 24h. (a) QRTPCR analysis of angiopoietin-1 (Ang1) and Tie2 mRNA expression. Data are normalized to GAPDH, mean ±SEM, n=3. (b) Western blot analysis of Ang1 and Tie2 protein levels in control and ethanol treated endothelial cells. Representative blots shown, together with cumulative data, mean ± SEM, n=3.
Ethanol stimulates Notch Signaling in endothelial cells
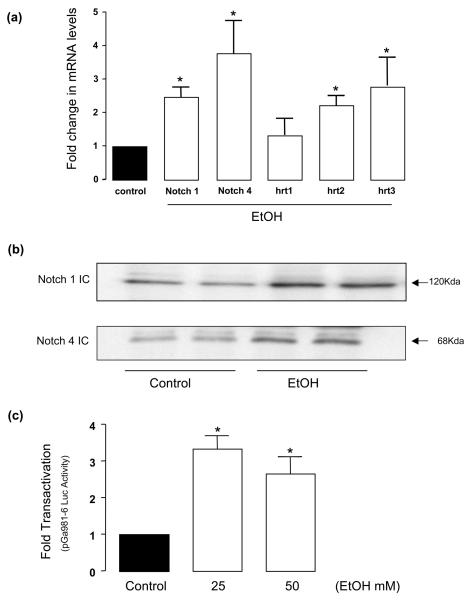
HUVEC were treated with ethanol (25 mM, 24 h) and Notch receptor mRNA and protein levels were determined by QRT-PCR and Western blot analysis, respectively. HUVEC treated with ethanol had significantly increased Notch receptor 1 and 4 mRNA and Notch 1 and 4 IC protein expression; 2.48±0.33 and 3.77±1 fold increase, respectively, for mRNA levels (Figure 4a) and 1.89±0.2 and 1.74±0.06 increase, respectively, for protein (Figure 4b). Moreover, HUVEC Notch target gene (hrt-1, 2 and 3) mRNA levels were increased following ethanol treatment (Figure 4a).
Figure 4. Ethanol stimulates Notch signaling in HUVEC.
Endothelial cells were treated without (control) or with EtOH (25mM) for 24h. (a) QRTPCR analysis of Notch 1 and 4 receptors and Notch target gene hrt-1, 2 and 3. Data were normalized to GAPDH and represent the mean ± SEM values from three independent experiments. (b) Representative Western blots of Notch 1 and 4 IC showing increase in protein expression following ethanol treatment. (c) The effect of ethanol on CBF-1/RBP-Jk promoter activity determined by luciferase assay as described in Methods. Data represent the mean ± SEM, n=3. *P<0.05 vs control.
Ethanol stimulates CBF-1/RBP-JK Promoter Activity
To determine the effect of ethanol at the level of transcriptional regulation, CBF-1/RBP-JK promoter activity was measured in HUVEC treated with or without ethanol for 24 h. In cells co-transfected with the luciferase reporter plasmid pGa981-6, which contains a CBF-1 regulated enhancer linked to the β-globin minimal promoter, there was a significant increase in CBF-1/RBP-Jk-dependent promoter activity of 3.35±.39 and 2.66±.49 fold for 25 mM and 50 mM Ethanol, respectively, when compared to control, untreated cells (Figure 4c).
Ethanol-induced angiogenic response is Notch-dependent
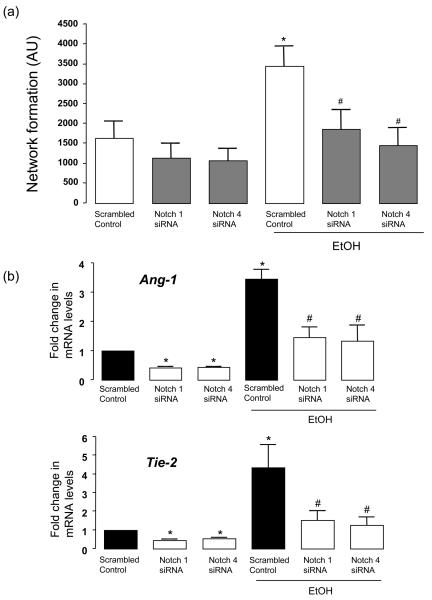
Gene silencing of Notch 1 or Notch 4 receptors with specific targeted siRNA duplexes was confirmed at the mRNA and protein level; >70% decrease in Notch 1 and Notch 4 mRNA and protein expression when compared to scrambled control transfected cells (data not shown). Moreover, knockdown of Notch 1 or Notch 4 with specific siRNA significantly attenuated ethanol-induced HUVEC network formation on matrigel in the absence of any significant effect on network formation by control HUVEC (Figure 5a); network length (AU) =3221±779 vs 1874±532 and 1468±479 for EtOH vs Notch 1 and Notch 4 siRNA, respectively. Moreover, Notch 1 and Notch 4 siRNA reduced Ang-1 and Tie-2 mRNA expression in control HUVEC and inhibited ethanol-induced Ang1/Tie2 mRNA expression (Figure 5b).
Figure 5. siRNA-directed knockdown of Notch 1 and Notch 4 inhibits EtOH-induced network formation and Ang1 and Tie2 mRNA.
HUVEC transfected with scrambled RNA (scrambled control) or with an siRNA targeted to Notch 1 or Notch 4 were treated without or with EtOH (25mM, 24h) before (a) Network formation on matrigel was assessed (cumulative data from 3 separate experiments conducted in triplicate shown. *P<0.05 vs scrambled control. #p<0.05 vs EtOH treated scrambled control) or (b) Ang1 and Tie2 mRNA levels were analyzed by QRTPCR. Data were normalized to GAPDH and represent the mean ± SEM values from three independent experiments. *P<0.05 vs scrambled control. #p<0.05 vs EtOH treated scrambled control.
Ethanol-induced angiogenic response is CBF-1/RBP-JK-dependent
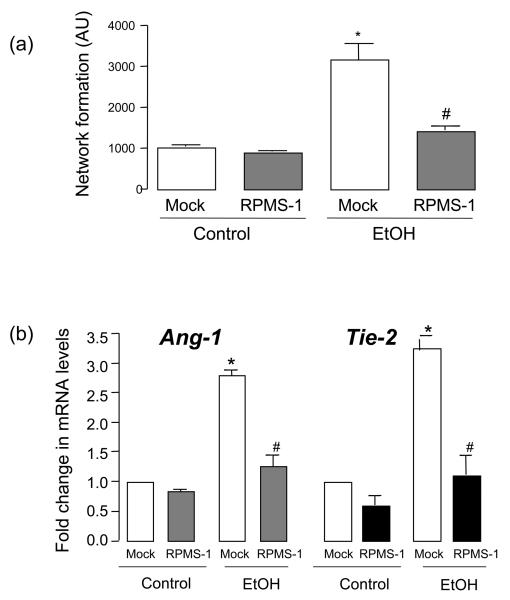
Blockade of endogenous Notch mediated CBF-1/RBP-JK regulated gene expression using Epstein-Barr virus encoded RPMS-1 resulted in a complete inhibition of ethanol-induced HUVEC network formation, in the absence of any effect on control cells (Figure 6a). Furthermore, RPMS-1 inhibited ethanol-stimulated Ang-1 and Tie-2 mRNA, in the absence of any effect in control HUVEC (Figure 6b).
Figure 6. Ethanol stimulated network formation and Ang1/Tie2 expression is CBF-1/RBP-JK-dependent.
HUVEC mock transfected (p7pcmv) or transfected with RPMS-1 (an Epstein Barr virus-encoded gene product that binds CBF-1) were treated with or without EtOH (25mM, 24 h) before their network formation on Matrigel and Ang1, Tie2 expression were determined. (a) Network formation data (cumulative data, n=3). (b) QRTPCR analysis of Ang1 and Tie2 mRNA. Data were normalized to GAPDH and represent the mean ± SEM values from three independent experiments. *P<0.05 vs mock transfected, #p<0.05 vs EtOH treated mock transfected. (c) siRNA-directed knockdown of Ang1 and Tie2 inhibits ethanol-stimulated network formation. Representative Western blots for Angiopoietin 1 (Ang1) and Tie2 protein following respective siRNA knockdown. HUVEC transfected with scrambled RNA (control) or with an siRNA targeted to Ang1 or Tie2 were treated with or without ethanol (EtOH, 25 mM, 24 h) before their network formation on Matrigel was determined. *P<0.05 vs control, #p<0.05 vs EtOH treated control.
siRNA-directed knockdown of Ang1 or Tie2 inhibits Ethanol-induced HUVEC network formation
Gene silencing of Ang1 or Tie2 with specific targeted siRNA duplexes was confirmed at the mRNA and protein level (data not shown, and Figure 6c). Inhibition of Ang1 or Tie2 with the respective siRNA had no effect on network formation by control HUVEC, but completely inhibited ethanol-induced HUVEC network formation (Figure 6c).
Discussion
Here we report for the first time that alcohol, at levels consistent with moderate consumption, stimulates a novel Notch-Angiopoietin1/Tie2 signaling pathway in endothelial cells, resulting in increased angiogenic activity.
Angiogenesis is associated with several pathologies including cardiovascular disease, chronic inflammation, cancer and wound healing and depending on the circumstance, can be beneficial or deleterious. Thus, manipulation of angiogenesis is an important clinical goal in many disease fields. With respect to cardiovascular disease, specifically, the development of new compensatory blood vessels involving both angiogenesis and arteriogenesis in response to occlusion ischemia, plays an important role in protecting tissues from ischemic damage and its stimulation has emerged as a principal therapeutic approach 34. Interestingly, clinical observations detail substantial differences in the extent of collateralization among patients with coronary artery disease, with some individuals demonstrating marked abundance and others showing nearly complete absence of these vessels 34 35. Factors responsible for the degree of collateralization are poorly understood but it is likely that genetic and lifestyle factors play a role.
A few studies have previously examined the relationship between ethanol and angiogenesis. Radek et al., recently reported that acute ethanol exposure inhibited angiogenesis in the context of wound healing 28. On the other hand and in agreement with our study, several groups report a stimulatory effect of ethanol on angiogenesis in a variety of in vivo and in vitro models 36 37 29 38 39. The mechanisms involved included ethanol stimulation of angiogenic growth factors such as VEGF 29 37, bFGF, 38 and TGF-beta1 38, while Qian et al., provided evidence of a signaling pathway linking ethanol-induced changes in Cdc42, H2O2, actin filaments and cell motility to in vitro angiogenesis 40. Endothelial cell proliferation and migration are central to the process of new blood vessel formation and our data also demonstrate a stimulatory effect of ethanol on HUVEC growth and migration. Of interest, we and others have previously demonstrated an inhibitory effect of ethanol on vascular smooth muscle cell proliferation 41 42 43. Taken with our current data this indicates a differential, cell specific effect of ethanol on vascular cell growth.
While it is well established that Notch receptors are important mediators of cell fate during embryogenesis 13 44, their role in adult physiology, and in particular in postnatal angiogenesis, is only beginning to be appreciated. The role of Notch signaling in angiogenesis has been assessed by manipulating the expression of different components in endothelial cells. Activation of Notch signaling by ectopic expression of Notch IC or Hes1 has been found to result in enhanced network formation of arterial endothelial cells 7. In that study, Notch-induced modulation occurred via an RBP-JK dependent mechanism resulting in the upregulation of several Notch target genes including Hes1. Moreover, inhibition of RBP-JK-dependent Notch signaling in human arterial endothelial cells resulted in attenuation of VEGF-driven network and cord formation in a 3D collagen angiogenesis model 7. Takeshita et al., recently elegantly demonstrated that endothelial Notch1 mediates the VEGF-induced angiogenic response to limb ischemia 10. They found an impaired angiogenic response in ecN1+/− mice and an inhibition of VEGF-induced endothelial proliferation, migration and survival by the γ-secretase inhibitor DAPT 10. Their data place Notch1 downstream of VEGF. These authors did not, however, provide any information as to the precise mechanism whereby Notch1 regulates angiogenesis. A recent study by Limbourg et al., also using the hind limb ischemia model, identified Notch ligand Delta-like 1 (Dll1) as an essential regulator of postnatal arteriogenesis 12. Notch signaling has also been implicated in the process of tumor angiogenesis 11 45 9.
Given that ethanol reportedly stimulates VEGF 29 38, and that there is evidence of an interaction between Notch signaling and VEGF 7 46, the possibility that stimulation of the Notch pathway in endothelial cells by ethanol is mediated via its effect on VEGF warrants investigation.
An increasing amount of attention has been directed toward the role of the angiopoietin family of growth factors in angiogenesis. Angiopoietin-1 (Ang1) is a ligand for the Tie2 (tyrosine kinase with immunoglobulin-like loop and EGF homology domains) receptor expressed mainly on endothelial cells 47. Ang1 is essentially involved in maturation, stabilization and remodeling of blood vessels through inducing tyrosine kinase receptor auto-phosphorylation, promoting endothelial cell migration and survival 47. In our study, ethanol treatment stimulated HUVEC Notch 1 and 4 mRNA and protein expression and downstream target gene hrt 2 and 3 mRNA levels. Ethanol also stimulated Ang1 and Tie2 mRNA and protein expression in these cells and increased angiogenic activity as assessed by network formation on Matrigel. Knockdown of Notch 1 or 4 by siRNA, or inhibition of Notch mediated CBF-1/RBP-Jk regulated gene expression by RPMS-1, which competes at the SKIP/SMART complex of CBF-1, inhibited both ethanol-induced Ang1/Tie2 expression and ethanol-induced HUVEC network formation. Moreover, knockdown of Ang1 or Tie2 by siRNA inhibited ethanol-induced endothelial network formation. Thus, collectively these data demonstrate that a pathway involving Ang1/Tie2 downstream of Notch mediates the ethanol-induced in vitro angiogenic response in these cells, and that Ang1/Tie2 may be a key mediator of Notch-induced angiogenesis in adult cells. Of note, we have recently reported that cyclic strain-stimulated angiogenesis is also mediated by a Notch-dependent, Ang1/Tie2 pathway 23, suggesting that alcohol and mechanical forces, two very different stimuli, may share common signaling mechanisms.
‘Moderate’ alcohol consumption is generally recognized to be one to three drinks per day giving rise to blood alcohol levels (BAL) of ~5-25 mM 48 2. A BAL of 0.1 g% is approximately equivalent to 25 mM ethanol. Thus, the concentration of ethanol used in the majority of our experiments can be considered in the moderate range. The precise mechanism whereby ethanol stimulates the Notch pathway in endothelial cells remains to be determined. Interaction of the Notch receptor with a ligand initiates proteolytic cleavage at the extracellular site by α-secretase, followed by cleavage at the intracellular site by γ-secretase, (which is dependent on the presence of presenilins), resulting in the release of Notch IC from the cytoplasmic side of the cell membrane. Notch-IC is then translocated into the nucleus where it interacts primarily with CSL and recruits coactivators to form a transcription–activating complex. Notch-IC can be polyubiquitylated and targeted for degradation in a proteosome-dependent manner. Thus, there are several potential regulatory points in this pathway that could be affected by ethanol and which warrant investigation. Nevertheless, our data demonstrate increased Notch IC and target gene expression as well as enhanced CBF-1/RBP-Jk promoter activity in HUVEC with ethanol treatment.
According to the American Heart Association (Cardiovascular Disease (CVD) Statistics 2004), heart attacks and other forms of cardiovascular disease result in approximately 800,000 deaths annually in the USA, accounting for 36% of the nations total mortality. Epidemiologic studies from more than 20 countries demonstrate a 20-40% lower CVD incidence among drinkers compared with non-drinkers. The precise cellular and signaling mechanisms mediating these cardiovascular protective effects of alcohol are not known. Our in vitro data, demonstrating an ethanol-induced increase in endothelial cell angiogenic activity mediated by a Notch/CBF-1/RBP-Jk - Ang1/Tie2-dependent pathway provides a possible mechanistic pathway whereby moderate alcohol consumption could potentially improve survival in the setting of atherosclerotic-induced arterial occlusion and ischemia. Of interest, in addition to CVD, included in the list of diseases that epidemiological studies indicate moderate alcohol as being either a positive or negative risk factor are rheumatoid arthritis 49 50, diabetes mellitus 51, Alzheimers disease 52 and certain cancers 5, all of which have an angiogenesis component. It is therefore tempting to speculate that the novel Notch-dependent effect of ethanol on in vitro angiogenesis reported here, may be relevant to the mechanism of the varied effects of alcohol consumption on the progression of these seemingly unrelated diseases. Further in vivo investigation is merited to establish whether or not this is the case.
Acknowledgments
Funding
This work was supported in parts by grants from the National Institutes of Health (AA-12610 to EMR), the American Heart Association (Grant-in-Aid 0555785T to EMR), and by grants from the Welcome Trust and the Health Research Board of Ireland (PAC). DM is the recipient of a Postdoctoral Fellowship from the American Heart Association (0625890T). JPC is the recipient of a Scientist Development grant from the American Heart Association (0435237N).
Footnotes
There are no conflicts of interest for any of the authors (DM, JPC, PAC, EMR).
References
- 1.Pearson TA. Alcohol and heart disease. Circulation. 1996;94:3023–3025. doi: 10.1161/01.cir.94.11.3023. [DOI] [PubMed] [Google Scholar]
- 2.Thun MJ, Peto R, Lopez AD, Monaco JH, Henley SJ, Heath CW, Jr., et al. Alcohol consumption and mortality among middle-aged and elderly U.S. adults. New Engl J Med. 1997;337:1705–1714. doi: 10.1056/NEJM199712113372401. [DOI] [PubMed] [Google Scholar]
- 3.Di Castelnuovo A, Costanzo S, Bagnardi V, Donati MB, Iacoviello L, de Gaetano G. Alcohol dosing and total mortality in men and women: an updated meta-analysis of 34 prospective studies. Arch Intern Med. 2006;166:2437–2445. doi: 10.1001/archinte.166.22.2437. [DOI] [PubMed] [Google Scholar]
- 4.Mukamal KJ, Maclure M, Muller JE, Mittleman MA. Binge drinking and mortality after acute myocardial infarction. Circulation. 2005;112:3839–3845. doi: 10.1161/CIRCULATIONAHA.105.574749. [DOI] [PubMed] [Google Scholar]
- 5.Poschl G, Seitz HK. Alcohol and cancer. Alcohol Alcoholism. 2004;39:155–165. doi: 10.1093/alcalc/agh057. [DOI] [PubMed] [Google Scholar]
- 6.Carmeliet P. Angiogenesis in life, disease and medicine. Nature. 2005;438:932–936. doi: 10.1038/nature04478. [DOI] [PubMed] [Google Scholar]
- 7.Liu ZJ, Shirakawa T, Li Y, Soma A, Oka M, Dotto GP, et al. Regulation of Notch1 and Dll4 by vascular endothelial growth factor in arterial endothelial cells: implications for modulating arteriogenesis and angiogenesis. Mol Cell Biol. 2003;23:14–25. doi: 10.1128/MCB.23.1.14-25.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ridgway J, Zhang G, Wu Y, Stawicki S, Liang WC, Chanthery Y, et al. Inhibition of Dll4 signalling inhibits tumour growth by deregulating angiogenesis. Nature. 2006;444:1083–1087. doi: 10.1038/nature05313. [DOI] [PubMed] [Google Scholar]
- 9.Hainaud P, Contreres JO, Villemain A, Liu LX, Plouet J, Tobelem G, et al. The Role of the Vascular Endothelial Growth Factor-Delta-like 4 Ligand/Notch4-Ephrin B2 Cascade in Tumor Vessel Remodeling and Endothelial Cell Functions. Cancer Res. 2006;66:8501–8510. doi: 10.1158/0008-5472.CAN-05-4226. [DOI] [PubMed] [Google Scholar]
- 10.Takeshita K, Satoh M, Ii M, Silver M, Limbourg FP, Mukai Y, et al. Critical role of endothelial Notch1 signaling in postnatal angiogenesis. Circ Res. 2007;100:70–78. doi: 10.1161/01.RES.0000254788.47304.6e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rehman AO, Wang CY. Notch signaling in the regulation of tumor angiogenesis. Trends Cell Biol. 2006;16:293–300. doi: 10.1016/j.tcb.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 12.Limbourg A, Ploom M, Elligsen D, Sorensen I, Ziegelhoeffer T, Gossler A, et al. Notch ligand Delta-like 1 is essential for postnatal arteriogenesis. Circ Res. 2007;100:363–371. doi: 10.1161/01.RES.0000258174.77370.2c. [DOI] [PubMed] [Google Scholar]
- 13.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 14.Shawber CJ, Kitajewski J. Notch function in the vasculature: insights from zebrafish, mouse and man. Bioessays. 2004;26:225–234. doi: 10.1002/bies.20004. [DOI] [PubMed] [Google Scholar]
- 15.Sweeney C, Morrow D, Birney YA, Coyle S, Hennessy C, Scheller A, et al. Notch 1 and 3 receptor signaling modulates vascular smooth muscle cell growth, apoptosis, and migration via a CBF-1/RBP-Jk dependent pathway. Faseb J. 2004;18:1421–1423. doi: 10.1096/fj.04-1700fje. [DOI] [PubMed] [Google Scholar]
- 16.Wang W, Campos AH, Prince CZ, Mou Y, Pollman MJ. Coordinate Notch3-hairy-related transcription factor pathway regulation in response to arterial injury. Mediator role of platelet-derived growth factor and ERK. J Biol Chem. 2002;277:23165–23171. doi: 10.1074/jbc.M201409200. [DOI] [PubMed] [Google Scholar]
- 17.Wang W, Prince CZ, Hu X, Pollman MJ. HRT1 modulates vascular smooth muscle cell proliferation and apoptosis. Biochem Biophs Res Co. 2003;308:596–601. doi: 10.1016/s0006-291x(03)01453-0. [DOI] [PubMed] [Google Scholar]
- 18.Lai EC. Keeping a good pathway down: transcriptional repression of Notch pathway target genes by CSL proteins. EMBO Rep. 2002;3:840–845. doi: 10.1093/embo-reports/kvf170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iso T, Kedes L, Hamamori Y. HES and HERP families: multiple effectors of the Notch signaling pathway. J Cell Physiol. 2003;194:237–255. doi: 10.1002/jcp.10208. [DOI] [PubMed] [Google Scholar]
- 20.Shutter JR, Scully S, Fan W, Richards WG, Kitajewski J, Deblandre GA, et al. Dll4, a novel Notch ligand expressed in arterial endothelium. Gene Dev. 2000;14:1313–1318. [PMC free article] [PubMed] [Google Scholar]
- 21.Uyttendaele H, Marazzi G, Wu G, Yan Q, Sassoon D, Kitajewski J. Notch4/int-3, a mammary proto-oncogene, is an endothelial cell-specific mammalian Notch gene. Development. 1996;122:2251–2259. doi: 10.1242/dev.122.7.2251. [DOI] [PubMed] [Google Scholar]
- 22.Tsai S, Fero J, Bartelmez S. Mouse Jagged2 is differentially expressed in hematopoietic progenitors and endothelial cells and promotes the survival and proliferation of hematopoietic progenitors by direct cell-to-cell contact. Blood. 2000;96:950–957. [PubMed] [Google Scholar]
- 23.Morrow D, Cullen JP, Cahill PA, Redmond EM. Cyclic strain regulates the Notch/CBF-1 signaling pathway in endothelial cells: role in angiogenic activity. Arterioscl Thromb Vas. 2007;27:1289–1296. doi: 10.1161/ATVBAHA.107.142778. [DOI] [PubMed] [Google Scholar]
- 24.Fiedler U, Augustin HG. Angiopoietins: a link between angiogenesis and inflammation. Trends Immunol. 2006;27:552–558. doi: 10.1016/j.it.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 25.Witzenbichler B, Maisonpierre PC, Jones P, Yancopoulos GD, Isner JM. Chemotactic properties of angiopoietin-1 and -2, ligands for the endothelial-specific receptor tyrosine kinase Tie2. J Biol Chem. 1998;273:18514–18521. doi: 10.1074/jbc.273.29.18514. [DOI] [PubMed] [Google Scholar]
- 26.Carlson TR, Feng Y, Maisonpierre PC, Mrksich M, Morla AO. Direct cell adhesion to the angiopoietins mediated by integrins. J Biol Chem. 2001;276:26516–26525. doi: 10.1074/jbc.M100282200. [DOI] [PubMed] [Google Scholar]
- 27.Harfouche R, Hassessian HM, Guo Y, Faivre V, Srikant CB, Yancopoulos GD, et al. Mechanisms which mediate the antiapoptotic effects of angiopoietin-1 on endothelial cells. Microvasc Res. 2002;64:135–147. doi: 10.1006/mvre.2002.2421. [DOI] [PubMed] [Google Scholar]
- 28.Radek KA, Matthies AM, Burns AL, Heinrich SA, Kovacs EJ, Dipietro LA. Acute ethanol exposure impairs angiogenesis and the proliferative phase of wound healing. Am J Physiol-Heart C. 2005;289:H1084–1090. doi: 10.1152/ajpheart.00080.2005. [DOI] [PubMed] [Google Scholar]
- 29.Gu JW, Bailey AP, Sartin A, Makey I, Brady AL. Ethanol stimulates tumor progression and expression of vascular endothelial growth factor in chick embryos. Cancer. 2005;103:422–431. doi: 10.1002/cncr.20781. [DOI] [PubMed] [Google Scholar]
- 30.Redmond EM, Cullen JP, Cahill PA, Sitzmann JV, Stefansson S, Lawrence DA, et al. Endothelial cells inhibit flow-induced smooth muscle cell migration: role of plasminogen activator inhibitor-1. Circulation. 2001;103:597–603. doi: 10.1161/01.cir.103.4.597. [DOI] [PubMed] [Google Scholar]
- 31.Cullen JP, Sayeed S, Sawai RS, Theodorakis NG, Cahill PA, Sitzmann JV, et al. Pulsatile flow-induced angiogenesis: role of G(i) subunits. Arterioscl Thromb Vas. 2002;22:1610–1616. doi: 10.1161/01.atv.0000034470.37007.58. [DOI] [PubMed] [Google Scholar]
- 32.Morrow D, Sweeney C, Birney YA, Cummins PM, Walls D, Redmond EM, et al. Cyclic strain inhibits Notch receptor signaling in vascular smooth muscle cells in vitro. Circulation Res. 2005;96:567–575. doi: 10.1161/01.RES.0000159182.98874.43. [DOI] [PubMed] [Google Scholar]
- 33.Dumont E, Fuchs KP, Bommer G, Christoph B, Kremmer E, Kempkes B. Neoplastic transformation by Notch is independent of transcriptional activation by RBP-J signalling. Oncogene. 2000;19:556–561. doi: 10.1038/sj.onc.1203352. [DOI] [PubMed] [Google Scholar]
- 34.Koerselman J, van der Graaf Y, de Jaegere PP, Grobbee DE. Coronary collaterals: an important and underexposed aspect of coronary artery disease. Circulation. 2003;107:2507–2511. doi: 10.1161/01.CIR.0000065118.99409.5F. [DOI] [PubMed] [Google Scholar]
- 35.Hansen JF. Coronary collateral circulation: clinical significance and influence on survival in patients with coronary artery occlusion. Am Heart J. 1989;117:290–295. doi: 10.1016/0002-8703(89)90771-0. [DOI] [PubMed] [Google Scholar]
- 36.Jones MK, Sarfeh IJ, Tarnawski AS. Induction of in vitro angiogenesis in the endothelial-derived cell line, EA hy926, by ethanol is mediated through PKC and MAPK. Biochem Bioph Res Co. 1998;249:118–123. doi: 10.1006/bbrc.1998.9095. [DOI] [PubMed] [Google Scholar]
- 37.Gu JW, Elam J, Sartin A, Li W, Roach R, Adair TH. Moderate levels of ethanol induce expression of vascular endothelial growth factor and stimulate angiogenesis. Am J Physiol-Reg I. 2001;281:R365–372. doi: 10.1152/ajpregu.2001.281.1.R365. [DOI] [PubMed] [Google Scholar]
- 38.Gavin TP, Wagner PD. Acute ethanol increases angiogenic growth factor gene expression in rat skeletal muscle. J Appl Physiol. 2002;92:1176–1182. doi: 10.1152/japplphysiol.00929.2001. [DOI] [PubMed] [Google Scholar]
- 39.Bora PS, Kaliappan S, Xu Q, Kumar S, Wang Y, Kaplan HJ, et al. Alcohol linked to enhanced angiogenesis in rat model of choroidal neovascularization. FEBS J. 2006;273:1403–1414. doi: 10.1111/j.1742-4658.2006.05163.x. [DOI] [PubMed] [Google Scholar]
- 40.Qian Y, Luo J, Leonard SS, Harris GK, Millecchia L, Flynn DC, et al. Hydrogen peroxide formation and actin filament reorganization by Cdc42 are essential for ethanol-induced in vitro angiogenesis. J Biol Chem. 2003;278:16189–16197. doi: 10.1074/jbc.M207517200. [DOI] [PubMed] [Google Scholar]
- 41.Hendrickson RJ, Cahill PA, McKillop IH, Sitzmann JV, Redmond EM. Ethanol inhibits mitogen activated protein kinase activity and growth of vascular smooth muscle cells in vitro. Eur J Pharmacol. 1998;362:251–259. doi: 10.1016/s0014-2999(98)00771-7. [DOI] [PubMed] [Google Scholar]
- 42.Sayeed S, Cullen JP, Coppage M, Sitzmann JV, Redmond EM. Ethanol differentially modulates the expression and activity of cell cycle regulatory proteins in rat aortic smooth muscle cells. Eur J Pharmacol. 2002;445:163–170. doi: 10.1016/s0014-2999(02)01761-2. [DOI] [PubMed] [Google Scholar]
- 43.Ghiselli G, Chen J, Kaou M, Hallak H, Rubin R. Ethanol inhibits fibroblast growth factor-induced proliferation of aortic smooth muscle cells. Arterioscl Thromb Vas. 2003;23:1808–1813. doi: 10.1161/01.ATV.0000090140.20291.CE. [DOI] [PubMed] [Google Scholar]
- 44.Krebs LT, Xue Y, Norton CR, Shutter JR, Maguire M, Sundberg JP, et al. Notch signaling is essential for vascular morphogenesis in mice. Gene Dev. 2000;14:1343–1352. [PMC free article] [PubMed] [Google Scholar]
- 45.Li JL, Harris AL. Notch signaling from tumor cells: a new mechanism of angiogenesis. Cancer cell. 2005;8:1–3. doi: 10.1016/j.ccr.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 46.Lawson ND, Vogel AM, Weinstein BM. sonic hedgehog and vascular endothelial growth factor act upstream of the Notch pathway during arterial endothelial differentiation. Dev Cell. 2002;3:127–136. doi: 10.1016/s1534-5807(02)00198-3. [DOI] [PubMed] [Google Scholar]
- 47.Eklund L, Olsen BR. Tie receptors and their angiopoietin ligands are context-dependent regulators of vascular remodeling. Exp Cell Res. 2006;312:630–641. doi: 10.1016/j.yexcr.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 48.Mukamal KJ, Conigrave KM, Mittleman MA, Camargo CA, Jr., Stampfer MJ, Willett WC, et al. Roles of drinking pattern and type of alcohol consumed in coronary heart disease in men. New Engl J Med. 2003;348:109–118. doi: 10.1056/NEJMoa022095. [DOI] [PubMed] [Google Scholar]
- 49.Hazes JM, Dijkmans BA, Vandenbroucke JP, de Vries RR, Cats A. Lifestyle and the risk of rheumatoid arthritis: cigarette smoking and alcohol consumption. Ann Rheum Dis. 1990;49:980–982. doi: 10.1136/ard.49.12.980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jonsson IM, Verdrengh M, Brisslert M, Lindblad S, Bokarewa M, Islander U, et al. Ethanol prevents development of destructive arthritis. P Natl Acad Sci USA. 2007;104:258–263. doi: 10.1073/pnas.0608620104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Conigrave KM, Rimm EB. Alcohol for the prevention of type 2 diabetes mellitus? Treat Endocrinol. 2003;2:145–152. doi: 10.2165/00024677-200302030-00001. [DOI] [PubMed] [Google Scholar]
- 52.Pinder RM, Sandler M. Alcohol, wine and mental health: focus on dementia and stroke. J Psychopharmacol. 2004;18:449–456. doi: 10.1177/0269881104047272. [DOI] [PubMed] [Google Scholar]