Abstract
Natural bacterial communities are extremely diverse and highly dynamic, but evidence is mounting that the compositions of these communities follow predictable temporal patterns. We investigated these patterns with a 3-year, circumpolar study of bacterioplankton communities in the six largest rivers of the pan-arctic watershed (Ob', Yenisey, Lena, Kolyma, Yukon, and Mackenzie), five of which are among Earth's 25 largest rivers. Communities in the six rivers shifted synchronously over time, correlating with seasonal shifts in hydrology and biogeochemistry and clustering into three groups: winter/spring, spring freshet, and summer/fall. This synchrony indicates that hemisphere-scale variation in seasonal climate sets the pace of variation in microbial diversity. Moreover, these seasonal communities reassembled each year in all six rivers, suggesting a long-term, predictable succession in the composition of big river bacterioplankton communities.
Keywords: 16S, DGGE, diversity, seasonality, succession
Bacterioplankton are essential components of freshwater and marine ecosystems, catalyzing critical biogeochemical reactions and serving as central members of aquatic microbial food webs (1). Natural communities of bacterioplankton are extremely diverse (2, 3) and highly dynamic, undergoing shifts in dominant phylotypes in response to spatial and temporal environmental gradients (4–6). Our current understanding of freshwater bacterioplankton diversity and dynamics comes almost entirely from studies of lakes (7). Several small rivers have been surveyed (6, 8); but of the world's 25 largest rivers, only four have bacterioplankton gene sequences entered in GenBank: the Columbia River in the United States (U.S.) (5), the ChangJiang River in China (9), the Mackenzie River in Canada (10), and the Paraná River in Brazil (11). This lack of basic information on big river bacterioplankton limits our understanding of global biogeochemical cycles and our ability to detect biological community responses to anthropogenic and climate change impacts to these critical ecosystems.
Recently, several studies have show evidence of predictable temporal patterns in the composition of these communities including seasonality (12, 13), annual reassembly (6, 14), and synchronous community shifts in nearby but non-attached systems (e.g., lakes in separate catchments) (6, 15). However, these phenomena have only been identified in a handful of small regional studies, and the scales at which these patterns exist remain unknown.
We investigated these patterns as part of the PARTNERS program, a large coordinated study of the six largest rivers of the pan-arctic watershed (Ob', Yenisey, Lena, Kolyma, Yukon, Mackenzie) (Fig. 1) by a collaborative team of researchers from the U.S., Canada, and Russia (16). This study included five of the world's 25 largest rivers, and provided a unique opportunity to investigate global-scale patterns in bacterial diversity. Our results demonstrate that synchrony, seasonality, and annual reassembly in bacterioplankton communities occur on global scales and that bacterioplankton communities in big arctic rivers shift predictably with circumpolar seasonal changes in environmental conditions. These predictable temporal patterns provide insights to Arctic biogeochemistry and may serve as sensitive indicators of climate change in the Arctic.
Fig. 1.
Watershed map of the six largest rivers in the pan-arctic watershed with red dots indicating sampling sites.
Results and Discussion
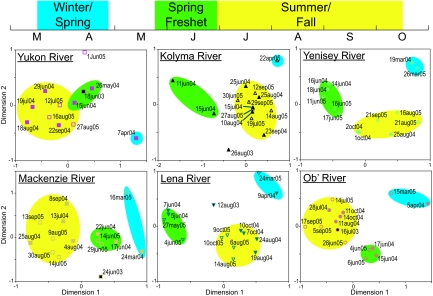
Bacterial communities displayed three patterns of change over the period of our study: synchrony, seasonality, and annual reassembly. They shifted synchronously over time in all six rivers, changing from winter to spring freshet communities during May and from spring freshet to summer/fall communities during late June (Fig. 2). During each of these seasons, bacterial communities remained relatively stable in all six rivers; community similarity values averaged 77% within season in each year compared with 51% between seasons (Table S1). Also, each of these three seasonal communities reassembled in each year of the study. In fact, community similarity values between years for each of the three seasons were only slightly lower than within year similarity values (72%). Moreover, seasonal clustering of bacterial communities was supported by Analysis of Similarity (ANOSIM), which showed higher similarity for communities collected during the same seasons than for communities collected during different seasons regardless of year (R >0.80, P < 0.05 for all comparisons except winter vs. spring in the Lena and Ob', for which R = 1, P < 0.07).
Fig. 2.
Multidimensional scaling diagrams showing synchronous seasonal variation and annual reassembly in bacterioplankton community composition over three years. A calendar bar at the top shows sampling periods. Circled clusters of samples contained similar bacterial communities within each river (ANOSIM R >0.8, P < 0.05). Closed symbols represent samples collected in 2004, open symbols 2005, and symbols with black fill 2003.
Spatial synchrony is defined as concurrent variation in time series data among ecosystems (17), and it has been identified in diverse physical, chemical, and biological data from freshwater ecosystems (18). Synchrony is an important signal of ecosystem structure because it indicates that governing exogenous forces such as climate impact independent ecosystems similarly and set the pace of temporal variation and ecosystem processes regionally. Lack of synchrony or asynchrony is thought to indicate the relative importance of internal intrinsic forces to drive distinct temporal variation within a single ecosystem and to cause divergence in ecosystem function across ecosystems. For example, Pace and Cole observed synchronous changes in dissolved organic carbon concentration and water color among 20 lakes in northern Michigan (U.S.) related to ice-out date and spring and summer precipitation, and found deviations from this pattern in lakes that lacked an outlet stream (17).
For biological populations, synchrony can be driven by exogenous factors (i.e., the Moran effect) (19, 20), but may also be influenced by dispersal of organisms among ecosystems (21). For example, spatial synchrony in bacterial community composition has been identified in a few small-scale studies on lake regions (15) and nearby rivers (6) among environments that are not hydraulically connected. In the relatively small Ipswich and Parker Rivers in Massachusetts (U.S.), bacterioplankton community composition shifted synchronously over 2.5 years, apparently driven by seasonal variation in water temperature and river flow (6). Kent et al. used a higher-resolution dataset (sampling twice weekly) to identify similar synchronous changes among six humic lakes in Michigan over a 3-month period (15).
Because they were not hydraulically connected, it is likely that local climate and weather patterns were the primary drivers of synchronous changes in these aquatic systems; but because they were in close proximity (within 8 km) it is possible that low-level dispersal or common inoculation via air, precipitation, or organisms such as birds contributed to this synchrony. In contrast, the watersheds of the six big arctic rivers described in this study drain more than 11 million square kilometers and are distributed all around the northern hemisphere, many extending far into the temperate zone. Thus, synchrony in the bacterioplankton communities of these rivers is likely driven by hemisphere-scale variations in seasonal climate and hydrology, and the influence of dispersal is limited to circumpolar transport via air currents (22). Divergence from this synchronous pattern may provide an early signal of climate change in some regions of the Arctic, and may result in changes to river microbial communities and the biogeochemical transformations that they carry out.
DNA sequences from clone libraries revealed typical freshwater bacterial assemblages dominated by Bacteroidetes, Betaproteobacteria, and Actinobacteria (Table 1). Diversity indices for these clone libraries were similar; Shannon's H' ranged from 3.1 to 3.5, and Simpson's index (1-d) ranged from 0.3 to 0.6. Comparisons of these assemblages showed that 72% of the sequences fell into phylogenetic groups (operational taxonomic units [OTUs], 97% sequence similarity) that included sequences from more than one river (Table S2). Moreover, nine of the 148 OTUs identified in this study accounted for 39% of all clone sequences and were present in at least five of the six rivers (Table 2). When these sequences were compared with the global database in GenBank, we found that these dominant bacterial groups are also found in freshwater systems around the world (Fig. 3 and Fig. S1). It appears that during the spring freshet, the six big arctic rivers contain a common and globally distributed community of freshwater bacteria. Differences in the communities of these rivers appear to be restricted to rare taxa (Table S2), but more DNA sequencing is necessary to identify river-specific taxa.
Table 1.
Taxonomic classification of 16S rRNA genes from each river collected June 7–17, 2004
| Phylum or class | KL | YE | YU | OB | MA | LE |
|---|---|---|---|---|---|---|
| Bacteroidetes | 42 | 28 | 18 | 25 | 31 | 22 |
| Betaproteobacteria | 16 | 27 | 23 | 16 | 16 | 30 |
| Actinobacteria | 9 | 16 | 19 | 20 | 20 | 12 |
| Alphaproteobacteria | 3 | 3 | 7 | 6 | ||
| Verrucomicrobia | 1 | 3 | 4 | 5 | 1 | 1 |
| Acidobacteria | 1 | 3 | 2 | 2 | ||
| Gammaproteobacteria | 3 | 1 | 1 | |||
| Deltaproteobacteria | 2 | 1 | ||||
| Chloroflexi | 1 | 1 | ||||
| Nitrospira | 1 | 1 | ||||
| Firmicutes | 1 | |||||
| Unclassified Proteobacteria | 1 | |||||
| Cyanobacteria | 1 | |||||
| OP10 | 1 | |||||
| Unclassified bacteria | 2 | 2 | 2 | 1 | 1 | 1 |
| Total clone sequences | 82 | 80 | 79 | 71 | 77 | 68 |
Table 2.
Bacterial OTUs that appeared in at least five of the six rivers, the fraction of all clones belonging to these OTUs, and taxonomic assignments
| Representative sequence for OTU | No. of rivers in which OTU appeared | All clones belonging to OTU, % | Phylum | Class | Order | Family | Genus |
|---|---|---|---|---|---|---|---|
| YU201A11 | 6 | 11.2 | Proteobacteria | Betaproteobacteria | Methylophilales | Methylophilaceae | Methylophilus |
| MA101D10 | 6 | 6.8 | Bacteroidetes | Sphingobacteria | Sphingobacteriales | Flexibacteraceae | Arcicella |
| YE201C08 | 6 | 5.3 | Actinobacteria | Actinobacteria | Actinomycetales | ||
| MA101F04 | 5 | 3.9 | Actinobacteria | Actinobacteria | Actinomycetales | ||
| YU201B06 | 5 | 3.3 | Actinobacteria | Actinobacteria | Actinomycetales | ||
| YU201A10 | 5 | 2.6 | Proteobacteria | Betaproteobacteria | Burkholderiales | Alcaligenaceae | |
| MA101A07 | 5 | 2.2 | Proteobacteria | Betaproteobacteria | Burkholderiales | Comamonadaceae | Rhodoferax |
| YE201E09 | 5 | 2.0 | Actinobacteria | Actinobacteria | Actinomycetales | ||
| YU201H04 | 5 | 1.3 | Actinobacteria | Actinobacteria | Actinomycetales | Microbacteriaceae |
Note that subclass and suborder information were omitted from the table.
Fig. 3.
Map of the 114 major watersheds of the world (35) with marks indicating rivers (red) and lakes (blue) where members of some of the nine dominant OTUs in Table 2 have been found. [Adapted with permission from ref. 36 (Copyright 1998, World Resources Institute).]
The second pattern that we identified in big river bacterioplankton communities is seasonality. In each river, bacterial communities were relatively stable during each of the three seasonal periods (winter, spring freshet, summer/fall), and changed rapidly between seasons. This pattern of seasonality in each river is linked to the overall pattern of synchrony across all rivers because seasonal shifts occurred at the same times in all systems. Taxonomic diversity estimated from the number of denaturing gradient gel electrophoresis (DGGE) bands also varied with season, being low in the winter (19 ± 3 SD), high during the freshet (29 ± 5), and intermediate during summer/fall (25 ± 4).
It is becoming clear that seasonality is a common feature of aquatic bacterial communities. In coastal and estuarine systems, bacterial communities change with regional conditions, shifting steadily throughout the year in the southwestern U.S. (14), and forming distinct winter and summer communities in the northeast U.S. (13). In the Sargasso Sea, communities shift seasonally and are greatly influenced by deep convective mixing in winter and spring (12). In lakes and rivers, communities shift according to seasonal environmental conditions and are affected by major seasonal events such as lake mixing (23) and the spring freshet (6). In our study, major aspects of seasonal environmental conditions (e.g., discharge, NO3− concentration, dissolved organic matter indices) were common among the six rivers. Winter bacterial communities were sampled before the spring freshet and were always collected through ice, spring communities were sampled during high river flow, and summer/fall communities were collected during extended periods of low river flow. These and other seasonal environmental conditions shared by the six rivers followed the same annual pattern (see below) and appeared to drive changes in the composition of bacterioplankton communities (Fig. 2), suggesting a strong link between climate, biogeochemistry, and communities.
In each river, bacterial community composition was strongly predicted by environmental variables (Table 3). We found that the best models for describing variability in bacterial communities included inconsistent sets of environmental variables; but that was not unexpected, given that there was a high degree of covariation among these variables. However, we found that most of these models included factors describing the concentration and character of dissolved organic matter (DOC; absorbance at 375 nm, lignin phenol concentration, specific UV absorbance [SUV], ΔDO14C) (24), the concentration of nitrate (NO3−), and the concentration of major ions (Mg2+, K+, Na+, Ca+, Cl−, SO42−, alkalinity). We selected variables representative of these three categories (a375, NO3−, Mg2+) and found that they alone described most of the variability in bacterial community composition in all six rivers including the Yukon River, for which we were not able to include winter samples in the analysis (Table 3). Many of these environmental variables correlated with seasonal variability in water temperature and river flow rate, which also explained a large portion of the variability in bacterial community composition.
Table 3.
Spearman's coefficients (r) describing rank correlations between microbial community composition and environmental factors for two select sets of factors calculated with Bio-Env, and for all factors calculated with the BV-Step forward selection procedure (Primer 6.0)
| River | r | r | r (BV-Step, all variables) |
|---|---|---|---|
| Mackenzie | 0.66 (a375, Mg, NO3) | 0.66 (Q, Temp) | 0.74 (a375, DO14C, NO3, SO4, pH, Temp, Si) |
| Yukon | 0.54 (a375, Mg)* | 0.74 (SUVA, DOC, Cl, TDP, Si)† | |
| Kolyma | 0.77 (a375, Mg, NO3) | 0.85 (a375, NO3, LP)‡ | |
| Lena | 0.80 (a375, Mg, NO3) | 0.59 (Q)§ | 0.88 (a375, LP, NO3, SO4, NH4, Temp) |
| Yenisey | 0.89 (a375, Mg, NO3) | 0.77 (Q, Temp) | 0.91 (a375, NO3, SO4, Na, K, pH, DOC) |
| Ob | 0.75 (a375, Mg, NO3) | 0.54 (Q, Temp) | 0.87 (NO3, Cl, Ca) |
Note that some samples were excluded from analyses because of missing environmental data. River flow (Q) was not determined for the Yukon or Kolyma.
*NO3 did not improve correlation.
†Q, TSS, ΔDO14C are not included.
‡Q are not included.
§Temp did not improve correlation.
Seasonal shifts in water chemistry are thought to be driven by differences in the flow paths of source waters feeding these rivers (25). During winter, elevated major ion and nutrient concentrations and reduced dissolved organic matter (DOM) result from relatively high groundwater contributions (26). During the spring freshet, major ions and nutrients are diluted, and DOM is elevated by inputs of relatively young DOM from organic-rich surface soil layers (24). As summer progresses and the ground thaws, major ions gradually increase and DOM decreases, reflecting deepening flow paths of source water. Nutrients remain low in summer because of increased biological uptake in the soils and river water. Shifts in these variables define common seasonal changes in large temperate and arctic rivers, and they correlate with predictable, global-scale patterns of change in bacterioplankton community composition. As global climate shifts in the future, the seasonal flow paths of water in these catchments will change, as will the interaction of this water with the vast stores of organic rich peat; consequently, river chemistry and microbial community composition will change as well.
Annual reassembly was the third pattern that we identified in these bacterioplankton communities. We observed that bacterial populations specific to individual seasons become too rare to detect in other seasons but then become dominant again the following year. This is not necessarily what one would expect, given the remarkable bacterial diversity in natural systems (2) and the sensitivity and limited resilience of microbial communities (36). However, early studies of individual populations using cell counting techniques show repeatable annual patterns in the abundance of SAR11 bacteria in the Sargasso Sea (27), and Polynucleobacter sp. in Lake Mondsee (28). Also, four different multiyear studies of bacterioplankton community composition show that whole assemblages of bacteria reappear each year and form predictable seasonal communities in the Ipswich and Parker Rivers in the U.S. (6), Lake Mendota in the U.S. (23), the coastal ocean off California in the U.S. (14), and the Sargasso Sea (12). Our study showed a similar pattern for big arctic rivers (Fig. 2), and, like these other studies, demonstrated a predictable succession for bacterioplankton communities that may be useful for interpreting biogeochemical cycles and identifying long-term changes to ecosystems.
Bacterial communities in most natural systems are extremely diverse and highly variable between systems and over time (2, 6). However, our study demonstrates predictable temporal patterns in the composition of these communities that occur on a global scale, and shows a common, globally distributed set of freshwater bacterial populations. Similar synchronous, phenological patterns in plant and animal communities can be used to document long-term changes in climate (29). However, unlike these “macroscopic” organisms, bacterial communities are much more diverse, grow more rapidly, and may be more sensitive to environmental change. It is therefore likely that signals of climate change in the Arctic and other parts of the world can be detected by monitoring the composition, successional cycles, and interecosystem synchrony of microbial communities.
Materials and Methods
Water samples were collected over a 3-year period near the river mouths using ultraclean sampling equipment and protocols based on the U.S. Geological Survey (USGS) sampling protocols (http://pubs.usgs.gov/twri) (24). Sampling locations, dates, and methods for collecting depth-integrated water samples are described elsewhere (24) except for the Koyma River, which was sampled near Chersky, Russia (68.75 N 161.30 E).
We used a community fingerprinting technique called denaturing gradient gel electrophoresis of PCR-amplified 16S rRNA genes (PCR-DGGE) (6, 30) to show shifts in dominant bacterioplankton communities within each river. Methods for DNA sample collection, extraction, and DGGE analysis are described elsewhere (6), with the exceptions that DGGE gel gradients contained 35–60% denaturants.
DGGE separates PCR-amplified 16S rRNA gene fragments by melting characteristics on acrylamide gels containing gradients of denaturing chemicals. DGGE is highly sensitive and can reveal differences between DNA fragments as small as a single base pair (31). DGGE bands are assumed to represent individual bacterial populations (i.e., phylotypes or operational taxonomic units), although they may represent more than one population when DNA fragments fall at the same position in the gel. Experiments indicate that DGGE is capable of detecting 99–99.9% of bacterioplankton in a sample, depending on copy number of rRNA operons per cell and PCR primer specificity (30, 32). So, although this method does not characterize the vast microdiversity contained in bacterial communities (2), it does identify and compare the dominant members of these communities.
To identify bacteria in the six rivers, we collected DNA sequences of PCR-amplified and cloned 16S rRNA genes from river samples collected in June 2004 (≈96 sequences per river). Methods for preparing clone libraries are described elsewhere (6) with the following exceptions. PCR primers amplified bacterial 16S rRNA genes and 16S-23S intergenic spacer regions (27f 5′-agagtttgatcctggctcag-3′, 23Sr 5′-gggttbccccattcrg-3′) (33). Initial amplifications were done in single tubes with 27 cycles of PCR. These PCR products were diluted 1 in 10 in eight tubes per sample, subjected to two cycles of reconditioning PCR (34), and then combined. Plasmids were extracted using Nunc plasmid purification plates following the manufacturer's instructions, and the first 900 bp of the 16S genes were sequenced bidirectionally.
We tested the relationship between bacterioplankton community composition (DGGE) and a suite of environmental variables using the BVStep procedure (Primer 6.0). This program uses Spearman's rank correlation method to determine the degree of association between similarity matrices of DGGE data (Sorensen's equation) and environmental data (Euclidean distance) with forward stepwise selection (Spearman's, rho >0.95, delta rho <0.001, random starting variables). We also calculate Spearman's rho for select sets of environmental variables using the Bio-Env procedure. Environmental variables were transformed if necessary to achieve a normal distribution. Variables included in the analysis were river flow rate (Q), pH, water temperature, alkalinity, dissolved organic carbon (DOC), ΔDO14C, specific UV absorbance of DOC (SUVA), absorbance at 375 nm (a375), lignin phenols (LP), total suspended solids (TSS), Cl−, SO42−, Na+, K+, Mg2+, Ca+, NH4+, NO3−, SiO2, dissolved organic nitrogen (DON), total dissolved nitrogen (TDN), total dissolved phosphorous (TDP), particulate organic nitrogen (PON), and particulate organic carbon (POC).
Aligned clone sequences were exported from ARB after applying a 50% base pair frequency filter to remove nonhomologous sequences (Escherichia coli positions 61–833, 646 base pairs). Phylogenetic distances were calculated with DNADIST using Jukes-Cantor model, and the DOTUR application was applied to calculate diversity indices and to assign sequences to operational taxonomic units (OTUs) based on 97% sequence similarity. Taxonomic assignments were made using the Ribosomal Database Project naive Bayesian rRNA classifier tool using a confidence threshold of 80% (http://rdp.cme.msu.edu).
Supplementary Material
Acknowledgments.
Sampling efforts by personnel from the United States Geological Survey National Research Program and Alaska Water Science Center, Canada's Department of Indian Affairs and Northern Development, the South Russian Centre for Preparation and Implementation of International Projects, and the Northeast Science Station in Russia were essential to the success of this project. We thank Erica Kiss and Suzanne Thomas for assistance in the laboratory, and John Hobbie and Jennifer Bowen for helpful comments on the manuscript. This work was supported by National Science Foundation Grants 0520480, 0229302, 0425582, and 0519840.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The DNA sequences reported in this paper have been deposited in the GenBank database (accession nos. FJ694228–FJ694684).
This article contains supporting information online at www.pnas.org/cgi/content/full/0906149106/DCSupplemental.
References
- 1.Azam F, Worden AZ. Microbes, molecules, and marine ecosystems. Science. 2004;303:1622–1624. doi: 10.1126/science.1093892. [DOI] [PubMed] [Google Scholar]
- 2.Sogin ML, et al. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc Natl Acad Sci USA. 2006;103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huber JA, et al. Microbial population structures in the deep marine biosphere. Science. 2007;318:97–100. doi: 10.1126/science.1146689. [DOI] [PubMed] [Google Scholar]
- 4.Fenchel T. Biogeography for bacteria. Science. 2003;301:925–926. doi: 10.1126/science.1089242. [DOI] [PubMed] [Google Scholar]
- 5.Crump BC, Armbrust EV, Baross JA. Phylogenetic analysis of particle-attached and free-living bacterial communities in the Columbia River, its estuary, and the adjacent coastal ocean. Appl Environ Microbiol. 1999;65:3192–3204. doi: 10.1128/aem.65.7.3192-3204.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crump BC, Hobbie JE. Synchrony and seasonality of bacterioplankton communities in two temperate rivers. Limnol Oceanogr. 2005;50:1718–1729. [Google Scholar]
- 7.Zwart G, Crump BC, Agterveld M, Hagen F, Han SK. Typical freshwater bacteria: An analysis of available 16S rRNA gene sequences from plankton of lakes and rivers. Aquat Microb Ecol. 2002;28:141–155. [Google Scholar]
- 8.Cottrell MT, Waidner LA, Yu LY, Kirchman DL. Bacterial diversity of metagenomic and PCR libraries from the Delaware River. Environ Microbiol. 2005;7:1883–1895. doi: 10.1111/j.1462-2920.2005.00762.x. [DOI] [PubMed] [Google Scholar]
- 9.Sekiguchi H, Watanabe M, Nakahara T, Xu BH, Uchiyama H. Succession of bacterial community structure along the Changjiang River determined by denaturing gradient gel electrophoresis and clone library analysis. Appl Environ Microbiol. 2002;68:5142–5150. doi: 10.1128/AEM.68.10.5142-5150.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galand PE, Lovejoy C, Pouliot J, Garneau ME, Vincent WF. Microbial community diversity and heterotrophic production in a coastal Arctic ecosystem: A Stamukhi lake and its source waters. Limnol Oceanogr. 2008;53:813–823. [Google Scholar]
- 11.Lemke MJ, et al. Description of freshwater bacterial assemblages from the Upper Parana River Floodpulse System, Brazil. Microb Ecol. 2009;57:94–103. doi: 10.1007/s00248-008-9398-3. [DOI] [PubMed] [Google Scholar]
- 12.Morris RM, et al. Temporal and spatial response of bacterioplankton lineages to annual convective overturn at the Bermuda Atlantic Time-series Study site. Limnol Oceanogr. 2005;50:1687–1696. [Google Scholar]
- 13.Kan JJ, Crump BC, Wang K, Chen F. Bacterioplankton community in Chesapeake Bay: Predictable or random assemblages. Limnol Oceanogr. 2006;51:2157–2169. [Google Scholar]
- 14.Fuhrman JA, et al. Annually reoccurring bacterial communities are predictable from ocean conditions. Proc Natl Acad Sci USA. 2006;103:13104–13109. doi: 10.1073/pnas.0602399103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kent AD, Yannarell AC, Rusak JA, Triplett EW, McMahon KD. Synchrony in aquatic microbial community dynamics. ISME J. 2007;1:38–47. doi: 10.1038/ismej.2007.6. [DOI] [PubMed] [Google Scholar]
- 16.McClelland JW, et al. Development of a pan-arctic database for river chemistry. EOS Trans Am Geophys Union. 2008;89:217–218. [Google Scholar]
- 17.Pace ML, Cole JJ. Synchronous variation of dissolved organic carbon and color in lakes. Limnol Oceanogr. 2002;47:333–342. [Google Scholar]
- 18.Magnuson JJ, Benson BJ, Kratz TK. Temporal coherence in the limnology of a suite of lakes in Wisconsin, USA. Freshw Biol. 1990;23:145–159. [Google Scholar]
- 19.Grenfell BT, et al. Noise and determinism in synchronized sheep dynamics. Nature. 1998;394:674–677. [Google Scholar]
- 20.Ranta E, Kaitala V, Lindstrom J, Helle E. The Moran effect and synchrony in population dynamics. Oikos. 1997;78:136–142. [Google Scholar]
- 21.Liebhold A, Koenig WD, Bjornstad ON. Spatial synchrony in population dynamics. Annu Rev Ecol Evol Syst. 2004;35:467–490. [Google Scholar]
- 22.Finlay BJ. Global dispersal of free-living microbial eukaryote species. Science. 2002;296:1061–1063. doi: 10.1126/science.1070710. [DOI] [PubMed] [Google Scholar]
- 23.Shade A, et al. Interannual dynamics and phenology of bacterial communities in a eutrophic lake. Limnol Oceanogr. 2007;52:487–494. [Google Scholar]
- 24.Raymond PA, et al. Flux and age of dissolved organic carbon exported to the Arctic Ocean: A carbon isotopic study of the five largest arctic rivers. Glob Biogeochem Cycle. 2007;21:GB4011. doi: 10.1029/2007GB002934. [DOI] [Google Scholar]
- 25.Striegl RG, Aiken GR, Dornblaser MM, Raymond PA, Wickland KP. A decrease in discharge-normalized DOC export by the Yukon River during summer through autumn. Geophys Res Lett. 2005;32:L21413. doi: 10.1029/2005GL024413. [DOI] [Google Scholar]
- 26.Gordeev VV, Martin JM, Sidorov IS, Sidorova MV. A reassessment of the Eurasian river input of water, sediment, major elements, and nutrients to the Arctic Ocean. Am J Sci. 1996;296:664–691. [Google Scholar]
- 27.Morris RM, et al. SAR11 clade dominates ocean surface bacterioplankton communities. Nature. 2002;420:806–810. doi: 10.1038/nature01240. [DOI] [PubMed] [Google Scholar]
- 28.Wu QLL, Hahn MW. High predictability of the seasonal dynamics of a species-like Polynucleobacter population in a freshwater lake. Environ Microbiol. 2006;8:1660–1666. doi: 10.1111/j.1462-2920.2006.01049.x. [DOI] [PubMed] [Google Scholar]
- 29.Menzel A, et al. European phenological response to climate change matches the warming pattern. Glob Change Biol. 2006;12:1969–1976. [Google Scholar]
- 30.Muyzer G, Dewaal EC, Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel-electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16s ribosomal-RNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sheffield VC, Cox DR, Lerman LS, Myers RM. Attachment of a 40-base-pair G+C-rich sequence (GC-clamp) to genomic DNA fragments by the polymerase chain-reaction results in improved detection of single-base changes. Proc Natl Acad Sci USA. 1989;86:232–236. doi: 10.1073/pnas.86.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kan JJ, Wang K, Chen F. Temporal variation and detection limit of an estuarine bacterioplankton community analyzed by denaturing gradient gel electrophoresis (DGGE) Aquat Microb Ecol. 2006;42:7–18. [Google Scholar]
- 33.Brown MV, Schwalbach MS, Hewson I, Fuhrman JA. Coupling 16S-ITS rDNA clone libraries and automated ribosomal intergenic spacer analysis to show marine microbial diversity: Development and application to a time series. Environ Microbiol. 2005;7:1466–1479. doi: 10.1111/j.1462-2920.2005.00835.x. [DOI] [PubMed] [Google Scholar]
- 34.Thompson JR, Marcelino LA, Polz MF. Heteroduplexes in mixed-template amplifications: Formation, consequence and elimination by ‘reconditioning PCR’. Nucleic Acids Res. 2002;30:2083–2088. doi: 10.1093/nar/30.9.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ravenga C, Murray S, Abramovitz J, Hammond A. Watersheds of the World: Ecological Value and Vulnerability. Washington, DC: World Resources Institute; 1998. [Google Scholar]
- 36.Allison SD, Martiny JBH. Resistance, resillience, and redundancy in microbial communities. Pro Natl Acad Sci USA. 2008;105:11512–11519. doi: 10.1073/pnas.0801925105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.