Abstract
Cyclooxygenase-2 (COX-2), a key mediator of inflammation, and its product, prostaglandin E2 (PGE2), enhance carcinogenesis, particularly in skin. Ultraviolet (UV) B is the most carcinogenic component of solar irradiation, and a crucial role of COX-2 in UVB-mediated skin carcinogenesis has been reported. Here, we investigated the effects of delphinidin, an abundant dietary anthocyanin, on UVB-induced COX-2 upregulation and the underlying molecular mechanism. We found that delphinidin suppressed UVB-induced COX-2 expression in JB6 P+ mouse epidermal cells. COX-2 promoter activity and PGE2 production were also suppressed by delphinidin treatment within non-cytotoxic concentrations. Activator protein-1 and nuclear factor-κB, crucial transcription factors involved in COX-2 expression, were activated by UVB and delphinidin abolished this activation. UVB-induced phosphorylation of c-Jun N-terminal kinase, p38 kinase and Akt was inhibited by delphinidin. The activities of mitogen-activated protein kinase kinase (MAPKK) 4 and phosphatidylinositol-3 kinase (PI-3K) were inhibited markedly by delphinidin. A pull-down assay using delphinidin–Sepharose beads revealed that delphinidin binds directly with MAPKK4 or PI-3K in a manner that was competitive with adenosine triphosphate. Moreover, in vivo investigations using mouse skin revealed that the upregulation of COX-2 expression, MAPKK4 activity and PI-3K activity induced by UVB was abolished with delphinidin treatment. Collectively, our results demonstrated that delphinidin targets MAPKK4 and PI-3K directly to suppress COX-2 overexpression, suggesting a potential protective role for delphinidin against UVB-mediated skin carcinogenesis.
Introduction
A strong link between inflammation and carcinogenesis has been reported (1,2). Cyclooxygenase-2 (COX-2), a major mediator of inflammation, and its product, prostaglandin E2 (PGE2), enhance carcinogenesis, particularly skin cancer (3,4). The pivotal role of COX-2 in ultraviolet (UV)-related skin carcinogenesis was reported (5). UVB (280–320 nm), a key component of solar UV radiation, is suggested as a major cause of skin cancer (5). UVB radiation activates a variety of cellular signaling pathways and molecular targets linked to photoaging and photocarcinogenesis. Chronic exposure to UVB irradiation leads to induction of COX-2 expression and inflammatory responses, resulting in the development of skin cancer (6). UVB irradiation stimulates activator protein-1 (AP-1) and nuclear factor-κB (NF-κB), which are crucial transcription factors involved in COX-2 expression and carcinogenesis (7,8), especially skin cancer development (9).
Major signaling pathways that are known to mediate UVB-induced biological responses involve mitogen-activated protein kinases (MAPKs) (10). MAPKs mediate a wide range of intracellular signaling molecules involved in biological processes, including cell proliferation, differentiation and apoptosis. Three types of MAPKs have been characterized, including extracellular signal-regulated kinases (ERKs), c-Jun N-terminal kinases (JNKs) and p38. They are activated by specific mitogen-activated protein kinase kinases (MAPKKs), including mitogen-activated protein kinase/extracellular signal-regulated kinase (MEK) 1/2, MAPKK4/7 and MAPKK3/6, respectively (11). Specific inhibitors of these kinase families cause a downregulation of MAPK activity and could be used to treat MAPKs-mediated diseases, including cancer. Another key mediator of the UVB-induced cellular response is the phosphatidylinositol-3 kinase (PI-3K) pathway. PI-3K is a major upstream kinase of Akt. This pathway regulates various cellular processes, such as apoptosis, proliferation and growth (12), and requires UVB-induced COX-2 expression (13,14). Therefore, MAPKKs and PI-3K are possible molecular target candidates for suppressing UVB-induced COX-2 expression.
Anthocyanins are abundant natural polyphenolic compounds that contribute intense color to fruits and vegetables, including berries, red grapes, purple sweet potato and red cabbage (15). Anthocyanins have been noted for their health-promoting effects and biological function associated with a lower risk of cancer (16,17). Among the anthocyanidins, the aglycon form of anthocyanin, delphinidin, has the most potent anticarcinogenic properties. Delphinidin was reported to exert a stronger inhibitory potency against cancer cell migration (18) and cell transformation (19) than other anthocyanidin compounds tested. However, direct protein targets of delphinidin and antitumor-promoting mechanisms remain largely unknown. In the present study, we investigated the chemopreventive effects of delphinidin on UVB-induced tumor promotion in vitro and in vivo and examined the underlying molecular mechanism. Here, we report that delphinidin suppresses UVB-induced COX-2 expression by acting as a potent inhibitor of MAPKK4 and PI-3K.
Materials and methods
Chemicals
Delphinidin was purchased from Indofine Chemical (Hillsborough, NJ) and the purity of the chemical was >99% according to the manufacturer's information. Dimethylsulfoxide was obtained from Sigma–Aldrich (St Louis, MO). Delphinidin was prepared as a 40 mM stock in dimethylsulfoxide, 40 μl aliquots was stored at −80°C and fresh aliquots were used for each experiment. Eagle's minimum essential medium (EMEM), basal medium Eagle, gentamicin, fetal bovine serum (FBS) and L-glutamine were from Gibco BRL (Carlsbad, CA). The antibodies against phosphorylated MEK1/2 (Ser217/221), phosphorylated MAPKK3/6 (Ser189/207), total MAPKK3/6, phosphorylated MAPKK4 (Ser257/Thr261) and total p90RSK were purchased from Cell Signaling Technology (Beverly, MA). The antibodies against total MEK1/2, total MAPKK4 phosphorylated ERKs (Thr202/Tyr204) and total ERKs were from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-β-actin was purchased from Sigma–Aldrich. The MAPKK4 and PI-3K assay kits were obtained from Upstate Biotechnology (Lake Placid, NY). Cyanogen bromide–Sepharose 4B, glutathione–Sepharose 4B, [γ-32P] adenosine triphosphate (ATP) and the chemiluminescence detection kit were purchased from Amersham Pharmacia Biotech (Piscataway, NJ), and the protein assay kit was acquired from Bio-Rad Laboratories (Hercules, CA). A PGE2 enzyme immunoassay kit was obtained from Cayman Chemical (Ann Arbor, MI). G418 and the luciferase assay substrate were purchased from Promega (Madison, WI).
Cell viability assay
To estimate cell viability, JB6 P+ cells were seeded (103 cells per well) in 96-well plates with 5% FBS and EMEM and incubated at 37°C in a 5% CO2 incubator. The cells were treated with delphinidin at the concentrations indicated (0, 5, 10 or 20 μM) for 4 h. After incubation, 20 μl of CellTiter 96 Aqueous One Solution (Promega) were added to each well, and the cells were then incubated for 1 h at 37°C in a 5% CO2 incubator. Absorbance was measured at 492 and 690 nm.
UVB irradiation
A UVB irradiation system was used to stimulate cells in serum-free media. The spectral peak from the UVB source (Bio-Link crosslinker, Vilber Lourmat, Cedex 1, France) was at 312 nm. Cells were exposed to UVB at a dose of 0.5 kJ/m2 and then cultured for various periods of time depending on the experiment.
Cell culture
JB6 P+ mouse epidermal (JB6 P+) cells were cultured in monolayers in EMEM containing 5% FBS, 2 mM L-glutamine and 100 u penicillin/100 μg/ml streptomycin at 37°C in a 5% CO2 incubator. JB6 P+ cells were stably transfected with a COX-2, AP-1 or NF-κB luciferase reporter plasmid and maintained in 5% FBS/EMEM and 200 μg/ml G418.
Animals
Female ICR mice (5 weeks of age; mean body weight at 25 g) were purchased from the Institute of Laboratory Animal Resources at Seoul National University (Seoul, Korea). Animals were acclimated for 1 week prior to the study and had free access to food and water. The animals were housed in climate-controlled quarters (24°C at 50% humidity) with a 12 h light–dark cycle.
Western blot analysis
JB6 P+ cells were cultured for 48 h and then incubated in EMEM containing 0.1% FBS for an additional 24 h. Media were changed and cells were incubated with or without delphinidin for 1 h before exposure to 0.5 kJ/m2 UVB and then harvested at the indicated time. Cell lysates were scraped and disrupted with lysis buffer [10 mM Tris (pH 7.5), 150 mM NaCl, 5 mM ethylenediaminetetraacetic acid (EDTA), 1% Triton X-100, 1 mM dithiothreitol (DTT), 0.1 mM phenylmethylsulfonyl fluoride (PMSF), 10% glycerol and a protease inhibitor cocktail tablet] for 40 min on ice and then centrifuged at 16 000g for 10 min. The protein concentration of the supernatant fraction was measured using a dye-binding protein assay kit as described in the manufacturer's manual. Lysate proteins (40 μg) were subjected to 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis and electrophoretically transferred to a polyvinylidene fluoride membrane (Millipore Corporation, Bedford, MA). After transfer, the membrane was blocked in 5% fat-free milk for 1 h and then incubated with the specific primary antibody for 2 h at room temperature. After hybridization with the horseradish peroxidase-conjugated secondary antibody, protein bands were detected using an enhanced chemiluminescence detection kit.
Luciferase assays for COX-2 promoter activity and AP-1 and NF-κB transcription activities
Confluent monolayers of JB6 P+ cells stably transfected with a COX-2, AP-1 or NF-κB luciferase reporter plasmid were trypsinized and harvested. Cells (8 × 103) suspended in 100 μl of 5% FBS/EMEM were added to each well of a 96-well plate. Plates were incubated at 37°C in a 5% CO2 incubator. At 80–90% confluence, cells were cultured in 0.1% FBS/EMEM for 24 h. The cells were then treated with various concentrations of delphinidin for 1 h and then exposed to 0.5 kJ/m2 UVB and harvested after 6 h to assess COX-2, AP-1 or NF-κB activities. After treatment, cells were disrupted with 100 μl of lysis buffer [0.1 M potassium phosphate buffer (pH 7.8), 1% Triton X-100, 1 mM DTT and 2 mM EDTA], and luciferase activity was measured using a luminometer (Microlumat Plus LB 96V, Berthold Technologies, Bad Wildbach, Germany).
PGE2 assay
JB6 P+ cells were plated in six-well dishes and grown to 80% confluence in 2 ml of growth medium, treated with delphinidin for 1 h before exposure to 0.5 kJ/m2 UVB and then harvested 18 h later. The amount of PGE2 released into the medium was measured using the PGE2 enzyme immunoassay kit.
In vitro MAPKK4 kinase assay
MAPKK4 activity was determined directly, according to the instructions provided by Upstate Biotechnology. In brief, each reaction contained 2.5 μl of assay buffer [500 mM Tris–HCl (pH 7.5), 1 mM ethyleneglycol-bis(aminoethylether)-tetraacetic acid, 1 mM Na3VO4, 1% 2-mercaptoethanol and 0.1% Brij-35] added to 10 ng of active MAPKK4 protein. MAPKK4 protein was incubated with delphinidin (0, 5, 10 or 20 μM) for 10 min at room temperature. Next, inactive JNK (250 ng) and activating transcription factor 2 (25 μg) were added with 10 μl of diluted [γ-32P]ATP in magnesium acetate–ATP cocktail buffer [2.5 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (pH 7.4), 50 mM magnesium acetate and 0.5 mM ATP]. The reactions were incubated at 30°C for 10 min and then 15 μl aliquots were transferred onto p81 paper and washed three times with 0.75% phosphoric acid for 5 min and once with acetone for 5 min. The radioactive incorporation was determined using a scintillation counter. Each experiment was performed three times.
Ex vivo MAPKK4 immunoprecipitation and kinase assays in JB6 P+ cells
For the ex vivo MAPKK4 kinase assay, JB6 P+ cells were cultured to 80% confluence and then serum starved in 0.1% FBS/EMEM for 24 h at 37°C. Cells were either treated or not treated with delphinidin (0–20 μM) for 1 h before being exposed to 0.5 kJ/m2 UVB and harvested after 15 min, disrupted with lysis buffer [20 mM Tris–HCl (pH 7.4), 1 mM EDTA, 150 mM NaCl, 1 mM ethyleneglycol-bis(aminoethylether)-tetraacetic acid, 1% Triton X-100, 1 mM β-glycerophosphate, 1 mg/ml leupeptin, 1 mM Na3VO4 and 1 mM PMSF], and finally centrifuged at 16 000g for 10 min in a microcentrifuge. Lysate samples containing 500 μg of protein were used for immunoprecipitation with an antibody against MAPKK4 and incubated at 4°C overnight. Protein A/G Plus agarose beads were then added, and the mixture was rotated continuously for another 3 h at 4°C. The beads were washed three times with kinase buffer [20 mM 3-(N-morpholino)propanesulfonic acid (pH 7.2) and 25 mM β-glycerol phosphate]. Radioactive incorporation was determined as for the in vitro MAPKK4 kinase assay described above.
In vivo MAPKK4 immunoprecipitation and kinase assay
Mice were treated with delphinidin (0, 40 or 200 nmol) applied topically in 200 μl of acetone, and dorsal skin was prepared 2 h after UVB exposure (0.5 kJ/m2). Proteins were extracted as described above and centrifuged at 16 000g for 15 min. In advance, 700 μg of mouse skin extract was mixed with protein A/G beads (20 ml) for 1 h at 4°C. The mixture was processed and radioactive incorporation was determined as for the in vitro MAPKK4 kinase assay described above. Data are presented as the mean of data points from three mice in each treatment group.
In vitro PI-3K assay
An active PI-3K protein (100 ng) was incubated with delphinidin (0, 10 and 20 μM) for 10 min at 30°C. The mixtures were then incubated with 20 μl of 0.5 mg/ml phosphatidylinositol (Avanti Polar Lipids, Alabaster, AL) for 5 min at room temperature, followed by incubation with reaction buffer [100 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (pH 7.6), 50 mM MgCl2 and 250 μM ATP containing 10 μCi of [γ-32P]ATP] for an additional 10 min at 30°C. The reaction was stopped by adding 15 μl of 4 N HCl and 130 μl of chloroform:methanol (1:1). After vortexing, 30 μl of the lower chloroform phase were spotted onto a 1% potassium oxalate-coated silica gel plate that was previously activated for 1 h at 110°C. The resulting 32P-labeled phosphatidylinositol-3-phosphate was separated by thin layer chromatography and radiolabeled spots were visualized by autoradiography.
Ex vivo PI-3K immunoprecipitation and kinase assay
JB6 P+ cells were cultured to 80% confluence and then serum starved in 0.1% FBS/EMEM for 24 h at 37°C. Cells were either treated or not treated with delphinidin (0–20 μM) for 1 h before being exposed to UVB (0.5 kJ/m2) and harvested after 15 min, disrupted with lysis buffer [20 mM Tris–HCl (pH 7.4), 1 mM EDTA, 150 mM NaCl, 1 mM ethyleneglycol-bis(aminoethylether)-tetraacetic acid, 1% Triton X-100, 1 mM β-glycerophosphate, 1 mg/ml leupeptin, 1 mM Na3VO4 and 1 mM PMSF] and finally centrifuged at 16 000g for 10 min in a microcentrifuge. The lysates containing 500 μg of protein were used for immunoprecipitation with an antibody against p110 (to detect PI-3K) and incubated at 4°C overnight. Protein A/G Plus agarose beads were then added, and the mixture was rotated continuously for another 3 h at 4°C. The beads were washed three times with kinase buffer. Radioactive incorporation was determined as for the in vitro PI-3K assay described above.
In vivo PI-3K immunoprecipitation and kinase assay
Mice were treated with delphinidin (0, 40 or 200 nmol) applied topically in 200 μl of acetone, and dorsal skin was prepared 2 h after UVB exposure (0.5 kJ/m2). Proteins were extracted as described above and centrifuged at 16 000g for 15 min. In advance, 700 μg of mouse skin extract was mixed with protein A/G beads (20 ml) for 1 h at 4°C. The mixture was processed and radioactive incorporation was determined as for the ex vivo PI-3K assay described above. The spots were quantified using the ImageJ software program (National Institutes of Health).
Direct, cell-based and in vivo pull-down assays
Active MAPKK4 protein (0.2 μg) or a JB6 P+ cellular supernatant fraction (500 μg) was incubated with the delphinidin–Sepharose 4B (or Sepharose 4B only as a control) (100 μl, 50% slurry) in a reaction buffer [50 mM Tris (pH 7.5), 5 mM EDTA, 150 mM NaCl, 1 mM DTT, 0.01% Nonidet P-40, 2 μg/ml bovine serum albumin, 0.02 mM PMSF and 1× protease inhibitor mixture]. After incubation with gentle rocking overnight at 4°C, the beads were washed five times with buffer [50 mM Tris (pH 7.5), 5 mM EDTA, 150 mM NaCl, 1 mM DTT, 0.01% Nonidet P-40 and 0.02 mM PMSF], and proteins bound to the beads were analyzed by western blotting. For the in vivo pull-down assay, mice received a topical application of 200 μl acetone alone 1 h before UVB irradiation (0.5 kJ/m2). Dorsal skin was prepared as described above for the in vivo western blotting assay and proteins were extracted as described above for the in vivo MAPKK4 and PI-3K immunoprecipitation and kinase assays. Then, 500 μg of mouse skin extract was incubated with delphinidin–Sepharose 4B (or Sepharose 4B alone as a control) beads (100 μl, 50% slurry) in reaction buffer as described for the cell-based pull-down assay. Beads were incubated and washed, and proteins bound to the beads were analyzed by western blotting as described above.
ATP and delphinidin competition assay
Recombinant MAPKK4 or p110 for PI-3K (0.2 μg each) was incubated with 100 μl of delphinidin–Sepharose 4B or 100 μl of Sepharose 4B in a reaction buffer (see direct and cell-based pull-down assays) for 12 h at 4°C, and ATP (10 or 100 μM) was added to a final volume of 500 μl and incubated for 30 min. The samples were washed, and proteins were then detected by western blotting.
Molecular modeling
The homology model structure of MAPKK4 was generated by Geno3D (http://geno3d-pbil.ibcp.fr) using the co-ordinates of MAPKK7 (Protein Data Bank code 2DYL) as a template. The co-ordinates of PI-3K in complex with ATP or myricetin (Protein Data Bank codes 1E8X or 1E90, respectively) were used for the docking of delphinidin to PI-3K. Insight II (Accelrys, San Diego, CA) was used for the modeling study and structure analysis.
Statistical analysis
When necessary, data are expressed as means ± SDs and the Student's t-test was used for single statistical comparisons. A probability value of P <0.05 was used as the criterion for significance.
Results
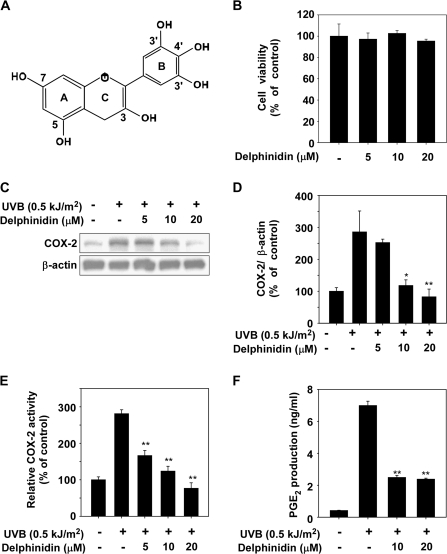
Delphinidin inhibits UVB-induced COX-2 protein expression and PGE2 production in JB6 P+ cells
Because abnormal upregulation of COX-2 and inflammation were reported to play an important role in skin cancer (6), we first investigated the effect of delphinidin on UVB-induced COX-2 upregulation. The optimal time for induction of COX-2 protein expression by UVB irradiation was determined by a time-course study (data not shown). The results of cell viability assay data indicated that delphinidin (Figure 1A) between 5 and 20 μM concentration had no effect on cell viability (Figure 1B). However, delphinidin at 5–20 μM suppressed UVB-induced COX-2 protein expression in JB6 P+ cells (Figure 1C and D). UVB-induced COX-2 promoter activity was dose-dependently suppressed by delphinidin treatment in JB6 P+ cells, which were stably transfected with a COX-2 luciferase plasmid (Figure 1E). The production of PGE2, which increased following exposure to UVB, was also suppressed by delphinidin treatment (Figure 1F). These results indicated that delphinidin could effectively repress UVB-induced COX-2 protein expression and PGE2 production in JB6 P+ cells.
Fig. 1.
Effect of delphinidin on UVB-induced COX-2 expression and PGE2 production in JB6 P+ cells. (A) Chemical structure of delphinidin. (B) Effect of delphinidin on viability of JB6 P+ cells. JB6 P+ cells were treated with delphinidin at the concentrations indicated (0, 5, 10 or 20 μM) for 4 h. After incubation, 20 μl of CellTiter 96 Aqueous One Solution was added to each well, and the cells were then incubated for 1 h. Absorbance was measured at 492 and 690 nm. (C) UVB-induced COX-2 expression in JB6 P+ cells is inhibited by delphinidin. JB6 P+ cells were pretreated with delphinidin at the concentrations indicated (0, 5, 10 or 20 μM) for 1 h before being exposed to 0.5 kJ/m2 UVB and harvested 4 h later. The cells were disrupted, and COX-2 protein level was determined by western blot analysis as described in Materials and Methods. β-Actin was detected to verify equal loading of proteins. Data are representative of three independent experiments. (D) COX-2 and β-actin bands were quantified by densitometric analysis using the ImageJ software program. (E) Delphinidin suppresses UVB-induced COX-2 promoter activity. JB6 P+ cells, which were stably transfected with a COX-2 luciferase reporter plasmid, were treated with delphinidin at the concentrations indicated (0, 5, 10 or 20 μM) for 1 h before being exposed to 0.5 kJ/m2 UVB and harvested 6 h later. Relative activities were determined using a luciferase assay as described in Materials and Methods. (F) Delphinidin suppresses UVB-induced PGE2 production. JB6 P+ cells were treated with delphinidin at the concentrations indicated (0, 10 or 20 μM) for 1 h before being exposed to 0.5 kJ/m2 UVB and harvested 18 h later. PGE2 production was measured using a PGE2 assay kit as described in Materials and Methods. For (B–D) and (E–F), data are shown as mean ± SD and asterisks indicate a significant inhibition by delphinidin compared with the group treated with UVB alone (*P < 0.05 and **P < 0.01).
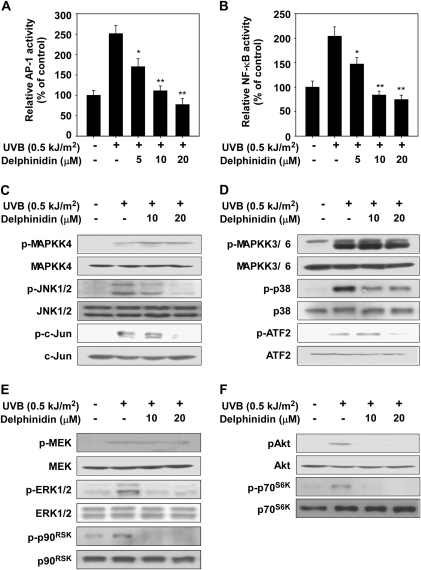
Delphinidin suppresses UVB-induced transactivation of AP-1 and NF-κB and phosphorylation of JNKs, p38 and Akt in JB6 P+ cells
Next, we measured the effect of delphinidin on transactivation of AP-1 and NF-κB using JB6 P+ cells stably transfected with an AP-1 or NF-κB luciferase reporter plasmid. Consistent with the results for COX-2 expression, delphinidin inhibited UVB-induced transactivation of AP-1 (Figure 2A) or NF-κB (Figure 2B) in a dose-dependent manner, which may contribute to the antitumor-promoting and anti-inflammatory activities of delphinidin. We found that delphinidin suppressed UVB-induced phosphorylation of JNKs, c-Jun, p38, activating transcription fractor 2, ERK1/2 and p90RSK but not MAPKK4, MAPKK3/6 or MEK1/2 (Figure 2C–E). We could not detect UVB-induced phosphorylation of MAPKK7. Delphinidin also strongly suppressed the phosphorylation of the PI-3K downstream kinases, including Akt and p70S6K (Figure 2F). These results suggested that the inhibition of the JNKs, p38, ERKs and Akt pathways by delphinidin leads to the suppression of AP-1 and NF-κB transactivation, resulting in decreased COX-2 expression.
Fig. 2.
Effect of delphinidin on UVB-induced signaling in JB6 P+ cells. (A and B) Delphinidin suppresses UVB-induced AP-1 (A) or NF-κB (B) transactivation. JB6 P+ cells, which were stably transfected with an AP-1 or NF-κB luciferase reporter plasmid, were treated with delphinidin at the concentrations indicated (0, 5, 10 or 20 μM) for 1 h before being exposed to 0.5 kJ/m2 UVB and harvested 6 h later. Relative activities were determined by luciferase assay as described in Materials and Methods. (C–F) JB6 P+ cells were treated with delphinidin at the concentrations indicated (0, 10 or 20 μM) for 1 h before being exposed to 0.5 kJ/m2 UVB and harvested after 30 min. Western blot analysis was conducted as described in Materials and Methods using specific antibodies as indicated. Data are presented as means ± SDs and asterisks for (A) and (B) indicate significant inhibition of luciferase activity by delphinidin compared with the group treated with UVB alone (*P < 0.05 and **P < 0.01).
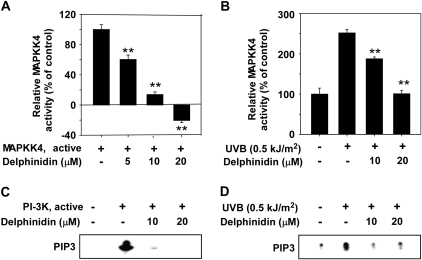
Delphinidin inhibits MAPKK4 or PI-3K activity in vitro and ex vivo
Because delphinidin strongly suppressed the JNKs and Akt signaling pathways, we investigated the effects of delphinidin on the kinase activity of MAPKK4 and PI-3K. Kinase assay data revealed that delphinidin strongly suppressed MAPKK4 and PI-3K activity in vitro (Figure 3A and C). However, delphinidin had no effect on MAPKK6 activity (data not shown). Further, ex vivo kinase assay data revealed that delphinidin inhibited MAPKK4 and PI-3K activity in UVB-treated cell lysates (Figure 3B and D).
Fig. 3.
Delphinidin inhibits MAPKK4 or PI-3K activity in vitro and ex vivo. (A and C) MAPKK4 (A) and PI-3K (C) activity assays were performed as described in Materials and Methods. (B and D) Delphinidin inhibits UVB-induced MAPKK4 or PI-3K activity. JB6 P+ cells were treated with delphinidin at the concentrations indicated (0, 10 or 20 μM) for 1 h before being exposed to 0.5 kJ/m2 UVB and harvested after 15 min. MAPKK4 (B) or PI-3K (D) activity was measured as described in Materials and Methods. For (A and B) data are shown as means ± SDs and asterisks indicate significant inhibition of MAPKK4 activity by delphinidin compared with the active MAPKK4 only group (A) or the group treated with UVB (B) alone (**P < 0.01).
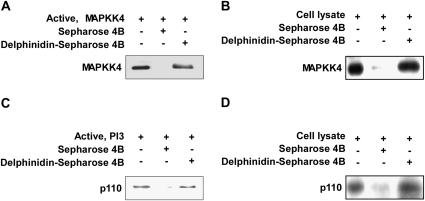
Delphinidin binds directly to MAPKK4 and PI-3K
We next determined whether delphinidin interacts directly with MAPKK4 or PI-3K. Direct binding of delphinidin to MAPKK4 or PI-3K was demonstrated by the in vitro pull-down assay (Figure 4A and C). We also found ex vivo binding between delphinidin and MAPKK4 or PI-3K in JB6 P+ cell lysates (Figure 4B and D).
Fig. 4.
Binding of delphinidin to MAPKK4 or PI-3K. (A and C) Delphinidin binds directly to the MAPKK4 (A) or PI-3K (C) protein. MAPKK4– or PI-3K–delphinidin binding was confirmed by immunoblotting using an antibody against MAPKK4 (A) or p110 for PI-3K (C): lane 1 (input control), MAPKK4 (A) or PI-3K (C) protein standard; lane 2 (control), Sepharose 4B was used for a pull-down assay as described in Materials and Methods and lanes 3 and 4, MAPKK4 (A) or PI-3K (C) was pulled down using delphinidin–Sepharose 4B beads as described in Materials and Methods. (B and D) Delphinidin binds to UVB-activated MAPKK4 (B) or PI-3K (D). The MAPKK4–delphinidin (B) or PI-3K–delphinidin (D) binding in UVB-exposed JB6 P+ cells was confirmed by immunoblotting using an antibody against MAPKK4 (B) or p110 for PI-3K (D): lane 1 (input control), whole-cell lysates from JB6 P+ cells; lane 2 (control), lysates of JB6 P+ cells were precipitated with Sepharose 4B beads as described in Materials and Methods and lane 3, whole-cell lysates from JB6 P+ cells were precipitated by delphinidin–Sepharose 4B as described in Materials and Methods.
Delphinidin suppresses UVB-induced COX-2, MAPKK4 and PI-3K activity in vivo
We examined the effect of delphinidin on UVB-treated mouse skin in vivo. UVB irradiation highly upregulated COX-2 expression in mouse skin, and this was suppressed by delphinidin (Figure 5A). We also found that UVB-induced MAPKK4 or PI-3K activity in mouse skin was suppressed significantly by delphinidin treatment (Figure 5B and C). The pull-down assay data indicated direct binding of delphinidin with MAPKK4 (Figure 5D, upper panel) or PI-3K (Figure 5D, lower panel) using in vivo skin lysates. Together, these findings provide evidence that delphinidin suppresses UVB-induced COX-2 expression mainly by targeting MAPKK4 and PI-3K.
Fig. 5.
Effect of delphinidin on UVB-induced COX-2 expression and MAPKK4 or PI-3K activity in mouse dorsal skin. (A) Delphinidin inhibits UVB-induced COX-2 expression in mouse skin extracts. The levels of COX-2 and β-actin were determined by western blot analysis using specific antibodies against the corresponding COX-2 and β-actin proteins. Each band was quantified using the ImageJ software program. Results are shown as mean ± SE (n = 3). (B and C) Delphinidin inhibits UVB-induced MAPKK4 (B) or PI-3K (C) activity in mouse skin extracts. For the MAPKK4 and PI-3K assays, dorsal skin protein lysates were prepared from the epidermis, and the assays were carried out as described in Materials and Methods. For (A–C) the asterisks indicates a significant difference between groups treated with delphinidin and irradiated with UVB and the group exposed to UVB alone (*P < 0.05 and **P < 0.01) and the # sign indicates a significant difference between untreated and UV treated (P < 0.01). (D) Delphinidin specifically binds with MAPKK4 and PI-3K in mouse skin extracts. The MAPKK4– and PI-3K–delphinidin binding in vivo was confirmed by immunoblotting using an antibody against MAPKK4 and p110 for PI-3K: lane 1 (input control), mouse dorsal skin lysate; lane 2 (control), a lysate of mouse dorsal skin precipitated with Sepharose 4B beads as described in the Materials and Methods and lane 3, mouse dorsal skin lysate precipitated by delphinidin–Sepharose 4B affinity beads.
Delphinidin binds directly to MAPKK4 and PI-3K in an ATP-competitive manner
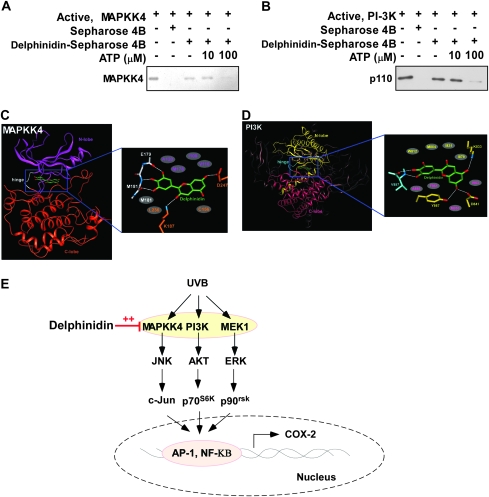
We next determined the means by which delphinidin interacts directly with MAPKK4 or PI-3K and results indicated that ATP competed with delphinidin for binding with MAPKK4 (Figure 6A) or PI-3K (Figure 6B). These results suggested that delphinidin inhibits MAPKK4 and PI-3K activity competitively with ATP. Using a computer modeling study, we found that delphinidin easily docked to the ATP-binding site of MAPKK4 (Figure 6C) or PI-3K (Figure 6D).
Fig. 6.
Delphinidin binds to MAPKK4 or PI-3K competitively with ATP. Active MAPKK4 (A) or PI-3K (B) (0.2 μg each) was incubated with ATP at the concentrations indicated (0, 10 or 100 μM) and 100 μl of delphinidin–Sepharose 4B or Sepharose 4B (as a negative control) in a reaction buffer to a final volume of 500 μl. The pulled-down proteins were detected by western blot analysis as described in Materials and Methods: lane 1 (input control), MAPKK4 (A) and PI-3K (B) protein standard; lane 2 is the negative control, indicating that neither MAPKK4 (A) nor PI-3K (B) binds with Sepharose 4B and lane 3 is the positive control, which indicates that MAPKK4 (A) or PI-3K (B) binds with delphinidin–Sepharose 4B. (C and D) Hypothetical model of MAPKK4 and PI-3K in complex with delphinidin. (C) Delphinidin binds to the ATP-binding site in the kinase domain of MAPKK4. The close-up view of the inhibitor interaction shows the hydrogen bonds depicted as white lines, and the residues in gray ellipses are the hydrophobic residues interacting with delphinidin. (D) Model structure of PI-3K in complex with delphinidin and its close-up view. The Ras-binding domain, C2 domain and helical domain of PI-3K are shown in gray. Delphinidin binds to the ATP-binding site in the catalytic domain of PI-3K. The residues in gray ellipses are the hydrophobic residues interacting with delphinidin. The hydrogen bonds are depicted as white lines. (E) Simplified depiction of the effects of delphinidin on UVB-induced COX-2 expression.
Discussion
Dietary anthocyanins or anthocyanin-rich fruits and extracts exhibit protective effects against a variety of chronic diseases. Previous studies have shown that anthocyanins possess a strong antioxidant capacity, preventing oxidative stress-induced apoptosis (20) and doxorubicin-induced cardiotoxicity (21). Anthocyanins possess strong antioxidant capacity and antiproliferative activity against cancer cell growth (22) and suppress tumor cell invasion and migration (23). Delphinidin was shown to inhibit cell transformation and migration most strongly among several anthocyanidins tested (18,19). The beneficial effects of anthocyanidins have been explained by their antioxidant effects (24). However, due to their low effective dose and specific signaling inhibition, antioxidant effects cannot account for all the beneficial effects (25). Therefore, our previous studies and other studies suggested that flavonoids, including anthocyanidins, can inhibit certain cellular kinase activities. Myricetin suppresses MEK1, Fyn and PI-3K activities and quercetin inhibits MEK1, Raf1 and PI-3K, whereas kaempferol attenuates p90RSK activity. However, the targets of delphinidin have not been fully elucidated (26–29).
Accumulating data suggest that inflammation is associated with cell transformation and the development of cancer. COX-2, a key player in the inflammatory response, is highly upregulated during carcinogenesis (30). Evidence suggests that high levels of COX-2 expression and PGE2 production enhance tumorigenesis of various cell types, particularly in skin (3,4,31). UVB, the most carcinogenic component of solar irradiation, is a major cause of skin cancer (32) and COX-2 plays a critical role in UVB-induced skin cancer. A recent study indicated that UVB-induced photocarcinogenesis could be abolished by COX-2 inhibition (33). A transgenic mouse study using COX-2 knockout or COX-2 overexpressing mice also revealed that COX-2 is required for UVB-induced skin carcinogenesis (34,35). AP-1 and NF-κB are the most important transcription factors involved in COX-2 expression. UVB activates AP-1 and NF-κB through the MAPKs and PI-3K/Akt pathways (10). Therefore, targeting these pathways could be effective to inhibit COX-2 expression and prevent skin carcinogenesis.
In this study, we found that delphinidin effectively suppressed the phosphorylation of JNKs but not MAPKK4. Instead, the kinase activity of MAPKK4 was inhibited substantially by delphinidin. MAPKK4 is well known for phosphorylating JNKs and p38. Phosphorylation of JNK decreases when activity of MAPKK4 is suppressed in most cell types. However, the effect of MAPKK4 on p38 differs by cell type and stimuli (36). UVB-induced phosphorylation of p38 did not change in MAPKK4−/− murine embryonic fibroblasts compared with wild-type murine embryonic fibroblasts (37). Therefore, inhibition of MAPKK4 activity by delphinidin causes a decrease in JNKs phosphorylation but not p38. A new molecular target of delphinidin that regulates p38 must be elucidated. To investigate the molecular basis of MAPKK4 inhibition by delphinidin, we carried out a docking study (Figure 6C) using a homology model structure of the MAPKK4 kinase domain derived from the crystal structure of MAPKK7, which has 66% amino acid sequence homology with MAPKK4. The kinase domain of MAPKK4 consists of an N-lobe and a C-lobe. These N- and C-lobes are linked through a loop called the ‘hinge region’. The backbone of this loop interacts with the adenine moiety of ATP by hydrogen bonding. Considering the experimental result indicating that delphinidin is an ATP-competitive inhibitor of MAPKK4, we docked the compound to the ATP-binding site of MAPKK4 and delphinidin was easily docked to this site of MAPKK4. Delphinidin can form hydrogen bonds with the backbone of the hinge region of MAPKK4 as do other ATP-competitive kinase inhibitors. The hydroxyl groups at the 3′ and 5′ positions can form hydrogen bonds with the backbone carbonyl groups of Glu179 and Met181. The hydroxyl group at the 4′ position forms a hydrogen bond with the backbone amide group of Met181. The hydroxyl groups at positions 3 and 7 form hydrogen bonds with the side chains of Lys187 and Asp247, respectively. In addition, the inhibitor would be sandwiched by the side chains of the hydrophobic residues in the ATP-binding site, including Ala120, Met178, Ile108, Val116, Cys156, Leu236 and Met181.
We suggest that another molecular target of delphinidin is PI-3K. The PI-3K/Akt pathway has been pointed out as another potential target for suppressing UVB-mediated COX-2 expression (13,14). Inhibition of Akt phosphorylation by a PI-3K inhibitor or dominant-negative Akt mutant suppressed UVB-induced COX-2 transcription in human keratinocytes (14). UVB-induced phosphorylation of Akt/p70S6K was attenuated effectively by delphinidin because PI-3K activity, a well-known upstream kinase of Akt, was inhibited by delphinidin. We also created a model structure (Figure 6D) of PI-3K in complex with delphinidin using the crystal structure of PI-3K in complex with ATP or delphinidin (38). PI-3K comprises four domains: a Ras-binding domain, a C2 domain, a helical domain and a catalytic domain. Although the substrate of PI-3K is not a protein, the catalytic domain of the enzyme consists of an N-lobe, a C-lobe and a hinge loop with a fold similar to protein kinases, and this structural similarity is also conserved in the ATP-binding site that is flanked by these two lobes. Consequently, ATP binds between these lobes in a manner similar to the ATP binding in protein kinases. Because delphinidin was an ATP-competitive inhibitor of PI-3K in our experiment, we docked the compound to the ATP-binding site of PI-3K. The hydroxyl groups at the 3′ and 4′ positions of delphinidin could form hydrogen bonds with the backbone atoms of Val887 in the hinge loop of PI-3K. The hydroxyl groups at positions 3 and 5 could also make hydrogen bonds with the side chains of Lys833, Asp841 and Tyr867. Delphinidin could hydrophobically interact with the side chains of Met804, Trp812, Ile831 and Ile879 from the N-lobe and Ala885, Phe961, Met953 and Ile963 from the C-lobe. The high inhibitory activity of delphinidin for PI-3K could be due to the hydrogen bonding and hydrophobic interactions.
MEK1 is another molecular target of delphinidin. Flavonoids that possess a hydroxyl group at the 3′ position (e.g. myricetin, quercetin but not kaempferol) inhibit MEK1 activity. The hydroxyl group at the 3′ position plays a key role in the formation of the hydrogen bond between flavonoids and the backbone amide group of Ser212 in MEK1. Because delphinidin has a hydroxyl group at the 3′ position, it also suppresses MEK1 activity. Delphinidin attenuated 12-O-tetradecanoylphorbol 13-acetates-induced neoplastic transformation by inhibiting the MEK/ERK signaling pathway. In the present study, the UVB-induced phosphorylation of ERKs was decreased by delphinidin treatment, which may be due to the inhibitory effects of delphinidin on MEK1 (Figure 6E).
Targeted inhibition of kinase signaling pathways by small molecules is an important strategy in chemoprevention and chemotherapy. Especially, multi-target kinase inhibitors are of interest with recent approval as anticancer drugs (39). Inhibition by highly specific inhibitors can cause drug resistance problems in cancer patients by activating an alternative pathway. Broad inhibition of multiple targets, rather than a single specific target, could be much more effective and applicable in treating cancer and other disorders. Sorafenib and Sunitinib are anticancer drugs that act as small molecular inhibitors of Raf, vascular endothelial growth factor receptor, platelet-derived growth factor receptor, FLT-3 and c-Kit (40,41). Both drugs are used to treat renal cell carcinoma but are also approved for the treatment of hepatocellular carcinoma and gastrointestinal stromal tumors, respectively. This suggests that the broad reactivity of multi-kinase inhibitors allows their multiple applications for a number of conditions. A potential drawback with the use of a multi-target kinase inhibitor is the appearance of undesirable side effects. Safety is a pivotal concern in using multi-target kinase inhibitors. Food components such as delphinidin are generally regarded as safe due to their longtime use. In this regard, delphinidin should be considered as a potential chemopreventive agent.
To summarize (Figure 6E), delphinidin inhibits UVB-induced COX-2 expression in JB6 P+ cells by blocking the MAPKK4 and PI-3K pathways and subsequently suppressing AP-1 and NF-κB activities. Our results suggest MAPKK4 and PI-3K as potent molecular targets of delphinidin in suppressing UVB-mediated skin carcinogenesis. Taken together, these results provide insight into the molecular action of delphinidin and indicate the potential of delphinidin as a novel chemopreventive agent. Further studies, including X-ray crystallography, to determine the inhibitor complex structure would elucidate the exact binding mode of delphinidin to targeted kinases.
Funding
The Hormel Foundation; National Institutes of Health (CA27502, CA120388, CA111536, CA88961, CA 81064); World Class University program (R31-2008-00-10056-0), research grants (Nos. R01-2007-000-11957-0 and M10510140004-08N1014-00410) through the Korea Science and Engineering Foundation funded by the Ministry of Education, Science and Technology; the BioGreen 21 Program (no. 20070301-034-042), Rural Development Administration; Technology Development Program for Agriculture and Forestry (no. 107055-02), Ministry for Food, Agriculture, Forestry and Fisheries, Republic of Korea.
Acknowledgments
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- AP-1
activator protein-1
- ATP
adenosine triphosphate
- COX-2
cyclooxygenase-2
- DTT
dithiothreitol
- EDTA
ethylenediaminetetraacetic acid
- EMEM
Eagle's minimum essential medium
- ERK
extracellular signal-regulated kinase
- FBS
fetal bovine serum
- JNK
c-Jun N-terminal kinase
- MAPK
mitogen-activated protein kinase
- MAPKK
mitogen-activated protein kinase kinase
- NF-κB
nuclear factor-κB
- PGE2
prostaglandin E2
- PI-3K
phosphatidylinositol-3 kinase
- PMSF
phenylmethylsulfonyl fluoride
- UV
ultraviolet
References
- 1.Balkwill F, et al. Inflammation and cancer: back to Virchow? Lancet. 2001;357:539–545. doi: 10.1016/S0140-6736(00)04046-0. [DOI] [PubMed] [Google Scholar]
- 2.Coussens LM, et al. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Higashi Y, et al. Enhanced expression of cyclooxygenase (COX)-2 in human skin epidermal cancer cells: evidence for growth suppression by inhibiting COX-2 expression. Int. J. Cancer. 2000;86:667–671. doi: 10.1002/(sici)1097-0215(20000601)86:5<667::aid-ijc10>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 4.Lee JL, et al. Cyclooxygenases in the skin: pharmacological and toxicological implications. Toxicol. Appl. Pharmacol. 2003;192:294–306. doi: 10.1016/s0041-008x(03)00301-6. [DOI] [PubMed] [Google Scholar]
- 5.Rundhaug JE, et al. Cyclo-oxygenase-2 plays a critical role in UV-induced skin carcinogenesis. Photochem. Photobiol. 2008;84:322–329. doi: 10.1111/j.1751-1097.2007.00261.x. [DOI] [PubMed] [Google Scholar]
- 6.Buckman SY, et al. COX-2 expression is induced by UVB exposure in human skin: implications for the development of skin cancer. Carcinogenesis. 1998;19:723–729. doi: 10.1093/carcin/19.5.723. [DOI] [PubMed] [Google Scholar]
- 7.Kang YJ, et al. Cyclooxygenase-2 gene transcription in a macrophage model of inflammation. J. Immunol. 2006;177:8111–8122. doi: 10.4049/jimmunol.177.11.8111. [DOI] [PubMed] [Google Scholar]
- 8.Konstantinopoulos PA, et al. NF-kappaB/PPAR gamma and/or AP-1/PPAR gamma ‘on/off’ switches and induction of CBP in colon adenocarcinomas: correlation with COX-2 expression. Int. J. Colorectal Dis. 2007;22:57–68. doi: 10.1007/s00384-006-0112-y. [DOI] [PubMed] [Google Scholar]
- 9.Cooper SJ, et al. Ultraviolet B regulation of transcription factor families: roles of nuclear factor-kappa B (NF-kappaB) and activator protein-1 (AP-1) in UVB-induced skin carcinogenesis. Curr. Cancer Drug Targets. 2007;7:325–334. doi: 10.2174/156800907780809714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bode AM, et al. Mitogen-activated protein kinase activation in UV-induced signal transduction. Sci. STKE. 2003;2003:RE2. doi: 10.1126/stke.2003.167.re2. [DOI] [PubMed] [Google Scholar]
- 11.Chang L, et al. Mammalian MAP kinase signalling cascades. Nature. 2001;410:37–40. doi: 10.1038/35065000. [DOI] [PubMed] [Google Scholar]
- 12.Vivanco I, et al. The phosphatidylinositol 3-kinase-AKT pathway in human cancer. Nat. Rev. Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 13.Bachelor MA, et al. Inhibition of p38 mitogen-activated protein kinase and phosphatidylinositol 3-kinase decreases UVB-induced activator protein-1 and cyclooxygenase-2 in a SKH-1 hairless mouse model. Mol. Cancer Res. 2005;3:90–99. doi: 10.1158/1541-7786.MCR-04-0065. [DOI] [PubMed] [Google Scholar]
- 14.Tang Q, et al. Roles of Akt and glycogen synthase kinase 3beta in the ultraviolet B induction of cyclooxygenase-2 transcription in human keratinocytes. Cancer Res. 2001;61:4329–4332. [PubMed] [Google Scholar]
- 15.Mazza G. Anthocyanins in grapes and grape products. Crit. Rev. Food Sci. Nutr. 1995;35:341–371. doi: 10.1080/10408399509527704. [DOI] [PubMed] [Google Scholar]
- 16.Bohm H, et al. [Flavonols, flavone and anthocyanins as natural antioxidants of food and their possible role in the prevention of chronic diseases] Z. Ernahrungswiss. 1998;37:147–163. doi: 10.1007/pl00007376. [DOI] [PubMed] [Google Scholar]
- 17.Zafra-Stone S, et al. Berry anthocyanins as novel antioxidants in human health and disease prevention. Mol. Nutr. Food Res. 2007;51:675–683. doi: 10.1002/mnfr.200700002. [DOI] [PubMed] [Google Scholar]
- 18.Lamy S, et al. Anthocyanidins inhibit migration of glioblastoma cells: structure-activity relationship and involvement of the plasminolytic system. J. Cell. Biochem. 2007;100:100–111. doi: 10.1002/jcb.21023. [DOI] [PubMed] [Google Scholar]
- 19.Hou DX, et al. Anthocyanidins inhibit activator protein 1 activity and cell transformation: structure-activity relationship and molecular mechanisms. Carcinogenesis. 2004;25:29–36. doi: 10.1093/carcin/bgg184. [DOI] [PubMed] [Google Scholar]
- 20.Shih PH, et al. Anthocyanins induce the activation of phase II enzymes through the antioxidant response element pathway against oxidative stress-induced apoptosis. J. Agric. Food Chem. 2007;55:9427–9435. doi: 10.1021/jf071933i. [DOI] [PubMed] [Google Scholar]
- 21.Choi EH, et al. Cytoprotective effect of anthocyanins against doxorubicin-induced toxicity in H9c2 cardiomyocytes in relation to their antioxidant activities. Food Chem. Toxicol. 2007;45:1873–1881. doi: 10.1016/j.fct.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y, et al. Human tumor cell growth inhibition by nontoxic anthocyanidins, the pigments in fruits and vegetables. Life Sci. 2005;76:1465–1472. doi: 10.1016/j.lfs.2004.08.025. [DOI] [PubMed] [Google Scholar]
- 23.Chen PN, et al. Black rice anthocyanins inhibit cancer cells invasion via repressions of MMPs and u-PA expression. Chem. Biol. Interact. 2006;163:218–229. doi: 10.1016/j.cbi.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 24.Rice-Evans CA, et al. Structure-antioxidant activity relationships of flavonoids and phenolic acids. Free Radic. Biol. Med. 1996;20:933–956. doi: 10.1016/0891-5849(95)02227-9. [DOI] [PubMed] [Google Scholar]
- 25.Williams RJ, et al. Flavonoids: antioxidants or signalling molecules? Free Radic. Biol. Med. 2004;36:838–849. doi: 10.1016/j.freeradbiomed.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 26.Cho YY, et al. Ribosomal S6 kinase 2 is a key regulator in tumor promoter-induced cell transformation. Cancer Res. 2007;67:8104–8112. doi: 10.1158/0008-5472.CAN-06-4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim JE, et al. MKK4 is a novel target for the inhibition of tumor necrosis factor-alpha-induced vascular endothelial growth factor expression by myricetin. Biochem. Pharmacol. 2009;77:412–421. doi: 10.1016/j.bcp.2008.10.027. [DOI] [PubMed] [Google Scholar]
- 28.Lee KW, et al. Raf and MEK protein kinases are direct molecular targets for the chemopreventive effect of quercetin, a major flavonol in red wine. Cancer Res. 2008;68:946–955. doi: 10.1158/0008-5472.CAN-07-3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee KW, et al. Myricetin is a novel natural inhibitor of neoplastic cell transformation and MEK1. Carcinogenesis. 2007;28:1918–1927. doi: 10.1093/carcin/bgm110. [DOI] [PubMed] [Google Scholar]
- 30.Dubois RN, et al. Cyclooxygenase in biology and disease. FASEB J. 1998;12:1063–1073. [PubMed] [Google Scholar]
- 31.Prosperi JR, et al. Invasive and angiogenic phenotype of MCF-7 human breast tumor cells expressing human cyclooxygenase-2. Prostaglandins Other Lipid Mediat. 2004;73:249–264. doi: 10.1016/j.prostaglandins.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 32.Ichihashi M, et al. UV-induced skin damage. Toxicology. 2003;189:21–39. doi: 10.1016/s0300-483x(03)00150-1. [DOI] [PubMed] [Google Scholar]
- 33.Tang X, et al. Cyclooxygenase-2 inhibitor nimesulide blocks ultraviolet B-induced photocarcinogenesis in SKH-1 hairless mice. Photochem. Photobiol. 2008;84:522–527. doi: 10.1111/j.1751-1097.2008.00303.x. [DOI] [PubMed] [Google Scholar]
- 34.Bol DK, et al. Cyclooxygenase-2 overexpression in the skin of transgenic mice results in suppression of tumor development. Cancer Res. 2002;62:2516–2521. [PubMed] [Google Scholar]
- 35.Fischer SM, et al. Cyclooxygenase-2 expression is critical for chronic UV-induced murine skin carcinogenesis. Mol. Carcinog. 2007;46:363–371. doi: 10.1002/mc.20284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Whitmarsh AJ, et al. Role of mitogen-activated protein kinase kinase 4 in cancer. Oncogene. 2007;26:3172–3184. doi: 10.1038/sj.onc.1210410. [DOI] [PubMed] [Google Scholar]
- 37.Brancho D, et al. Mechanism of p38 MAP kinase activation in vivo. Genes Dev. 2003;17:1969–1978. doi: 10.1101/gad.1107303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walker EH, et al. Structural determinants of phosphoinositide 3-kinase inhibition by wortmannin, LY294002, quercetin, myricetin, and staurosporine. Mol. Cell. 2000;6:909–919. doi: 10.1016/s1097-2765(05)00089-4. [DOI] [PubMed] [Google Scholar]
- 39.Petrelli A, et al. From single- to multi-target drugs in cancer therapy: when aspecificity becomes an advantage. Curr. Med. Chem. 2008;15:422–432. doi: 10.2174/092986708783503212. [DOI] [PubMed] [Google Scholar]
- 40.Grandinetti CA, et al. Sorafenib and sunitinib: novel targeted therapies for renal cell cancer. Pharmacotherapy. 2007;27:1125–1144. doi: 10.1592/phco.27.8.1125. [DOI] [PubMed] [Google Scholar]
- 41.Hiles JJ, et al. Role of sunitinib and sorafenib in the treatment of metastatic renal cell carcinoma. Am. J. Health Syst. Pharm. 2008;65:123–131. doi: 10.2146/ajhp060661. [DOI] [PubMed] [Google Scholar]