Abstract
Prostate cancer has been shown to undergo unique metabolic changes associated with neoplastic transformation, with associated changes in citrate, alanine, and lactate concentrations. 13C HR-MAS spectroscopy provides an opportunity to simultaneously investigate the metabolic pathways implicated in these changes by using 13C labeled substrates as metabolic probes. In this work, a method to reproducibly interrogate metabolism in prostate cancer cells in primary culture was developed using HR-MAS spectroscopy. Optimization of cell culture protocols, labeling parameters, harvesting, storage, and transfer was performed. Using [3-13C] pyruvate as a metabolic probe, 1H and 13C HR-MAS spectroscopy were used to quantify the net amount and fractional enrichment of several labeled metabolites that evolved in multiple cell samples from each of five different prostate cancers. Average enrichment across all cancers was 32.4±5.4% for [3-13C] alanine, 24.5±5.4% for [4-13C] glutamate, 9.1±2.5% for [3-13C] glutamate, 25.2±5.7% for [3-13C] aspartate, and 4.2±1.0% for [3-13C] lactate. Cell samples from the same parent population demonstrated reproducible fractional enrichments of alanine, glutamate, and aspartate to within 12%, 10%, and 10%, respectively. Furthermore, the cells produced a significant amount of [4-13C] glutamate, which supports the bioenergetic theory for prostate cancer. These methods will allow further characterization of metabolic properties of prostate cancer cells in the future.
INTRODUCTION
Prostate cancer, has been shown in in vivo and in vitro studies to undergo unique metabolic changes as the healthy prostate tissue undergoes neoplastic transformation. A number of studies have demonstrated diminished citrate in prostate cancer (1-3), as well as higher levels of lactate and alanine compared to normal prostate tissue (4-6). Several explanations have been proposed for these metabolic alterations, including changes in membrane synthesis, ATP production, and modification of the extracellular environment to promote invasion and metastasis (7-9).
The goal of this study was to develop a method for studying metabolism of intact primary cultured prostate cancer cells (10,11) using 13C HR-MAS and to evaluate how well the cells incorporated [3-13C] pyruvate into the metabolic pathways implicated in the citrate, lactate, and alanine changes that occur in prostate cancer. The study of pure human prostate cancer cells avoids the complexity introduced by the presence of multiple cell types that exist in tissue specimens; while the use of primary culture rather than immortalized cell lines avoids alterations in metabolism that can occur in immortalized cells. The measurement of 13C labeled metabolites requires very sensitive NMR techniques for several reasons. The NMR measurements must be capable of detecting the 13C labeling in approximately 2 × 106 cells because the primary cultured prostate cells are mortal and have a limited lifespan. While NMR extraction protocols often require 108 cells for a single sample (12), HR-MAS spectroscopy has the ability to record spectra from smaller samples. The technique also allows solution-like NMR spectra to be obtained from intact tissue and cell specimens while preserving the tissue for further pathologic and genetic study (5,13-16). Since the HR-MAS experiments are performed on intact samples, the measurements are unaffected by any artifacts the extraction process may introduce.
As part of the study, a cell culture protocol was developed that successfully incorporated the label from [3-13C] pyruvate into the metabolites of interest. In addition, the culture protocol was optimized to permit measurement of 13C labeling in intact cells with HR-MAS spectroscopy. Finally, an initial comparison was made between the metabolic adaptations proposed to occur in prostate cancer, and a preliminary comparison was made between the metabolism of cells cultured from Gleason grade 3+3 and 4+3 cancers.
METHODS
Cell Culture and Labeling Procedure
In this study, prostate cancer cells from 5 different patients were studied. Two of the cancers were of Gleason grade 4+3, while three were of Gleason grade 3+3. The primary cultured prostate cancer cells were isolated and cultured as previously described (11). Briefly, tissues were harvested from radical prostatectomy specimens, minced, enzymatically disaggregated and then incubated in specialized medium. Cells from primary cultures were passaged twice then grown to 80% confluency, yielding 5 to 8 HR-MAS samples from each cancer culture. A modification to the cell culture protocol described in (10) was required because the PFMR-4A medium resulted in contamination of the proton spectrum even after washing the cells three times in phosphate buffered saline (PBS). Instead, Dulbecco's Modified Eagle's (DME) medium (Sigma-Aldrich, St. Louis, MO), which resulted in less contamination, was used for the final 24 hours of cell culture. The culture supplements used with the PFMR-4A medium (11) were added to the DME medium along with 1mM glucose and 1mM pyruvate. Additionally, threonine was excluded from the DME medium because the threonine resonance at 1.33 ppm significantly overlaps the lactate resonance at 1.31 ppm and was detected in the 1H spectra when it was present in the medium.
Following incubation in DME for 24 hours, the medium was aspirated from the cell culture plate. DME medium containing 5 mM [3-13C] pyruvate (Cambridge Isotopes, Andover, MA) and 1 mM unlabelled glucose was then added to the cells, and they were returned to the incubator. For one set of cells, several different labeling durations were used: 30 min, 60 min, 90 min. After 60 minutes of labeling, the glutamate C-4 enrichment had reached a maximum and the other metabolites of interests were approaching a maximum. Therefore, the remaining cell cultures used a 60-minute labeling duration.
Preparation of Sample Tubes
A specialized apparatus was constructed that allowed the cells to be directly loaded into a 3 mm diameter HR-MAS rotor (sample holder) from a storage tube at −80° C; this was done in order to minimize metabolic activity and breakdown of the cells during the transfer.. The narrow end of a shortened Pasteur pipette was inserted into a modified blood Caraway Tube (2.8 mm inner diameter, Chase Scientific Glass, Inc., Rockwood, TN). The two glass components were fused using an epoxy. A rubber cap was placed at the bottom of the blood capillary tube, and the tube was scored with a metal file approximately 3 cm from the bottom so that it could later be broken. A fully assembled sample tube is shown in Figure 1.
Figure 1.
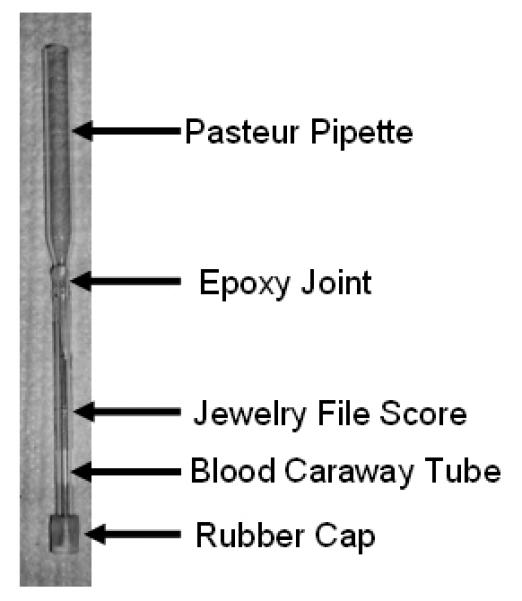
Photograph of a sample tube just after fabrication. The sample tube was assembled from a modified Pasteur pipette and a modified blood caraway tube.
Harvesting the Cells
Following incubation in labeled medium, the conditioned medium was aspirated from the plate. Plates were washed with 25 mL of PBS, which was then aspirated. TrypLE Express (Invitrogen, Carlsbad, CA) was added to the plate, and the plate was returned to the incubator for 6 minutes until the cells detached. Following detachment, the cells were maintained at a maximum of 1°C in order to minimize further metabolism and were washed by adding 25 mL of PBS, previously cooled to 1°C, to the plate. The cell suspension was transferred into a 50 mL conical tube and centrifugated at 1°C and 400 RCF for 5 minutes and the supernatant was then aspirated.
Cells were washed a second time by resuspending in 2 mL of 1°C PBS. The suspension was pipetted into a sample tube as described above and centrifuged for 5 minutes at 1°C and 400 RCF again. The sample tube was broken at the point at which it had been scored, and the top part of the apparatus was discarded. The supernatant was aspirated from the remaining portion of the tube. The tube was capped at the top, transferred to a cryo-container, and cooled to −80°C for storage. Based on cell count estimates performed on a few representative culture plates, each sample contained approximately 2×106 cells.
HR-MAS Experiments, Data Acquisition and Processing
A sample tube containing frozen cells was removed from the freezer and placed on dry ice. A 35-μL leakproof zirconium HR-MAS rotor (Varian Inc, Palo Alto, CA) was weighed using a balance capable of 0.01 mg accuracy. Three microliters of D2O+TSP were pipetted into the rotor and provided a frequency reference. Three microliters of 101.24 mM [2-13C] glycine was then added to the rotor to enable calibration of the 13C pulses and provide a quantification reference. The sample tube was removed from the dry ice, and its two rubber caps were cut off. The frozen cell sample was transferred to the HR-MAS rotor by holding the sample tube above the rotor and pushing the cells out the sample tube into the rotor using a small plastic rod. Following the insertion of each solution or cell sample, the rotor was weighed in order to determine the mass of the most recently added substance. The HR-MAS rotor was then assembled and closed as described in (5).
The cell sample was analyzed using a Varian INOVA 11.7T spectrometer equipped with a 4 mm gHX nanoprobe, previously cooled to 1°C. Once the rotor was loaded into the HR-MAS nanoprobe, the sample was spun at 2,250 Hz and maintained at 1°C. A pulse-acquire sequence was used to obtain a 1H[13C] spectrum with a 90° flip angle, 30,000 complex points, 20 kHz bandwidth, TR = 7 s, 128 averages, and 10 dummy scans for a total scan time of 16.1 minutes. The carbon decoupling was implemented using the GARP scheme with a pulse power of 43dB. The resulting spectrum was zero-filled to 131,072 points and Fourier transformed.
To obtain information from 13C labeled metabolites, a heteronuclear single quantum coherence (HSQC) experiment was performed, allowing detection of signal only from those protons attached to 13C nuclei. The evolution times were set to a single bond scalar coupling (JCH) of 143 Hz, which corresponds to the J-coupling constant for the C-2 labeled glycine reference. This choice necessarily resulted in incomplete polarization transfer for 13C labeled metabolites of the pyruvate substrate, including glutamate C-4 (JCH = 127 Hz), glutamate C-3 (JCH = 130 Hz), aspartate C-3 (JCH = 129 Hz, JCH = 130 Hz), alanine C-3 (JCH = 130 Hz), and lactate C-3 (JCH = 128 Hz). However, correction was made for the incomplete transfer, as described below. HSQC spectra were acquired with 4,096 points and a 10 kHz spectral width along the proton dimension, 256 complex increments and a 15 kHz spectral width along the carbon dimension, GARP 13C decoupling during the data acquisition, a TR of 2.41 s, 32 dummy scans, and 8 averages for each increment leading to a total scan time of 2.78 hrs. Linear prediction was used along the 13C dimension to produce an additional 512 points resulting in a matrix size of 4,096 × 768 points. This matrix was then zero-filled to 16,384 × 4,096 points and multiplied by Gaussian apodizations in both dimensions before Fourier transformation. Following the HSQC experiment, a second 1H[13C] spectrum was recorded to determine whether the metabolite concentrations had changed during the course of the HR-MAS experiment.
Quantitation
Metabolites were quantified by integrating the peaks in the 1D and 2D spectra. For the 1H data, the spectra were baseline corrected and integrated using 5% of peak height to determine limits of integration. The amount of each metabolite (MolesX) was calculated using the [2-13C] glycine reference as follows:
| Eq. [1] |
where AreaX and AreaGly are the peak areas in the 1H spectrum for the metabolite of interest (X) and glycine, respectively; NEPX and NEPGly are the number of equivalent protons for X and the glycine reference; MolesGly is the number of moles of the glycine reference that were added to the HR-MAS sample. A similar method was used to calculate the amount of 13C labeled metabolite (Moles13Cx) from volumes of the cross peaks in the HSQC data, with a small correction factor included to account for incomplete polarization transfer:
| Eq. [2] |
where Vol13Cx and Vol13C-Gly are the peak volumes in the HSQC spectrum for the metabolite of interest (X) and [2-13C] glycine, respectively; JCHx is the J-coupling in hertz for X; Moles13C-Gly is the number of moles of the [2-13C] glycine reference that were added to the HR-MAS sample. The concentration of the 13C labeled metabolites was calculated by dividing Moles13Cx from Eq. [2] by the mass of the HR-MAS sample.
The fractional enrichment of the metabolite (FEX) was calculated by computing the ratio of the two quantities calculated in Eq. [1] and Eq. [2]:
| Eq. [3] |
For this calculation, the MolesX was determined from the first 1H[13C] spectrum recorded for each cell sample.
RESULTS
Reducing Contamination
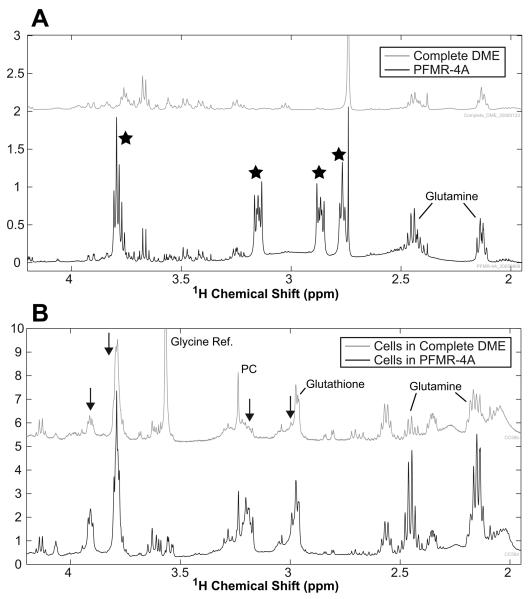
The 1H spectra in Figure 2 illustrate that the PFMR-4A medium produced several large peaks. While the glutamine concentration was much higher in the PFMR-4A than in DME medium, it was present in both media (Figure 2), resulting in uncertainty in the quantitation of glutamine in the proton cell spectra. Most of the other large peaks (highlighted with black stars at 2.6, 2.87, 3.15, and 3.75 ppm in Figure 2A) present in the PFMR-4A medium spectrum were absent from the DME medium and were produced by the HEPES component of the PFMR-4A medium. When the cells were cultured in PFMR-4A medium and washed three times with PBS, high concentrations of glutamine were observed in the proton spectra along with spectral contaminants at ~3.0, ~3.2, ~3.8, and ~3.9 ppm (Figure 2B). Because of these observations, the medium was switched to DME for the final 24 hours of the experiment. The proton spectra in Figure 2B from the cells cultured in DME for the final 24 hours demonstrated a reduction in glutamine and all of the spectral contaminants.
Figure 2.
(A) Proton HR-MAS spectra of DME medium (TOP) and PFMR-4A medium (BOTTOM). The ) that ultimately contaminated PFMR-4A spectrum contains several large peaks ( the spectra of the cells cultured in this medium prior to harvesting the cells as shown in the bottom of (B). By culturing the cells in DME for the last 24 hours (TOP of B), the spectral contamination caused by the PFMR-4A medium was significantly reduced as indicated by the black arrows. In addition, the glutamine levels were significantly reduced because the DME medium contained less glutamine. Note the large glycine peak in the DME cell spectrum was produced by the reference compound added for the HR-MAS experiment.
The Proton Spectrum
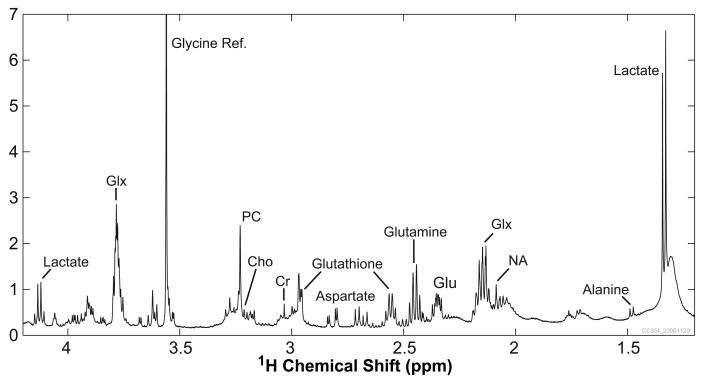
Similar to the published spectral profiles for prostate cancer tissue, the representative 1H[13C] HR-MAS spectrum in Figure 3 contains high levels of PC and lactate and no citrate. This spectral profile was consistent across the cell cultures derived from the five patients studied. In addition, the high spectral resolution of these intact cell samples readily permits identification of alanine, creatine, choline, glycerophosphocholine (GPC), and myo-inositol (mI) in the spectra, many of which are the subject of current prostate cancer research. Very important to the 13C labeling experiments included in this study, aspartate and glutamate have prominent resonances in the spectra, along with glutamine, glutathione, and an N-acetyl moiety. The strong glycine resonance was produced by the [2-13C] glycine reference added to the HR-MAS sample. While there are still contamination peaks present in the 1H[13C] spectra, they are small enough and isolated enough that most of the metabolites are easily identified and quantified. The high PC levels suggest that the cells were actively growing and proliferating when they were harvested. Also, it is unknown if the recorded glutamine levels are representative of the intrinsic cellular concentrations or if they were biased by the glutamine that was present in the modified DME medium.
Figure 3.
Representative 1H[13C] HR-MAS spectrum from primary cultured prostate cancer cells. The spectrum was acquired from 25.6 mg of cells in 16.1 min. Prominent peaks include the glycine reference, lactate, phosphocholine (PC), aspartate, glutathione, glutamine, glutamate (Glu), and Glx (composite of glutamate, glutamine, and glutathione). Cho: choline; Cr: creatine; NA: N-acetyl peak
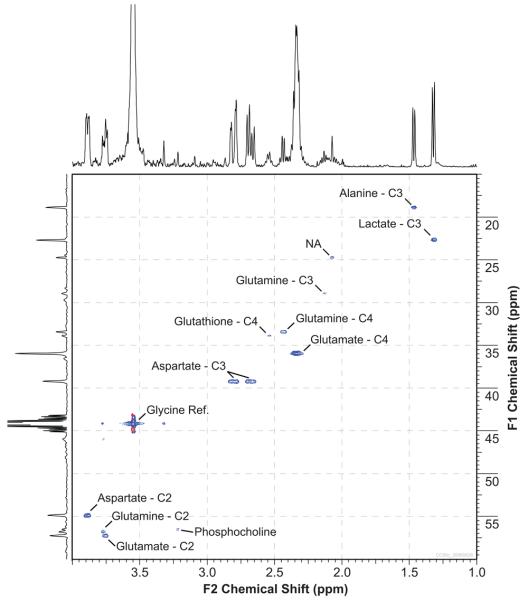
The HSQC Spectrum
The representative 13C HSQC spectrum presented in Figure 4 demonstrates that it is possible to measure the 13C enrichment of metabolites in intact prostate cancer cells using HR-MAS. The most prominently labeled metabolites were C-3 lactate, C-3 alanine, C-4 glutamate, C-2 glutamate, C-3 aspartate, and C-2 aspartate. Also labeled were an N-acetyl peak, C-2 glutamine, C-3 glutamine, C-4 glutamine, and C-4 glutathione. While PC was observed in the HSQC spectrum, its peak represents natural abundance enrichment. The large peak at 3.54 ppm was from the [2-13C] glycine reference compound. The spectrum in Figure 4 was graphically modified by removing the T1 noise from the [2-13C] glycine peak in the carbon dimension. This was done to facilitate easy viewing of the 2D spectrum and its projections.
Figure 4.
Representative 13C C HSQC spectrum from primary cultured prostate cancer cells using HR-MAS. Maximum intensity projections along 1Hand 13C dimensions are shown at the top and left, respectively. The spectrum was acquired from 26.5 mg of cells in 2.78 hrs.
Comparison of 1H[13C] spectra obtained before and after the HSQC experiment demonstrated that lactate was the only metabolite that changed during the HSQC experiment. Thus, with the exception of lactate, the duration of the HSQC experiment had no effect on the results of the metabolic measurements because the metabolism reflected in these measurements took place during the cell-labeling portion of the experiment rather than during the HR-MAS measurements. Lactate, however, increased in concentration by approximately 80% in the 2.78 hours between the first and second one-dimensional acquisitions. The significant evolution of lactate during the HR-MAS experiment indicated that there was significant lactate dehydrogenase activity even at 1°C and precluded precise quantitation of lactate in these experiments. Nonetheless, the lactate fractional enrichment computed using the first 1H[13C] experiment provided an estimate of the upper limit for the fractional enrichment of the primary cultured prostate cancer cells.
Quantitative Results
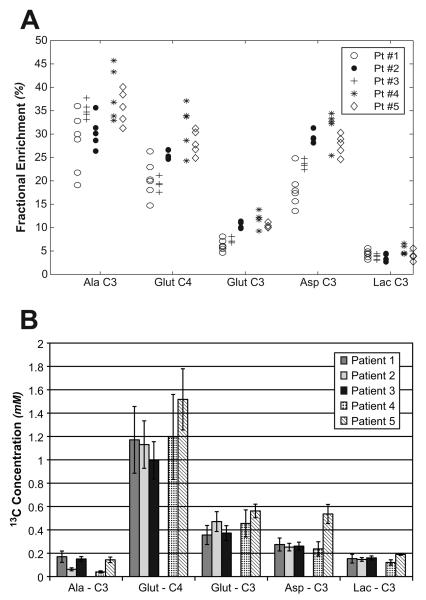
As demonstrated by the fractional enrichment plot in Figure 5A, the C-3 of alanine, the C-4 of glutamate, C-3 of glutamate, and the C-3 of aspartate were enriched well above the natural enrichment of 1.1% for all of the prostate cancer cell cultures. The average relative standard deviations for the fractional enrichments for multiple samples from each cancer cell culture were 12.6%, 11%, 9.4%, and 22% for C-3 alanine, C-4 glutamate, C-3 aspartate, and C-3 lactate, respectively. While there were no statistical differences in the fractional enrichment between Gleason grade 3+3 and 4+3 primary cultures, there was a trend toward higher fractional enrichments in the 4+3 cultures (17% higher for C-3 alanine, 33% higher for C-4 glutamate, 38% higher for C-3 glutamate, 25% higher for C-3 aspartate, and 14% higher for C-3 lactate). Average enrichments across all cancers were 32.4 ± 5.4% for C-3 alanine, 24.5 ± 5.4% for C-4 glutamate, 9.1 ± 2.5% for C-3 glutamate, 25.2 ± 5.7% for C-3 aspartate, and 4.2 ± 1.0% for C-3 lactate. Additionally, there was a substantial difference in the amount of 13C labeled metabolite that was produced for each of the metabolites following the one hour incubation of the cells with medium containing [3-13C] pyruvate (Figure 5B). In particular, the amount of [4-13C] glutamate produced during the labeling experiment was substantially more than the amount of 13C compound produced for the other metabolites. Combining data from all of the samples, the prostate cancer cells produced an average of 0.115 ± 0.061 mM of C-3 alanine, 1.21 ± 0.29 mM of C-4 glutamate, 0.440 ± 0.11 mM of C-3 glutamate, 0.308 ± 0.12 mM of C-3 aspartate, and 0.154 ± 0.031 mM of C-3 lactate.
Figure 5.
(A) Fractional enrichment of the several metabolites following the delivery of [3-13C] pyruvate to primary cultured prostate cancer cells derived from five different prostate cancer patients. The prostate cancers from patients 1-3 had a Gleason score of 3+3 and the patients 4 and 5 had a Gleason score of 4+3. (B) The average and standard deviation of the concentration of the 13C pool for each of the metabolites is also presented. While all the metabolites were enriched above natural abundance, the [3-13C] pyruvate was converted to glutamate more than any of the other metabolites during a 2 hour exposure to the pyruvate. Pt: Patient; Ala: Alanine; Glut: Glutamate; Asp: Aspartate; Lac: Lactate.
DISCUSSION
This work describes a new method for performing HR-MAS spectroscopy on intact human prostate cancer cells grown in primary culture, and provides a method to analyze their metabolism using information obtained from both 1H and 13C spectra. By performing the experiment with intact cells rather than extracts, fewer cells were required to produce a single HR-MAS sample. However, a culture protocol had to be developed for primary cultured cells that optimized the harvesting procedure used to prepare the cells for subsequent HR-MAS experiments.
The results of the one-dimensional experiments yielded several new conclusions, which clarify and expand upon prior published work. First, in a previous study of high resolution NMR of prostate cancer cells in primary culture by Yacoe et al (12) using PFMR-4A medium and cell extracts, the large complex peaks that were found at 2.87 and 3.15 ppm were assigned to α-ketoglutarate. In the present study, it was found that these peaks, as well as the large unidentified triplet at 2.73 ppm, were in fact not from the cells but rather from the medium. The absence of these spurious peaks in the cell spectra described in this work enabled observation of aspartate in the 1H[13C] spectra. This is of interest because aspartate is hypothesized to play a significant role in the metabolism of both normal and diseased prostate (17,18), and we confirmed that although the peaks are relatively small, they are nonetheless present. Furthermore, aspartate is significantly enriched upon incubation with [3-13C] pyruvate.
The 13C HSQC spectra recorded from intact primary cultured cells under HR-MAS conditions provided a robust measurement of 13C labeling in the metabolites observed in the 1H[13C] spectra. Acquisition of 1H[13C] spectra before and after the HSQC data demonstrated that, with the exception of lactate, the other metabolites of interest were stable throughout the HR-MAS experiment. This supports the conclusion that the quantitation of these metabolites reflects metabolism that occurred during the incubation of cells in medium rather than during the HR-MAS measurements. Although lactate enrichment cannot be absolutely quantified because of the lactate synthesis that occurred while the cells were in the HR-MAS spectrometer, an upper bound on the lactate 13C enrichment can be determined using the HSQC peak volume and the peak area of the first 1H[13C] experiment. Finally, the method used for quantifying enrichment demonstrates sufficient reproducibility that enrichments of glutamate and aspartate can be measured with 10% precision, and alanine within 12%.
By virtue of the excellent reproducibility of the fractional enrichment calculations between multiple samples taken from the same cancer culture, the results of the experiments indicate that the percent enrichment calculated for a single sample is in fact representative of the cells from that primary culture. The metabolites that show the greatest degree of enrichment in our combined proton and HSQC experiments are alanine, glutamate C-4, and aspartate C-3. Comparing the Gleason grade 3+3 and 4+3 cancers, there was a trend for higher fractional enrichment of C-3 alanine, C-3 and C-4 glutamate, C-3 aspartate , and C-3 lactate in cells with a higher Gleason score as compared to those with a lower Gleason score. The lack of a statistically significant difference in the fractional enrichment was possibly due to the small sample size.
In contrast to other 13C studies of metabolism in cancer cells (19,20) which showed significant evolution of labeled lactate from [1-13C] glucose tracer, the present study demonstrated far greater evolution of labeled glutamate than lactate. Since it was not possible to accurately quantify the fractional enrichment of the lactate exported into medium by these cells, the cells may have produced significantly more labeled lactate than reported. Ignoring the lactate in the medium for the moment, the apparent high rate of citric acid cycle metabolism would be consistent with a large body of work by Costello and Franklin (8,21-26), who have studied the loss of citrate production in secretory prostatic cells which occurs when the cells undergo neoplastic transformation. Another plausible explanation for this difference lies in the choice of substrate. The pyruvate used in this study requires an independent source of NADH for conversion to lactate, whereas the labeled glucose used in the other studies does not require an independent NADH source. Since NADH was not present in the primary prostate cell culture medium in excess concentration, it is possible that the production of lactate was artificially low because of the absence of the cofactor.
One shortcoming of this method is the inability to culture normal secretory prostate epithelial cells capable of synthesizing citrate. While metabolic differences between neoplastic cells and normal secretory prostatic cells would likely be apparent in cell cultures, the inability to culture normal cells that produce citrate prevents this observation. Primary cultures of normal prostatic epithelial cells consist of basal and transit amplifying cells which typically do not differentiate into citrate synthesizing secretory cells (27). However, culture conditions that promote differentiation have recently been reported (28), which will permit future comparative metabolic studies of normal versus malignant prostate cancer cells.
Conclusion
The method presented in this work for studying metabolism of prostate cancer cells in primary culture enabled quantification of the fractional enrichment and the concentration of the 13C labeled pool for the metabolic products of [3-13C] pyruvate. Fractional enrichment of these metabolites was consistent to within approximately 10% for multiple samples obtained from the same parent population. While alanine demonstrated the highest fractional enrichment in these cells, much of the [3-13C] pyruvate was converted to glutamate, indicating that citric acid cycle metabolism may be the significant consumer of pyruvate in human prostate cancer cells.
Acknowledgements
We would like to acknowledge Dr. Samson Jarso, Dr. Tracy McKnight, and Christopher Sundberg for developing the original cell culture protocols that served as a starting point for this study. We want to thank Bao Nguyen from Varian, Inc. for continually maintaining and repairing our Nanoprobe. We are also grateful to Lawrence Hooser and Robert Sellers for culturing the cells used in this study.
Grant Sponsors:
NIH R21 EB005363
NIH R01 CA102751
References
- 1.Kurhanewicz J, Vigneron DB, Nelson SJ, Hricak H, MacDonald JM, Konety B, Narayan P. Citrate as an in vivo marker to discriminate prostate cancer from benign prostatic hyperplasia and normal prostate peripheral zone: detection via localized proton spectroscopy. Urology. 1995;45(3):459–466. doi: 10.1016/S0090-4295(99)80016-8. [DOI] [PubMed] [Google Scholar]
- 2.Kurhanewicz J, Dahiya R, Macdonald JM, Chang LH, James TL, Narayan P. Citrate alterations in primary and metastatic human prostatic adenocarcinomas: 1H magnetic resonance spectroscopy and biochemical study. Magn Reson Med. 1993;29(2):149–157. doi: 10.1002/mrm.1910290202. [DOI] [PubMed] [Google Scholar]
- 3.Schick F, Bongers H, Kurz S, Jung WI, Pfeffer M, Lutz O. Localized proton MR spectroscopy of citrate in vitro and of the human prostate in vivo at 1.5 T. Magn Reson Med. 1993;29(1):38–43. doi: 10.1002/mrm.1910290109. [DOI] [PubMed] [Google Scholar]
- 4.Cornel EB, Smits GA, Oosterhof GO, Karthaus HF, Deburyne FM, Schalken JA, Heerschap A. Characterization of human prostate cancer, benign prostatic hyperplasia and normal prostate by in vitro 1H and 31P magnetic resonance spectroscopy. J Urol. 1993;150(6):2019–2024. doi: 10.1016/s0022-5347(17)35957-8. [DOI] [PubMed] [Google Scholar]
- 5.Swanson MG, Zektzer AS, Tabatabai ZL, Simko J, Jarso S, Keshari KR, Schmitt L, Carroll PR, Shinohara K, Vigneron DB, Kurhanewicz J. Quantitative analysis of prostate metabolites using 1H HR-MAS spectroscopy. Magn Reson Med. 2006;55(6):1257–1264. doi: 10.1002/mrm.20909. [DOI] [PubMed] [Google Scholar]
- 6.Tessem MB, Swanson MG, Keshari KR, Albers MJ, Joun D, Tabatabai ZL, Simko JP, Shinohara K, Nelson SJ, Vigneron DB, Gribbestad IS, Kurhanewicz J. Evaluation of lactate and alanine as metabolic biomarkers of prostate cancer using 1H HR-MAS spectroscopy of biopsy tissues. Magn Reson Med. 2008;60(3):510–516. doi: 10.1002/mrm.21694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Costello LC, Franklin RB. Bioenergetic theory of prostate malignancy. Prostate. 1994;25(3):162–166. doi: 10.1002/pros.2990250308. [DOI] [PubMed] [Google Scholar]
- 8.Costello LC, Franklin RB. ‘Why do tumour cells glycolyse?’: from glycolysis through citrate to lipogenesis. Mol Cell Biochem. 2005;280(1-2):1–8. doi: 10.1007/s11010-005-8841-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4(11):891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 10.Peehl DM. Primary cell cultures as models of prostate cancer development. Endocr Relat Cancer. 2005;12(1):19–47. doi: 10.1677/erc.1.00795. [DOI] [PubMed] [Google Scholar]
- 11.Peehl DM. Human prostatic epithelial cells. In: Freshney RI, Freshney MG, editors. Culture of Epithelial Cells. 2nd ed. Wiley-Liss, Inc.; New York: 2002. pp. 171–194. [Google Scholar]
- 12.Yacoe ME, Sommer G, Peehl D. In vitro proton spectroscopy of normal and abnormal prostate. Magn Reson Med. 1991;19(2):429–438. doi: 10.1002/mrm.1910190234. [DOI] [PubMed] [Google Scholar]
- 13.Swanson MG, Vigneron DB, Tabatabai ZL, Males RG, Schmitt L, Carroll PR, James JK, Hurd RE, Kurhanewicz J. Proton HR-MAS spectroscopy and quantitative pathologic analysis of MRI/3D-MRSI-targeted postsurgical prostate tissues. Magn Reson Med. 2003;50(5):944–954. doi: 10.1002/mrm.10614. [DOI] [PubMed] [Google Scholar]
- 14.Cheng LL, Wu C, Smith MR, Gonzalez RG. Non-destructive quantitation of spermine in human prostate tissue samples using HRMAS 1H NMR spectroscopy at 9.4 T. FEBS Lett. 2001;494(1-2):112–116. doi: 10.1016/s0014-5793(01)02329-8. [DOI] [PubMed] [Google Scholar]
- 15.Griffin JL, Blenkiron C, Valonen PK, Caldas C, Kauppinen RA. High-resolution magic angle spinning 1H NMR spectroscopy and reverse transcription-PCR analysis of apoptosis in a rat glioma. Anal Chem. 2006;78(5):1546–1552. doi: 10.1021/ac051418o. [DOI] [PubMed] [Google Scholar]
- 16.Santos CF, Kurhanewicz J, Tabatabai ZL, Simko JP, Keshari KR, Gbegnon A, DeLos Santos R, Federman S, Shinohara K, Carroll PR, Haqq CM. Swanson MG Metabolic, pathologic, and genetic analysis of prostate tissues: Quantitative evaluation of histopathologic and mRNA integrity after HR-MAS spectroscopy. NMR Biomed. 2009 doi: 10.1002/nbm.1474. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Costello LC, Akuffo V, Franklin RB. Testosterone stimulates net citrate production from aspartate by prostate epithelial cells. Horm Metab Res. 1988;20(4):252–253. doi: 10.1055/s-2007-1010807. [DOI] [PubMed] [Google Scholar]
- 18.Franklin RB, Lao LX, Costello LC. Evidence for two aspartate transport systems in prostate epithelial cells. Prostate. 1990;16(2):137–145. doi: 10.1002/pros.2990160205. [DOI] [PubMed] [Google Scholar]
- 19.Singer S, Okunieff P, Gostin C, Thilly WG, Chen LB, Neuringer LJ. 13C- and 31P-NMR studies of human colon cancer in-vitro and in-vivo. Surg Oncol. 1993;2(1):7–18. doi: 10.1016/0960-7404(93)90039-2. [DOI] [PubMed] [Google Scholar]
- 20.Wehrle JP, Ng CE, McGovern KA, Aiken NR, Shungu DC, Chance EM, Glickson JD. Metabolism of alternative substrates and the bioenergetic status of EMT6 tumor cell spheroids. NMR Biomed. 2000;13(6):349–360. doi: 10.1002/1099-1492(200010)13:6<349::aid-nbm652>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 21.Costello LC, Franklin RB. The clinical relevance of the metabolism of prostate cancer; zinc and tumor suppression: connecting the dots. Mol Cancer. 2006;5:17. doi: 10.1186/1476-4598-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Costello LC, Franklin RB. Tumor cell metabolism: the marriage of molecular genetics and proteomics with cellular intermediary metabolism; proceed with caution! Mol Cancer. 2006;5:59. doi: 10.1186/1476-4598-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Costello LC, Franklin RB, Feng P. Mitochondrial function, zinc, and intermediary metabolism relationships in normal prostate and prostate cancer. Mitochondrion. 2005;5(3):143–153. doi: 10.1016/j.mito.2005.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Costello LC, Guan Z, Kukoyi B, Feng P, Franklin RB. Terminal oxidation and the effects of zinc in prostate versus liver mitochondria. Mitochondrion. 2004;4(4):331–338. doi: 10.1016/j.mito.2004.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dakubo GD, Parr RL, Costello LC, Franklin RB, Thayer RE. Altered metabolism and mitochondrial genome in prostate cancer. J Clin Pathol. 2006;59(1):10–16. doi: 10.1136/jcp.2005.027664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh KK, Desouki MM, Franklin RB, Costello LC. Mitochondrial aconitase and citrate metabolism in malignant and nonmalignant human prostate tissues. Mol Cancer. 2006;5:14. doi: 10.1186/1476-4598-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peehl DM. Primary cell cultures as models of prostate cancer development. Endocrine-related Cancer. 2005;12:19–47. doi: 10.1677/erc.1.00795. [DOI] [PubMed] [Google Scholar]
- 28.Zhao H, Nolley R, Chen Z, Reese SW, Peehl DM. Inhibition of monoamine oxidase A promotes secretory differentiation in basal prostatic epithelial cells. Differentiation. 2008;76(7):820–830. doi: 10.1111/j.1432-0436.2007.00263.x. [DOI] [PMC free article] [PubMed] [Google Scholar]