Abstract
Recently, we identified in adult tissues a population of Oct4+SSEA-1+Sca-1+lin-CD45- very small embryonic-like stem cells (VSELs). First, to address recent controversies on Oct4 expression in cells isolated from adult organs we show here an evidence that Oct4 promoter in bone marrow (BM)-derived VSELs has an open chromatin structure and is actively transcribed. Next, to explain VSELs quiescence and lack of teratoma formation we demonstrate a unique DNA methylation pattern at some developmentally crucial, imprinted-genes, showing hypomethylation/erasure of imprints in paternally methylated and hypermethylation of imprints in maternally methylated ones. These epigenetic characteristics leading to upregulation in VSELs of H19 and p57KIP2 (also known as Cdkn1c) and repression of Igf2 and Rasgrf1 explain VSEL's quiescent status. Interestingly, this unique pattern in imprinted-genes methylation is reverted in co-cultures with a C2C12 supportive cell-line when VSELs are induced to form VSEL-derived spheres (VSEL-DSs) enriched for stem cells able to differentiate into all three germ layers. Therefore, we suggest that the proliferative/developmental potential of Oct4+ VSELs is epigenetically regulated by expression of Oct4 and some imprinted-genes, and postulate that restoring the proper methylation pattern of imprinted-genes will be crucial step for employing these cells in regenerative medicine.
Keywords: VSELs, Genomic imprinting, Oct4, Epiblast/Germ line
Introduction
Pluripotent stem cells (PSCs) are able to differentiate in in vitro cultures into cells from all three germ layers and in vivo they grow teratomas and complement blastocyst development. PSCs are derived from early embryos i) as embryonic stem cells (ESCs) or epiblast stem cells (EpiSCs) or ii) from migrating primordial germ cells (PGCs) as embryonic germ cells (EGCs).1-3 In addition, recently a novel type of PSCs, known as inducible PSCs (iPSCs) was obtained after transduction of somatic cells with genes encoding embryonic transcription factors.4,5 Some investigators postulated presence of Oct4+ PSCs in postnatal adult tissues that are able to differentiate in vitro into cells from all three germ layers,6,7 however their in vivo pluripotentiality was not demonstrated. Nevertheless, some recent reports, cast some doubts if Oct4 is truly expressed in cells isolated from adult tissues.8,9
Recently, our group identified a population of very small embryonic-like stem cells (VSELs) in adult murine tissues including bone marrow (BM)10 and human cord blood.11 These cells isolated from BM by multiparameter fluorescence activated cell sorter (FACS) as a population of Sca-1+lin-CD45- are: i) very small in size (∼3-6 μm); ii) express pluripotent markers such as Oct4, Nanog, Rex-1, and SSEA-1 and iii) possess large nuclei containing unorganized chromatin (euchromatin). We have shown that VSELs are mobilized into peripheral blood during organ injuries (e.g., heart infarct, stroke),12-14 what suggests that they may contribute in regeneration of the damaged tissues.
Unlike ESCs, highly purified BM-derived Oct4+ VSELs do not proliferate in vitro if cultured alone and do not grow teratomas in vivo. However, in co-cultures with myoblastic C2C12 cells, VSELs form EB-like structures, VSEL-derived spheres (VSEL-DSs) that contain primitive stem cells able to differentiate into cells from all three germ layers.10 On one hand, this suggests that VSELs are a quiescent cell population and that some mechanisms must exist to prevent their unleashed proliferation and teratoma formation. On the other, the ability of VSELs to change quiescent fate in co-cultures with C2C12 cells demonstrates that their quiescent status could be modulated. This supports the overall concept that VSELs may contribute to rejuvenation of organs and tissue repair.15 However, molecular mechanisms that modulate both pluripotency and quiescence of these cells remain unclear. Identification of these mechanisms is important to not only to understand better the biological significance of VSELs presence in adult tissues, but also will be crucial for future use of these cells in regenerative medicine.
We have hypothesized, that quiescence of VSELs could be modulated similarly as in PGCs, by status of DNA methylation on some developmentally important imprinted-genes.15 Genomic imprinting is epigenetic process responsible for paternal-specific, mono-allelic expression of so-called imprinted-genes.16 There are ∼80 imprinted-genes (expressed from maternal or paternal chromosomes only) for which proper mono-allelic expression regulates totipotential status of the zygote, and pluripotency of developmentally early stem cells.17-19 In addition, most imprinted-genes such as insulin-like growth factor 2 (Igf2), H19, Igf2 receptor (Igf2R), p57Kip2 (also known as Cdkn1c) play a direct role in embryo development.16 The majority of imprinted-genes exist as gene clusters enriched for CpG islands and their expression is coordinately regulated by DNA methylation status on CpG-rich cis-elements known as differently methylated regions (DMRs). The DMRs are differentially methylated on CpG sites by DNA methyltransferase (Dnmts), depending on the parental allele origin.20 In addition depending on developmental period of methylation “primary DMRs” are differentially methylated during gametogenesis, and “secondary DMRs” acquire allelic specific methylation after fertilization.21 So far, 15 primary DMRs have been identified in mouse genome. Interestingly, most DMRs are methylated at maternal allele and only three DMRs (Igf2-H19, Rasgrf1, Meg3 loci) are paternally methylated.22 Although DMR methylation is of primary importance, histone modifications also contribute to monoallelic expression of these genes.23,24
In present study, we demonstrate that the proliferative quiescence of VSELs could be epigenetically controlled by DNA methylation on some developmentally important imprinted-genes. Moreover, for the first time, we provide molecular evidence for an open/active chromatin structure in Oct4 promoter in VSELs, supporting that Oct4 could be truly expressed in stem cells isolated from adult tissues.
Materials and Methods
Animals and preparation of BM cells for FACS
The present study was performed in accordance with the guidelines of the Animal Care and Use Committee of the University of Louisville (UofL) School of Medicine and with the Guide for the Care and Use of Laboratory Animals (Department of Health and Human Services, Publication No. NIH 86–23). Murine MNCs were isolated from BM of pathogen-free, 4 to 6 week-old female and male C57BL/6 or C57BL/6-Tg(ACTB-EGFP)1Osb/J mice (Jackson Laboratory, Bar Harbor, ME, USA). Isolated by flushing bones, BM cell suspensions were lysed in BD lysing buffer (BD Biosciences, San Jose, CA, USA) for 15 min in room temperature (rt) and washed twice in phosphate buffered saline (PBS).
Isolation of VSELs from BM by FACS
VSELs (Lin-Sca-1+CD45-) and HSCs (Lin-Sca-1+CD45+) cells were isolated from 4 to 6 week-old mice BM cells by multiparameter, live cell sorting (FACSVantage SE; Becton Dickinson, Mountainview, CA, USA or MoFlo, Dako A/S, Fort Collins, CO, USA).10 Briefly, BM-MNCs (10×107 cells/ml) were resuspended in cell-sort medium (CSM) containing 1× Hank's Balanced Salt Solution without phenol red (GIBCO, Grand Island, NY, USA), 2% heat-inactivated fetal calf serum (FCS; GIBCO), 10mM HEPES buffer (GIBCO), and 30 U/ml of Gentamicin (GIBCO). The following monoclonal antibodies (mAbs) were employed for cell staining: biotinconjugated rat anti-mouse Ly-6A/E (Sca-1) (clone E13-161.7); streptavidin-PE-Cy5 conjugate; anti-CD45-APC-Cy7 (clone 30-F11); anti-CD45R/B220-PE (clone RA3-6B2); anti-Gr-1-PE (clone RB6-8C5); anti-TCRab PE (clone H57-597); anti-TCRgz PE (clone GL3); anti-CD11b PE (clone M1/70); and anti-Ter-119 PE (clone TER-119). All mAbs were added at saturating concentrations and the cells were incubated for 30 min on ice and washed twice, then resuspended for sort in CSM at a concentration of 5×106 cells/ml. The double-sorted populations of cells were employed.
Formation of VSEL-DSs and cell culture
The VSEL-DSs were cultured as previously described10 and cells isolated from VSEL-DSs at days 5, 7, and 11 were employed in our studies. Murine ESC-D3 cells were purchased from ATCC (Rockville, MD, USA) and grown in Dulbecco's modified Eagle's medium (DMEM; GIBCO) containing 4 mM L-glutamine, 1.5 g/L sodium bicarbonate, 4.5 g/L glucose, 0.1 mM β-mercaptoethanol (Sigma, St Louis, MO, USA), 15% heat-inactivated fetal bovine serum (FBS; GIBCO), 100 IU/ml penicillin, 100 μg/ml streptomycin (Invitrogen, Carlsbad, CA, USA), and 5 ng/ml of recombinant mouse LIF (Chemicon-Millipore, Billerica, MA, USA) without a feeder layer. EB formation was performed by the hanging drop method. The human hematopoietic cell line, THP-1, and murine BM STs were maintained in RPMI1640 (GIBCO) and DMEM medium, respectively, supplemented with 10% FBS, 100 IU/ml penicillin, 100 μg/ml streptomycin, and 2 mM L-glutamine.
Carrier Chromatin-Immunoprecipitation (Carrier-ChIP)
Carrier-ChIP analysis was performed as previously described with some modifications.25 Instead of Drosophila melanogaster SL2 cells, THP-1 cells were used as a source of carrier chromatin. The ChIP assay was performed using Magna ChIP™ G kit (Upstate-Millipore, Billerica, MA, USA) according to the manufacturer's instructions. In brief, 5×106 THP-1 cells were resuspended in culture media and mixed with 2×104 freshly isolated VSELs, HSCs, BM-MNCs, ESC-D3s, and EB-derived cells. The cell mixtures were subsequently fixed with 1% formaldehyde in culture media for 10 min at rt with rotation. The excess of formaldehyde was quenched by adding 10×glycine stock followed by incubation for 5 min at rt. The crosslinked chromatins of cell mixtures were subsequently sheared by sonication (Model 150T, Fisher Scientific, Pittsburgh, PA, USA) at 40% amplitude (four times 15 sec pulse on with incubation at ice for 1 min at intervals) in 200 μl of Nuclear Lysis Buffer. After centrifugation at 10,000×g at 4°C for 10 min, sheared chromatin was immunoprecipitated using Protein G magnetic beads, conjugated with 3 μg of ChIP grade antibodies against H3Ac (Upstate-Millipore), H3K9me2 (Abcam, Cambridge, MA, USA), or rabbit immunoglobulin (Ig)G control antibodies (Sigma). The bound and unbound sheared crosslinked chromatins were subsequently eluted according to the manufacturer's instructions. PCR reactions were performed using Amplitaq Gold Taq (Applied Biosystems, Foster City, CA, USA), primers for murine sequence specific Oct4, Nanog, or β-Actin promoter (Supplementary Table 1) at 1st cycle of 8 min at 95°C, 2nd cycle of 2 min at 95°C, 1 min at the annealing temperature (AT), and 1 min at 72°C. Subsequently, the reaction was set as follows: indicated cycles of 30 sec at 95°C; 1 min at AT°C; and 1 min at 72°C; followed by 1 cycle of 10 min at 72°C. The AT was 62°C for Oct4, 60°C for Nanog, and 65°C for β-Actin. Finally, PCR products were visualized by electrophoresis on 2% agarose gel. To quantify the enrichment of each histone modification, we applied RQ-PCR using qChIP primer sets (Supplementary Table 1). The copy number of bound or unbound PCR products was calculated by the absolute quantification method. The enrichment of each histone modification was calculated as the ratio of amplicon amounts from bound (B) to unbound (UB) fractions and fold differences are shown as mean ± S.D. from at least four independent experiments. All the clones obtained by employing these ChIP primers were subsequently sequenced to rule out the possibility of amplification of Oct4 pseudogenes or nonspecific sequences (data not shown).
Bisulfite-sequencing and combined bisulfite-restriction analysis (COBRA)
The DNA methylation status of the promoters of pluripotent regulators (Oct4, Nanog) and DMRs of imprinted-genes were investigated using bisulfite DNA modification followed by sequencing as well as by COBRA assay. In brief, genomic DNA were prepared from double-sorted VSELs, HSCs, STs, ESC-D3s, and cells derived from VSEL-DSs (2×104) using the DNeasy Blood & Tissue Kit (Qiagen Inc, Valencia, CA, USA). Next, 100 ng of gDNA were used in bisulfite modification, performed using the EpiTect Bisulfite Kit (Qiagen Inc) according to the manufacturer's instructions. DMRs of imprinted-genes were amplified by nested PCR using bisulfite treated gDNA and specific primers (Supplementary Table 1). Both first and second round PCR were performed at 2 cycles of 2 min at 95°C, 1 min at 55°C, 1 min at 72°C, and subsequent 35 cycles of 30 sec at 95°C, 1 min at 55°C, 1 min at 72°C, and 1 cycle of 10 min at 72°C. After agarose gel electrophoresis, amplicons were eluted using QIAquick Gel Extraction Kits (Qiagen Inc). Eluted amplicons were subsequently ligated into pCR®2.1-TOPO® vector and transformed into TOP10 bacteria using a TOPO TA Cloning Kit (Invitrogen). The plasmids were prepared using a QIAprep Spin Miniprep Kit (Qiagen Inc) and sequenced with M13 forward and reverse primers. The methylation pattern in DMRs was analyzed using CpGviewer software.26 The COBRA assay was performed by cutting amplicons of DMRs with TaqI or BstUI restriction enzyme for 2 h and subsequent agarose gel electrophoresis, as previously described.18 All experiments were conducted with three independent isolations of all the cell populations and two independent PCRs of each isolated cell.
Reverse transcriptase- polymerase chain reaction (RT-PCR)
Total RNA from various cells was isolated using the RNeasy Mini Kit (Qiagen Inc) including treatment of DNase I (Qiagen Inc). The mRNA (10 ng) was reverse-transcribed with Taqman Reverse Transcription Reagents (Applied Biosystems) according to the manufacturer's instructions. The resulting cDNA fragments were amplified using Amplitaq Gold at 1 cycle of 8 min at 95°C, 2 cycles of 2 min at 95°C, 1 min at 60°C, 1 min at 72°C, and subsequent 35 cycles of 30 sec at 95°C, 1 min at 60°C, 1 min at 72°C, and 1 cycle of 10 min at 72°C by employing sequence specific primers (Supplementary Table 2). All primers were designed with Primer Express software (Applied Biosystems), as at least one primer included an exon-intron boundary.
Real-time quantitative PCR (RQ-PCR)
Quantitative assessment of mRNA levels of target genes was performed by RQ-PCR using an ABI Prism 7500 Sequence Detection System (Applied Biosystems). The cDNA templates from each cell were amplified using SYBR Green PCR Master Mix (Applied Biosystems) and specific primers (Supplementary Table 2). All primers were designed with Primer Express software (Applied Biosystems), as at least one primer included an exon-intron boundary. In case of Oct4 expression analysis, the already published primer set was used,9 and by sequencing the PCR products, we exclude the possibility of amplification of Oct4 pseudogenes or nonspecific sequences (data not shown). The threshold cycle (Ct), the cycle number at which the fluorescence of amplified gene reached a fixed threshold, was subsequently determined and relative quantification of the expression level of target genes was performed with the 2-ΔΔCt method, using the mRNA level of β2-microglobulin as an endogenous control gene and that of ST as a calibrator.
Immunocytochemistry
Immunocytochemistry for Oct4, SSEA-1, p57KIP2 (polyclonal, Abcam), Dnmt1 (C-17, polyclonal, Santa Cruz, Santa Cruz, CA, USA), and Dnmt3b (N-19, polyclonal, Santa Cruz) proteins was performed as previously described.10
Statistic Analysis
All the data in quantitative ChIP and gene expression analysis were analyzed using one factor ANOVA with Bonferroni's Multiple Comparison Test. We used Instat1.14 program (GraphPad, La Jolla, CA, USA) and statistical significance was defined as p<0.05 or p<0.01.
Results
The open chromatin structure of Oct4 promoter in VSELs
A few recent reports cast some doubt on whether Oct4+ cells are truly present in adult tissues.8,9 These reports suggested that Oct4 expression in putative candidates of PSCs could merely be a result of detection of Oct4 pseudogenes by RT-PCR or unspecific staining. Therefore, it becomes important to reappraise these data and to see if Oct4 is truly expressed in candidate PSCs isolated from adult tissues. To prove that VSELs express the Oct4 gene, we investigated the epigenetic status of Oct4 promoter in these cells. To do this, Sca-1+Lin-CD45- VSELs were double purified along with Sca-1+Lin-CD45+ hematopoietic stem cells (HSCs) by FACS (Figure 1a). First, we reconfirmed that highly purified VSELs, similarly to ESC cell-line ESC-D3, express Oct4 both at the mRNA and protein levels (Figure 1b and c). Next, since expression of Oct4 is repressed in differentiated cells by a mechanism involving promoter methylation,27 we examined the DNA methylation status of the Oct4 promoter (Figure 1d) by employing bisulfite-sequencing in murine VSELs, HSCs, BM-derived stroma-cells (STs), and cells isolated from ESC-D3 derived 1-day embryoid bodies (EBs) (Figure 1e). We noticed that the Oct4 promoter in VSELs, similar to EBs, is hypomethylated (28% and 13.2%, respectively). In contrast, Oct4 promoter was hypermethylated in adult HSCs (63.4%) and STs (60.4%), as expected.
Figure 1. The Oct4 promoter in VSELs is transcriptionally active.

(a) The strategy of isolation of VSELs (Lin-Sca-1+CD45-) and HSCs (Lin-Sca-1+CD45+) from murine BM. Note that the lymphocyte gate (R1) was extended to the left to include small sized SCs. (b) Expression of Oct4 mRNA in indicated cells. β-Actin was used as an endogenous housekeeping gene. Control reaction were performed without RTase (-) (c) Immunostaining of Oct4 (Red) and SSEA-1 (Green) protein in VSELs. Note that Oct4 was localized in the nucleus (DAPI blue staining). (d) CpG sites (open-circles) in Oct4 promoter and the target sites of primers (Oct4-S1 and Oct4-S2) employed for the ChIP assay. (e) Bisulfite-sequencing results of DNA methylation of the Oct4 promoter in indicated cells. Methylated and unmethylated CpG sites are shown in filled and open circles, respectively. The numbers under the bisulfite-sequencing profiles indicate the percentage of methylated CpG sites. Regular (f) and quantitative (g) ChIP analyses for the Oct4 promoter in indicated cells to evaluate its association with H3Ac and H3K9me2 histones. In regular ChIP analysis, the amplification of the β-Actin promoter was performed as a control reaction of the endogenous housekeeping gene. Rabbit IgG antibody was used as negative control of immunoprecipitation. In the quantitative ChIP assay, the enrichment of each histone modification was calculated as the ratio of the value from the bound fraction (B) to that from the unbound fraction (UB). Fold differences are shown as the mean ± S.D. from at least four independent experiments. *p<0.05, **p<0.01 compared to BMMNC. IgG; Rabbit IgG antibody
To provide additional direct evidence that the Oct4 promoter in VSELs is in an active/open state, we performed the chromatin-immunoprecipitation (ChIP) assay to evaluate its association with acetylated-histone3 (H3Ac) and dimethylated-lysine-9 of histone-3 (H3K9me2), the molecular features for open- and closed-type chromatin, respectively.28 To overcome the problem of low VSELs number, we performed the Carrier-ChIP assay using human hematopoietic cell-line THP-1 as carrier. As shown in Figure 1f, Oct4 promoter chromatin association with H3Ac was detected in both VSELs and ESC-D3, but not in primary HSCs, BM mononuclear cells (MNCs), and THP-1 cells, even in PCR reactions after employing high cycle numbers (Supplementary Figure 1). Furthermore, RQ-PCR analysis of the ChIP products revealed that the Oct4 promoter in VSELs was highly enriched for H3Ac similarly to ESC-D3 and EB (1-day) cells and its association with H3K9me2 was relatively very low (Figure 1g). Interestingly, in contrast to BM-derived MNCs, Oct4 promoter in HSCs showed a weak association with both H3Ac and H3K9me2. Since VSELs also express Nanog, we evaluated the epigenetic status of the Nanog promoter in these cells as well. We found that the Nanog promoter was methylated (∼50%) however, quantitative ChIP data confirmed that the H3Ac/H3K9me2 ratio supports the active status of the Nanog promoter in VSELs (Supplementary Figure 2).
Thus, this part of our experiments strongly supported our previous data that VSELs truly express Oct4 and Nanog.
The unique genomic imprinting patterns result in quiescent transcriptome of VSELs
As mentioned above, highly purified BM-derived VSELs do not proliferate in vitro if cultured alone. Based on the expression of PSCs markers and primitive morphology, we hypothesized that the quiescence of VSELs could be controlled in a manner similar to epiblast-derived PGCs by erasure/modification of methylation on some developmentally important imprinted-genes.15 To test whether VSELs similarly as PGCs undergo epigenetic reprogramming/modification of genomic imprinting, we first investigated the DNA methylation status on DMRs of paternally methylated imprinted-genes (Igf2-H19, Rasgrf1, and Meg3) (Figure 2a). The rationale was that paternally imprinted-genes are much less frequent22 and, more importantly, the proper mono-allelic imprint of the Igf2-H19 genes was crucial for obtaining viable parthenogenetic mice derived from a reconstructed oocyte containing two haploid sets of maternal genomes.29 We noticed that VSELs showed significant hypomethylation (∼10%) of the DMR for Igf2-H19 locus (Figure 2b). In contrast, this region was normally methylated (∼50%) in HSCs and STs and even slightly hypermethylated in ESC-D3 (Figure 2b). These bisulfite-sequencing results were subsequently confirmed by combined bisulfite-restriction analysis (COBRA) (Figure 2e). Next, we investigated the methylation status of DMRs for Rasgrf1 and Meg3 and found that VSELs in contrast to other cells erase imprinting on the DMR for Rasgrf1 (Figure 2c). However, the DMR for Meg3 is properly methylated (Figure 2d). These results indicate that VSELs erase the genomic imprinting on DMRs for paternally imprinted Igf2-H19 and Rasgrf1 loci similarly to PGCs,30 but not at the DMR for Meg3.
Figure 2. Erasure of genomic imprinting for paternally methylated imprinted-genes in VSELs.
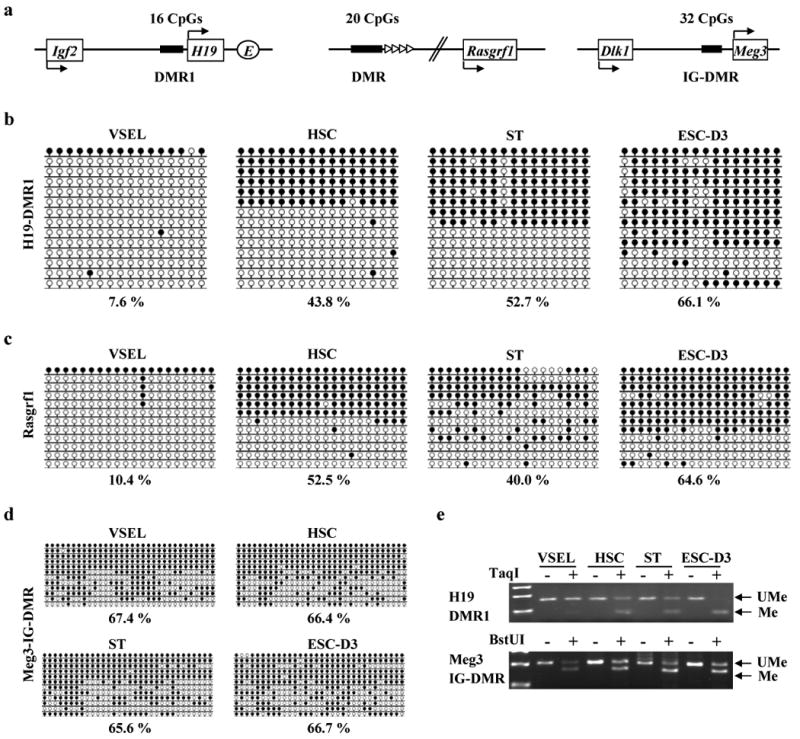
(a) The schematic diagram of DMRs for Igf2-H19, Rasgrf1, and Meg3 loci. Upper and bottom arrows represent the maternally and paternally initiated transcription of indicated genes respectively. E: Enhancer. Bisulfite-sequencing profiles of DNA methylation of DMRs for Igf2-H19 (b), Rasgrf1 (c), and Meg3 (d) loci. The percentage of methylated CpG sites was shown by employing bisulfite modification and sequencing results. Unlike DMRs of Igf2-H19 and Rasgrf1, there was little difference in DNA methylation for intergenic (IG)-DMR for Meg3 locus. (e) COBRA assay of Igf2-H19 DMR1 by TaqI restriction enzyme (upper panel) and IG-DMR for Meg3 locus by cleaved BstUI restriction enzyme (lower panel). The unmethylated DNA (UMe) was not cleaved in contrast to methylated DNA (Me) because of a sequence change in the site recognized by a restriction enzyme after bisulfite reaction.
Next, we examined DMRs for selected maternally methylated (Kcnq1, Igf2R) loci that are most well-studied for regulation of embryo growth (Figure 3a). We found that DMRs for both maternally imprinted loci, Kcnq1 (Figure 3b) and Igf2R (Figure 3c) were both hypermethylated in VSELs. At the same time, all these regions were normally methylated (∼50%) in adult HSCs and STs. Highly proliferative ESC-D3 cells showed opposite methylation patterns in DMRs for Kcnq1 (Figure 3b), Igf2-H19 (Figure 2b), and Rasgrf1 (Figure 2c) loci, compared with VSELs. These bisulfite-sequencing results were subsequently confirmed by COBRA assay (Figure 3d). When we investigated other maternally methylated (Peg1, SNRPN) genes, we found that the DMR for Peg1 was hypermethylated (Figure 3e), however the DMR for SNRPN was slightly hypomethylated (Figure 3f) in VSELs as compared to other cells.
Figure 3. VSELs show the hypermethylated status of DMRs in maternally methylated imprinted-genes.

(a) The schematic diagram of DMRs for Kcnq1 and Igf2R loci. Note that DMRs for Kcnq1 and Igf2R loci are located in promoter for antisense-transcripts, Lit1 and Air, respectively. Bisulfite-sequencing results of DNA methylation of DMRs for Kcnq1 (b), Igf2R (c), Peg1 (e), and SNRPN (f) loci. The percentage of methylated CpG sites was shown under the bisulfite-sequencing results. (d) COBRA assay of Igf2R DMR2 cleaved by TaqI restriction enzyme (upper panel) and KvDMR by cleaved BstUI restriction enzyme (lower panel). The UMe was not cleaved in contrast to Me because of a sequence change in the site recognized by a restriction enzyme after bisulfite reaction.
Thus, our DMR methylation results revealed a unique genomic imprinting pattern in VSELs showing the tendency of erasure in paternally methylated DMRs but hypermethylation of the maternally methylated DMRs. It is accepted that while paternally expressed imprinted-genes (Igf2, Rasgrf1) enhance the embryo growth, maternally expressed ones (H19, p57KIP2, Igf2R) inhibit cell proliferation.16 Therefore, the differences observed on VSELs demonstrate growth-repressive imprints in these cells.
To confirm our DMR methylation results, we performed RQ-PCR analysis of expression of imprinted-genes that should presumably be affected by the observed changes in genomic imprinting. As expected, according to chromatin insulator (CTCF) model,31,32 VSELs downregulate mRNA for Igf2 (Figure 4a) and Rasgrf1 (Figure 4b) while simultaneously highly upregulate H19 (Figure 4a). However, H19 and Igf2 were highly expressed in ESC-D3 cells (Figure 4a), the ratio of H19/Igf2 mRNA for VSELs and ESC-D3 was ∼400 vs. ∼1, respectively. This suggests that a proper balance in expression of both these genes may regulate proliferation of PSCs as seen in i) oocyte-derived parthenogenetic mice,29 ii) PGC-derived EGCs,33 iii) nuclear transfer clonotes,17 and iv) iPSCs.5 Rasgrf1 is one of Ras specific guanine nucleotide exchange factors (GEFs) that activate Ras protein and is highly upregulated in several human malignancies.34 Therefore, selective downregulation of this gene in VSELs is also intriguing and requires further study.
Figure 4. The unique genomic imprinting patterns in VSELs affect the expression level of imprinted-genes.
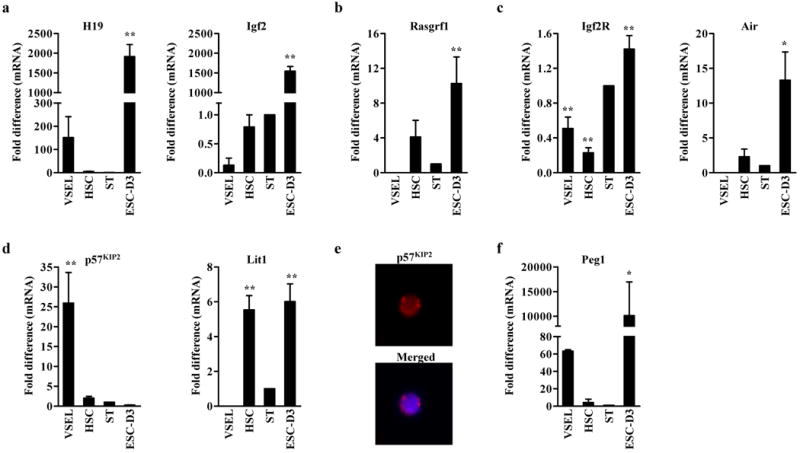
RQ-PCR analysis of Igf2-H19 (a) and Rasgrf1 (b), which DMRs were hypomethylated in VSELs, and maternally methylated imprinted-genes, Igf2R (c), p57KIP2 (d), and Peg1 (f). Of note, VSELs express little antisense-transcripts, Air (c) and Lit1 (d) for Igf2R and Kcnq1 loci, respectively. The relative expression level is represented as the fold difference to the value of STs and shown as the mean ± S.D. from at least three independent experiments on different samples of double-sorted VSELs, HSCs, STs, and ESC-D3. *p<0.05, **p<0.01 compared to ST. (e) The immunostaining of p57KIP2 in VSELs. Note that p57KIP2 protein was localized in the nucleus (merged).
In contrast to CTCF-regulated genes (Igf2-H19, Rasgrf1), different mechanism regulates expression of Igf2R and Kcnq1.20 DMRs for these loci are located in promoters of antisense-transcripts (Air and Lit1 respectively) that coordinately repress expression of clustered imprinted-genes (Figure 3a). As expected to hypermethylation of DMRs for Igf2R and Kcnq1 (Figure 3), VSELs downregulate expression of Air and Lit1 (Figure 4c and D). As result of this, VSELs highly express Ig2R (Figure 4c) and, more importantly, highly upregulate p57KIP2, a known negative regulator of the cell cycle (Figure 4d and e). In addition to p57KIP2, we also evaluated other cyclin-dependent kinases (Cdks) or inhibitors (CDKIs). We found high expression of p21Cip1, but no significant differences in the expression level of Cdks2, 4, and 6, which suggest that CDKIs could play more important roles in VSELs quiescence than Cdks (Supplementary Figure 3).
Some of the DMRs of imprinted genes are located directly in their promoters (e.g., Peg1). Although the Peg1 DMR was hypermethylated in VSELs, these cells highly express mRNA for this gene (Figure 4f) similarly to ESC-D3 cells. The high level of Peg1 expression in VSELs, similar to that observed in embryonic cells,35 further supports their embryonic-like character.
Based on our data, all the epigenetic reprogramming of genomic imprinting in VSELs results in quiescent transcriptome by upregulation of growth-repressive genes (H19, p57KIP2, Igf2R) and downregulation of growth-promoting genes (Igf2, Rasgrf1). Therefore, these unique changes in methylation patterns reveal a potentially novel mechanism involved in regulating the pluripotency of early developmental stem cells deposited in adult tissues.
VSELs highly express Dnmts
Since the methylation status of imprinted genes and their expression is regulated by Dnmts (Dnmt1, Dnmt3b, Dnmt3a, Dnmt3L), we focused on expression of these genes in VSELs. Dnmt1 is believed to be responsible for the maintenance of DNA methylation and Dnmt3a and 3b are accountable for de novo DNA methylation.36 Furthermore, Dnmt3L, a gene that shares homology with the Dnmt3 family methyltransferases despite the lack of enzymatic activity, is also essential for DNA methylation of imprinted genes.37 As reported, undifferentiated ESCs highly express Dnmts and Dnmt-deficient ESCs die during differentiation, indicating that Dnmts perform essential roles in maintaining pluripotency and differentiation of ESCs.36 We noticed that VSELs highly express all Dnmts similarly to ESCs and, in particular, are highly enriched for mRNA for Dnmt3L (Figure 5a). The intranuclear expression of Dnmt1 and Dnmt3b in VSELs was additionally confirmed by immunostaining (Figure 5b). Because the expression of de novo Dnmts and Dnmt3L is low in differentiated somatic cells, high expression of these Dnmts in VSELs suggests their high epigenetic plasticity, similarly as observed in ESCs.
Figure 5. VSELs express a high level of Dnmts.

(a) RQ-PCR analysis of Dnmt1, 3b, 3a, and related protein Dnmt3L. The relative expression level was represented as the fold difference to the value of STs and shown as the mean ± S.D. from at least three independent experiments performed on double-sorted VSELs, HSCs, STs, and ESC-D3 cells. *p<0.05, **p<0.01 compared to ST. (b) The immunostaining of Dnmt1 and Dnmt3b proteins in VSELs. DAPI staining was performed to visualize the nucleus and the image merged with DAPI (merged) is shown in right panel. Note that both Dnmts were located in the nucleus (merged).
The reprogramming of genomic imprinting during VSEL-DSs formation
Finally, although purified VSELs remain quiescent if cultured alone in vitro, they grow VSEL-DSs in co-cultures with C2C12 cells as previously reported.10 This suggests that their quiescent status can be modulated by epigenetic reprogramming in VSELs after cell-to-cell contact with the C2C12 supportive cell line. To address this better, we analyzed the DNA methylation of the Oct4 promoter and selected imprinted-genes in cells isolated from VSEL-DSs at days 5, 7, and 11 (Figure 6a). We observed both gradual hypermethylation of the Oct4 promoter and occurrence of somatic methylation-pattern on DMRs for Igf2-H19, Rasgrf1, Igf2R, Kcnq1, and Peg1 (Figure 6b and Supplementary Figure 4). These results summarized in Figure 6b suggest that growth-repressive genomic imprinting in VSELs is gradually restored in stem cells that form VSEL-DS, which suggests that VSELs may show dynamic epigenetic plasticity potential similarly to ESCs. However, restoration of genomic imprints in cells isolated from VSEL-DSs is paralleled by hypermethylation of the Oct4 promoter (Figure 6b), which suggests that cells present in these spheres gradually lose their pluripotency. Therefore, these results demonstrate that the DNA methylation of Oct4 promoter and DMRs in certain imprinted-genes both orchestrate the pluripotent and quiescent status of VSELs (Figure 6c).
Figure 6. The repressive genomic imprinting is recovered during VSEL-DS formation.

(a) Experimental strategy to isolate cells from VSEL-DSs. VSELs freshly isolated from the BM of GFP transgenic mice (GFP-Tg) were used to grow VSEL-DSs. GFP+ cells were sorted from the cultures. (b) Summary for bisulfite-sequencing results (Supplementary Figure 4) of DNA methylation in the imprinted-genes DMRs and the Oct4 promoter in freshly isolated VSELs and VSEL-DSs (5, 7, 11 days). The paternally imprinted DMRs (H19, Rasgrf1) were marked as blue lines and the maternally imprinted DMRs (Igf2R, KvDMR, Peg1) were marked as red lines. The dashed red line indicates the normal methylation status (50%). (c) Proposed model for epigenetic reprogramming of VSELs deposited in adult tissues during development and their potential activation in response to tissue and organ injury. The proliferative potential of VSELs was indicated in blue (means quiescent status) or red (means proliferative status).
Discussion
Our present study for the first time provides molecular evidence at chromatin level that Oct4 gene is actively transcribed in VSELs isolated from adult BM. In addition, our methylation studies of the Oct4 promoter and crucial imprinted-genes reveal novel mechanisms that may prevent “unleashed” proliferation of developmentally early stem cells deposited in adult tissues. VSELs show some similarities in methylation pattern to PGCs, which suggests their close relationship to epiblast/germ line cells (Figure 6c).
Some recent reports cast some doubts if Oct4 could be truly expressed in cells isolated from adult tissues and prompt us to reappraise expression of Oct4 in VSELs. To rule out an argument that Oct4 expression could be result of misinterpretation of RT-PCR and immunostaining results we employed i) Oct4 specific primers that do not amplify pseudogenes, ii) removed during RNA isolation contaminating genomic DNA by deoxyribonuclease I (DNase I) treatment, iii) confirmed the PCR products by DNA sequencing, and iv) show the intranuclear localization of Oct4 protein. More importantly we performed epigenetic analysis (DNA methylation and histone modifications) of Oct4 promoter (Figure 1). Our data provide strong molecular evidence at chromatin level that Oct4 gene is truly transcribed in VSELs isolated from adult BM. Similarly, by performing similar approach we also provided evidence for an open status of Nanog promoter.
Furthermore, in the present study, we confirmed our hypothesis that VSELs show some unique genomic imprinting patterns that regulate their quiescent status. The role of status of genomic imprinting in regulation of pluripotentiality was described for another epiblast-derived stem cell population, PGCs. These germ-line committed stem cells gradually reprogram/erase their genomic imprinting during migration to genital ridges (8.5-12.5 dpc).30 Erasure of genomic imprint in PGCs result that these pluripotent cells i) are quiescent, ii) do not complete blastocyst development and iii) PGCs-derived nuclei, in contrast to any other somatic cell nuclei are ineffective as DNA donors for nuclear transfer.17 We noticed that in VSELs (Figure 2 and 3), paternally methylated DMRs (Igf-2-H19 and Rasgrf1) similarly as in PGCs are hypomethylated, in contrast to maternally methylated ones (Kcnq1, Igf2R, and Peg1) that remain hypermethylated in VSELs.
The methylation status of DMRs is regulated by Dnmts. While PGCs express Dnmt1, both Dnmt3a and Dnmt3L are not expressed in these cells.30,38. Furthermore, VSELs in contrast to PGCs highly express all types of Dnmts (Figure 5), particularly Dnmt3L that is important for establishing maternal methylated imprints.37 Thus, the high expression of DNA methylation machinery in VSELs could explain hypermethylation status of maternally imprinted DMRs. However, despite of potential high capacity of DNA methylation in these cells, we noticed that paternally methylated DMRs for Igf2-H19, and Rasgrf1 loci were erased. To explain this striking discrepancy, we found that VSELs highly express chromatin insulator CTCF gene that prevents methylation of paternally imprinted DMRs (data not shown). Therefore, the high CTCF expression could protect the methylation of paternally methylated DMRs in these cells. This notion could be further supported by our additional observation showing that VSELs show a slight hypomethylation in DMR for SNRPN, which is also regulated by CTCF.31 Based on this we postulate that the balance between Dnmts and CTCF expression influences a final outcome of DMRs methylation patterns in VSELs.
It is well known that imprinted-genes play crucial roles in fetal growth, development, pluripotency of stem cell, and tumorigenesis. As result of unique reprogramming of genomic imprinting VSELs show upregulation of growth-repressive imprinted-genes (H19, p57KIP2, Igf2R) and downregulation of growth-promoting genes (Igf2, Rasgrf1) (Figure 4). Since Igf2 has been described as an important autocrine growth-factor that promotes expansion of several cell types39 and, on other hand H19 regulatory mRNA was found to inhibit cell proliferation40, the changes in expression of both these genes are probably responsible for a quiescent status of VSELs. Importantly, the significance of a proper mono-allelic imprint of Igf2-H19 gene was demonstrated to be crucial in generation of viable parthenogenetic mice using two haploid sets of maternal genomes.29 In addition, since non-signaling receptor Igf2R functions as decoy receptor for Igf2, that prevents its availability for Igf1R,41 upregulation of Igf2R in VSELs may additionally protect VSELs from autocrine/paracrine stimulation by Igf2. Another important gene that is downregulated by changes in DMRs methylation is Rasgrf1, protein involved in Igf1R signaling transduction.42 Thus, our preliminary data suggest that VSELs show some changes in the expression of genes that are related to Igf signaling machinery. The elucidation of a potential role of these factors in quiescence/activation of VSELs, however requires further more complex studies.
Furthermore, we noticed that VSELs highly express p57KIP2 transcript as result of hypermethylation of DMR in Kcnq1 locus. Since the specific upregulation of p57KIP2 is responsible for transforming growth factor β (TGFβ)-induced quiescence of primitive human HSCs43 and murine BM side population (SP) cells,44 these results indicate that p57KIP2 could also play a significant role in maintaining VSELs quiescence. More importantly our data also demonstrated that all these changes in genomic imprinting that affect pluripotent and quiescent status of VSELs become reverted when these cells expand/differentiate into VSEL-DSs.
In conclusion, herein we provide molecular evidence that some rare Oct4+ VSELs are present in adult tissues. We hypothesize, that these cells could serve as a backup for tissue committed monopotent SCs and that their proliferative potential is tightly regulated by both i) Oct4 expression and the ii) methylation status of some imprinted-genes that are directly involved in regulation of cell proliferation (e.g., Igf2, H19, Igf2R, p57KIP2, Rasgrf1). Therefore, potential modulation of mechanisms that control genomic imprinting in VSELs would be crucial for developing more powerful strategies to unleash the regenerative potential of these cells to employ them efficiently in clinic.
Supplementary Material
Acknowledgments
We thank Chris Worth, Rui Liu, and Izabela Klich for technical support; Dr. Hal Broxmeyer, Dr. Jan Nolta, Dr. Miodrag Stojkovic, and Dr. Mervin Yoder for critical comments. This work was supported by NIH P20RR018733 from the National Center for Research Resources to MK and NIH R01 CA106281-01, NIH R01 DK074720, and Stella and Henry Endowment to MZR.
This study was supported by NIH P20RR018733 from the National Center for Research Resources to MK and NIH R01 CA106281-01, NIH R01 DK074720, and Stella and Henry Endowment to MZR
Footnotes
Conflicts of Interest Statement: The authors declare that they have no competing financial interests.
Supplementary Information: The Supplementary Information includes four figures, two tables and Supplementary References.
The Supplementary Information is available at the Leukemia website (http://www.nature.com/leu)
References
- 1.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 2.Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, et al. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- 3.Matsui Y, Zsebo K, Hogan BLM. Derivation of pluripotential embryonic stem cells from murine primordial germ cells in culture. Cell. 1992;70:841–847. doi: 10.1016/0092-8674(92)90317-6. [DOI] [PubMed] [Google Scholar]
- 4.Takahashi K, Yamanaka S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 5.Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 6.Jiang Y, Jahagirdar BN, Reinhardt RL, Schwartz RE, Keene CD, Ortiz-Gonzalez XR, et al. Pluripotency of mesenchymal stem cells derived from adult marrow. Nature. 2002;418:41–49. doi: 10.1038/nature00870. [DOI] [PubMed] [Google Scholar]
- 7.D'Ippolito G, Diabira S, Howard GA, Menei P, Roos BA, Schiller PC. Marrow-isolated adult multilineage inducible (MIAMI) cells, a unique population of postnatal young and old human cells with extensive expansion and differentiation potential. J Cell Sci. 2004;117:2971–2981. doi: 10.1242/jcs.01103. [DOI] [PubMed] [Google Scholar]
- 8.Liedtke S, Enczmann Jg, Waclawczyk S, Wernet P, Kogler G. Oct4 and Its Pseudogenes Confuse Stem Cell Research. Cell Stem Cell. 2007;1:364–366. doi: 10.1016/j.stem.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 9.Lengner CJ, Camargo FD, Hochedlinger K, Welstead GG, Zaidi S, Gokhale S, et al. Oct4 Expression Is Not Required for Mouse Somatic Stem Cell Self-Renewal. Cell Stem Cell. 2007;1:403–415. doi: 10.1016/j.stem.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kucia M, Reca R, Campbell FR, Zuba-Surma E, Majka M, Ratajczak J, et al. A population of very small embryonic-like (VSEL) CXCR4+SSEA-1+Oct-4+ stem cells identified in adult bone marrow. Leukemia. 2006;20:857–869. doi: 10.1038/sj.leu.2404171. [DOI] [PubMed] [Google Scholar]
- 11.Kucia M, Halasa M, Wysoczynski M, Baskiewicz-Masiuk M, Moldenhawer S, Zuba-Surma E, et al. Morphological and molecular characterization of novel population of CXCR4+ SSEA-4+ Oct-4+ very small embryonic-like cells purified from human cord blood - preliminary report. Leukemia. 2006;21:297–303. doi: 10.1038/sj.leu.2404470. [DOI] [PubMed] [Google Scholar]
- 12.Kucia M, Zhang YP, Reca R, Wysoczynski M, Machalinski B, Majka M, et al. Cells enriched in markers of neural tissue-committed stem cells reside in the bone marrow and are mobilized into the peripheral blood following stroke. Leukemia. 2005;20:18–28. doi: 10.1038/sj.leu.2404011. [DOI] [PubMed] [Google Scholar]
- 13.Wojakowski W, Tendera M, Kucia M, Zuba-Surma E, Paczkowska E, Ciosek J, et al. Mobilization of Bone Marrow-Derived Oct-4+ SSEA-4+ Very Small Embryonic-Like Stem Cells in Patients With Acute Myocardial Infarction. J Am Coll Cardio. 2009;53:1–9. doi: 10.1016/j.jacc.2008.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paczkowska E, Kucia M, Koziarska D, Halasa M, Safranow K, Masiuk M, et al. Clinical Evidence That Very Small Embryonic-Like Stem Cells Are Mobilized Into Peripheral Blood in Patients After Stroke. Stroke. 2009;40:1237–1244. doi: 10.1161/STROKEAHA.108.535062. [DOI] [PubMed] [Google Scholar]
- 15.Ratajczak MZ, Machalinski B, Wojakowski W, Ratajczak J, Kucia M. A hypothesis for an embryonic origin of pluripotent Oct-4+ stem cells in adult bone marrow and other tissues. Leukemia. 2007;21:860–867. doi: 10.1038/sj.leu.2404630. [DOI] [PubMed] [Google Scholar]
- 16.Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat Rev Genet. 2001;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- 17.Yamazaki Y, Mann MR, Lee SS, Marh J, McCarrey JR, Yanagimachi R, et al. Reprogramming of primordial germ cells begins before migration into the genital ridge, making these cells inadequate donors for reproductive cloning. Proc Natl Acad Sci U S A. 2003;100:12207–12212. doi: 10.1073/pnas.2035119100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horii T, Kimura M, Morita S, Nagao Y, Hatada I. Loss of Genomic Imprinting in Mouse Parthenogenetic Embryonic Stem Cells. Stem Cells. 2008;26:79–88. doi: 10.1634/stemcells.2006-0635. [DOI] [PubMed] [Google Scholar]
- 19.Pannetier Ml, Feil R. Epigenetic stability of embryonic stem cells and developmental potential. Trends in Biotechnology. 2007;25:556–562. doi: 10.1016/j.tibtech.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 20.Delaval K, Feil R. Epigenetic regulation of mammalian genomic imprinting. Curr Opin Genet Dev. 2004;14:188–195. doi: 10.1016/j.gde.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 21.Lopes S, Lewis A, Hajkova P, Dean W, Oswald J, Forne T, et al. Epigenetic modifications in an imprinting cluster are controlled by a hierarchy of DMRs suggesting long-range chromatin interactions. Hum Mol Genet. 2003;12:295–305. doi: 10.1093/hmg/ddg022. [DOI] [PubMed] [Google Scholar]
- 22.Kobayashi H, Suda C, Abe T, Kohara Y, Ikemura T, Sasaki H. Bisulfite sequencing and dinucleotide content analysis of 15 imprinted mouse differentially methylated regions (DMRs): paternally methylated DMRs contain less CpGs than maternally methylated DMRs. Cytogenet Genome Res. 2006;113:130–137. doi: 10.1159/000090824. [DOI] [PubMed] [Google Scholar]
- 23.Mager J, Montgomery ND, de Villena FPM, Magnuson T. Genome imprinting regulated by the mouse Polycomb group protein Eed. Nat Genet. 2003;33:502–507. doi: 10.1038/ng1125. [DOI] [PubMed] [Google Scholar]
- 24.Fournier C, Goto Y, Ballestar E, Delaval K, Hever AM, Esteller M, et al. Allele-specific histone lysine methylation marks regulatory regions at imprinted mouse genes. EMBO J. 2002;21:6560–6570. doi: 10.1093/emboj/cdf655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Neill LP, VerMilyea MD, Turner BM. Epigenetic characterization of the early embryo with a chromatin immunoprecipitation protocol applicable to small cell populations. Nat Genet. 2006;38:835–841. doi: 10.1038/ng1820. [DOI] [PubMed] [Google Scholar]
- 26.Carr IM, Valleley EMA, Cordery SF, Markham AF, Bonthron DT. Sequence analysis and editing for bisulphite genomic sequencing projects. Nucl Acids Res. 2007 May 11;35(10):e79. doi: 10.1093/nar/gkm330. 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feldman N, Gerson A, Fang J, Li E, Zhang Y, Shinkai Y, et al. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat Cell Biol. 2006;8:188–194. doi: 10.1038/ncb1353. [DOI] [PubMed] [Google Scholar]
- 28.Margueron R, Trojer P, Reinberg D. The key to development: interpreting the histone code? Curr Opin Genet Dev. 2005;15:163–176. doi: 10.1016/j.gde.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 29.Kono T, Obata Y, Wu Q, Niwa K, Ono Y, Yamamoto Y, et al. Birth of parthenogenetic mice that can develop to adulthood. Nature. 2004;428:860–864. doi: 10.1038/nature02402. [DOI] [PubMed] [Google Scholar]
- 30.Hajkova P, Erhardt S, Lane N, Haaf T, El-Maarri O, Reik W, et al. Epigenetic reprogramming in mouse primordial germ cells. Mech Dev. 2002;117:15–23. doi: 10.1016/s0925-4773(02)00181-8. [DOI] [PubMed] [Google Scholar]
- 31.Pant V, Mariano P, Kanduri C, Mattsson A, Lobanenkov V, Heuchel R, et al. The nucleotides responsible for the direct physical contact between the chromatin insulator protein CTCF and the H19 imprinting control region manifest parent of origin-specific long-distance insulation and methylation-free domains. Genes Dev. 2003;17:586–590. doi: 10.1101/gad.254903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoon B, Herman H, Hu B, Park YJ, Lindroth A, Bell A, et al. Rasgrf1 Imprinting Is Regulated by a CTCF-Dependent Methylation-Sensitive Enhancer Blocker. Mol Cell Biol. 2005;25:11184–11190. doi: 10.1128/MCB.25.24.11184-11190.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shovlin TC, Durcova-Hills G, Surani A, McLaren A. Heterogeneity in imprinted methylation patterns of pluripotent embryonic germ cells derived from pre-migratory mouse germ cells. Dev Biol. 2008;313:674–681. doi: 10.1016/j.ydbio.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 34.Zhu TN, He HJ, Kole S, D'Souza T, Agarwal R, Morin PJ, et al. Filamin A-mediated Down-regulation of the Exchange Factor Ras-GRF1 Correlates with Decreased Matrix Metalloproteinase-9 Expression in Human Melanoma Cells. J Biol Chem. 2007;282:14816–14826. doi: 10.1074/jbc.M611430200. [DOI] [PubMed] [Google Scholar]
- 35.Lefebvre L, Viville S, Barton SC, Ishino F, Keverne EB, Surani MA. Abnormal maternal behaviour and growth retardation associated with loss of the imprinted gene Mest. Nat Genet. 1998;20:163–169. doi: 10.1038/2464. [DOI] [PubMed] [Google Scholar]
- 36.Chen T, Ueda Y, Dodge JE, Wang Z, Li E. Establishment and Maintenance of Genomic Methylation Patterns in Mouse Embryonic Stem Cells by Dnmt3a and Dnmt3b. Mol Cell Biol. 2003;23:5594–5605. doi: 10.1128/MCB.23.16.5594-5605.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bourc'his D, Xu GL, Lin CS, Bollman B, Bestor TH. Dnmt3L and the Establishment of Maternal Genomic Imprints. Science. 2001;294:2536–2539. doi: 10.1126/science.1065848. [DOI] [PubMed] [Google Scholar]
- 38.Durcova-Hills G, Tang F, Doody G, Tooze R, Surani MA. Reprogramming Primordial Germ Cells into Pluripotent Stem Cells. PLoS ONE. 2008;3:e3531. doi: 10.1371/journal.pone.0003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eggenschwiler J, Ludwig T, Fisher P, Leighton PA, Tilghman SM, Efstratiadis A. Mouse mutant embryos overexpressing IGF-II exhibit phenotypic features of the Beckwith-Wiedemann and Simpson-Golabi-Behmel syndromes. Genes Dev. 1997;11:3128–3142. doi: 10.1101/gad.11.23.3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hao Y, Crenshaw T, Moulton T, Newcomb E, Tycko B. Tumour-suppressor activity of H19 RNA. Nature. 1993;365:764–767. doi: 10.1038/365764a0. [DOI] [PubMed] [Google Scholar]
- 41.Ludwig T, Eggenschwiler J, Fisher P, D'Ercole AJ, Davenport ML, Efstratiadis A. Mouse Mutants Lacking the Type 2 IGF Receptor (IGF2R) Are Rescued from Perinatal Lethality inIgf2 and Igf1r Null Backgrounds. Dev Biol. 1996;177:517–535. doi: 10.1006/dbio.1996.0182. [DOI] [PubMed] [Google Scholar]
- 42.Font de Mora J, Esteban LM, Burks DJ, Nunez A, Garces C, Garcia-Barrado MJ, et al. Ras-GRF1 signaling is required for normal beta-cell development and glucose homeostasis. EMBO J. 2003;22:3039–3049. doi: 10.1093/emboj/cdg280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scandura JM, Boccuni P, Massague J, Nimer SD. Transforming growth factor β-induced cell cycle arrest of human hematopoietic cells requires p57KIP2 upregulation. Proc Natl Acad Sci U S A. 2004;101:15231–15236. doi: 10.1073/pnas.0406771101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Umemoto T, Yamato M, Nishida K, Yang J, Tano Y, Okano T. p57Kip2 is expressed in quiescent mouse bone marrow side population cells. Biochem Biophys Res Commun. 2005;337:14–21. doi: 10.1016/j.bbrc.2005.09.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


