SUMMARY
The C. elegans Dosage Compensation Complex (DCC) associates with both X chromosomes of XX animals to reduce X-linked transcript levels. Five DCC members are homologous to subunits of the evolutionarily conserved condensin complex, while two non-condensin subunits are required for DCC recruitment to X. Here, we investigated the molecular mechanism of DCC recruitment and spreading along X by examining gene expression and the binding patterns of DCC subunits in different stages of development, and in strains harboring X;autosome fusions. We show that DCC binding is dynamically specified according to gene activity during development, and that the mechanism of DCC spreading is independent of X-chromosome DNA sequence. Accordingly, in X;A fusion strains DCC spreading propagates from X-linked recruitment sites onto autosomal promoters as a function of distance. Quantitative analysis of spreading suggests that the condensin-like subunits spread from recruitment sites to promoters more readily than subunits involved in initial X-targeting. Via these mechanisms, a highly conserved chromatin complex is appropriated to accomplish domain-scale transcriptional regulation during development. Similarities to the X-recognition and spreading strategies used by the Drosophila DCC suggest mechanisms fundamental to chromosome-scale gene regulation.
INTRODUCTION
In many animal species, sex is determined by how many copies of a particular chromosome is inherited from the parental gametes. One consequence of such a mechanism is that the two sexes will have a potentially lethal imbalance in the dosage of one chromosome. Mechanisms to correct for this imbalance have evolved, and are referred to as “dosage compensation”. Most dosage compensation mechanisms studied to date involve specific changes to the chromatin of the sex chromosome, which ultimately act to balance sex chromosome gene expression between males and females [1]. In C. elegans, XX hermaphrodites reduce transcript levels from each X chromosome by a factor of two to match the expression of XO males [2]. This is fascinating in many respects, among which is that the compensation must somehow be “tuned” to each locus so that genes expressed over a wide dynamic range are all subtly repressed by approximately two-fold.
The C. elegans Dosage Compensation Complex (DCC) is composed of proteins encoded by the genes sdc-1, sdc-2, sdc-3, dpy-21, dpy-26, dpy-27, dpy-28, capg-1 and mix-1 (Figure 1A) [3, 4]. DPY-30, a 13-kDa protein homologous to a subunit of a protein complex that methylates histone H3 at Lysine 4 (H3K4), is also required for dosage compensation [5, 6]. CAPG-1, DPY-26, DPY-28, DPY-27 and MIX-1 are homologous to the members of the condensin complex, which functions during chromosome condensation and segregation in organisms ranging from bacteria to humans [7]. Except for DPY-27, which is specific to the DCC, all of the condensin-like subunits also function as part of more typical mitotic and meiotic condensin complexes on all chromosomes [3].
Figure 1. DCC binding is dynamically specified according to transcriptional activity during development.
(A) A schematic representation of the DCC inferred from condensin homology (in parenthesis) and co-immunoprecipitation experiments. Members of the complex homologous to condensin subunits are shown in orange. SDC-2 and SDC-3 (dark blue) are involved in X-specific recruitment. DPY-30 is homologous to a subunit of an H3K4 methyltransferase complex. (B) Average z scores of ChIPs performed in embryos or in L4 worms are plotted. F22A3.1 and F22A3.6 are expressed in L4s but not in embryos. Both RNA Polymerase II and DPY-27 binding are higher in L4s compared to embryos. In embryos, C25B8.4 and C25B8.1 are expressed, and correspondingly are bound by high levels of DPY-27. In L4s, C25B8.4 and C25B8.1 transcription and DPY-27 binding are reduced. (C) RNA abundance for the four genes highlighted in panel B [35]. (D) Change in DPY-27 and RNA Polymerase II binding at promoters was calculated by subtracting the ChIP value in embryos from that of L4s. A moving average of values is plotted. Change in DPY-27 level (y axis) correlates positively with change in RNA Polymerase II binding (x axis) on the X (blue), but not on chromosome III (gray). (E) Average DPY-27 and RNA Polymerase II binding in embryos and L4s at two distinct rex sites.
During C. elegans embryogenesis, the DCC recognizes and associates specifically with each of the X chromosomes in XX embryos, but does not bind to X in XO embryos [8–13]. Current hypotheses posit two distinct modes of DCC association with the X. The first involves initial recognition and recruitment of the DCC by discrete sites along the X called “rex” sites (for recruitment element on X). Immunofluorescence microscopy revealed at least thirty-eight rex sites, which were defined by their ability recruit the DCC to multiple-copy extra-chromosomal transgenic DNA [14–16]. The immunofluorescence studies [15, 16] and two genomewide ChIP-chip studies [15, 17] identified a DNA sequence motif with a 10-bp core (TCGCGCAGGG) that occurs at many sites of DCC recruitment. Mutating the motif at a rex site reduces DCC binding, suggesting that the motif is critical for recruitment [15, 16]. However, the DNA sequence motifs do not fully account for X-specificity, because many perfect matches to the motif occur on autosomes but are not bound by the DCC [15]. The second mode of DCC association involves spreading of the DCC from the recruitment elements to adjacent chromatin.
In this manuscript, we test current hypotheses regarding DCC spreading and present two key findings. First, we show that DCC binding is dynamically specified according to gene activity during development, providing insight regarding how the process might be tuned to gene activity. Second, we show that the mechanism of DCC spreading is independent of X-chromosome DNA sequence and that spreading propagates from X-linked recruitment sites onto autosomal promoters as a function of distance. Additionally, quantitative comparison of binding data at rex-1 and the nearby dpy-23 promoter indicates that the condensin-like subunits of the DCC spread from recruitment sites to active promoters more readily than the SDC subunits involved in initial X-targeting, suggesting a DCC sub-complex involved in spreading.
RESULTS
Along the X, DCC binding is dynamically specified according to gene activity during development
Previously published ChIP experiments performed in C. elegans embryos established two modes of binding on X [17]. The first mode is represented by high-amplitude signals termed “foci” (defined empirically as being more than two standard deviations greater than the mean peak amplitude). These foci were associated with a specific DNA motif and hypothesized to be involved in initial X-recognition. The second mode of binding was a lower-amplitude accumulation of the DCC at gene promoters. Unlike foci, DCC accumulation at promoters was correlated with transcriptional activity, and was not specified by a stereotypic DNA sequence motif. This led to the hypothesis that while recruitment is governed at least in part by DNA sequence, DCC association with promoters is specified chiefly by transcriptional activity [17]. The hypothesis predicts that the DCC would be redistributed to a new set of gene promoters in the context of a different transcriptional program.
To test this prediction, we performed DPY-27 and RNA Polymerase II ChIPs from animals in the fourth larval stage of development (L4) (Figure 1B, Supplemental Figure 1A). Loci that are transcriptionally silent in embryos but expressed in L4 animals are bound by DPY-27 specifically in L4 (Figure 1B–C). The converse is also true; the DCC disengages from loci that are transcribed in embryos but silent in L4s (Figure 1B–C). DCC disengagement from repressed genes and recruitment to active genes during development occurs across the entire length of the X chromosome (Figure 1D).
In contrast to the dramatic changes in DPY-27 localization observed at gene promoters (P=5.7 e-32), DPY-27 binding at rex sites [15] remained constant between the embryo and L4 stages of growth (P=0.474 Figure 1E).
The condensin-like members of the DCC spread to adjacent chromatin more efficiently than non-condensin members
The zinc-finger containing protein SDC-3 functions during the early steps of DCC recruitment and is important for X-recognition [10]. While both DPY-27 (an SMC4 homolog) and SDC-3 bind strongly to foci thought to be involved in recruitment [17], the binding of SDC-3 to adjacent chromatin decreases sharply, while DPY-27 binding decreases more gradually [17]. Furthermore, DPY-27 binding at promoters was higher than that of SDC-3 [15, 17]. This led us to hypothesize that following recruitment a condensin-like sub-complex spreads more efficiently onto gene promoters. This hypothesis makes two predictions. First, other DCC members that are homologous to condensin subunits should behave similarly to DPY-27, and spread to gene promoters more readily. Second, SDC-2 should accumulate at the recruitment regions and spread less readily to gene promoters, because among the DCC subunits only SDC-2 can localize to X chromosome autonomously [11], and only SDC-2 is required for the localization of all DCC members [9, 10].
We tested these predictions by determining the binding patterns of the condensin-subunit homologs DPY-26 and MIX-1, and the non-condensin protein SDC-2 (Supplemental Figure 1B). The new ChIP data were analyzed jointly with previously published DPY-27 and SDC-3 data (Methods) [17]. The binding patterns of the condensin-like subunits were consistent with the DPY-27 pattern, with 92% (DPY-26) and 98% (MIX-1) of the peaks being located on the X chromosome (P<10−55 for X specificity; Supplemental Figure 2A). Furthermore, like DPY-27, the binding of both proteins accumulated preferentially at the 5′ end of genes (Figure 2A, Supplemental Figure 2B) and was positively correlated with RNA Pol II localization (Figure 2B). However, SDC-2 exhibited a unique binding pattern indicative of association with recruitment sites. Two independent antibodies revealed that SDC-2 binding did not accumulate strongly at promoters or scale extensively with expression (Figure 2B; Supplemental Figure 2A, C–D). Fewer than 200 SDC-2 peaks were observed on X. In contrast, using the same peak-finding criteria, 1358 MIX-1 and 1976 DPY-26 peaks were found. Furthermore, SDC-2 peaks were coincident with 26 of the 38 known rex sites (P<10−100; Figure 2C, Supplemental Figure 2E–F, Discussion). Therefore, SDC-2 exhibited even greater specificity than SDC-3 to recruitment sites.
Figure 2. The binding of DCC components occurs preferentially at promoters and correlates positively with RNA Polymerase II occupancy.
(A) Data were centered at the translation start sites of each X-linked gene, and the average z score of the probes in a sliding window was plotted. Data from members of the DCC that are homologous to the subunits of condensin are shown in orange. SDC-2 and SDC-3 (in blue) are involved in X-specific recruitment. (B) A moving average of ChIP enrichment at promoters of X-linked genes is plotted as a function of RNA Polymerase II occupancy [36]. (C) The locations of the 38 known rex sites (red bars), sites of SDC-2 binding (Antibody 1), and DCC foci are indicated along the X chromosome. Green bars indicate overlap with rex sites.
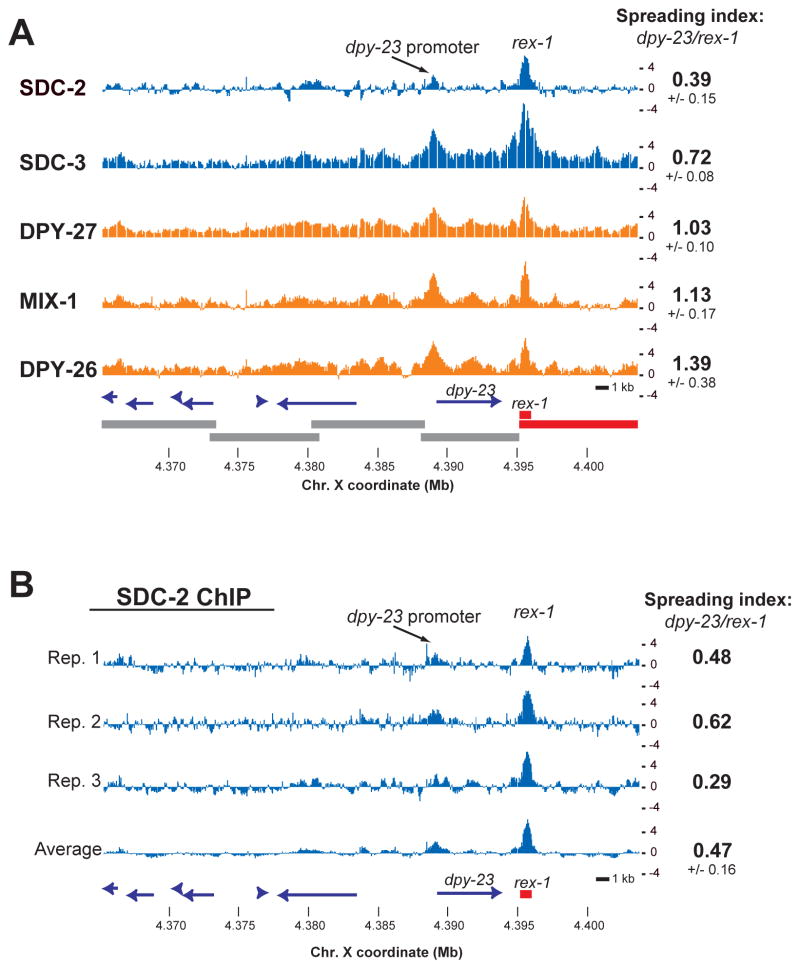
To quantitatively assess the recruitment and spreading properties of different DCC subunits, we studied a well-characterized region containing rex-1 [16] and DNA fragments that cannot recruit the complex on their own but are bound by the DCC in the context of the natural chromosome (e.g. the promoter of dpy-23) [14–17] (Figure 3A). We calculated a “spreading index” by measuring the average amplitude of binding at rex-1 relative to the average amplitude of binding at the dpy-23 promoter. This analysis was repeated for each biological replicate to calculate the average and standard deviation (Figure 3A). DPY-27, DPY-26 and MIX-1 have similar spreading indices (1.03 +/−0.10, 1.39 +/−0.38 and 1.13 +/−0.17), all of which are higher than those of SDC-2 and SDC-3 (0.39 +/−0.15 (Ab 1) and 0.72 +/−0.08).
Figure 3. The condensin-like portion of the C. elegans Dosage Compensation Complex (DCC) spreads more efficiently relative to subunits involved in early steps of recruitment.
(A) Average z scores of ChIP enrichment are plotted at the rex-1 dpy-23 locus [14, 16]. Data from members of the DCC that are homologous to the subunits of condensin are shown in orange. SDC-2 and SDC-3 (in blue) are involved in X-specific recruitment. DNA fragments shown in red recruit the DCC onto extra-chromosomal arrays [14]. Fragments in grey fail to recruit but are bound in the context of the natural chromosome. The spreading index is calculated by dividing average ChIP score at the dpy-23 promoter by the corresponding value at rex-1. SDC-2 data are from antibody 1. (B) A second polyclonal SDC-2 antibody raised against a different portion of the protein (antibody 2) in two different rabbits (SDQ3146 for rep1 and 2 and SDQ3148 for rep3) was used to generate the ChIP-chip profiles.
Spreading index calculated with antibodies raised to a completely independent SDC-2 epitope (Ab 2) (0.47 +/−0.16) was similar to the first (0.39 +/−0.15). Each of the individual SDC-2 replicates performed with either antibody had a lower spreading index than the corresponding condensin-like DCC members (Figure 3B). Furthermore, data published independently by others [15] showed that the spreading indices for the condensin-subunit homologs DPY-27 (1.39) and MIX-1 (1.61) were greater than that of SDC-2 (0.79) and SDC-3 (0.87). The spreading indices at the dpy-23-rex-1 locus quantify what we observe throughout genome: strong binding of both condensins and non-condensins to recruitment sites, followed by more efficient spreading of the condensin-like portion of the complex to surrounding genes.
DCC spreading does not require X-chromosome DNA sequence
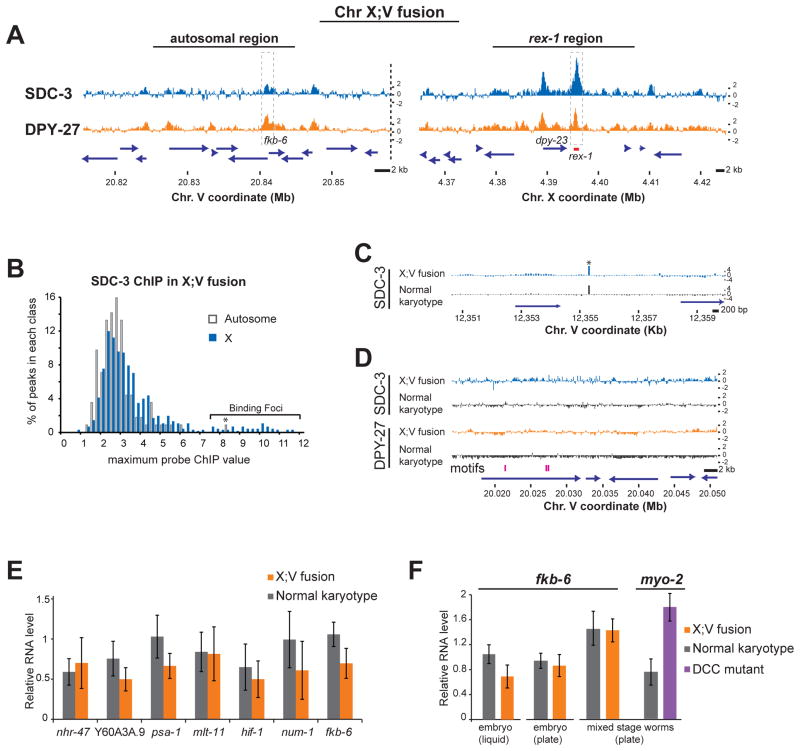
We hypothesized that the mechanism of DCC spreading was not directly dependent on X-chromosome DNA sequence, but instead could operate on any transcriptionally active sequence near a recruitment site. This hypothesis was based on observations that DCC binding at promoters is dynamically correlated with transcriptional activity (Figure 1), and that the accumulation of the DCC at gene promoters does not seem to correspond to any particular DNA sequence motif [17]. We tested our hypothesis by examining DCC binding at high resolution in three strains harboring precisely defined X;autosome fusion chromosomes [18]. In these strains, the right end of the X is fused to the right end of chromosome V (X;V), the left end of chromosome II (X;II), or the right of chromosome I (X;I) respectively [18]. DPY-27 ChIP-chip from mixed-stage embryos was compared to the binding pattern of the wild-type strain with a normal karyotype. In all three strains, DPY-27 spreads across the X;A junction and into the autosomal sequences of the fused chromosome (Figure 4). Replicates of the individual ChIPs reveal a highly reproducible binding pattern (Supplemental Figure 3A).
Figure 4. DCC binding spreads into the autosomal regions of an X;autosome fusion chromosome.
Average z scores of DPY-27 ChIP performed in embryos of three strains that contain an X;V, X;II or X;I chromosome are plotted around each fusion site, which is indicated with a dashed line. Dashed boxes highlight DPY-27 binding at promoters. DPY-27 accumulates at the promoters of both autosomal and X-liked genes to a similar degree near the fusion site.
DCC spreading decreases as a function of distance from the nearest recruitment site
Earlier low-resolution studies with X-chromosome DNA attached to an autosome showed that DCC did not spread such that the autosome was engulfed [14, 16, 19, 20]. We investigated different hypotheses for how DCC spreading onto the whole autosome is limited (Figure 5A). If spreading continues from the nearest recruitment site until it encounters an autosomal blocking element, one would expect uniform levels of DCC binding until the block was encountered. Alternatively, if there were no special autosomal blocking elements, the levels of DCC association with active gene promoters might decrease continuously as a function of distance from the nearest recruitment site.
Figure 5. DPY-27 spreading diminishes as a function of distance from the X.
(A) Possible modes of spreading are represented along a virtual X;autosome fusion chromosome. (B) DPY-27 ChIP z score moving average with a window size of 100 kb and a step size of 10 kb is plotted along the coordinates of fused chromosome in X;V. (C) Same as B, for a normal karyotype. (C) An X;II fusion. (D) An X;I fusion.
To distinguish these alternatives, DPY-27 binding was quantified as a sliding window of 100 kb along the fused X;A chromosome (Figure 5B) and compared to data from a normal-karyotype strain (Figure 5C). In all three strains harboring a chromosomal fusion, DPY-27 association with the autosomal regions diminishes continuously with increasing distance from the X (Figure 5B,D,E). This suggests that there are not discrete elements on autosomes that block DCC spreading, and that spreading decreases as a function of chromosomal distance from the nearest recruitment site.
DCC binding extends greater than two million bases from the nearest X recruitment element
To estimate the extent of DPY-27 spreading onto the autosome, we identified autosomal peaks that occurred in the fusion strains, but not in wild-type animals. We identified 82 peaks that occurred on chromosome V specifically in the X;V fusion strain. A similar number of autosomal peaks, nearly all of which were specific to the fusion autosome, were found in each of the other fusion strains (Supplemental Figure 3B). Ninety-three percent of the autosomal DPY-27 peaks in the fusion strains were within 2 Mb of a fusion site, with a few peaks present up to 3.5 Mb into the autosome (Figure 6A, Supplemental Figure 3C,D).
Figure 6. DCC binding extends ~2 Mb into autosomal regions and is determined by distance from the X and transcriptional activity.
(A) Each point represents a site of DPY-27 binding that occurs in the X;V fusion strain but not in a wild-type strain (Methods). The z score of the maximum value probe value is plotted as a function of distance from the X;V fusion site. (B) Distribution of all DPY-27 peaks in X;V fusion (p value E-10) with respect to underlying genes. The maximum value probe within each peak was assigned to a region by CEAS [32]. P values were obtained by chi-square test using the observed and expected distribution of probes among the annotation classes. Expected value was calculated from the CEAS assignment of all microarray probes. The most commonly bound region is highlighted with an asterisk. (C) A scatter plot of average DPY-27 ChIP z scores at promoters (y axis) and RNA abundance (log2RMA-normalized intensity, x axis) of X-linked genes and genes located within 1 Mb of the fusion site on chromosome V. (D) Average DPY-27 ChIP z scores at chromosome V gene promoters is plotted as a function of distance from the fusion site. The outliers at ~12Mb are histone genes which may cross-hybridize to other histone loci in the genome.
The functionally characterized rex site nearest to the right end of X is 2 MB from the chromosome terminus (rex-35), but it is possible that other rex sites nearer to the end await characterization. For example, there is a ChIP-defined DCC focus 173 kb from the end. Regardless, the gradual decline of binding from the fusion site explains the need to have multiple recruitment elements or “way stations” [19] spaced along the X chromosome to maintain the required levels of DCC association with X.
The mode of DCC association with autosomal genes near the X;A fusion boundary is indistinguishable from the mode of DCC association with natural X-linked genes
If the mechanism underlying the spreading onto the autosomes is the same as the spreading on natural X chromosomes, the manner in which the autosomal genes near the fusion site are bound by the DCC would be similar to that of X-linked genes. On the X, the DCC associates with promoters (Figure 4, dashed boxes, Figure 2A, Supplemental Figure 2B), with an amplitude that correlates with the polymerase occupancy and transcription rate of the downstream gene (Figure 2B).
Like DCC binding on X, the autosomal sites of DCC binding in the fusion strains exhibited a strong preference for the 5′ regions of genes (Figure 4, dashed boxes, Figure 6B, Supplemental Figure 3E). Furthermore, DPY-27 binding at promoters of autosomal genes near the fusion site correlated with the transcriptional activity of the downstream gene, just as is observed for DCC association with gene promoters on X (Figure 6C, Supplemental Figure 3F). As we noted previously, binding at the promoters decreases with the distance from the fusion site (Figure 6D, Supplemental Figure 4A). Therefore, the degree of DCC association with promoters appears to be governed by two factors: distance from the nearest recruitment site and transcriptional activity. The autosomal fusions provide a unique window on this process, since the close spacing of recruitment sites on the natural X make the dependence of binding amplitude on the distance from a recruitment site difficult to observe.
As is observed on X, the condensin-like members of the DCC spread to adjacent autosomal chromatin more efficiently than non-condensin members
To test whether the propagation of DPY-27 binding onto the autosomal regions is more efficient than that of SDC-3, we determined the ratio of binding between the X-linked rex-1 and the autosomal fkb-6 promoter (Figure 7A). This analysis indicated that SDC-3 spread less efficiently (0.26) than DPY-27 (0.82 +/−0.09).
Figure 7. In fusion strains, DPY-27 spreads more efficiently than SDC-3 onto autosomal genes, and autosomal genes bound by the DCC are not strongly repressed.
(A) SDC-3 and DPY-27 binding in the X;V fusion strain, at autosomal loci near the fusion site and at rex-1 on X. The fusion site is indicated with a dashed line. At the recruitment site (rex-1) SDC-3 binds more strongly than DPY-27, but in the same sample, DPY-27 binding is stronger within autosomal regions near the fusion site (compare peaks highlighted with dashed boxes). (B) The distribution of maximum probe values within SDC-3 ChIP peaks in the X;V fusion strain, segregated according to their chromosomal location. Binding foci are shown with a bracket. One autosomal peak (indicated with an asterisk) was identified as a focus. (C) However, this peak was created by a single probe that is identical to a focus on the X. (D) SDC-3 and DPY-27 ChIP profiles shown at an autosomal region that contains three copies of the 10 bp sequence motif (pink, bottom). (E) Real-time PCR analysis of the expression of seven genes in the X:V fusion strain embryos grown in liquid media. Six genes are within 2 megabases of the fusion site on chrV. nhr-47 is more than 15 Mb away. (F) Real time PCR analysis of fkb-6 and myo-2 (a dosage compensated gene on the X). RNA was isolated from embryos obtained from worms grown in liquid or plates, and from mixed stage worms grown on plates. The DCC mutant is strain CB428, dpy-21(e428).
This observation also suggests that the DCC binding on autosomes was indeed due to the nearby natural recruitment elements on X, rather than to new recruitment elements that may have arisen on the autosomes as a result of the chromosomal fusion. In fact, all of the high-amplitude binding foci in the fusion chromosome strains were located on the X, arguing that the autosomal sequences on the fused chromosome failed to recruit DCC on their own (Supplemental Figure 4B). Additionally, no SDC-3 binding foci were located on autosomal sequences, with one exception (Figure 7B). This single autosomal peak included a probe identical to a sequence located in an X focus, indicating cross-hybridization as the likely cause (Figure 7C). Finally, a region of chromosome V 870 kb from the fusion site contained three 10-bp core DCC motifs located in close proximity to each other, but failed to bind SDC-3 or DPY-27, reinforcing the likelihood that X-specific factors other than DNA sequence are involved in specifying recruitment sites (Figure 7D).
Autosomal genes located near the X;A fusion boundary are not strongly repressed by the DCC
We asked whether the DCC represses autosomal genes bound by DPY-27 in the fusion strains. We performed gene expression microarray analysis of normal karyotype (N2), X;V, and X;II fusion strains. We identified 25 genes in X;V strains and 80 in X;II strains whose expression differed significantly (Supplemental Table 1). These genes were not significantly overrepresented on the X chromosome, or within the autosomal regions one mega base from the fusion site (Supplemental Figure 5A). Additionally, the transcript level of all genes within one megabase of the fusion site was not significantly different between fusion and normal karyotype strains (Supplemental Figure 5B).
We also analyzed the transcription of seven autosomal genes bound by DPY-27 in the X;V fusion strain with real-time PCR, using RNA prepared from embryos (Figure 7E). We established that changes in transcript levels due to dosage compensation can be detected with this platform by showing that the dosage-compensated gene myo-2 [2] is de-repressed in a DCC mutant strain (Figure 7F). Indeed, all six genes that are within two megabases of the fusion site were expressed at lower levels in X;V fusion embryos relative to the wild-type strain (Figure 7E). However, the change was significant only for the gene closest to the fusion site, fkb-6 (Figure 7E, Supplemental Figure 5C). It is not clear whether the difference in fkb-6 RNA level is due to dosage compensation or other factors, because reduction of fkb-6 transcript was not observed in mixed-stage cultures or in embryos prepared from adults grown on solid media (Figure 7E). Our results are consistent with the lack of correlation between the binding location of the DCC and changes in RNA abundance upon the disruption of dosage compensation [15]. However, many explanations for this apparent lack of concordance are possible (Discussion).
DISCUSSION
The advantages of distinct subcomplexes for recruitment and spreading
The DCC is composed of three functionally distinct groups of proteins. The first group contains the condensin subunit homologs DPY-27, MIX-1, DPY-26, DPY-28, and CAPG-1. Among these, only DPY-27 is specific to DCC. The other members are also part of canonical condensins [3]. The second group, comprised of SDC-2 and SDC-3, operates during early steps of X-specific recruitment and has limited homology in other species, [9, 10]. The role of the third group (SDC-1 and DPY-21) is not clear. Both SDC-1 and DPY-21 are required for dosage compensation and co-immunoprecipitate with the other DCC subunits, but are not required for binding of other DCC subunits to X, and are not essential for viability of XX animals [21]. Targeting of the DCC to X through relatively few recruitment sites by the action of a set of proteins dedicated specifically to targeting (SDC-2 and SDC-3) would provide a means of delivering the condensins (DPY-27, MIX-1, DPY-26, DPY-28 and CAPG-1) to their sites of action without hard-wiring DCC targeting at each site. Critically, it would also provide a means for dynamic binding of the DCC, allowing binding to be responsive to changes in transcription due to developmental cues or environmental perturbation.
The function of SDC-2 in X chromosome recognition
SDC-2 binding sites are associated with a 12 bp motif, which contains the previously identified 10-bp core motif and differs slightly from 12 bp motif identified by analysis of rex sites [15] (Supplemental Figure 6A). Much like the 10-bp motif, the 12-bp motif is overrepresented only 2.8-fold on X and is more clustered on X than the autosomes, suggesting that motif proximity contributes to X specificity (Supplemental Figure 6B). At the most strict definition of the motif, 75% of the motifs on X are within an SDC-2 peak and 38% of the SDC-2 peaks contain a motif (Ab 1; Methods). Although the sequence motif is critical for recruitment [15, 16], it clearly does not explain all recruitment and X-specificity, suggesting that other chromatin factors are involved in specifying the recruitment sites on the X. The distinction between recruitment and spreading mechanisms is further highlighted by the fact that although spreading onto the autosomes was observed in X;A fusion strains, autosomal regions with the DNA sequence properties of recruitment elements do not become recruitment elements even when physically attached to the X.
We found that the SDC-2 spreading index (0.39 +/−0.15, Ab 1) was lower than that of SDC-3 (0.72 +/−0.08), which suggests that the spreading dynamics of SDC-2 and SDC-3 may differ significantly. The SDC-2 and SDC-3 spreading indices calculated from data produced by others [15] were more similar to each other (0.79 and 0.87, respectively). In those studies, epitope-tagged SDC-2 was produced by a multi-copy transgene, which may result in overexpression.
Dynamic propagation of the DCC to promoters does not depend on the DNA sequence of X-linked genes
Comparison of DPY-27 binding in embryos and L4 animals indicated that DCC binding at promoters is dynamically targeted to active genes, and with equal specificity dissociates from inactivated genes. Our study reveals that after recruitment, the mode of DCC propagation does not involve any special aspect of X chromosome DNA sequence. DCC spreading into the autosomal regions in X;A fusion strains also accumulates at promoters and is directly correlated with transcriptional activity. Therefore this mode of binding is specified by a general marker of transcriptional activity, perhaps involving the DPY-30 protein and H3K4 methylation, a ubiquitous molecular marker of active promoters [22].
Does transcriptional repression by the DCC act locally or globally?
There are two possibilities for the mechanism of DCC action, neither of which excludes the other. On one hand, the DCC may act locally, repressing individual genes by binding specifically to promoters in proportion to transcription rate. Such local action would help explain how two-fold repression could be effected on genes that are transcribed over a wide range of levels. On the other hand, expression data gathered in dosage compensation deficient embryos reveal disconnect between DCC localization and resulting transcriptional changes, which argues for a global mechanism of repression. X-linked genes bound by the DCC and those not bound are equally likely to experience transcriptional changes upon disruption of dosage compensation [15]. This apparent discordance may be explained by a global mechanism for repression, but an alternative interpretation is that the expression changes reflect the secondary effects of disrupting dosage compensation. For example, the deletion of regulatory factors is known to cause transcriptional changes in many genes that are not direct binding targets [25]. Taken at face value, our analysis of fusion strains would appear to support a global repression model, since we did not observe repression of autosomal genes at which DCC localization was observed. However, it is possible that the levels of association at autosomal promoters were not normal, or that some other component required for the normal function of the DCC was absent on autosomes.
DCC subunits are recruited to few autosomal loci
In this study we concentrated on the vast majority of DCC binding sites that occur on X, but we also observed some autosomal sites of binding (fewer than 260, Supplemental Figure 2A), many fewer than was previously reported by others [15]. The function of DCC at these autosomal sites is not clear. Although expression of several autosomal genes was altered in dosage compensation mutants, these genes were not specifically associated with DCC peaks. Conversely, transcript levels of genes near sites of autosomal DCC binding were not significantly altered in dosage compensation mutants [15].
Similarities in C. elegans and Drosophila dosage compensation complex targeting
Drosophila males (XO) increase transcription of their single X approximately two-fold as a means of dosage compensation. The MSL (male-specific lethal) complex is directed to the X by DNA-sequence based recruitment motifs [26, 27], and similar to our results for the C. elegans DCC, MSL spreading is sequence independent [28]. Targeting of the MSL complex to active genes is aided by the chromodomain of MSL-3, which recognizes H3K36 tri-methylation [29].
It is remarkable that two arguably converse strategies (modest upregulation of X in files and modest downregulation of two Xs in worms) share many aspects of how the complexes recognize and spread along the X. These include a recruitment mechanism specified in part by DNA sequence but also requiring other cis-acting factors, followed by a spreading mechanism that is generic in nature, but directed by the initial recruitment. These strategies are held in common despite the use of two completely different molecular machines (histone acetyltransferase vs condensin) that likely act to modulate different parts of the transcription cycle (elongation vs initiation). These shared properties of chromosome recognition and spreading represent a convergent evolution of strategy that is likely fundamental to the nature of chromosome-scale gene regulation.
EXPERIMENTAL PROCEDURES
Antibodies and Strains
Polyclonal antibodies against DPY-27 and SDC-3 were described previously [17]. Anti-RNA Polymerase II (CTD) antibodies were clone 8WG16 (Millipore 05-952). Recombinant proteins SDC-2 (1-455 aa), DPY-26 (740-1262 aa), MIX-1 (837-1244 aa) were prepared with Novagen’s Pet30-EkLIC system. Rabbit polyclonal antibodies were produced at Covance Immunology Services. The epitopes were cloned into pGEX-5X-2 vector (GE Healthcare) and a GST tag was utilized for affinity purification. SDQ3146 and SDQ4148 antibodies against SDC-2 (1749-1848 aa) were produced by genetic immunization at Strategic Diagnostics Inc. Antibodies against SDC-2 regions 1–455 aa and 1749–1848 aa are referred to as antibody 1 (Ab 1) and antibody 2 (Ab 2), respectively.
N2 (normal karyotype) and dpy-21 mutant (e428) (CB428) strains were obtained from The Caenorhabditis Genetics Center (CGC). The X::autosome fusion strains (X;V aka. YPT47 or 15eh#1; X;II aka. YTP41; X;I aka. YPT40 or 11dh) were provided by Shawn Ahmed [18].
Worm growth and ChIP-chip
Standard worm growth techniques were used to obtain embryos from worms grown in S-liquid media [30]. The embryo ChIP protocol was described previously [17]. L4 worms were obtained by synchronizing at L1 by allowing embryos to hatch on NGM plates or in M9 buffer without food for 24 hours. The starved L1s were filtered through a 20 micron nylon mesh and grown with food for ~40 hours at 20°C until the worm population was predominantly L4s, with few L3s and young adults not harboring any embryos. Worms were collected, washed with M9 and re-suspended in equal volume of M9 with protease inhibitors (Calbiochem). This mixture was drizzled into liquid nitrogen to form frozen “popcorn” and stored at −80°C. Approximately 2 grams of frozen sample was processed with Biopulverizer (BioSpec Products, Inc.) and further ground to a fine powder with a cryo-mortar and pestle. The powder was fixed in 1% formaldehyde in 5 volumes of M9 at room temperature for 5 (L4 ChIP replicates 1, 2 and 4) or 10 minutes (L4 ChIP replicate 3). 125 mM glycine was added and incubated for 5 minutes. Samples were collected by centrifugation at 4000g for 5 minutes at 4°C, washed once with 20 ml M9+protease inhibitors and 10 ml of FA (50 mM HEPES/KOH pH 7.5, 1 mM EDTA, 1% Triton X-100, 0.1 % sodium deoxycholate; 150 mM NaCl) with protease inhibitors, and re-suspended in FA buffer for sonication. 2 ml sample were sonicated with a Branson sonifier 7 to 10 cycles of 12 pulses (0.9 s on, 0.1 s off) at 35% amplitude in 15 ml conical tubes, with cooling in dry ice-ethanol bath for 2 s between cycles. Cellular debris was removed by centrifugation at 13,000 g for 15 min at 4°C. 6–8 mg of extract was brought to 1 ml volume, sarkosyl was added to 1% final concentration, samples were centrifuged for 5 minutes and supernatant was taken. Prior to adding antibody, 10% of the volume was taken for input sample. The remaining sample was incubated with 3–5 μg affinity purified antibodies overnight at 4°C. Collection of immunocomplexes were performed as described [17] except in SDC-2 ChIPs, MIX-1 ChIP1, and DPY-26 ChIP1, 40 ul Dynabeads M280 Sheep anti-Rabbit IgG (Invitrogen) were used for collection. ChIP DNA was amplified by LM-PCR as described [17].
Microarrays, data extraction and processing
DPY-27, SDC-3 and No antibody (NoAb) control data are available from GEO, GSE6739 [17]. All microarrays were designed and manufactured by Roche NimbleGen. Data from X;V fusion strain was obtained from the C. elegans ChIP-chip array based on WS120 (C4533-03-01), which contains 385K probes that span chrV (~5MB-end) and all of X at 86-bp resolution. All other data was produced from a single array containing 1.9 million probes that spans the entire genome at 50 bp resolution and is based on WS170. Sample hybridization and data extraction for X;V fusion strain was performed as per the manufacturer’s protocols. The remaining hybridizations and data collection were done by NimbleGen Service Laboratory as described [31]. All ChIPs were performed at least in triplicate except X;II, X;I DPY-27 and X;V SDC-3 ChIPs, which were done in duplicate. DPY-26, MIX-1, SDC-2, X;V DPY-27 ChIPs 1 and 3, L4 ChIP1 and 4, X;II, X;I DPY-27 ChIPs 1, X;V SDC-3 ChIP1 were labeled with Cy5 and input DNA was labeled with Cy3. In the remaining replicates, the dyes were swapped.
For each data set, log2 ratio of intensity from Cy5/Cy3 is obtained from NimbleScan. Log ratios were transformed to z scores and ChIP enrichment of each data point was calculated by taking the average of replicates. L4 DPY-27 and RNA Polymerase II ChIP replicates 2 and 3 were from same worm collection, thus these two were averaged before combining with the remaining replicates. To directly compare normal karyotype data to that of X;V fusion, probes that were present in the C4533-03-01 microarray were identified and normalized for comparison. Data were visualized by UCSC genome browser (http://genome.ucsc.edu/).
Spreading index and Peak analysis
Enrichment within a 1 kb window centered at the SDC-3 ChIP maximum value probe at the rex-1 region was calculated. Promoter binding at the dpy-23 and fkb-6 gene was calculated by average binding between 500 bp upstream and 200 bp downstream of the translation start site.
The ChIPOTle peak finding algorithm was used on the average z score data sets. We ran ChIPOTle (http://sourceforge.net/projects/chipotle-2) with 500 bp window and 100 bp step size at the indicated p value cutoffs. SDC-2, SDC-3, MIX-1, DPY-27 and DPY-26 peaks (p value e-60) were refined by eliminating those that were within 100 bp of a NoAb peak. For distribution of peaks among chromosomes, p values were obtained by a chi-square test between observed and expected distribution that was calculated by allocation of peaks based on chromosome length.
Differentially bound peaks in the X;V fusion (p value e-40) were identified by running ChIPOTle on the normalized data in which ChIP score of each probe from N2 strain was subtracted from that of X;V. This was not possible for the X;II and X;I strains because the microarray platform was different. Therefore, differentially bound peaks in X;II and X;I fusions (p value e-10) were determined by eliminating those peaks that were within 250 bp of another peak in the N2 or the other strain’s peak set.
Annotation analysis
The coordinate corresponding to the center of the maximum value probe within a peak was used for annotation by CEAS [32]. A coordinate was assigned hierarchically to an annotation class: exon, intron, 5′, 3′ or more than 1 kb away from any gene. For distribution of peaks with respect to underlying genes, p values were obtained by a chi-square test between observed and expected distribution that was calculated by assignment of all probes on the microarray.
RNA extraction and expression analysis
The NimbleGen expression microarrays employed include three probes for each gene and are based on WS170. RNA was extracted by using Trizol reagent with the manufacturer’s protocol (Invitrogen). Total RNA was used for sample preparation and hybridization to microarrays by Roche NimbleGen service laboratories. The resulting single-channel data was normalized by RMA (Robust Multichip Average) using NimbleScan. The RMA calls were log2 transformed and average of three replicates were used.
2 μg of total RNA was used to prepare cDNA in 20 μl total volume using Invitrogen Superscript III kit. 1 μl of the reaction was used in 20 μl volume of real-time PCR using Maxima™ SYBR Green qPCR Master Mix (2×) (Fermentas). Primer sequences are provided in supplementary methods. The relative amount of RNA was calculated by using a standard curve and normalizing to the average level of ben-1 (tubulin) and fasn-1.
Data accession
All data are publically available at Gene Expression Omnibus, accession #GSE16621. Detailed protocols are at the modENCODE website (http://www.modencode.org/).
Supplementary Material
Acknowledgments
Sevinc Ercan was supported by National Institutes of Health under Ruth L. Kirschstein National Research Service Award GM084471. This work was partly supported by the NHGRI modENCODE project, grant U01 HG004270. We thank Mia Lowden and Shawn Ahmed for fusion strains, Paul Giresi and Christina Whittle for discussion on data analysis, Kirsten Hagstrom for help with characterization of the DPY-26 and MIX-1 antibodies. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Straub T, Becker PB. Dosage compensation: the beginning and end of generalization. Nat Rev Genet. 2007;8:47–57. doi: 10.1038/nrg2013. [DOI] [PubMed] [Google Scholar]
- 2.Meyer BJ, Casson LP. Caenorhabditis elegans compensates for the difference in X chromosome dosage between the sexes by regulating transcript levels. Cell. 1986;47:871–881. doi: 10.1016/0092-8674(86)90802-0. [DOI] [PubMed] [Google Scholar]
- 3.Csankovszki G, Collette K, Spahl K, Carey J, Snyder M, Petty E, Patel U, Tabuchi T, Liu H, McLeod I, et al. Three distinct condensin complexes control C. elegans chromosome dynamics. Curr Biol. 2009;19:9–19. doi: 10.1016/j.cub.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meyer BJ. Sex in the wormcounting and compensating X-chromosome dose. Trends Genet. 2000;16:247–253. doi: 10.1016/s0168-9525(00)02004-7. [DOI] [PubMed] [Google Scholar]
- 5.Hsu DR, Meyer BJ. The dpy-30 gene encodes an essential component of the Caenorhabditis elegans dosage compensation machinery. Genetics. 1994;137:999–1018. doi: 10.1093/genetics/137.4.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nagy PL, Griesenbeck J, Kornberg RD, Cleary ML. A trithorax-group complex purified from Saccharomyces cerevisiae is required for methylation of histone H3. Proc Natl Acad Sci U S A. 2002;99:90–94. doi: 10.1073/pnas.221596698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Losada A, Hirano T. Dynamic molecular linkers of the genome: the first decade of SMC proteins. Genes Dev. 2005;19:1269–1287. doi: 10.1101/gad.1320505. [DOI] [PubMed] [Google Scholar]
- 8.Chuang PT, Albertson DG, Meyer BJ. DPY-27:a chromosome condensation protein homolog that regulates C. elegans dosage compensation through association with the X chromosome. Cell. 1994;79:459–474. doi: 10.1016/0092-8674(94)90255-0. [DOI] [PubMed] [Google Scholar]
- 9.Chuang PT, Lieb JD, Meyer BJ. Sex-specific assembly of a dosage compensation complex on the nematode X chromosome. Science. 1996;274:1736–1739. doi: 10.1126/science.274.5293.1736. [DOI] [PubMed] [Google Scholar]
- 10.Davis TL, Meyer BJ. SDC-3 coordinates the assembly of a dosage compensation complex on the nematode X chromosome. Development. 1997;124:1019–1031. doi: 10.1242/dev.124.5.1019. [DOI] [PubMed] [Google Scholar]
- 11.Dawes HE, Berlin DS, Lapidus DM, Nusbaum C, Davis TL, Meyer BJ. Dosage compensation proteins targeted to X chromosomes by a determinant of hermaphrodite fate. Science. 1999;284:1800–1804. doi: 10.1126/science.284.5421.1800. [DOI] [PubMed] [Google Scholar]
- 12.Lieb JD, Albrecht MR, Chuang PT, Meyer BJ. MIX-1: an essential component of the C. elegans mitotic machinery executes X chromosome dosage compensation. Cell. 1998;92:265–277. doi: 10.1016/s0092-8674(00)80920-4. [DOI] [PubMed] [Google Scholar]
- 13.Lieb JD, Capowski EE, Meneely P, Meyer BJ. DPY-26, a link between dosage compensation and meiotic chromosome segregation in the nematode. Science. 1996;274:1732–1736. doi: 10.1126/science.274.5293.1732. [DOI] [PubMed] [Google Scholar]
- 14.Csankovszki G, McDonel P, Meyer BJ. Recruitment and spreading of the C. elegans dosage compensation complex along X chromosomes. Science. 2004;303:1182–1185. doi: 10.1126/science.1092938. [DOI] [PubMed] [Google Scholar]
- 15.Jans J, Gladden JM, Ralston EJ, Pickle CS, Michel AH, Pferdehirt RR, Eisen MB, Meyer BJ. A condensin-like dosage compensation complex acts at a distance to control expression throughout the genome. Genes Dev. 2009;23:602–618. doi: 10.1101/gad.1751109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McDonel P, Jans J, Peterson BK, Meyer BJ. Clustered DNA motifs mark X chromosomes for repression by a dosage compensation complex. Nature. 2006;444:614–618. doi: 10.1038/nature05338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ercan S, Giresi PG, Whittle CM, Zhang X, Green RD, Lieb JD. X chromosome repression by localization of the C. elegans dosage compensation machinery to sites of transcription initiation. Nat Genet. 2007;39:403–408. doi: 10.1038/ng1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lowden MR, Meier B, Lee TW, Hall J, Ahmed S. End joining at Caenorhabditis elegans telomeres. Genetics. 2008;180:741–754. doi: 10.1534/genetics.108.089920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blauwkamp TA, Csankovszki G. Two classes of dosage compensation complex binding elements along Caenorhabditis elegans X chromosomes. Mol Cell Biol. 2009;29:2023–2031. doi: 10.1128/MCB.01448-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lieb JD, de Solorzano CO, Rodriguez EG, Jones A, Angelo M, Lockett S, Meyer BJ. The Caenorhabditis elegans dosage compensation machinery is recruited to X chromosome DNA attached to an autosome. Genetics. 2000;156:1603–1621. doi: 10.1093/genetics/156.4.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yonker SA, Meyer BJ. Recruitment of C. elegans dosage compensation proteins for gene-specific versus chromosome-wide repression. Development. 2003;130:6519–6532. doi: 10.1242/dev.00886. [DOI] [PubMed] [Google Scholar]
- 22.Shilatifard A. Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr Opin Cell Biol. 2008;20:341–348. doi: 10.1016/j.ceb.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsu DR, Chuang PT, Meyer BJ. DPY-30, a nuclear protein essential early in embryogenesis for Caenorhabditis elegans dosage compensation. Development. 1995;121:3323–3334. doi: 10.1242/dev.121.10.3323. [DOI] [PubMed] [Google Scholar]
- 24.Simonet T, Dulermo R, Schott S, Palladino F. Antagonistic functions of SET-2/SET1 and HPL/HP1 proteins in C. elegans development. Dev Biol. 2007;312:367–383. doi: 10.1016/j.ydbio.2007.09.035. [DOI] [PubMed] [Google Scholar]
- 25.Chua G, Morris QD, Sopko R, Robinson MD, Ryan O, Chan ET, Frey BJ, Andrews BJ, Boone C, Hughes TR. Identifying transcription factor functions and targets by phenotypic activation. Proc Natl Acad Sci U S A. 2006;103:12045–12050. doi: 10.1073/pnas.0605140103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alekseyenko AA, Peng S, Larschan E, Gorchakov AA, Lee OK, Kharchenko P, McGrath SD, Wang CI, Mardis ER, Park PJ, et al. A sequence motif within chromatin entry sites directs MSL establishment on the Drosophila X chromosome. Cell. 2008;134:599–609. doi: 10.1016/j.cell.2008.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Straub T, Grimaud C, Gilfillan GD, Mitterweger A, Becker PB. The chromosomal high-affinity binding sites for the Drosophila dosage compensation complex. PLoS Genet. 2008;4:e1000302. doi: 10.1371/journal.pgen.1000302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larschan E, Alekseyenko AA, Gortchakov AA, Peng S, Li B, Yang P, Workman JL, Park PJ, Kuroda MI. MSL complex is attracted to genes marked by H3K36 trimethylation using a sequence-independent mechanism. Mol Cell. 2007;28:121–133. doi: 10.1016/j.molcel.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 29.Sural TH, Peng S, Li B, Workman JL, Park PJ, Kuroda MI. The MSL3 chromodomain directs a key targeting step for dosage compensation of the Drosophila melanogaster X chromosome. Nat Struct Mol Biol. 2008;15:1318–1325. doi: 10.1038/nsmb.1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stiernagle T WormBook, T.C.e.R. Community. Maintenance of C. elegans. 2006 doi: 10.1895/wormbook.1.101.1. (WormBook doi/10.1895/wormbook.1.8.1, http://www.wormbook.org) [DOI] [PMC free article] [PubMed]
- 31.Selzer RR, Richmond TA, Pofahl NJ, Green RD, Eis PS, Nair P, Brothman AR, Stallings RL. Analysis of chromosome breakpoints in neuroblastoma at sub-kilobase resolution using fine-tiling oligonucleotide array CGH. Genes Chromosomes Cancer. 2005;44:305–319. doi: 10.1002/gcc.20243. [DOI] [PubMed] [Google Scholar]
- 32.Ji X, Li W, Song J, Wei L, Liu XS. CEAS: cis-regulatory element annotation system. Nucleic Acids Res. 2006;34:W551–554. doi: 10.1093/nar/gkl322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu XS, Brutlag DL, Liu JS. An algorithm for finding protein-DNA binding sites with applications to chromatin-immunoprecipitation microarray experiments. Nat Biotechnol. 2002;20:835–839. doi: 10.1038/nbt717. [DOI] [PubMed] [Google Scholar]
- 34.Workman CT, Yin Y, Corcoran DL, Ideker T, Stormo GD, Benos PV. enoLOGOS: a versatile web tool for energy normalized sequence logos. Nucleic Acids Res. 2005;33:W389–392. doi: 10.1093/nar/gki439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang M, Ryu J, Kiraly M, Duke K, Reinke V, Kim SK. Genome-wide analysis of developmental and sex-regulated gene expression profiles in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2001;98:218–223. doi: 10.1073/pnas.011520898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Whittle CM, McClinic KN, Ercan S, Zhang X, Green RD, Kelly WG, Lieb JD. The genomic distribution and function of histone variant HTZ-1 during C. elegans embryogenesis. PLoS Genet. 2008;4:e1000187. doi: 10.1371/journal.pgen.1000187. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.