Abstract
Background
The Brassica species, related to Arabidopsis thaliana, include an important group of crops and represent an excellent system for studying the evolutionary consequences of polyploidy. Previous studies have led to a proposed structure for an ancestral karyotype and models for the evolution of the B. rapa genome by triplication and segmental rearrangement, but these have not been validated at the sequence level.
Results
We developed computational tools to analyse the public collection of B. rapa BAC end sequence, in order to identify candidates for representing collinearity discontinuities between the genomes of B. rapa and A. thaliana. For each putative discontinuity, one of the BACs was sequenced and analysed for collinearity with the genome of A. thaliana. Additional BAC clones were identified and sequenced as part of ongoing efforts to sequence four chromosomes of B. rapa. Strikingly few of the 19 inter-chromosomal rearrangements corresponded to the set of collinearity discontinuities anticipated on the basis of previous studies. Our analyses revealed numerous instances of newly detected collinearity blocks. For B. rapa linkage group A8, we were able to develop a model for the derivation of the chromosome from the ancestral karyotype. We were also able to identify a rearrangement event in the ancestor of B. rapa that was not shared with the ancestor of A. thaliana, and is represented in triplicate in the B. rapa genome. In addition to inter-chromosomal rearrangements, we identified and analysed 32 BACs containing the end points of segmental inversion events.
Conclusion
Our results show that previous studies of segmental collinearity between the A. thaliana, Brassica and ancestral karyotype genomes, although very useful, represent over-simplifications of their true relationships. The presence of numerous cryptic collinear genome segments and the frequent occurrence of segmental inversions mean that inference of the positions of genes in B. rapa based on the locations of orthologues in A. thaliana can be misleading. Our results will be of relevance to a wide range of plants that have polyploid genomes, many of which are being considered according to a paradigm of comprising conserved synteny blocks with respect to sequenced, related genomes.
Background
The cultivated Brassica species, like Arabidopsis thaliana, are members of the Brassicaceae family [1]. Brassica rapa (n = 10) contains the Brassica A genome, which is the smallest, at ca. 500 Mb [2]. A genome sequencing project is underway http://brassica.bbsrc.ac.uk/. A number of genome analysis studies have shown that the Brassica genomes contain extensive triplication, consistent with their having evolved from a hexaploid ancestor [3-5]. Two sequence-level studies, one in B. oleracea [6] and one in B. rapa [7] have provided further support for the hypothesis of hexaploid ancestry for the Brassica species. Recent cytogenetic studies have shown that a distinctive feature of the Brassiceae tribe, of which the Brassica species are members but A. thaliana is not, is that they contain extensively triplicated genomes [8].
An elegant study using sequenced RFLP markers demonstrated that 21 segments of the genome of A. thaliana, representing almost its entirety, could be replicated and rearranged to generate a structure approximating that of the B. napus genome [9]. In a similarly ground-breaking study, an ancestral karyotype (AK) of n = 8 was proposed for the Brassicaceae, which has been related to the A. thaliana genome sequence and the structure of the B. rapa genome derived by linkage mapping [10]. Thus the genome sequence of A. thaliana is being used, either directly via the B. napus comparative analysis or indirectly via the AK inferred genome, to inform studies in the Brassica species. A complication in such comparative studies is that there are typically multiple orthologues in Brassica species for each gene represented in A. thaliana, although interspersed gene loss has reduced the number that might be expected in paleohexaploids such as the Brassica species [6].
Brassica species have been used to study the early responses of genomes to the induction of polyploidy, via resynthesis of B. napus by hybridization of B. rapa with B. oleracea. Such lines display genome instability, which can persist for many generations [11]. Although this is hypothesised to involve homoeologous non-reciprocal translocations, such evolutionary events have not been studied at the sequence level. Indeed, sequence-level studies in Brassica to date have focussed on regions that show collinearity between the Brassica genome studied and that of A. thaliana. Similarly, in comparative studies in grass genomes, which are considered very much in terms of rearranged collinear blocks [12], little attention has been paid to the regions of collinearity breakdown.
We aimed to test the veracity of our present understanding of the evolution of the Brassica and Arabidopsis genomes from the AK genome by identifying and sequencing BAC clones containing genomic DNA of B. rapa that represent a sample of collinearity discontinuities (CDs) relative to the A. thaliana genome. This involved the development of bioinformatics tools and accessing data arising from ongoing activities to sequence the first four of the ten chromosomes of B. rapa.
Results
Identification of BAC clones putatively containing collinearity discontinuities
We developed a method by which candidate B. rapa BAC clones spanning CDs with the Arabidopsis genome could be identified and selected for sequencing. Our starting point was the set of BAC end sequence (BES) data available for the combined libraries from the 'Chiifu' cultivar, which is the subject of the multinational genome sequencing project. Using a strategy opposite to that employed for selection of seed BACs for that programme, we analysed the mate-pairs within the BES data primarily for inferred disruptions in short- to medium-range synteny (up to five-fold of an average BAC insert, i.e. 500 kb). We first conducted a BLASTN similarity search against the Arabidopsis genome sequence with all 200,031 individual BES available from 106,144 B. rapa BAC clones, these sequences comprising 93,887 mate-pairs and 12,257 singletons. For each BES we recorded the pseudochromosome coordinates of the most significant alignment above a threshold E-value of 10-30. Of the clones with both mate-pair BES available, 26,574 (28%) gave mappings with each E-value above this threshold and were therefore amenable to further analysis.
We loaded these pseudochromosome mappings into our own copy of the ATIDB Arabidopsis genome database [13] to enable a programmatic analysis. A Perl script was developed to interrogate the database and to identify associations between non-contiguous regions of the Arabidopsis genome that are linked by a number of disjoined mate-pair mappings and thus produce a list of cognate B. rapa BAC clones that might contain discontinuities. The algorithm we used is described in more detail in Methods. Our initial approach took into account several factors; filtering out instances of clone duplications and discounting mate-pair mappings whose DNA strand dispositions differed from the majority. We experimented with a threshold number, over the range of 2-5, of independent mate-pair mappings linking any given pair of bins required to signal an association.
BAC clones potentially representing CDs with the A. thaliana genome were thus selected, one from each association identified by three or more BAC clones. These BAC clones were sequenced and annotated, inter alia, for similarity to A. thaliana gene models and to B. rapa BES using BLASTN. Of the 68 sequenced BACs, 38 were found not to contain CDs. In the majority of these (25), the BACs show alignment of multiple gene models from two regions of the A. thaliana genome. These pairs of regions of the A. thaliana genome are related to each other, representing paralogous segments. The sequences at one end of each B. rapa BAC shows the highest similarity to the corresponding gene model from one of the A. thaliana genome segments, whereas the sequences at the other end of the B. rapa BAC shows the highest similarity to the corresponding gene model from the other A. thaliana genome segment. We termed this paralogue conflation. In the remaining cases, there appears to be at one end of the clone a small stretch of inverted sequence or a single gene (or gene fragment) with similarity elsewhere in the A. thaliana genome. The remaining 30 B. rapa BAC clones contain similarity to two or more collinear runs of multiple A. thaliana gene models.
These findings enabled us to substantially improve the algorithm and to reduce the false positive rate, using the sequenced BACs as a training set. Graphical representations of the results are shown in Figure 1 and a summary is given in Table 1. A majority proportion of the CDs we were seeking would be ancestral to the divergence of the B. rapa A genome and B. oleracea C genome lineages and so we experimented with adding supplementary data obtained from a B. oleracea BAC library. Using identical methods we analysed 85,317 BES derived from 43,691 B. oleracea clones, these sequences comprising 41,626 mate-pairs and 2,065 singletons. Of the clones with both mate-pair BES available, 6,623 clones (15.9%) yielded significant mappings at the same E-value threshold as before. This proportion was almost half that obtained from the B. rapa dataset, probably due to the higher repeat content in B. oleracea reducing the probability of each BES in a mate-pair containing conserved (genic) sequence. When these B. oleracea mappings were added and the database re-analysed, the number of associations revealed was increased significantly at each threshold value (Table 1).
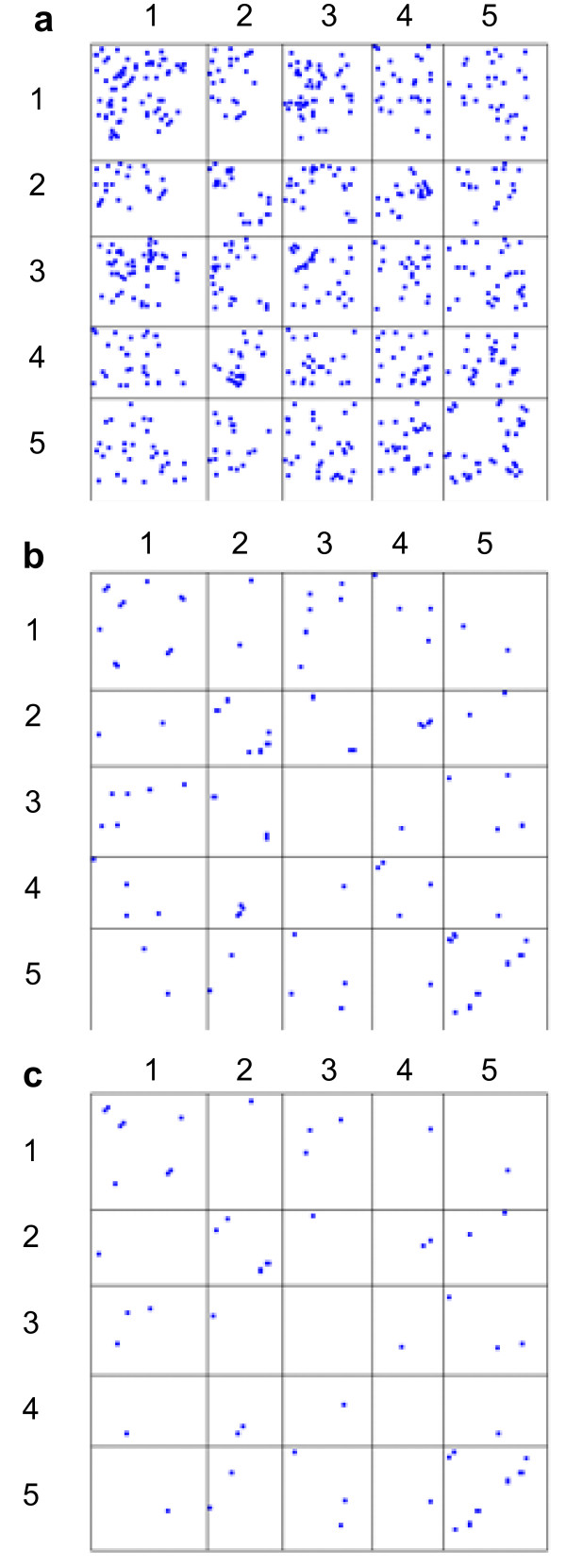
Figure 1.
Representation of candidate collinearity discontinuities within the A. thaliana genome with respect to B. rapa. The plot is divided into cells with dimensions proportional to the physical lengths of the five Arabidopsis pseudochromosomes (numbered on axes). A blue square within a cell signifies an association between 500 kb bins requiring (a.) 2; (b.) 3 or (c.) 4 distinct instances of disjoined B. rapa BES mate-pair mappings linking them.
Table 1.
Results of database interrogation for collinearity discontinuities
| B. rapa BES | B. rapa + B. oleracea BES | |||
|---|---|---|---|---|
| Thresholda | Associationsb | Clonesc | Associations | Clones |
| 2 | 380 | 893 | 719 | 1614 |
| 3 | 54 | 241 | 74 | 324 |
| 4 | 30 | 169 | 39 | 219 |
| 5 | 18 | 121 | 23 | 155 |
aNumber of independent BES mate-pair mappings required for an association.
bNumber of non-contiguous pairs of 500 kb bins linked by mate-pair mappings.
cNumber of clones linking bins.
Sequence validation of putative collinearity discontinuities
A genome sequencing project is underway for B. rapa, with sequences derived using a BAC-by-BAC strategy and with annotation being made publicly available http://brassica.bbsrc.ac.uk/. During the course of this effort, we sequenced a number of BAC clones (and sets of overlapping clones) containing CDs relative to the A. thaliana genome. Several of these had not had both mate-pair BES available previously and hence would have been undetectable by our computational analysis. These clones, along with those identified from the genome-wide analysis, were then assessed rigorously in order to exclude any that might not be representative of the genome (e.g. chimaeric ligation or the result of rearrangement in E. coli). This was done by using the sequence annotation to assess whether there are additional B. rapa BAC clones overlapping and confirming the discontinuity. This process involved the identification, based on congruous BAC end sequence alignments, of additional BAC clones that confirm the CD, and is illustrated in Figure 2. This resulted in the confirmation of a further 20 discontinuities represented in sequenced BAC clones, making a total of 50, as listed in Table 2. Relative to the A. thaliana chromosome structure, 19 of these represent inter-chromosomal rearrangements, with the remaining 31 representing the end points of intra-chromosomal rearrangements (segmental inversions).
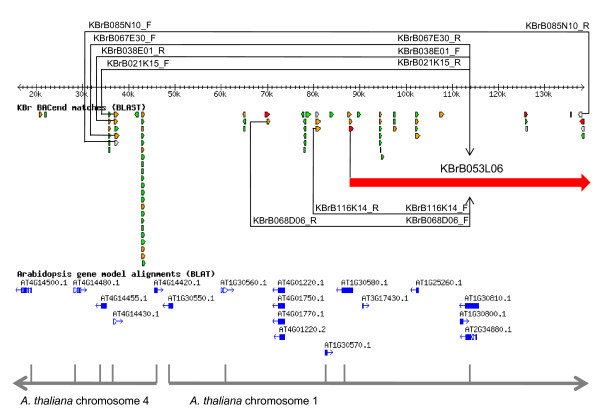
Figure 2.
Validation of collinearity discontinuities. Annotation of the sequence of BAC KBrB026E16 showing aligned BAC end sequences and homologous A. thaliana gene models. Sequence similarities with A. thaliana gene models are shown with blue arrows. Sequence similarity with end sequences derived from other B. rapa BAC clones are colour-coded: red for the end sequences of fully sequenced BACs, white for the end sequences of BACs for which both can be found within the annotated BAC, orange for end sequences which may bridge to another fully sequenced BAC, and green for other cases. The two inferred collinearity blocks are indicated by grey arrows. The collinearity discontinuity (CD) is confirmed by congruous end sequence alignments of KBrB085N10, KBrB067E30, KBrB038E01 and KBrB021K15. Both end sequences of KBrB085N10 align to KBrB026E16, with the expected orientation of the reads (one in each collinearity block). For each of the remaining three, one end aligns to the A. thaliana chromosome 4-related region and the other to regions of KBrB053L06 (shown in red) that extend beyond KBrB026E16, as do one end of each of KBrB068D06 and KBrB116K14 (from which the other ends align to the chromosome 1-related region. Thus multiple BACs (for which only end sequence is available) confirm the CD identified in the sequenced clone, in addition to the fully-contained BAC (KBrB085N10).
Table 2.
Characterization of B. rapa BAC clones containing discontinuities in collinearity with the A. thaliana genome
| BAC | Sequence Accession |
Collinearity with A. thaliana* | Linkage group | No. confirming BACsd |
|---|---|---|---|---|
| KBrH106N09 | CU915572 | 4g03430-4g03630| 4g05460-4g05430| 3g26280-3g26570 | A9b | 3 |
| KBrB089J13 | CU695271 | 4g38560-4g38350| 4g14145-4g14350| 1g30480-1g30400 | A8a | 7 |
| KBrH110M01 | AC237306 | 2g22840-2g20920| 2g41990-2g42005| 5g49760-5g49900 | A3b | 2 |
| KBrH013F08 | CU695290 | 3g52930-3g52770| 1g02080-1g01980| 5g42100-5g42020 | 4 | |
| KBrB028F11 | AC237302 | 2g20440-2g20900| 3g24620-3g25290| 1g62200-1g63390 | A9b | 2 |
| KBrB026E16 | CU914552 | 4g14550-4g14420| 1g30550-1g30810 | A8b | 4 |
| KBrH109L07 | CU695323 | 3g49370-3g49660| 5g61770-5g61780 | A6a | 5 |
| KBrH010M06 | AC237305 | 3g49870-3g49670| 5g61760-5g61580 | A3b | 3 |
| KBrH004M24 | AC232396 | 2g01210-2g01060| 5g47810-5g47540 | A6b | 4 |
| KBrB026A12 | AC237301 | 5g47680-5g47760| 2g01110-2g01410 | A9b | 5 |
| KBrH131P10 | FP245487 | 3g24350-3g24495| 1g62120-1g61890 | A1b | 4 |
| KBrH001J23 | CU695282 | 3g49700-3g49730| 1g21900-1g21650 | A6a | 5 |
| KBrH034P16 | CU740088 | 3g52490-3g52500| 4g15230-4g15410 | A3a | 8 |
| KBrH071G03 | FP236364 | 5g42920-5g42470| 4g39770-4g39480 | 7 | |
| KBrH024O16 | CU695293 | 5g49960-5g49810| 1g64970-1g65200 | 5 | |
| KBrB055E21 | AC232500 | 3g01590-3g02000| 4g00050-4g00370 | A3b | 14 |
| KBrH004I22 | AC237303 | 2g04740-2g04050| 3g25520-3g25550 | A3b | 5 |
| KBrH006I08 | AC237304 | 3g63240-3g63460| 2g26540-2g26260 | A9b | 3 |
| KBrH026A01 | CU928183 | 2g25290-2g26170| 4g00040-4g00080| 2g24690-2g24450 | 2 | |
| KBrH066L21 | CU695309 | 5g28060-5g28150| 5g30510-5g28490| 5g49570-5g49360 | A9a | 12 |
| KBrB129C20 | CU695281 | 5g22750-5g22980| 5g60800-5g60460 | A3a | 18 |
| KBrH055O17 | FP565592 | 5g23100-5g23080| 5g60830-5g61380 | 10 | |
| KBrB011D06 | AC232452 | 2g05540-2g05760| 2g07690-2g05840| 2g11890-2g12480 | A3c | 5 |
| KBrB022J01 | CU695257 | 4g35450-4g35335| 4g23620-4g24120| 4g36130-4g36140 | A8b | 3 |
| KBrH108B16 | CU984565 | 5g59700-5g59650| 5g60110-5g59320| 5g59030-5g59130 | A3a | 8 |
| KBrH064I20 | CU695308 | 4g36760-4g36870| 4g37240-4g36880| 4g37260-4g37410 | A8a | 7 |
| KBrH001C16 | CU928050 | 4g37410-4g37250| 4g36880-4g36910 | A1a | 9 |
| KBrH027B04 | CU915252 | 4g37170-4g37240| 4g36870-4g36660 | A1b | 5 |
| KBrH073M17 | CU914555 | 3g08930-3g08600| 3g10330-3g10650 | A3a | 10 |
| KBrH073M07 | CU695313 | 3g09760-3g10180| 3g08530-3g07980 | A3a | 11 |
| KBrH088K13 | CU984562 | 4g00170-4g00770| 4g01370-4g01270 | A3a | 4 |
| KBrH108F05 | CU695322 | 4g01040-4g00800| 4g01400-4g01450 | A3a | 2 |
| KBrB034C23 | AC229604 | 5g26850-5g27020| 5g27930-5g27520 | A9a | 22 |
| KBrH106B05 | CU915253 | 5g28080-5g27950| 5g27110-5g27380 | A6a | 3 |
| KBrH089D10 | FP085567 | 1g08450-1g07860| 1g07705-1g07720 | A8b | 1 |
| KBrH056E23 | CU695304 | 1g13380-1g13400| 1g11860-1g11490 | A6a | 3 |
| KBrB026M01 | CU695258 | 1g23820-1g24180| 1g27210-1g26970 | A8b | 6 |
| KBrB122B06 | FP236659 | 1g49630-1g49590| 1g49880-1g50220 | 2 | |
| KBrB100G10 | CU695273 | 1g50380-1g50980| 1g51420-1g51380 | 8 | |
| KBrB092C03 | CU984555 | 1g65580-1g64750| 1g67220-1g67330 | A7a | 7 |
| KBrH061M05 | CU695307 | 2g32870-2g32830| 2g33120-2g33710 | A3a | 4 |
| KBrH127P20 | CU695332 | 2g46300-2g46140| 2g40430-2g40700 | A5a | 8 |
| KBrH083O14 | FP325102 | 2g47250-2g47950| 2g29950-2g29320 | 8 | |
| KBrB037J21 | CU984548 | 3g24040-3g23940| 3g24300-3g24310 | A1a | 2 |
| KBrB008F10 | CU695255 | 4g17460-4g17480| 4g28380-4g28070 | A1b | 2 |
| KBrB045C11 | FP236657 | 4g20200-4g20890| 4g21710-4g21680 | 1 | |
| KBrB090F01 | CU915574 | 4g39980-4g40100| 4g38060-4g37870 | A1a | 9 |
| KBrB107K13 | CU695275 | 5g02800-5g02030| 5g01040-5g01075 | A2a | 11 |
| KBrB073M23 | CU915573 | 5g02880-5g03370| 5g02580-5g02310 | 5 | |
| KBrH100O23 | FP017268 | 4g03240-4g03610| 4g05460-4g04940 | 5 | |
* Collinear runs of gene model homologies indicated "-"; discontinuities in collinearity indicated "| "
a Determined by linkage mapping
b Identified during chromosome sequencing activities
c Determined by linkage mapping of an overlapping BAC
d In addition to the sequenced BAC
Inter-chromosomal rearrangements
Fifteen of the 19 inter-chromosomal CDs were genetically mapped in the B. rapa genome, either by direct linkage mapping or by sequence overlap with a BAC mapped by linkage mapping described elsewhere [14], as summarised in Table 2. These could be related to the position in the Brassica A genome of CDs previously inferred by linkage mapping-, defined relative to A. thaliana chromosomes [9] and subsequently to the AK [10]. We will use the nomenclature At(chromosome number, letter) to refer to the previously described A. thaliana chromosome blocks (e.g. At1A refers to A. thaliana chromosome 1, block A) as described in [9] and AK(letter) to refer to the ancestral karyotype blocks (e.g. AKA refers to ancestral karyotype block A), as described in [10].
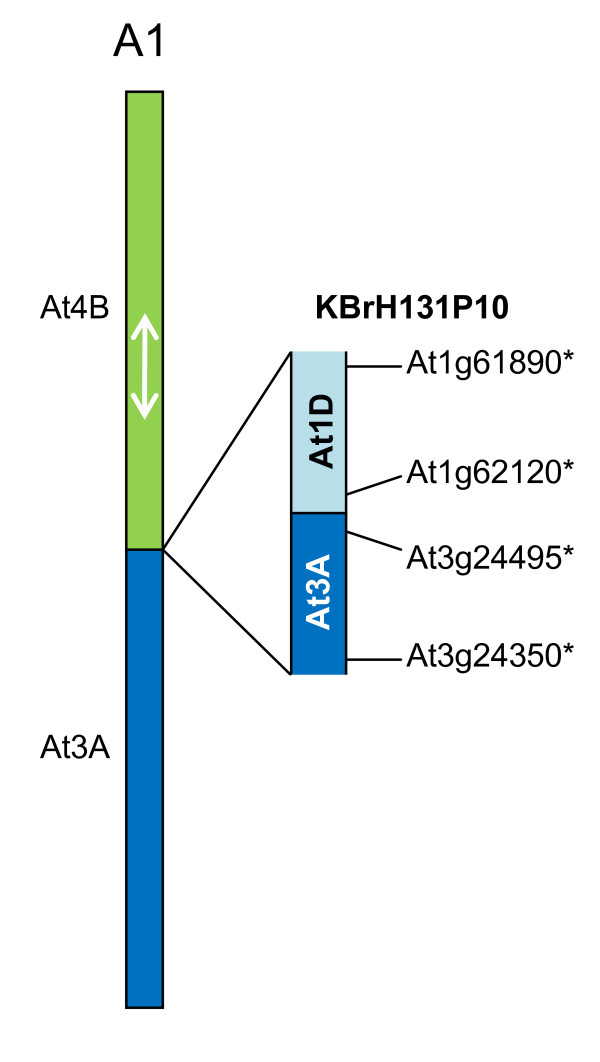
One sequenced CD (represented by BAC KBrH131P10) mapped to linkage group A1 and is consistent with the position of the inferred CD between blocks At3A and At4B (AKF and AKT), as illustrated in Figure 3. However, the transition revealed by the BAC sequence is between the expected part of At3A (AK block F) but with At1D (AKD). The linkage mapping study [9] had identified markers on A1 with similarity to this region of the A. thaliana genome, but there was insufficient evidence to call the block. Only one copy of AKD had been identified previously [10], so our study has identified the position of one of the "missing" two blocks in the B. rapa genome that would be expected from its paleohexaploid ancestry.
Figure 3.
Collinearity discontinuities mapped to B. rapa linkage group A1. The sequenced BAC clones are illustrated in relation to the CDs with which their mapping is consistent, as represented in [9]. Orthologues delineating the boundaries of recombination events and the end-most aligned gene models within the BACs are designated by the name of the A. thaliana gene model suffixed "*".
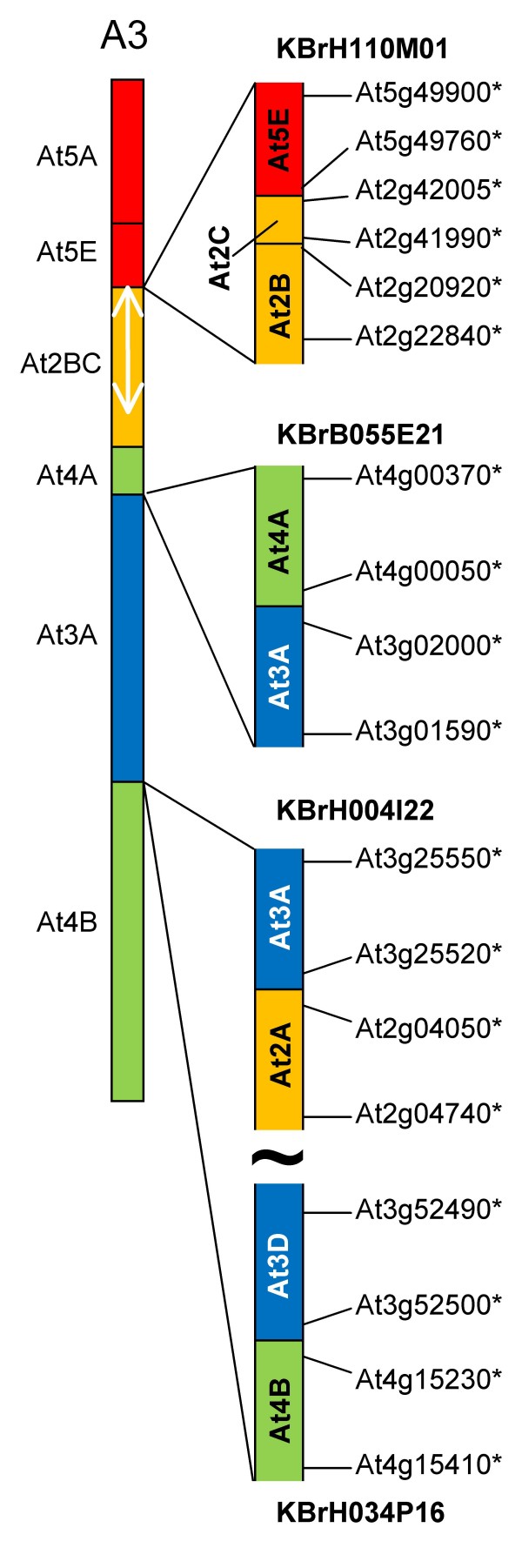
Five sequenced CDs (represented by BACs KBrH110M01, KBrH010M06, KBrH034P16, KBrB055E21 and KBrH004I22) mapped to linkage group A3, as illustrated in Figure 4. The position of KBrH110M01 is consistent with the position of the inferred CD between At5E and At2B/C (AKW and AK J). However, the transition revealed by the BAC sequence shows an additional small segment in between these, with collinearity to a more distant (internal) part of At2B/C (AKJ). This structure may be the result of rearrangements within this collinearity block.
Figure 4.
Collinearity discontinuities mapped to B. rapa linkage group A3. The sequenced BAC clones are illustrated in relation to the CDs with which their mapping is consistent, as represented in [9]. Orthologues delineating the boundaries of recombination events and the end-most aligned gene models within the BACs are designated by the name of the A. thaliana gene model suffixed "*".
The sequences within KBrH010M06 represent the end of collinearity block At3C (AKM) and sequences internal to collinearity blocks At5F (AKX), but neither had been identified previously on linkage group A3.
The sequences within KBrH034P16 are internal to collinearity blocks At3D and At4B (AKN and AKT). Although At4B (AKT) had been identified previously on B. rapa linkage group A3, At3D (AKN) had not. Therefore the transition previously inferred on this linkage group between collinearity blocks At4B (AKT) and At3A (AKF) [9,10] may be more complex than anticipated. This is supported by the results of analysis of the structure of the Brassica A genome as represented in B. juncea, in which AKN and AKG-H were identified between AKT and AKF [15].
The sequences within KBrB055E21 represent the ends of collinearity blocks At3A and At4A (AKF and AKO). This is consistent with the transition inferred on the basis of linkage mapping [9], but is not consistent with the inferred interpolation of AKP between AKF and AKO that has been proposed [10].
The sequences within KBrH004I22 correspond to the end of At3A (AKF) and sequences internal to At2A (or at the end of AKK). Although At3A (AKF) had been identified previously on linkage group A3, At2A (AKK) had not. Only two copies of AKK had been identified previously [10], so our study may have identified the position of the "missing" third block in the B. rapa genome that would be expected from its paleohexaploid ancestry. The linkage mapping study [9] had identified markers on A3 that have similarity to this region of the A. thaliana genome, but there was insufficient evidence to call the block.
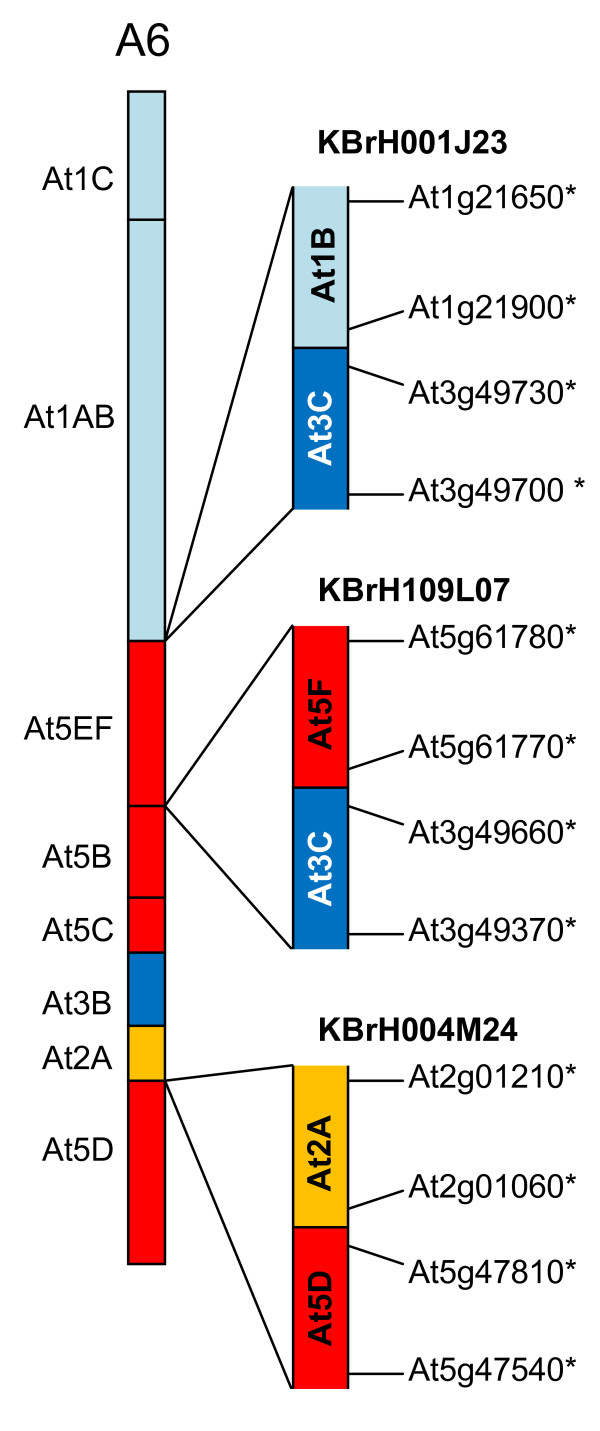
Three sequenced CDs (represented by BACs KBrH109L07, KBrH004M24 and KBrH001J23) mapped to linkage group A6, as illustrated in Figure 5. The sequences in KBrH109L07 represent the end of collinearity block At3C (AKM) and sequences internal to collinearity blocks At5F (AKX). Block At5F (AKX) had been position on linkage group A6 previously, but no copy of At3C (AKM) had been positioned previously on this linkage group. Thus this BAC represents the position of an additional copy of this segment (along with that identified in BAC KBrH010M06). The linkage mapping study [9] had identified markers on A6 that have similarity to this region of the A. thaliana genome, but there was insufficient evidence to call the block.
Figure 5.
Collinearity discontinuities mapped to B. rapa linkage group A6. The sequenced BAC clones are illustrated in relation to the CDs with which their mapping is consistent, as represented in [9]. Orthologues delineating the boundaries of recombination events and the end-most aligned gene models within the BACs are designated by the name of the A. thaliana gene model suffixed "*".
The sequences within KBrH004M24 correspond to the end of At2A (AKK) and sequences internal to At5D (or at the end of AKV) and confirm one of the CDs on linkage group A6 previously inferred [10].
The sequences within KBrH001J23 correspond to the end of collinearity block At3C (AKM) and sequences internal to collinearity blocks At1B (AKB). Block At1B (AKB) had been position on linkage group A6 previously, but no copy of At3C (AKM) had been positioned previously on this linkage group. Thus this BAC represents the position of the third copy of this segment (along with those identified in BACs KBrH010M06 and KBrH109L07).
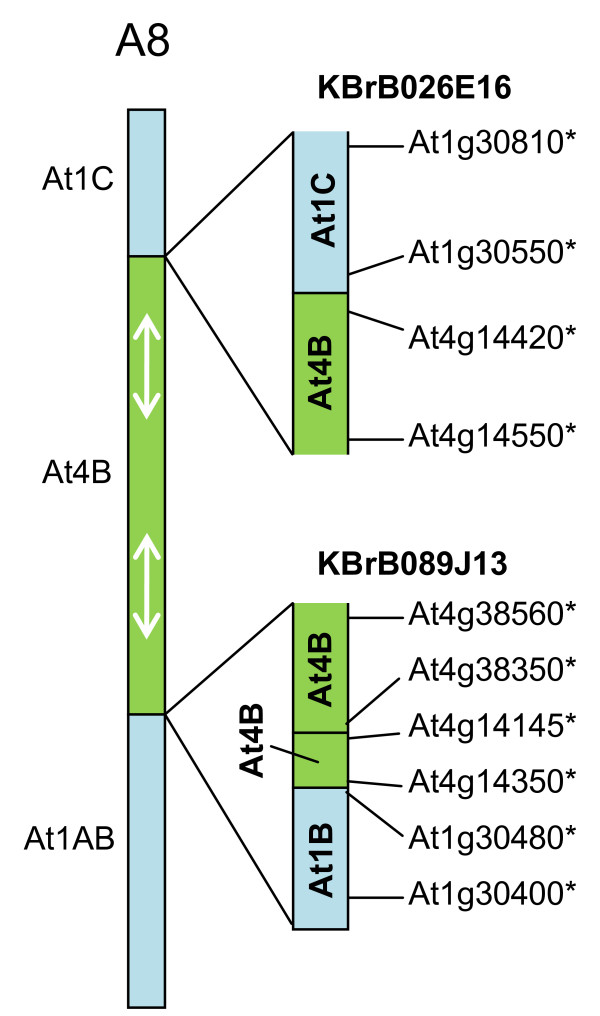
Two sequenced CDs (represented by BACs KBrB089J13 and KBrB026E16) mapped to linkage group A8, as illustrated in Figure 6. The sequences in both correspond to those close to the ends of collinearity blocks At4B (AKT) and At1B (AKB). Thus they are both candidates for representing the CDs on linkage group A8 previously inferred [9,10]. However, the transition revealed by the sequence of KBrB089J13 shows an additional small segment in between these, with collinearity to a more distant (internal) part of At4B (AKT). This structure may be the result of rearrangements within this collinearity block.
Figure 6.
Collinearity discontinuities mapped to B. rapa linkage group A8. The sequenced BAC clones are illustrated in relation to the CDs with which their mapping is consistent, as represented in [9]. Orthologues delineating the boundaries of recombination events and the end-most aligned gene models within the BACs are designated by the name of the A. thaliana gene model suffixed "*".
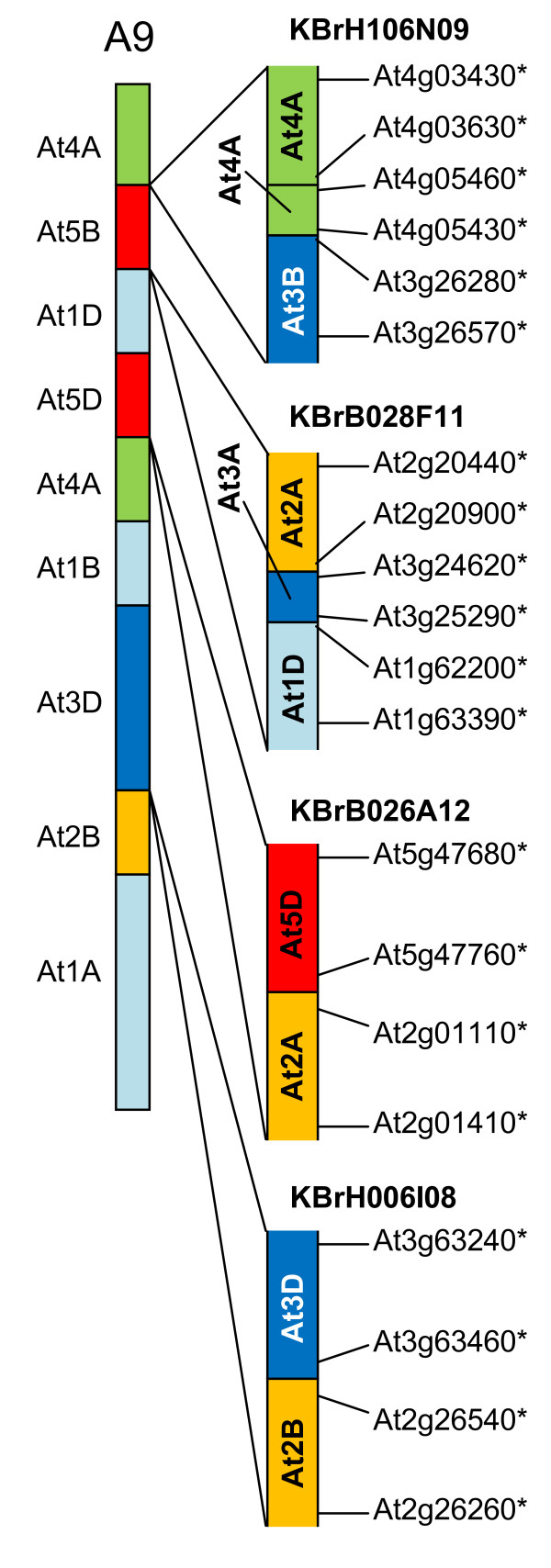
Four sequenced CDs (represented by BACs KBrH106N09, KBrB028F11, KBrB026A12 and KBrH006I08) mapped to linkage group A9, as illustrated in Figure 7. The sequences in KBrH106N09 represent sequences within collinearity block At4A (AKO) and within At3B (AKL). Although At4A (AKO) had been identified previously on linkage group A9, At3B (AKL) had not. Only two copies of AKL had been identified previously [10], so our study has identified the position of the "missing" third block in the B. rapa genome that would be expected from its paleohexaploid ancestry. There is an additional small collinear segment between AKO and AKL, corresponding to sequences anticipated to have been positioned between AKO and AKP, suggesting that the previously defined boundary of AKO (corresponding to an orthologue of At4g04995) [10] may have been incorrect, with this block extending to an orthologue of A4g05460.
Figure 7.
Collinearity discontinuities mapped to B. rapalinkage group A9. The sequenced BAC clones are illustrated in relation to the CDs with which their mapping is consistent, as represented in [9]. Orthologues delineating the boundaries of recombination events and the end-most aligned gene models within the BACs are designated by the name of the A. thaliana gene model suffixed "*".
The sequences in KBrB028F11 represent sequences at the end of collinearity blocks At2A (AKH) and within At1D (AKD). Although At1D (AKD) had been identified previously on B. rapa linkage group A9, At2A (AKH) had not. Therefore the transitions previously inferred on this linkage group bordering block At1D (AKD) [9,10] may be more complex than anticipated. This is supported by the results of analysis of the structure of the Brassica A genome as represented in B. juncea, in which AKH was identified as being adjacent to AKD [15]. The linkage mapping study [9] had identified markers on A9 that have similarity to this region of the A. thaliana genome, but there was insufficient evidence to call the block. The transition revealed by the BAC sequence shows an additional small segment in between At2A (AKH) and At1D (AKD), with collinearity to the end of At3A (AKF).
The sequences in KBrB026A12 represent sequences at the end collinearity block At2A (AKK) and within At5D (AKV). Although At5D (AKV) had been identified previously on linkage group A9, the part of At2A corresponding to AKK had not. Therefore the transitions previously inferred on this linkage group bordering block At5D (AKV) [9,10] may be more complex than anticipated. Only two copies of AKK had been identified previously [10], so our study has identified the position of the "missing" third block in the B. rapa genome that would be expected from its paleohexaploid ancestry. The linkage mapping study [9] had identified markers on A9 that have similarity to this region of the A. thaliana genome, but there was insufficient evidence to call the block.
The sequences in KBrH006I08 represent sequences at the end of collinearity blocks At3D (AKN) and within At2B (AKI). They confirm one of the CDs on linkage group A9 previously inferred [9,10], but indicate that At2B (AKI), as represented on linkage group A9, may be truncated.
Segmental inversions
Twenty four of the 31 CDs representing the end points of intra-chromosomal rearrangements (segmental inversions) were mapped in the B. rapa genome, either by direct linkage mapping or by overlap with a BAC mapped by linkage mapping. Their occurrence appears genome-wide, as summarised in Table 2. Few such rearrangements had been inferred previously, and these had not been clearly defined. The positions on linkage group A8 of BAC clones KBrB022J01 and KBrH064I20, and on linkage group A1 of BAC clones KBrH001C16, KBrH027B04, KBrB008F10 and KBrB090F01 are consistent with those expected for the inversions noted in At4B segments on these chromosomes [9].
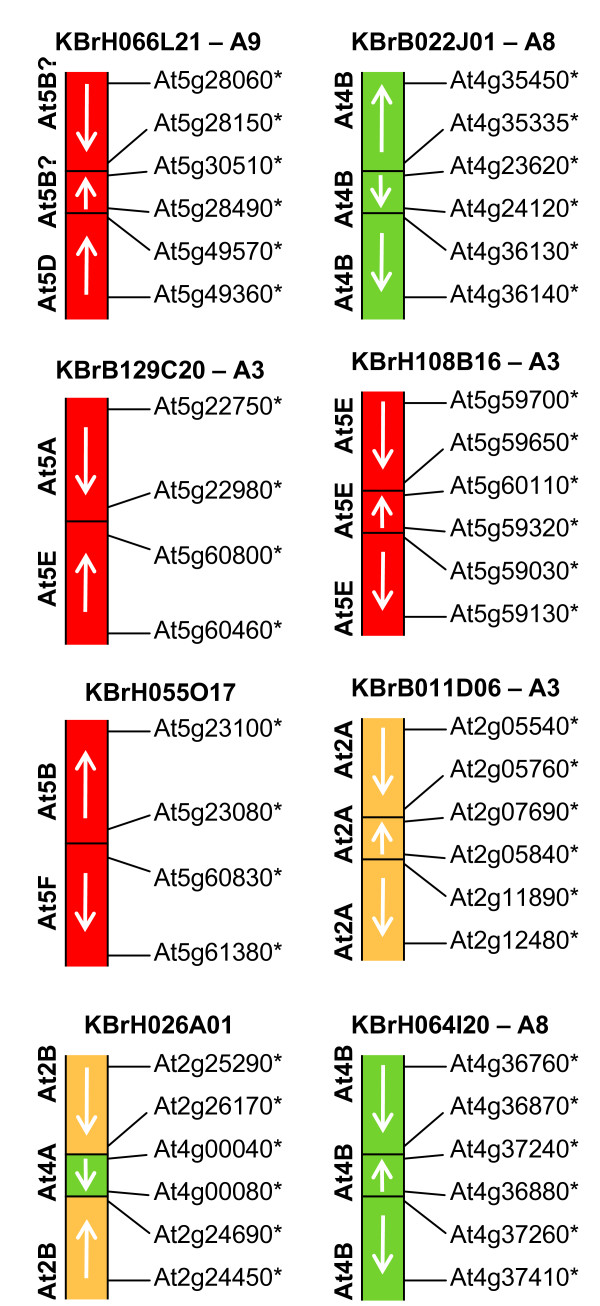
Some of the CDs classified as segmental inversions on the basis of the relationships of genes represented in the BACs to their orthologues in A. thaliana may have actually represented inter-chromosomal rearrangements, but segments of those ancestral chromosomes have subsequently come together in the A. thaliana genome. This is most notably the case for A. thaliana chromosome 5, for which we have three "segmental inversion" CDs spanning relatively large regions of the chromosome. These three are represented by KBrH066L21, KBrB129C20 and KBrB055O17, and are illustrated in Figure 8. The sequences in KBrH066L21, which has been mapped to linkage group A9, represent sequences near the end of collinearity block At5D (AKV) and between At5B and At5C (AKQ and AKS). Both At5D (AKV) and At5B (AKQ) had been identified on this linkage group previously, but not adjacent to each other. This suggests that the transitions previously inferred on this linkage group bordering these collinearity blocks [9,10] may be more complex than anticipated. The identification of sequences corresponding to a region between AKQ and AKS suggests that the previously defined boundary of AKQ (corresponding to an orthologue of At5g28897) [10] may have been incorrect, with this block extending to an orthologue of A5g30510. The distal part of this extended collinearity block AKQ contains a small segmental inversion (relative to the A. thaliana genome) that is wholly contained within the BAC. The sequences in KBrB129C20 represents a CD between the ends of collinearity blocks At5E and At5A (AKW and AKR) and is consistent with the position of a previously described CD on linkage group A3 [9,10], to which the clone maps. The sequences within KBrB055O17 represents a CD between At5F and At5B (AKX and AKQ), respectively, but we were unable to determine its position in the genome.
Figure 8.
Collinearity discontinuities involving segmental inversions. Orthologues delineating the boundaries of recombination events and the end-most aligned gene models within the BACs are designated by the name of the A. thaliana gene model suffixed "*". The linkage group to which each BAC maps, where determined, is shown next to the BAC name.
In addition to the small segmental inversion (relative to the A. thaliana genome) contained within KBrH066L21, we found further examples of secondary rearrangements at the points of CDs in KBrB022J01 and KBrH108B16, and wholly contained inversions within KBrB011D06 and KBrH064I20, as illustrated in Figure 8. We identified one example of a CD apparently representing intra-chromosomal rearrangements that were separated by sequences from elsewhere in the genome. The sequences in KBrH026A01 represent a small segment from the end of At4A (AKO) at one end of a segmental inversion within At2B (AKI), as illustrated in Figure 8.
Analysis of collinearity discontinuity sequences
None of the BAC clones containing the CDs was found to contain B. rapa satellite repeat sequences characteristic of centromeres [16,17], nor was any found with tandem tracts of TTTAGGG repeats that are associated with telomeres [18], although clone KBrH108B16 did have a cluster of 31 such repeats interspersed over 926 bp, but some 17 kb from the CD.
Minimal sequences defining the confirmed CDs were then selected from these BACs by manual inspection, aided by annotated ab initio gene predictions and Brassica EST alignments. These CD regions were then compared with the complete set of annotated B. rapa genome sequence currently available (approximately 120 Mb of gene space). The results are given in Table 3. The CD sequences appear to have nucleotide compositions intermediate to those of genic and intergenic regions in terms of microsatellite (simple sequence repeat) density. Although the CDs show densities of apparent gene features typical of the averaged genome sequence, detailed analysis reveals that these features are often gene fragments with homology to Arabidopsis gene models from regions outside those brought together at the discontinuities. Furthermore, they are relatively transcriptionally inactive, with the proportion of models with EST support (obtained from alignments with transcript assemblies derived from all available EST sequences) being about half that of the average for predicted genes. The CD regions also appear to be relatively transcriptionally silent when analysed with the very sensitive but targeted method of alignment with Brassica A genome Solexa reads derived from the B. napus leaf transcriptome.
Table 3.
Summary of features over identified collinearity discontinuities and all annotated B. rapa sequence
| All sequence | Genic | Intergenic | All CDs | Inter-chromosomal CDs | Intra-chromosomal CDs | |
|---|---|---|---|---|---|---|
| %GC content | 35.4 | 40.9 | 30.1 | 34.0 | 35.2 | 33.3 |
| SSR density/kb | 0.22 | 0.17 | 0.27 | 0.24 | 0.28 | 0.21 |
| Gene model density/kb | 0.23 | n/a | n/a | 0.20 | 0.23 | 0.18 |
| Models with EST support/kb | 0.13 | n/a | n/a | 0.09 | 0.11 | 0.08 |
| Mapped Solexa leaf read density/kb | 40.8 | 64.6 | 12.6 | 13.7 | 15.6 | 12.5 |
Discussion
Computational methods were used to successfully identify BAC clones representing verified CDs between the genomes of B. rapa and A. thaliana, and relative to an ancestral karyotype. Along with CDs identified during the ongoing chromosome sequencing project, these represent a substantial (but incomplete) sampling of the CDs in the genome of B. rapa. Previous studies had defined a segmental structure for the paleohexaploid Brassica genome based largely on genetic linkage of markers with similarity to sequences in the A. thaliana genome [9,10]. Whereas the seminal study in this area [9] compared the arrangements of the B. napus genome with that of A. thaliana, and ours compared the arrangement of the B. rapa genome with that of A. thaliana, we anticipate that the results should be directly comparable as there seems to be little difference in the organization of the A genome in these two Brassica species [19]. Remarkably few of our CDs correspond to those expected from this structure: 3 of the 18 representing inter-chromosomal rearrangements and 6 of the 32 representing intra-chromosomal rearrangements. The relatively high "noise" inherent to comparative genomics studies in Brassica species, which is a consequence of the widespread occurrence of apparently transduplicated fragments of genes [6], means that multiple instance of collinear alignments are required to correctly identify collinear genome segments. This requirement limits the ability to identify relatively small segments using, for example, comparative linkage mapping based on RFLP markers. Although the paleopolyploid ancestry of Brassica species is now widely accepted, the lack of discernable triplication throughout the genome has not been fully explained. The hypothesised segments that have not been identified had been assumed to have been deleted. However, we have found evidence for the existence of numerous additional copies of genome segments, bringing the count of many of these to (or closer to) the predicted three. In one case (At3C/AKM) we identified and mapped onto the B. rapa genome three copies where none had been identified previously in that species, with only one copy having been identified in the Brassica A genome as represented in B. juncea [15].
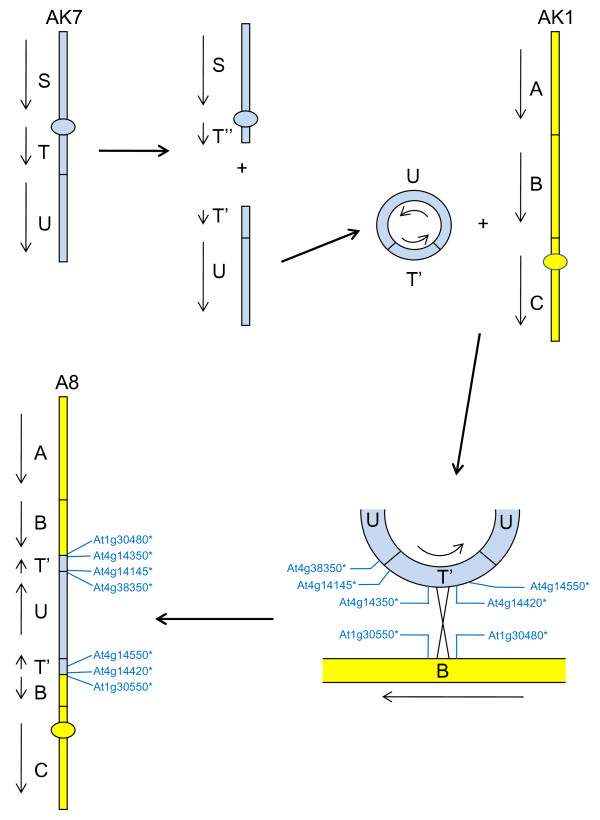
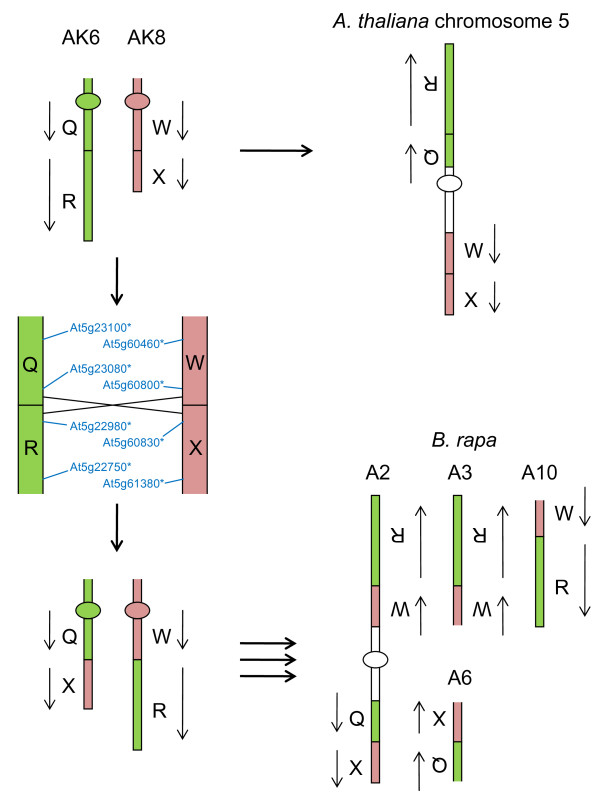
Our analyses of CDs enable us to hypothesise how parts of the genomes of A. thaliana and B. rapa have evolved from the AK. For B. rapa linkage group A8, which contains one copy of AK blocks T-U, our data enable the development of a model for the derivation of the chromosome from the AK, as shown in Figure 9. The detection of sequences from AKT at both ends of an AKU collinearity block indicates that there may have been a circular intermediate derived from linkage group AK7 which was integrated into AK1 to form the progenitor of B. rapa A8. Some rearrangements of the AK seem to have taken place in the Brassica lineage before genome triplication, but not in the A. thaliana lineage. For example, as shown in Figure 10, A. thaliana chromosome 5 contains the pairs of AK blocks AKQ-AKR and AKW-AKX (derived from linkage groups AK6 and AK8, respectively). Whereas these AK blocks appear to have been recombined in the ancestor of B. rapa. This recombination is represented in B. rapa three times [10], on linkage groups A2, A3 and A10 for AKW-AKR, and at least twice for the reciprocal outcome of the recombination, AKQ-AKX (on A2 and A6).
Figure 9.
Hypothetical derivation of B. rapa linkage group A8. Ancestral karyotype segments are labelled (A, B, C, S, T, U) and oriented by arrows. Orthologues delineating the boundaries of recombination events are designated by the name of the A. thaliana gene model suffixed "*".
Figure 10.
Hypothetical derivation of A. thaliana and B. rapa linkage groups from the ancestral karyotype. Ancestral karyotype segments are labelled (Q, R, W, X) and oriented by arrows. Orthologues delineating the boundaries of recombination events are designated by the name of the A. thaliana gene model suffixed "*".
Conclusion
Our results show that previous studies of segmental collinearity between A. thaliana, Brassica and AK genomes, although very useful, represent over-simplifications of the true inter-relationships of the genomes. In addition to the occurrence of individual genes in non-collinear regions of the genomes previously noted [6], the presence of numerous cryptic collinear genome segments and the frequent occurrence of segmental inversions mean that inference of the positions of genes based on the locations of orthologues in A. thaliana can be misleading. Indeed, excessive reliance on collinearity with the genome of A. thaliana may prove problematic for the ongoing efforts to sequence the B. rapa genome. Polyploidy is common in plants, and there is no reason to conclude that the greater complexity of segmental rearrangement and evolution that we have observed is unusual. Therefore, our results will be of relevance to studies in a wide range of polyploid plant genomes, many of which are being considered as having blocks of conserved synteny with respect to the genomes of model species, and studies relating to evolutionary breakpoints and their relation to genome organisation [20].
Methods
Computational analysis
The 200,031 publicly available BAC end sequence reads (BES) from the combined KBrH, KBrB and KBrS Brassica rapa ssp. pekinensis cv. Chiifu libraries, which were provided by the Korea Brassica Genome Resource Bank, were used in a WU-BLASTN search [21] versus the TAIR v6 Arabidopsis pseudomolecule sequences, using 1E-30 as the E-value cutoff. Supplementary BLAST parameters used were application of the Dust simple sequence filter and setting hspsepsmax = 1000, appropriate for use against very large subject sequences. Coordinates and scores for individual HSPs from the significant hits were then parsed into GFF format and loaded as features into a local copy of the ATIDB genome database [13] which is built on the GBrowse platform using a MySQL adaptor [22]. An identical exercise was performed with a set of 85,317 B. oleracea BES obtained from line TO 1434 and these data added incrementally.
A Perl CGI script was developed to interrogate the ATIDB database using the Bio::DB::GFF applications programming interface and methods. The five Arabidopsis reference chromosome sequences (pseudomolecules) were divided into bins of a selectable size (250 kb - 1 Mb) and the B. rapa (and B. oleracea) BES features mapping within each were extracted and loaded into a hashed array structure, keyed by chromosome and bin. Each bin was then systematically compared with every other bin, with the algorithm exploiting mirror symmetry for efficiency. Text string comparisons of feature object names (e.g. KBrH088K13_F and KBrH088K13_R) were used to identify mate-pairs amongst the BES mappings linking any given pair of bins. The raw mate-pair associations between bins identified from this initial process are inherently noisy and so the algorithm goes on to filter them on a combination of theoretical and empirical criteria. These can be summarised as follows: (1) any mate-pair mappings between neighbouring bins on the same chromosome were discounted if their physical separation in Arabidopsis pseudomolecule space was less than a set threshold of 500 kb, reflecting our estimate of the conserved microsynteny range; (2) duplicate instances of the mate-pair mappings, indicating either simple duplications of clones within libraries or multiple cloning events of the same DNA fragment during library construction, were eliminated; (3) DNA strand dispositions of mate-pair BES mappings (e.g. "Chr3:Bin20:plus" vs. "Chr5:Bin10:minus") were analysed to eliminate minority variants as it was reasoned that independent physical correlates of CDs should reflect a consistent pattern (unlike chimaeric clones generated by in vitro recombination events), and finally, in a development of the algorithm prompted by analysis of false positives; (4) the raw BES mappings in pseudomolecule space were used to locate the nearest annotated gene models and mate-pair mappings were eliminated if these conflicted at either end with the results of direct BLASTN query of the BES against annotated Arabidopsis genes - countering what we termed paralogue conflation;. We imposed an arbitrary threshold for the number of independent mate-pair mappings to annotated gene regions required to trigger a significant association. We varied this threshold between 2 and 5 in order to experiment with the signal to noise ratio in the dataset.
The final output of the script was directed through two routes, a graphical dot-plot style representation of the mate-pair associations using an interface to the GridMap Java applet [23] and also a spreadsheet format summary of the details underlying each significant association (clone identifiers, HSP coordinates and gene models, strand dispositions). The implementation is available from additional file 1 and additional file 2.
Sequence annotation and analysis
Our automated annotation pipeline [24] was used to analyse the sequences at the CDs. All annotated B. rapa BAC sequences were stored in a GBrowse MySQL database [22]. Minimal regions for each CD were manually selected by identifying the sequence flanked by the Arabidopsis gene models listed in Table 2, supplemented (where informative) either by annotated Brassica gene predictions from SNAP [25] post-processed with PASA [26] using raw EST data or by BLAT alignments [27] of Brassica transcript assemblies [28]. The CD sequences were analysed for the presence of characterised telomeric or centromeric repeats and then scanned for various annotated features with a Perl script using Bio::DB::GFF methods. This was repeated for extracted subsets of the entire annotated sequence defined as genic or intergenic by the gene predictions. Solexa leaf transcriptome reads obtained from B. napus [29] were aligned with MAQ [30] onto the B. rapa BAC sequences.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
IB conceived of the study, participated in its design and coordination, and helped to draft the manuscript. MT conceived and implemented the CD identification tool and helped to draft the manuscript. ND helped to implement the BAC annotation pipeline. SJK conceived BAC selection, sequencing and identification for CDs of A09 and A03. FF, ES, TJY, JHM and BSP identified and validated the BACs for sequencing. SRC, ZW, SYL and YPL anchored sequenced BACs to the B. rapa linkage map. CDT, AHP and JCP contributed to the conception of aspects of the study and contributed for analysis the end sequences of B. oleracea BAC clones prior to public release. All authors read and approved the final manuscript.
Supplementary Material
Simple HTML page used to select parameters and options for launching accompanying Perl CGI script (additional file 2).
Perl CGI script used to identify putative collinearity discontinuities (CDs) from BAC end mapping data. Output is directed to both graphical (requires GridMap applet [23]) and spreadsheet streams. There are numerous code dependencies, mostly commented in the script. The prerequisite is a Bio::DB::GFF database handle to a MySQL (or other) database storing the end mappings defined with respect to the reference sequence. The first author (MT) can assist with technical questions on implementation.
Contributor Information
Martin Trick, Email: martin.trick@bbsrc.ac.uk.
Soo-Jin Kwon, Email: sjkwon67@korea.kr.
Su Ryun Choi, Email: srchoi@cnu.ac.kr.
Fiona Fraser, Email: fiona.fraser@bbsrc.ac.uk.
Eleni Soumpourou, Email: eleni.soumpourou@bbsrc.ac.uk.
Nizar Drou, Email: nizar.drou@bbsrc.ac.uk.
Zhi Wang, Email: andy712@yahoo.cn.
Seo Yeon Lee, Email: phose84@hanmail.net.
Tae-Jin Yang, Email: tjyang@snu.ac.kr.
Jeong-Hwan Mun, Email: munjh1@korea.kr.
Andrew H Paterson, Email: paterson@dogwood.botany.uga.edu.
Christopher D Town, Email: cdtown@jcvi.org.
J Chris Pires, Email: piresjc@missouri.edu.
Yong Pyo Lim, Email: yplim@cnu.ac.kr.
Beom-Seok Park, Email: pbeom@korea.kr.
Ian Bancroft, Email: ian.bancroft@bbsrc.ac.uk.
Acknowledgements
We would like to thank the Beijing Genomics Institute for BAC sequencing. This work was funded by the UK Biotechnology and Biological Sciences Research Council (BB/E017363) and Rural Development Administration (BioGreen 21 Program 20050301034438, NAAS project No. 2007139062200001502 and 200901FHT020710397), and the Technology Development Program for Agriculture and Forestry, Ministry for Food, Agriculture, Forestry and Fisheries (Project No. 607003-05), Korea. CDT, AHP and JCP were supported by the U.S. National Science Foundation (DBI-0638536).
References
- Warwick SI, Black LD. Molecular systematics of Brassica and allied genera (Subtribe Brassicinae, Brassiceae) - Chloroplast genome and cytodeme congruence. Theor Appl Genet. 1991;82:81–92. doi: 10.1007/BF00231281. [DOI] [PubMed] [Google Scholar]
- Arumuganthan K, Earle ED. Nuclear DNA content of some important plant species. Plant Mol Biol Report. 1991;9:208–218. doi: 10.1007/BF02672069. [DOI] [Google Scholar]
- O'Neill CM, Bancroft I. Comparative physical mapping of segments of the genome of Brassica oleracea var alboglabra that are homoeologous to sequenced regions of the chromosomes 4 and 5 of Arabidopsis thaliana. Plant Journal. 2000;23:233–243. doi: 10.1046/j.1365-313x.2000.00781.x. [DOI] [PubMed] [Google Scholar]
- Rana D, Boogaart T van den, O'Neill CM, Hynes L, Bent E, Macpherson L, Park JY, Lim YP, Bancroft I. Conservation of the microstructure of genome segments in Brassica napus and its diploid relatives. Plant J. 2004;40:725–733. doi: 10.1111/j.1365-313X.2004.02244.x. [DOI] [PubMed] [Google Scholar]
- Park JY, Koo DH, Hong CP, Lee SJ, Jeon JW, Lee SH, Yun PY, Park BS, Kim HR, Bang JW, Plaha P, Bancroft I, Lim YP. Physical mapping and microsynteny of Brassica rapa ssp. pekinensis genome corresponding to a 222 kb gene-rich region of Arabidopsis chromosome 4 and partially duplicated on chromosome 5. Mol Gen Genomics. 2005;274:579–588. doi: 10.1007/s00438-005-0041-4. [DOI] [PubMed] [Google Scholar]
- Town CD, Cheung F, Maiti R, Crabtree J, Haas BJ, Wortman JR, Hine EE, Althoff R, Arbogast TS, Tallon LJ, Vigouroux M, Trick M, Bancroft I. Comparative genomics of Brassica oleracea and Arabidopsis thaliana reveals gene loss, fragmentation and dispersal following polyploidy. Plant Cell. 2006;18:1348–1359. doi: 10.1105/tpc.106.041665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang TJ, Kim JS, Kwon SJ, Lim KB, Choi BS, Kim JA, Jin M, Park JY, Lim MH, Kim HI, Lee MC, Lim YP, Kang JJ, Hong JH, Kim CB, Bhak J, Bancroft I, Park BS. Sequence-level analysis of the diploidization process in the triplicated FLC region of Brassica rapa. Plant Cell. 2006;18:1339–1347. doi: 10.1105/tpc.105.040535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysak MA, Koch MA, Pecinka A, Schubert I. Chromosome triplication found across the tribe Brassiceae. Genome Res. 2005;15:516–525. doi: 10.1101/gr.3531105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin IAP, Gulden SM, Sharpe AG, Lukens L, Trick M, Osborn TC, Lydiate DJ. Segmental Structure of the Brassica napus Genome Based on Comparative Analysis With Arabidopsis thaliana. Genetics. 2005;171:765–781. doi: 10.1534/genetics.105.042093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schranz ME, Lysak MA, Mitchell-Olds T. The ABC's of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends in Plant Sci. 2006;11:535–542. doi: 10.1016/j.tplants.2006.09.002. [DOI] [PubMed] [Google Scholar]
- Song K, Lu P, Tang K, Osborn TC. Rapid genome change in synthetic polyploids of Brassica and its implications for polyploid evolution. Proc Natl Acad Sci USA. 1995;92:7719–7723. doi: 10.1073/pnas.92.17.7719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore G. Grasses, line up and form a circle. Curr Biol. 1995;5:737–739. doi: 10.1016/S0960-9822(95)00148-5. [DOI] [PubMed] [Google Scholar]
- Pan X, Liu H, Clarke J, Jones J, Bevan M, Stein L. ATIDB: Arabidopsis thaliana insertion database. Nucl Acids Res. 2003;31:1245–1251. doi: 10.1093/nar/gkg222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Choi SR, Bae J, Hong CP, Lee SY, Hossain MJ, Van Nguyen D, Jin M, Park B-S, Bang J-W, Bancroft I, Lim Y-P. Sequenced BAC anchored reference genetic map that reconciles the ten individual chromosomes of Brassica rapa. BMC Genomics. 2009;10:432. doi: 10.1186/1471-2164-10-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panjabi P, Jagannath A, Bisht NC, Padmaja KL, Sharma S, Gupta V, Pradhan AK, Pental D. Comparative mapping of Brassica juncea and Arabidopsis thaliana using Intron Polymorphism (IP) markers: homoeologous relationships, diversification and evolution of the A, B and C Brassica genomes. BMC Genomics. 2008;9:113. doi: 10.1186/1471-2164-9-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison GE, Heslop-Harrison JS. Centromeric repetitive DNA sequences in the genus Brassica. Theor Appl Genet. 1995;90:157–165. doi: 10.1007/BF00222197. [DOI] [PubMed] [Google Scholar]
- Lim KB, Yang TJ, Hwang YJ, Kim JS, Park JY, Kwon SJ, Kim JA, Choi BS, Lim MH, Jin M, Kim HI, de Jong H, Bancroft I, Lim Y, Park BS. Characterization of the centromere and peri-centromere retrotransposons in Brassica rapa and their distribution in related Brassica species. Plant Journal. 2007;49:173–183. doi: 10.1111/j.1365-313X.2006.02952.x. [DOI] [PubMed] [Google Scholar]
- Richards EJ, Ausubel FM. Isolation of a higher eukaryotic telomere from Arabidopsis thaliana. Cell. 1988;53:127–136. doi: 10.1016/0092-8674(88)90494-1. [DOI] [PubMed] [Google Scholar]
- Cheung F, Trick M, Drou N, Lim Y-P, Park J-Y, Kwon S-J, Kim J-A, Scott R, Pires JC, Paterson AH, Town C, Bancroft I. Comparative analysis between homoeologous genome segments of Brassica napus and its progenitor species reveals extensive sequence-level divergence. Plant Cell. 2009;21:1912–1928. doi: 10.1105/tpc.108.060376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre C, Zaghloul L, Sagot MF, Gautier C, Arneodo A, Tannier E, Audit B. Analysis of fine-scale mammalian evolutionary breakpoints provides new insight into their relation to genome organization. BMC Genomics. 2009;10:335. doi: 10.1186/1471-2164-10-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gish W. WU-BLAST. 1996. http://blast.advbiocomp.com
- Stein LD. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priestly M Dickson J Dicks J Grid Map 2002http://cbr.jic.ac.uk/dicks/software/Grid_Map/12081263
- Trick M, Drou N. Brassica genome annotation pipeline. 2006. http://brassica.bbsrc.ac.uk
- Korf I. Gene finding in novel genomes. BMC Bioinformatics. 2004;5:59–68. doi: 10.1186/1471-2105-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Delcher AL, Mount SM, Wortman JR, Smith RK Jr, Hannick LI, Maiti R, Ronning CM, Rusch DB, Town CD, Salzberg SL, White O. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucl Acids Res. 2003;31:5654–5666. doi: 10.1093/nar/gkg770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. BLAT - The BLAST-Like Alignment Tool. Genome Res. 2002;4:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trick M, Cheung F, Drou N, Fraser F, Lobenhofer EK, Hurban P, Magusin A, Town CD, Bancroft I. A newly-developed community microarray resource for transcriptome profiling in Brassica species enables the confirmation of Brassica-specific expressed sequences. BMC Plant Biology. 2009;9:50–60. doi: 10.1186/1471-2229-9-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trick M, Long Y, Meng J, Bancroft I. Single nucleotide polymorphism (SNP) discovery in the polyploid Brassica napus using Solexa transcriptome sequencing. Plant Biotech J. 2009;7:334–346. doi: 10.1111/j.1467-7652.2008.00396.x. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Mapping short DNA reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Simple HTML page used to select parameters and options for launching accompanying Perl CGI script (additional file 2).
Perl CGI script used to identify putative collinearity discontinuities (CDs) from BAC end mapping data. Output is directed to both graphical (requires GridMap applet [23]) and spreadsheet streams. There are numerous code dependencies, mostly commented in the script. The prerequisite is a Bio::DB::GFF database handle to a MySQL (or other) database storing the end mappings defined with respect to the reference sequence. The first author (MT) can assist with technical questions on implementation.