Abstract
We show that the combination of spatially restricted uracil phosphoribosyltransferase (UPRT) expression with 4-thiouracil (4TU) delivery can be used to label and purify cell type specific RNA from intact complex tissues in Drosophila melanogaster. This method is useful for isolating RNA from cell types that are difficult to isolate by dissection or dissociation methods and should work in many organisms, including mammals and other vertebrates.
Cell type specific gene expression is a defining feature of multicellular organisms1. The analysis of cell type specific transcriptomes can provide insight into the mechanisms used to generate cellular diversity2, as well as help determine the underlying cause of disease3. Although a few methods are available for cell type specific RNA isolation4-7, each has constraints and researchers are often limited by their ability to isolate RNA from cell types of interest8. Thus, developing new methods for cell type specific RNA isolation is an important goal for genomic analysis of development and disease.
We previously showed that the Toxoplasma gondii nucleotide salvage enzyme uracil phosphoribosyltransferase (UPRT) can be used to biosynthetically label newly synthesized RNA in vivo9. Under natural conditions, UPRT couples ribose-5-phosphate to the N1 nitrogen of uracil to yield uridine monophosphate (UMP) which is subsequently incorporated into RNA. When the modified uracil analog 4-thiouracil (4TU) is provided to UPRT as a substrate, the resultant product is also incorporated into RNA, and this incorporation has little effect of cellular physiology9. Thio-substituted nucleotides are not a natural component of nucleic acids, and the resulting thio-labeled RNA can be readily tagged and purified using commercially available reagents. Due to the ability of this method to separate newly synthesized RNA from bulk cellular RNA, we and others have used it to measure RNA synthesis and decay rates9,10.
Here we describe a different use for 4TU/UPRT-based biosynthetic labeling that we call “TU-tagging”. We reasoned that by spatially restricting UPRT expression in a multicellular organism, 4TU will be modified and subsequently incorporated into newly synthesized RNA only in cells expressing UPRT. Thus, even if RNA is isolated from the whole organism, RNA from the cells expressing UPRT can be recovered by purifying labeled RNA (Fig. 1a). This method would be particularly useful for isolating RNA from cell types that are difficult to isolate by dissection or dissociation methods, such as subsets of neurons or glia in the CNS.
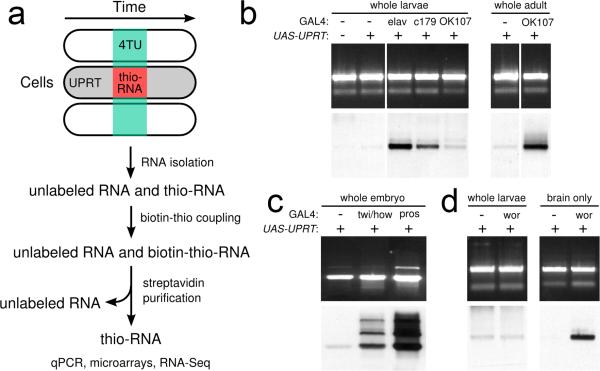
Figure 1. TU-tagging: overview and cell type specific labeling.
(A) TU-tagging procedure. (B) TU-tagging in larvae, and adults. RNA from larvae expressing GAL4 in no cell type (−), all neurons (elav), muscle cells (c179) or the mushroom body (OK 107) and either with (+) or without (−) UAS-UPRT, and adults of the indicated genotypes were electrophoresed and stained with ethidium bromide to detect all RNA (top) and streptavidinhorseradish peroxidase to detect thio-RNA (bottom).
(C) TU-tagging in embryos. 0−16h embryos of the indicated genotypes were treated with 4TU for 2 hours. RNA was analyzed as indicated above.
(D). Comparison of RNA labeling before and after tissue isolation. wor, neuroblasts.
To test the ability to biosynthetically label RNA in Drosophila, we delivered 4TU and monitored its incorporation into RNA by purifying total RNA, performing thio-biotin coupling, and using streptavidin-HRP to detect labeled RNA (see methods). Wild type or UAS-UPRT larvae or adults fed 4TU had very low levels of labeled RNA (Fig. 1b). In contrast, 4TU-fed larvae or adults containing both GAL4 and UAS-UPRT transgenes expressed UPRT in the correct cell types (data not shown) and showed robust RNA labeling (Fig. 1b,). Similarly, embryos soaked in 4TU-containing media showed robust RNA labeling only when both UAS-UPRT and GAL4 were present (Fig. 1c,). We conclude that the combination of UPRT and 4TU can be used to biosynthetically label RNA in Drosophila embryos, larvae, and adults.
To determine the limits of sensitivity, we fed larvae 4TU and expressed UPRT in different subsets of the larval brain. When UPRT is expressed in about 2,000 neurons within the entire larva we detect clear but low levels of labeled RNA (Fig. 1b,). When we reduce the number of UPRT expressing cells to about 250 neural progenitors per larva we see no detectable signal over background (Fig. 1d,). However, a simple dissection of the intact larval brain prior to RNA purification yielded excellent signal and dramatically reduced background levels of RNA labeling (Fig. 1d,). We conclude that the amount of TU-tagged RNA correlates well with the number of cells expressing UPRT; that simple tissue isolation can reduce background and increase sensitivity; and that non-CNS tissue contributes to nearly all of the low level of background RNA labeling in larvae.
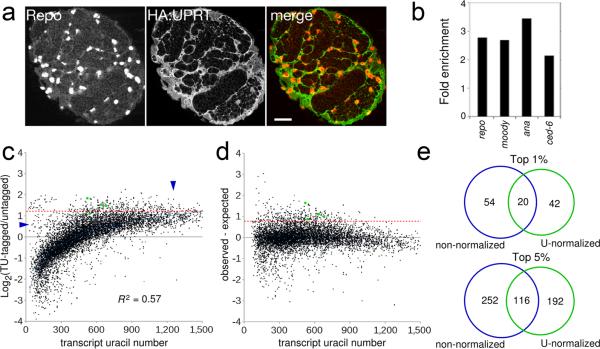
To confirm that thio-labeled RNA was from UPRT-expressing cells and demonstrate the utility of TU-tagging for cell type-specific RNA isolation, we purified TU-tagged and untagged RNA and compared them using microarrays. We chose to isolate RNA from larval glia, which are present in low number, are highly dispersed, and have a complex cell morphology (Fig. 2a), making them one of the most difficult cell types to isolate by dissection or dissociation methods. We used reversed polarity (repo)-GAL4 to drive expression of UAS-UPRT specifically in glial cells of the larval brain (Fig. 2a). We purified TU-tagged and untagged RNA from 72−96h larval brains, and hybridized them to custom Agilent microarrays (see methods). We detected signal for 7354 of the 14141 genes present on the microarray. If TU-tagged RNA was primarily from UPRT-expressing cells, known larval glia-specific genes should be among the most enriched in the TU-tagged relative to untagged RNA. There are four genes known to be expressed specifically in larval glia but not neurons or trachea of the brain. All four were enriched greater than 2-fold in the TU-tagged RNA (Fig. 2b) and three of the four were among the 5% most enriched genes: anachronism, 3.54-fold (top 0.3%, 19/7354); repo, 2.85-fold (top 1.6%, 115/7354); and moody, 2.75-fold (top 1.9%, 141/7354) which are expressed in all larval glia11,12 and ced-6 (2.13-fold, top 7.6%, 561/7354), which is expressed in a subset of larval glia13. Thus, known larval glia-specific genes were enriched in the TU-tagged RNA. We conclude that TU-tagging can effectively isolate glia-specific RNA from whole brain tissue without prior cell dissociation.
Figure 2. Cell-type specific RNA isolation and analysis.
(A) Single confocal section through a brain lobe from a 96h ALH larvae expressing HA:UPRT in all glia (repo-GAL4 UAS-HA:UPRT) stained for HA:UPRT (detected with an HA antibody) and glial nuclei (Repo). Scale bar, 20 μM.(B) fold-enrichment of selected larval glia-specific genes. See supplemental table 1 for all enriched genes.
(C) Average microarray ratios from two glia TU-tagging experiments plotted against transcript uracil number. Dashed red line indicates cutoff for top 5% enriched genes. Green dots, previously known larval glia-specific genes; vertical arrowhead, possible false positives; horizontal arrowhead, possible false negatives.
(D) TU-tagging microarray ratios after normalization and removal of transcripts with missing UTR annotations (see methods).
(E) Comparison of top 1% and 5% enriched genes before and after normalization.
We next tested if the number of uracils in a transcript influenced the levels of enrichment we observed, because long transcripts containing many uracils are expected to be labeled at a higher frequency than short transcripts with few uracils. To determine if transcript uracil number influenced our data, we plotted it against the observed microarray ratio for each transcript (see methods). There was a striking positive correlation (R2 = 0.57; Fig. 2c). Transcripts with many uracils are often among the most enriched transcripts overall even when they are not enriched relative to other transcripts with a similar uracil number (Fig. 2c,), whereas transcripts with few uracils that are clearly enriched relative to other transcripts with a similar uracil number are unlikely to be among the most enriched transcripts overall (Fig. 2c,). We conclude that transcript uracil number is a source of bias in TU-tagging experiments.
To remove the bias of transcript uracil number on the microarray results, we used the regression equation to calculate the expected ratio and subtracted it from the observed ratio for each transcript (see methods and Supplemental Table 1). This normalization procedure successfully removed the bias in data due to transcript uracil number (Fig. 2d), and had a large impact on which genes are present in the top 1% or 5% (Fig. 2e). We conclude that enrichment bias due to transcript uracil number can be removed using a simple normalization procedure, and that normalization has a large impact on which genes are considered most enriched.
We next determined how the normalization affected the enrichment levels of the known larval glia specific genes. Without normalization three out of four previously known larval-glia specific genes were within the top 5% of enriched genes (Fig. 2c). Strikingly, after normalization all four were within the top 3.2% of enriched genes: anachronism, top 0.2% (12/6167); repo, top 1.6% (99/6167); moody, top 2.4% (150/6167); and ced-6, top 3.2% (235/6167) (Figure 2D, green dots). Although the characterization of novel glia genes is beyond the scope of this paper, these results suggest that the other highly enriched genes in this dataset are excellent candidates for regulating aspects of larval glia biology. We conclude that normalizing for transcript uracil number improves TU-tagging data analysis, and that normalization should be useful for other 4TU/UPRT-based methods.
An important property of the TU-tagging method is that only newly synthesized RNAs are labeled. Thus, the percentage of labeled cellular RNA will depend on the length of time for which labeling is carried out. Although long labeling periods should work well for isolating the majority of RNA present in a particular cell type, short labeling periods could be used to detect changes in gene expression at successive timepoints in specific cell types, because newly synthesized RNA could be separated from bulk cellular RNA. This would be useful for studying rapid changes in gene expression following a particular developmental or physiological event.
The TU-tagging method is likely to work well in other systems including vertebrates. There is very little UPRT-independent “background” incorporation of 4TU into RNA in mouse or human cell culture lines9,14. Spatial control of UPRT in vertebrates could be achieved by using transgenes, as in Drosophila, or by retroviral delivery, electroporation, or mRNA injection. Thus, at least Drosophila, mice, and humans appear suitable for TU-tagging experiments. It is likely that TU-tagging can be used for cell type specific RNA isolation in many multicellular organisms and should be particularly useful for the study of development, neurobiology, and disease.
Online Methods
UPRT and 4TU toxicity tests
We crossed UAS-UPRT lines to various GAL4 lines to express UPRT in several cell types (see below) and ubiquitously (using tubulin-GAL4). In no case did we observe lethality, developmental defects, or delay in the time to pupariation and eclosion (data not shown). We next monitored development of larvae ubiquitously expressing UPRT when grown on food containing 4TU. We observed no effect on CNS development (developmental timing or neuroanatomy) in larvae grown on 4TU for 48h or less; we subsequently used an 8h or less feeding interval for all experiments. Longer exposure to 4TU (with or without UPRT expression) produced slight developmental delays but did not affect viability.
Fly Stocks
We used standard methods to clone the T. gondii UPRT coding sequence into pUAST to generate the UAS-UPRT and UAS-HA:UPRT plasmids and obtained independent viable insertions on the X, II or III chromosome. Three UAS-UPRT lines consistently showed low levels of background expression and high levels of GAL4-induced expression: UAS-HA:UPRT2.1 (chromosome II), UAS-UPRT3.1 (chromosome III), and UAS-HA:UPRT3.2 (chromosome III). We crossed one of these three lines to the following previously described GAL4 lines for all experiments (chromosome in parentheses):
worniu-GAL4(II) × UAS-HA:UPRT3.2
OK107-GAL4(IV) × UAS-HA:UPRT3.2
c179-GAL4(II) × UAS-HA:UPRT3.2
elav-GAL4(III) × UAS-HA:UPRT3.2
tubulin-GAL4(III) × UAS-HA-UPRT3.2
prospero-GAL4(III) × UAS-UPRT3.1
twist-GAL4(II);how24B-GAL4(III) × UAS-UPRT3.1
repo-GAL4(III) × UAS-HA:UPRT3.2
4TU treatment and RNA extraction
To treat embryos with 4TU, the embryos were dechorionated in bleach, washed, rinsed with isopropanol, blotted dry, and submerged in octane for 3 minutes (all within the basket). Embryos were blotted and air dried for ∼3 minutes until soft to touch. Embryos were transferred to Schneider's media containing 4-thiouracil (1.0 mM 4TU; Sigma-Aldrich 440736) for 2 hours at 25°C or 30°C, blotted dry, moved using a paintbrush to an eppendorf tube containing 1 × PBS with 1% tween (PT), and centrifuged at 4000 rpm for 30 seconds. PT was removed and the embryos were homogenized in Trizol and stored at −80C until RNA purification. To treat larvae with 4TU, larvae of the desired stage were placed on mocha food caps (20 ml H20, 0.4 g sucrose, 0.18 g agar, 1 g dextrose, and 0.5 g brewers yeast) containing 0.5 mM 4TU for the indicated time at 30°C, homogenized in Trizol, and stored at −80C until RNA purification. To treat adults with 4TU, adults were starved for ∼16 hours, placed on mocha food containing 1.0 mM 4TU for 6−8 hours at 25°C, homogenized in Trizol, and stored at −80C until RNA purification. A more detailed protocol is available on request.
Total RNA extraction from Trizol was performed using standard methods with the following additional steps: an initial centrifugation at 12,000 × g for 10 minutes at 4°C to remove insoluble material followed by a 5 minute incubation at room temperature to ensure complete dissociation of RNA-bound proteins. Only RNA samples with A260/280 ratios of ≥ 2.0 were used for subsequent biotinylation and purification steps. In all cases, RNA samples were resuspended at a final concentration of ≥ 0.4 μg/μl.
Purification of TU-tagged RNA
Detailed methods for biotinylating and purifying thio-tagged RNAs have been published9,15, and detailed protocols [AU correct as edited?] including the most recent improvements are available upon request. Relevant changes to previous protocols are summarized here. Biotinylation of RNA was performed using EZ-Link Biotin-HPDP (N-(6-(Biotinamido)hexyl)-3'-(2'-pyridyldithio)-propionamide; Pierce), as previously described. Biotinylation reactions contained 10 mM Tris (pH 7.4), 1 mM EDTA, and 2 μl of a 1 mg/ml Biotin-HPDP solution (in dimethylformamide) per 2 μg of RNA. The reaction volume was adjusted with water so that the concentration of Biotin-HPDP was equal to 30% of the final reaction volume. Biotinylation reactions were incubated in the dark for 3 hrs at 25°C prior to RNA precipitation. Biotinylated RNA was detected by blotting and probing with streptavidin-HRP as previously described. Purification of biotinylated TU-tagged RNA was performed as previously described with the following modifications: 2 μl of MPG streptavidin beads (PureBiotech) were used per μg of input RNA. The input RNA was always at a concentration of 0.5 μg/μl. After blocking with yeast tRNA and washing, beads were resuspended in the input RNA sample plus a volume of MPG buffer (1M NaCl, 10 mM EDTA, 100 mM Tris pH 7.4, in RNAse free H2O) equal to 1/3 the input RNA volume. Beads plus RNA were incubated at room temperature for 20 minutes prior to collecting the non-bound sample and performing washes with MPG buffer (one five minute wash at room temperature, two one minute washes at room temperature, a one minute wash in 65°C MPG buffer, and a final one minute wash at room temperature). After the removal of as much MPG buffer as possible, TU-tagged RNA was eluted by incubating the beads for ten minutes in freshly prepared 5% 2-mercaptoethanol. RNA was precipitated using isopropanol and linear acrylamide. After resuspending RNA in water, samples were placed in the magnetic stand again to remove any remaining MPG beads. For the purification procedures, input amounts of biotinylated RNA ranged from 14 μg to 20 μg.
Microarray Analysis
We used the Agilent (Palo Alto, CA) eArray platform to design a custom D. melanogaster oligonucleotide microarray representing 14,141 unique genes from the Flybase release 5.4 genome. 50 ng of TU-tagged and 200 ng untagged RNA were fluorescently labeled and hybridized directly against each other to Agilent microarrays. The microarray experiments were performed according to Agilent's protocol (Version 5.5, February 2007) and scanned using an Axon GenePix 4000B scanner. Fluorescent ratios for each microarray element were recovered and normalized using GenePix Pro 6.0.
Normalizing for transcript uracil number
We downloaded the dmel-all-transcript-r5.4.fasta file from Flybase which contains the sequences of every predicted transcript from genome release 5.4. We wrote a Perl script to count the transcript uracils number for each transcript. In cases where there was alternative splicing, we averaged the uracil number over the multiple isoforms. This was necessary because, for the most part, our microarray did not distinguish between different isoforms of the same gene. To normalize the data, these uracil counts were plotted against the observed microarray ratios using OpenOffice.org Spreadsheet and the regression equation was determined. This equation was used to calculate the expected ratio for each transcript based on the transcript's uracil number. For each transcript, the normalized ratio was calculated by subtracting the expected ratio from the observed ratio. After plotting the initial normalized ratios, we noticed a group of transcripts, all with low uracil numbers, that had very high normalized ratios. Upon investigation we noticed that these transcripts were missing annotations for either or both UTR regions. Thus, our transcript uracil counts for these transcripts was lower than their actual uracil number, leading to incorrect normalization. Therefore, transcripts with missing UTR annotations were excluded from normalization and further analysis. We note that normalization errors due to transcript mis-annotation will decrease as transcriptome annotations improve.
Supplementary Material
Acknowledgments
We thank Cristian Canestro, Tory Herman, Zack Lewis, and Sean O'Rourke for comments on the manuscript, and Clemens Cabernard for help with IMARIS. M.R.M. was supported by an NSF predoctoral fellowship, M.D.C. was supported by an NIH NRSA postdoctoral fellowship, and C.Q.D. was supported by NIH HD27056 and the Howard Hughes Medical Institute.
Footnotes
EdSumm
Expressing uracil phosphoribosyltransferase in specific tissues in the fly allows the incorporation of 4-thiouracil into newly synthesized RNA in vivo. The thio-labeled RNA can then be isolated and analyzed by routine procedures allowing the cell type specific measure of RNA sythesis and decay rates.
References
- 1.Tomancak P, Berman BP, Beaton A, et al. Genome Biol. 2007;8(7):R145. doi: 10.1186/gb-2007-8-7-r145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Z, Gerstein M, Snyder M. Nat Rev Genet. 2009;10(1):57. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schadt EE, Monks SA, Friend SH. Biochem Soc Trans. 2003;31(2):437. doi: 10.1042/bst0310437. [DOI] [PubMed] [Google Scholar]
- 4.Roy PJ, Stuart JM, Lund J, et al. Nature. 2002;418(6901):975. doi: 10.1038/nature01012. [DOI] [PubMed] [Google Scholar]
- 5.Tanke HJ, van der Keur M. Trends Biotechnol. 1993;11(2):55. doi: 10.1016/0167-7799(93)90123-Q. [DOI] [PubMed] [Google Scholar]
- 6.Doyle JP, Dougherty JD, Heiman M, et al. Cell. 2008;135(4):749. doi: 10.1016/j.cell.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heiman M, Schaefer A, Gong S, et al. Cell. 2008;135(4):738. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nelson SB, Hempel C, Sugino K. Curr Opin Neurobiol. 2006;16(5):571. doi: 10.1016/j.conb.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 9.Cleary MD, Meiering CD, Jan E, et al. Nat Biotechnol. 2005;23(2):232. doi: 10.1038/nbt1061. [DOI] [PubMed] [Google Scholar]
- 10.Dolken L, Ruzsics Z, Radle B, et al. RNA. 2008;14(9):1959. doi: 10.1261/rna.1136108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bainton RJ, Tsai LT, Schwabe T, et al. Cell. 2005;123(1):145. doi: 10.1016/j.cell.2005.07.029. [DOI] [PubMed] [Google Scholar]
- 12.Ebens AJ, Garren H, Cheyette BN, et al. Cell. 1993;74(1):15. doi: 10.1016/0092-8674(93)90291-w. [DOI] [PubMed] [Google Scholar]
- 13.Awasaki T, Tatsumi R, Takahashi K, et al. Neuron. 2006;50(6):855. doi: 10.1016/j.neuron.2006.04.027. [DOI] [PubMed] [Google Scholar]
- 14.Rinn JL, Wang JK, Allen N, et al. Genes Dev. 2008;22(3):303. doi: 10.1101/gad.1610508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeiner GM, Cleary MD, Fouts AE, et al. Methods Mol Biol. 2008;419:135. doi: 10.1007/978-1-59745-033-1_9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.