Abstract
Background
Transfer RNA (tRNA) gene predictions are complicated by challenges such as structural variation, limited sequence conservation and the presence of highly reiterated short interspersed sequences (SINEs) that originally derived from tRNA genes or tRNA-like transcription units. Annotation of “tRNA genes” in sequenced genomes generally have not been accompanied by experimental verification of the expression status of predicted sequences.
Results
To address this for mouse tRNA genes, we have employed two programs, tRNAScan-SE and ARAGORN, to predict the tRNA genes in the nuclear genome, resulting in diverse but overlapping predicted gene sets. From these, we removed known SINE repeats and sorted the genes into predicted families and single-copy genes. In particular, four families of intron-containing tRNA genes were predicted for the first time in mouse, with introns in positions and structures similar to the well characterized intron-containing tRNA genes in yeast. We verified the expression of the predicted tRNA genes by microarray analysis. We then confirmed the expression of appropriately sized RNA for the four intron-containing tRNA gene families, as well as the other 30 tRNA gene families creating an index of expression-verified mouse tRNAs.
Conclusions
These confirmed tRNA genes represent all anticodons and all known mammalian tRNA structural groups, as well as a variety of predicted “rogue” tRNA genes within families with altered anticodon identities.
Keywords: mammalian tRNA, SINE, tRNA intron, tRNA families, nuclear tRNA, rogue tRNAs
Transfer RNAs (tRNAs) are essential molecules responsible for decoding messenger RNAs (mRNAs) by delivering the proper amino acid into a growing peptide chain at the ribosome. Since a tRNA is required for each amino acid incorporated into every protein, tRNAs are one of the most abundant molecules in all living organisms. In order to make the large quantities of tRNAs needed, tRNA genes are replicated throughout the eukaryotic genomes through mechanisms such as retrotransposition. In some cases multiple copies of certain tRNA genes have been shown to be essential for a normal growth rate.1 The tRNAs are duplicated by creating cDNA copies of the primary transcript, which include the internal promoter sequences, and the copies then re-insert at distant locations in the genome.2,3 Thus, the duplicated tRNA genes in yeast retain limited (<20 base pairs) conservation of upstream and downstream flanking sequences, as well as their intron sequences, when present. Introns are found in tRNAs in bacteria, archaea and eukarya, although the structure of the intron and the splicing process is specific to the domain of life.4 In yeast, where the expression of individual gene copies has been verified, the tRNA genes' flanking sequences and introns are not as tightly conserved as the mature coding regions, consistent with greater selection pressure for retaining the mature domains of the tRNAs intact.
The expression of tRNAs has only been thoroughly studied in a few bacterial, archaeal and eukaryotic species. In eukaryotes, the synthesis, processing and utilization of tRNAs has been most extensively studied in the budding yeast, Saccharomyces cerevisiae. These studies have yielded important information on tRNA synthesis by RNA polymerase III (pol III), post-transcriptional modifications, RNA transport and gene organization, but there is a general lack of information on tRNA genes in mammalian genomes. For example, in mouse there are only 11 tRNA sequences verified as expressed in the tRNA database.2 Although there is a database of tRNA genes predicted by tRNAscan-SE available, so far there has been little confirmation of the predictive power of tRNA scanning programs in the mammalian genome.3 This is an especially difficult analysis in most vertebrate genomes since they contain many tRNA-derived short interspersed elements, or SINEs.5 Recent work from the Pan lab using microarray analysis confirmed the expression of expressed tRNAs that correspond to 374 predicted human tRNA genes, as identified by tRNAscan-SE.6,7
In an effort to comprehensively identify mouse tRNAs, we used two tRNA scanning programs, the commonly used tRNAscan-SE and more recently developed ARAGORN, to predict the probable tRNA genes in the mouse genome.3,8,9 tRNAscan-SE employs both heuristic algorithms and covariance models and is used extensively as the definitive tRNA gene identification program. Initial tRNA gene candidates are first identified by either tRNAscan, identifying the A and B box promoters and cloverleaf structure, or the Pavesi algorithm, identifying tRNAs on promoter and terminator sequences independent of a predicted secondary structure.10,11 tRNAscan-SE feeds the initial predictions into a third program that ranks the prediction based on a covariance model.12 ARAGORN identifies candidate tRNA genes with a heuristic algorithm exclusively, identifying portions of the B box sequence and then attempting to construct a cloverleaf with the neighboring sequences. In each case, the features common to all tRNAs are the characteristic “cloverleaf” secondary structures and very limited patches of sequence conservation, termed the A box and B box, used as common recognition elements in both the transcription of the genes and structural and recognition elements in the tRNA transcripts.
The predictions from scanning the Mus musculus genome with tRNAscan-SE and ARAGORN were strikingly different, although there was significant overlap.9 After combining the predictions and removing known SINE sequences, we sorted the genes into families and singly-occurring (“orphan”) genes based on sequence similarity, and we experimentally verified expression of the predicted gene families. Mouse RNA from embryos and several tissues was hybridized onto a custom microarray with oligonucleotide probes tiled against each of the tRNA gene families and orphan tRNA genes to test expression. We also confirmed the expression of the tRNA families by northern blot, especially focusing on whether pre-tRNAs the size of those predicted for intron-containing genes were expressed. The results show that all of the predicted families and several orphan tRNA genes are expressed in all tissues tested.
Results and Discussion
Scanning the mouse genome for predicted tRNA genes
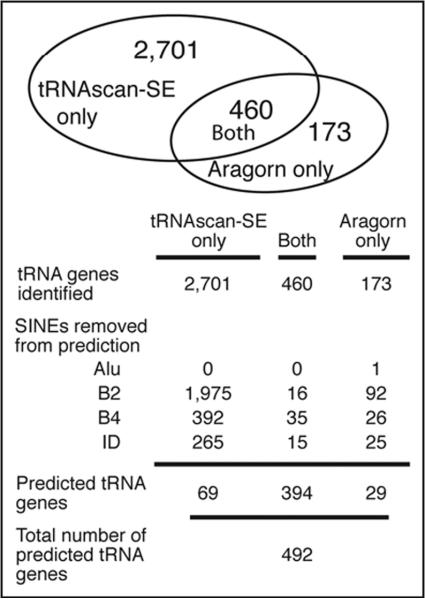
In an attempt to predict the functional tRNA genes in mouse, both tRNAscan-SE and ARAGORN were used to scan the May 2004 release of the Mus musculus genome (Fig. 1). tRNAscan-SE identified 3,161 putative tRNA genes and ARAGORN predicted 633 genes. Comparing the two sets, 460 of the putative tRNA genes identified by ARAGORN were also identified by tRNAscan-SE. Both programs identified all 11 of the verified mouse tRNA gene sequences and tRNA genes corresponding to all anticodons (Fig. 2). However, a large number of genes were predicted by only one of the programs, either tRNAscan-SE or ARAGORN, so we focused on removing potential artifacts from the set of tRNA gene predictions. The predicted tRNA genes were named in accordance with the tRNA naming convention established in yeast tX(anticodon)Y. X corresponds with the single character abbreviation for the encoded amino acid. Next, the anticodon is shown parenthetically. Y corresponds to the one letter designation of the chromosome where the gene is located (A for chromosome 1, B for chromosome 2, etc.), and a number follows if there are multiple tRNA genes with the same anticodon on an individual chromosome.
Figure 1.
Different tRNAs and SINEs are identified by both scanning programs. tRNAscan-SE and ARAGORN identify different sequences as tRNA genes. However, the majority of the genes identified by a single program are tRNA-derived SINE elements.
Figure 2.
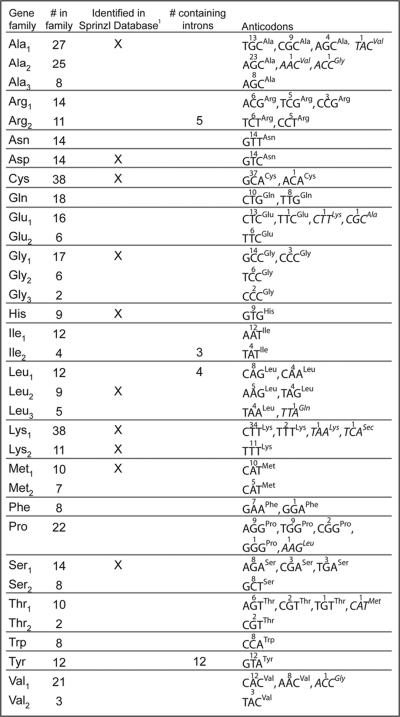
Mouse tRNA gene families. Mouse tRNA genes were separated into families based on sequence similarity. The number of genes within families and for any particular anticodon varies greatly. Genes that were previously identified in the Sprinzl database are marked, since their expression has been previously demonstrated. None of the intron-containing tRNA gene families were previously shown to be expressed.1 www.trna.uni-bayreuth.de/
Removing SINE elements from the predicted tRNA genes
One of the anticipated problems in this study was the abundance and variety of tRNA-derived SINE elements. tRNAscan-SE employs a scoring system to identify tRNA genes with predefined cut off levels to distinguish a `real gene' versus a pseudogene. tRNAscan-SE identified 22,027 sequences as pseudogenes, sequences that shared some features with tRNA genes but scored too low to be considered an actual tRNA gene. ARAGORN does not identify weak scoring sequences as pseudogenes. Examination of the pseudogene sequences identified by tRNAscan-SE revealed many to be similar to the B2 SINE consensus sequence. These pseudogenes were not considered in the rest of the analysis.
Given the high number of B2 SINEs identified as pseudo-tRNA genes by ARAGORN and to determine if any B2 SINEs were classified as tRNA genes, we used the B2 consensus sequence and ran a BLAST search against tRNA genes predicted by both tRNAscan-SE and ARAGORN.13 We used the entire B2 consensus sequence, both upstream and downstream of the tRNA-like domain, to avoid identifying the functional tRNA family from which B2 SINEs are derived. The tRNA genes identified by both predictive programs were the least likely to be similar to B2 SINEs, as only 16 sequences (3.5%) predicted by both tRNAscan-SE and ARAGORN were >70% similar to the B2 SINE consensus sequence. Ninety-two tRNA genes (53%) predicted only by ARAGORN and 1,975 tRNA genes (73%) predicted only by tRNAscan-SE were similar to the B2 SINE consensus sequence. The remaining predicted tRNA genes were compared to SINEs annotated on the UCSC Genome Browser by RepeatMasker, which identified B1, B3 and ID repeat elements (Fig. 1). Removing the SINEs from the predicted tRNA genes eliminated 97% of genes predicted by just tRNAscan-SE, 83% of genes predicted by just ARAGORN, and only 14% of the tRNA genes predicted by both programs. The high number of SINEs that were predicted by both programs indicates the close relationship between tRNAs and SINEs.
Assigning mouse tRNA genes into families
After removing the SINEs and merging the predicted tRNAs from tRNAscan-SE and ARAGORN, there are a total of 492 predicted tRNAs (Suppl. Fig. 3). These sequences were aligned using ClustalW and then manually sorted into families based on sequence similarity. The sequence of 440 predicted tRNA genes were highly similar to at least one other predicted tRNA gene. These were sorted into 34 tRNA families (Fig. 2) representing all 20 essential amino acid anticodons. Genes encoding the 11 previously verified mouse tRNAs from the tRNA database are all included in the tRNA families.2 The number of genes per family ranged from 2 to 38. In the Saccharomyces cerevisiae genome the number of copies of a single tRNA gene range from 1 to 16 copies. Most of these yeast tRNA gene copies have identical sequences in the mature tRNA coding regions, but this is not true with the predicted mouse genes; in fact, the sequence similarity between members of mouse tRNA families is strikingly less than the similarity between yeast tRNA family members. Most of the tRNA gene copies in the mouse genome have multiple nucleotides different among family members, even though the majority of the tRNA gene sequence is conserved (Suppl. Fig. 2). However, there must still be pressure to maintain sequence identity in these mouse tRNA genes, since the introns and flanking sequences of the intron-containing tRNA families diverge much more rapidly.
Over half (19) of the tRNA gene families contain genes with a single anticodon sequence (Fig. 2). The remaining 15 tRNA families contain genes with different anticodon sequences, eight of which include either one or two `rogue' tRNA genes that have an anticodon for a different amino acid than the majority of the family members (Suppl. Fig. 2—Ala1, Ala2, Glu1, Leu3, Lys1, Pro, Thr1, Val1). This was of concern, since “improper” charging of a tRNA relative to its anticodon would seem to be counter selected. We confirmed the sequence of several of these genes by amplifying the genomic DNA locus by PCR and directly sequencing the PCR products (data not shown).
Comparisons between mouse and human tRNA gene families
We examined the predicted tRNA genes identified by tRNAscan-SE in the human genome to see if rogue tRNA genes existed within human tRNA gene families. There are 36 tRNA families in the human genome, based on sequence similarity of the predicted human tRNA genes (Suppl. Fig. 4). Thirty-two of predicted human tRNA families have sequences similar to tRNA families in the mouse genome. The human Hs_Arg2, Hs_Leu3, Hs_Thr2 and Hs_Gln2 tRNA gene families do not have an identifiable ortholog in the mouse genome. Eight of the predicted human tRNA gene families contain at least one rogue tRNA gene (Hs_Ala1, Hs_Arg1, Hs_Arg2, Hs_Cys, Hs_Glu, Hs_Gly1, Hs_ Met2, Hs_Lys2), showing that the existence of rogue tRNAs is not unique to mouse. Neither the families that contains the rogue tRNA gene nor the rogue anticodons are conserved between the human and mouse genomes, indicating that the specific anticodon variants are not conserved in mammals.
Expression of predicted tRNAs
Custom microarrays were designed to test the expression of the predicted tRNA genes. RNA from mouse embryos and several different tissues were tested in case there was some substantially different pattern of expression for some families. Although most tRNAs are expressed constitutively, there are also examples in some eukaryotes where special tRNA families are transcribed in response to high demand for protein. In the silkworm Bombyx mori, an alanine tRNA is exclusively expressed in the silk gland and a glycine tRNA is overexpressed in the silk gland.14 This tissue-specific tRNA expression allows production of the glycine and alanine-rich silk protein, fibroin. In Xenopus, an entire set of highly reiterated tRNA genes are transcribed during oogenesis as part of a process to store large quantities of translational machinery for the upcoming high protein production in developing embryos.15 However, since the majority of tRNA studies have been performed on single cellular organisms, the possibility of tissue-specific or developmentally regulated tRNAs in mammals is largely unexplored. In humans, a subset of tRNAs have been examined for tissue specific expression of tRNA genes revealing modest variations in relative tRNA expression.7
Total RNA from different mouse developmental stages (7 day embryo, 10-12 day embryo) and different tissues (muscle, spleen, mammary gland, brain, ovary, thymus, liver, heart) was directly fluorescently labeled and used to probe the microarrays (see Methods). Probes unique to the predicted tRNA gene or gene family whose signal was greater than 99% of the negative controls were considered to be expressed. Using these criteria, all of the predicted tRNA gene families were detected as expressed by the microarray. However, because of the sequence similarity between family members we can only conclude that some subset of the tRNA gene copies are being expressed. While no families are exclusively expressed in a particular developmental stage or tissue, there is a five-fold greater expression of tRNAs in brain and ovary relative to muscle and liver. Overall tRNA expression ranges from Brain >Ovary >Heart >10-12 day embryo >Mammary Gland >Testis >Placenta >Thymus >7 day embryo >Spleen >Skeletal Muscle >Liver.
Each tRNA family was also confirmed by probing northern blots in a subset of mouse RNA samples to complement the microarray results, ensuring the signal detected was not due to cross hybridization of another type (size) RNA transcript. We were particularly interested in confirming for the first time the expression of intron-containing families in this work (see below), but confirmed the size and expression of all of the tRNA gene families in skeletal muscle, mammary gland, placenta and 7-day embryo (Suppl. Fig. 5) as well as the presence of the orthologous human tRNA family in RNA from HeLa cells. The northern blot results confirmed all of the tRNA gene families that were identified and found to be expressed by microarray analysis.
The microarray analysis identified 29 `orphan' tRNA genes are also expressed in the various RNA samples. Only a single copy of these `orphan' tRNA genes are found throughout the genome. One of the 29 orphan tRNAs, tSeC(TCA)G, is a known selenocysteine tRNA, Trsp.16 The microarray analysis detected the selenocysteine tRNA is expressed in all tissues types as well as in the 7 day and 10-12 day embryo, which is consistent with Trsp expression being essential for mouse embryogenesis.17 In addition to selenocysteine tRNA, which has two orthologs in the human genome, the tyrosine orphan tRNA, tY(GTA)B, has at least 20 orthologs (>90% sequence similarity) in the human genome. This tRNA gene was only identified by ARAGORN in the mouse genome and none of the human orthologs are identified by tRNAscan-SE. The predicted structure of tY(GTA)B is tRNA-like and the sequences are not tagged as a repetitive element by RepeatMasker in the mouse or human genomes. However, the sequences are missing a T in the conserved TTC sequence of the B box promoter and an oligo-T sequence (T5) is present in the aminoacyl acceptor stem of both the mouse and human copies of the gene, which could result in early pol III transcription termination. However, all of the probes on the microarray targeted to the mature region of tY(GTA)B gave signal well above background (data available on GEO GSE8224).
Intron-containing tRNA genes confirmed by northern blot
Introns are found in four previously uncharacterized mouse tRNA gene families: Arg3, Tyr, Leu1 and Ile2. Each of the orthologous families in the human tRNA set also contains introns. There is also a single intron containing tRNA gene in the 21-member human Pro family, which is not seen in the 22-member mouse Pro family. In S. cerevisiae there are introns in eight tRNA types: Phe, Ile, Lys, Leu, Pro, Ser, Trp and Tyr. The presence of introns in tyrosine, leucine and isoleucine tRNAs in organisms as divergent as yeast, mouse and humans might indicate that some tRNAs are more tolerant of introns than other tRNA types. Alternatively, the introns might be functional, such as the yeast tyrosine tRNA where the presence of the intron is required for proper folding of the tRNA.18
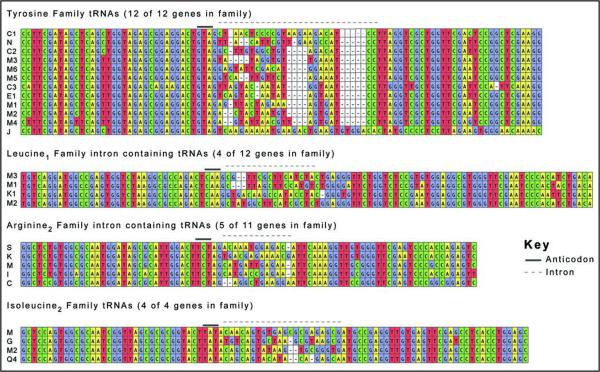
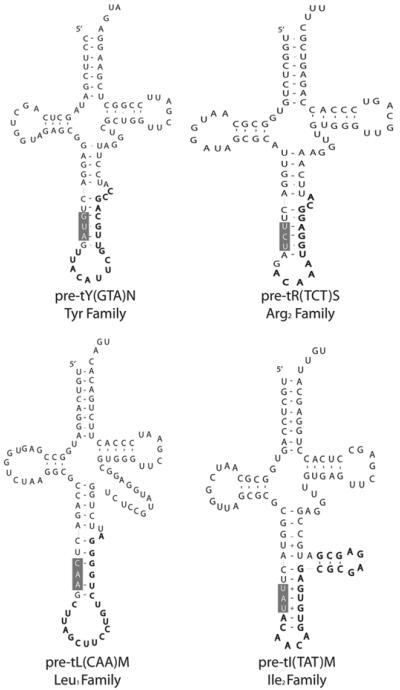
Expression of all intron-containing mouse families was confirmed by northern blot (Fig. 5). All of the genes in the mouse families Arg3, Ile2 and Tyr contain introns, while only four of the 12 tRNA genes in the Leu1 family contain introns (Fig. 2). There appears to be little selective pressure to maintain the sequences of the introns in duplicated genes relative to the greater conservation of mature tRNA sequence (Fig. 3), although the intron sequences are more conserved than the sequences immediately upstream or downstream of the mature domain, predicted to be in the pre-tRNA primary transcript. The predicted structures of the intron containing tRNA genes are consistent with the intron location and structure in yeast (Fig. 4).19 However, any new intron integration strategies would probably be excluded from the computer predictions.
Figure 5.
Northern blot confirmation of intron-containing tRNA genes. Northern blot analysis confirms the expression of intron-containing tRNA gene families in all mouse tissues tested. Separate panels for precursor tRNAs and mature tRNAs allow for sufficient contrast of the precursors, since precursor tRNAs exist at a small fraction of the mature levels relative to more rapidly growing organisms. Precursor sizes were consistent with the predicted intron sizes. It is not known whether the multiple bands in the mature tRNA are due to variations in “mature” domain length of family members, incomplete removal of amino acids from the 3' termini, or trimming of the 3' CCA residues. In Arg2, Ile2 and Leu1, families with both intron containing and non-intron containing tRNA genes, this method is unable to distinguish between the expression of the non-intron containing tRNA genes and matured intron containing tRNAs. While the expression levels vary between tissue types, the intron-containing tRNA gene families are ubiquitously expressed in all tissues tested.
Figure 3.
Sequence alignment of intron-containing tRNA genes. The four families of intron-containing genes are aligned. The solid line above the sequence indicates anticodon position and the introns are indicated with a dashed line. The letters and numbers (e.g., C1, N, C2 for Tyrosine family) correspond to the tRNA gene names in Supplemental Figure 3.
Figure 4.
Predicted structure of intron-containing tRNA genes. One representative structure is shown for each of the intron-containing tRNA gene families. The anticodon position and general helix-bulge-helix structure is consistent with yeast intron-containing tRNA structures.
There are also 10 intron-containing orphan tRNAs that were detected as expressed by the microarray analysis. Multiple probes unique to the predicted tRNA sequence gave signal above the 99% confidence level based on the negative control probes. However, the predicted intron containing phenylalanine tRNA, tF(GAA)O, has a 478 nt long intron and tA(GGC)O1 has a 2nt intron. Neither of these intron lengths are consistent with intron lengths or splicing mechanism in yeast (Suppl. Fig. 6) and there are no identifiable orthologs in the human genome. The intron in yeast isoleucine tRNA genes is 58 nt, but the remaining yeast introns are all between 13-32 nt long. The remaining intron-containing mouse orphans include: tA(AGC)Q4, tI(TAT)G, tI(TAT)M, tL(CAA)K, tP(AGG)P, tT(TGT)E1, tT(TGT)M2 and tV(TAC)G, the four genes that are predicted by only ARAGORN are indicated with italics (Suppl. Fig. 3).
Materials and Methods
Identifying potential tRNA genes
tRNA genes were predicted from the May 2004 release of the mouse genome using two publicly available computer programs: tRNAscan-SE and ARAGORN.3,8 The default settings were used with tRNAscan-SE, but ARAGORN was run with intron detection enabled. The resulting list of tRNA genes were merged based on genomic location. The tRNA genes were aligned by sequence similarity using Clustal X and then assigned to families based on high degrees of similarity.23 A Clustal X alignment of the 5'-end of the predicted tRNA genes revealed many (1,804) predicted tRNAs were similar to the tRNA region of B2 SINEs. Comparison of predicted mouse tRNAs with the SINEs predicted by RepeatMasker, as annotated on the UCSC Genome Browser, identified additional B2 SINEs as well as Alu, B4 and ID SINEs.24 tRNA genes that did not have at least one similar sequence were not assigned to a family and are designated `orphan' tRNA genes. The structure of intron containing tRNA gene sequences were predicted using Mfold and then refined by hand.25
Microarray design
Twenty probe sequences were allotted for each ncRNA prediction. Complementary DNA probes were designed to maximize spatial coverage of each predicted sequence while avoiding probes that have high self-folding potential as described previously (See 26) and were normalized by length (i.e., probe lengths were adjusted) to a uniform DNA-RNA melting temperature of 70°C. Probe sequences were on average 25.5 nt and were concatenated to 60 nucleotides. Probe sequences were submitted to Agilent Technologies for microarray production (Palo Alto, California). The designs included 1,000 60-mer probes of random sequence, which were used as negative controls, and 696 positive control probes tiled across U4 and U5 snRNAs and 18S and 28S rRNAs. The design is accessible at NCBIs Gene Expression Omnibus (GEO, www.ncbi.nlm.nih.gov/geo/) database under platform accession GPL5420.27
RNA extraction, labeling and hybridizations
Total RNAs from various mouse tissues were purchased from Ambion and Clontech. Integrity of rRNA was confirmed on 1% agarose-formaldehyde gels. 7 mg of total RNA was chemically labeled with Ulysis Alexa Fluor 546 or Ulysis Alexa Fluor 647 (Invitrogen) according to manufacturer's instructions. This protocol labels G residues, and there were no predicted RNAs that lacked G residues.28 Samples were resuspended in 0.5 mL of hybridization buffer (1 M NaCl, 0.5% sodium sarcosine, 50 mM N-morpholino ethane sulfonate, pH 6.5, 33% formamide and 40 mg salmon sperm DNA), denatured by heating at 65°C for 5 minutes, and snap-cooled on ice prior to hybridization. Hybridizations were carried out for 16-24 h at 42°C in a rotating hybridization oven. Slides were then washed (rocking ~30 seconds in 6x SSPE, 0.005% sarcosine, then rocking ~30 seconds in 0.06x SSPE) and scanned with a 4000A microarray scanner (Axon Instruments, Union City, CA).
Microarray data processing and normalization
TIFF images were quantified with GenePix 3.0 (Axon Instruments, Union City, CA). Individual channels were spatially detrended (i.e., overall correlations between spot intensity and position on the slide removed) by high-pass filtering using 5% outliers. The individual channels were then normalized using Variance Stabilization that allows for comparison across channels.29 All data is accessible at the GEO database under series accession GSE8224.27
Northern blots
Ten micrograms of RNA from each mouse tissue type was electrophoresed on an 8% polyacrlamide gel (SequaGel) and then electroblotted onto a Nytran SuperCharge membrane (Schleicher & Schuell Bioscience). Blots were probed with oligonucleotide probes to mature regions of the predicted tRNAs. In the case of genes predicted to have introns, intron probes were also used. Blots were reprobed for 5.8S ribosomal RNA (rRNA) as a loading control. The probe sequences can be found in online supplemental S_1. Blots were exposed to a phosphocapture screen, detected on a PhosphorImager (Molecular Dynamics 445 SI), and quantified with IPlab Gel software (Signal Analytics).
Conclusions
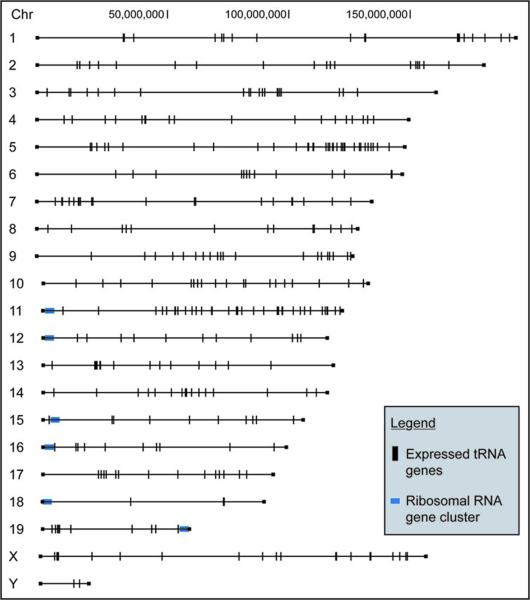
This work provides experimental confirmation of the predicted tRNAs in mouse, with comparisons to human genes that suggest conservation. It was necessary to first screen the predicted mouse tRNA genes for known SINE elements, since most of these highly repetitive sequences in most non-human vertebrates derive originally from tRNA genes. This eliminated 85% of the predictions by tRNAscan-SE and 33% from ARAGORN, although it should be noted that most of the ~900,000 tRNA-derived SINE elements are not identified by either program. Both of these programs are susceptible to identifying tRNA-derived SINE elements as functional tRNA genes, however overlapping predictions by both tRNAscan-SE and ARAGORN had the lowest likelihood of being SINEs. The predicted tRNA genes were then sorted into families based on sequence similarity. The sequence variations of mouse tRNA gene copies within families are much greater than in yeast or bacteria, but similar to human tRNA genes.6 tRNA genes were found on every chromosome (Fig. 6). This is in marked contrast to ribosomal genes, which are found on only a few chromosomes. However, there are clusters of genes of tRNA genes such as the 26 Cysteine family tRNA genes located within 400,000 nt on Chromosome 6.
Figure 6.
Map of tRNA gene locations in the mouse genome. Vertical lines indicate the location of verified tRNA genes throughout the mouse genome. The map illustrates the dispersed nature of tRNA genes with genes on every chromosome, compared with the ribosomal RNA gene clusters that are found on only a few chromosomes. However, there are instances of tRNA gene clustering, which appear as a thick line. One striking example of clustering is 26 of the 38 genes in the Cysteine family located within 400,000 nt on Chromosome 6 (Suppl. Fig. 3).
Analysis of the gene families identified several “rogue” tRNA genes, defined as having an anticodon for a different amino acid than the majority of the family members. In both mouse and human there are eight “rogue” tRNA genes. However, neither the anticodon nor the gene family of the rogue tRNA are conserved, the presence of rogue tRNAs in both the mouse and human genomes suggests the possibility that these tRNA genes are functional. These rogue tRNAs might facilitate anticodon variations, similar to the ambiguous intermediate hypothesis.20,21 The ambiguous intermediate hypothesis states that a particular codon can be made more free for the incorporation of alternative amino acids, such as selenocysteine, by weakening the fidelity between a particular codon/anticodon.
This is the first extensive examination of tRNAs that are found in mouse. Northern blot analysis of the 34 tRNA gene families confirms that tRNA-sized RNAs from all predicted families are expressed in mouse skeletal muscle, mammary gland, placenta and 7-day embryo. However, because of the sequence similarity between family members we can only conclude that some subset of the members of the tRNA gene families are being expressed. There are four intron-containing tRNA gene families that are expressed in mouse. The intron-containing tRNA families are conserved between mouse and humans and similar to the intron-containing tRNA types found in yeast. It is noteworthy that the intron-containing precursors can be arranged into structures similar to the intron precursor structures found in yeast.19 Since the structure is implicated in recognition by the tRNA splicing endonuclease, this conservation would be consistent with the conservation of the splicing machinery, SEN2 in yeast and tSEN2 in mouse.22 The conservation of intron structures would also be consistent with any functions contributed by the introns. For example, it has been suggested that introns in certain tRNAs provide an additional driver for folding of the precursors to allow greater latitude in the permitted mature domain sequences.
In addition to the 440 mouse tRNA genes found in families, there are 23 expressed orphan tRNAs, which were found as a single gene copy. One of the orphans is the well studied selenocysteine tRNA, TRSP, while another, tY(GTA)B, has 20 orthologs in the human genome (Suppl. Fig. 7). This tyrosine tRNA was only identified by ARAGORN and none of the eight orthologs in the human genome have been identified in the tRNAscan-SE database for the human genome. The sequence conservation and gene copy expansion in the human genome strongly argue that this is a functional RNA. Assignment of the remaining orphan tRNA genes, including the six with predicted non-canonical introns, remains tentative at this time.
All of the mouse tRNA gene families are also found in multiple copies in the human genome, however while the number of gene copies per gene family is often similar it can vary significantly. The asparagine tRNA family has 29 gene copies in the human genome compared with only 14 in mouse. However, it is not a general trend that humans have more tRNA gene copies than mouse, as mouse families Ala2 and Lys1 contain twice as many gene copies as in humans. The presence of introns in a family also varies, as the 26-member proline family in humans contains one member that has an intron but the 22-member mouse family does not. We detected appropriate sized transcripts for each of the tRNA gene families in both mouse and human RNA samples, which indicates at least some of the gene copies are active in both organisms. The rogue tRNA genes and orphan tRNA genes in both mouse and human genomes require further study to determine whether they are active and producing functional tRNAs.
Supplementary Material
Acknowledgements
We thank Paul Good for his contributions to Supplemental Figure 5, Tom Glover for providing mouse DNA, and Todd Lowe for feedback on the manuscript. Funding for this research was provided by NIH grants RO1 GM034869 and GM082875 to D.R.E., NIH Pharmacological Sciences predoctoral training grant (T32 GM007767) fellowship to D.J.C., CIHR Grant MOP-49451 to T.R.H., and an NSERC predoctoral fellowship to T.B.
Footnotes
Note Supplementary materials can be found at: www.landesbioscience.com/supplement/CoughlinRNA6-2-Sup.pdf
References
- 1.Bystrom AS, Fink GR. A functional analysis of the repeated methionine initiator tRNA genes (IMT) in yeast. Mol Gen Genet. 1989;216:276–86. doi: 10.1007/BF00334366. [DOI] [PubMed] [Google Scholar]
- 2.Sprinzl M, Vassilenko KS. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 2005;33:139–40. doi: 10.1093/nar/gki012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–64. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abelson J, Trotta CR, Li H. tRNA splicing. J Biol Chem. 1998;273:12685–8. doi: 10.1074/jbc.273.21.12685. [DOI] [PubMed] [Google Scholar]
- 5.Weiner AM. SINEs and LINEs: the art of biting the hand that feeds you. Current opinion in cell biology. 2002;14:343–50. doi: 10.1016/s0955-0674(02)00338-1. [DOI] [PubMed] [Google Scholar]
- 6.Goodenbour JM, Pan T. Diversity of tRNA genes in eukaryotes. Nucleic Acids Res. 2006;34:6137–46. doi: 10.1093/nar/gkl725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dittmar KA, Goodenbour JM, Pan T. Tissue-Specific Differences in Human Transfer RNA Expression. PLoS Genet. 2006:2–221. doi: 10.1371/journal.pgen.0020221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laslett D, Canback B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004;32:11–6. doi: 10.1093/nar/gkh152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–62. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 10.Pavesi A, Conterio F, Bolchi A, Dieci G, Ottonello S. Identification of new eukaryotic tRNA genes in genomic DNA databases by a multistep weight matrix analysis of transcriptional control regions. Nucleic Acids Res. 1994;22:1247–56. doi: 10.1093/nar/22.7.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fichant GA, Burks C. Identifying potential tRNA genes in genomic DNA sequences. J Mol Biol. 1991;220:659–71. doi: 10.1016/0022-2836(91)90108-i. [DOI] [PubMed] [Google Scholar]
- 12.Eddy SR, Durbin R. RNA sequence analysis using covariance models. Nucleic Acids Res. 1994;22:2079–88. doi: 10.1093/nar/22.11.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krayev AS, Markusheva TV, Kramerov DA, Ryskov AP, Skryabin KG, Bayev AA, Georgiev GP. Ubiquitous transposon-like repeats B1 and B2 of the mouse genome: B2 sequencing. Nucleic Acids Res. 1982;10:7461–75. doi: 10.1093/nar/10.23.7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patel CV, Gopinathan KP. Development stage-specific expression of fibroin in the silk worm Bombyx mori is regulated translationally. Indian journal of biochemistry & biophysics. 1991;28:521–30. [PubMed] [Google Scholar]
- 15.Stutz F, Gouilloud E, Clarkson SG. Oocyte and somatic tyrosine tRNA genes in Xenopus laevis. Genes & development. 1989;3:1190–8. doi: 10.1101/gad.3.8.1190. [DOI] [PubMed] [Google Scholar]
- 16.Ohama T, Choi IS, Hatfield DL, Johnson KR. Mouse selenocysteine tRNA([Ser]Sec) gene (Trsp) and its localization on chromosome 7. Genomics. 1994;19:595–6. doi: 10.1006/geno.1994.1116. [DOI] [PubMed] [Google Scholar]
- 17.Kelly VP, Suzuki T, Nakajima O, Arai T, Tamai Y, Takahashi S, et al. The distal sequence element of the selenocysteine tRNA gene is a tissue-dependent enhancer essential for mouse embryogenesis. Molecular and cellular biology. 2005;25:3658–69. doi: 10.1128/MCB.25.9.3658-3669.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leontis N, DaLio A, Strobel M, Engelke D. Effects of tRNA-intron structure on cleavage of precursor tRNAs by RNase P from Saccharomyces cerevisiae. Nucleic Acids Res. 1988;16:2537–52. doi: 10.1093/nar/16.6.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hopper AK, Phizicky EM. tRNA transfers to the limelight. Genes & development. 2003;17:162–80. doi: 10.1101/gad.1049103. [DOI] [PubMed] [Google Scholar]
- 20.Soll D, RajBhandary UL. The genetic code—thawing the `frozen accident'. Journal of biosciences. 2006;31:459–63. doi: 10.1007/BF02705185. [DOI] [PubMed] [Google Scholar]
- 21.Schultz DW, Yarus M. Transfer RNA mutation and the malleability of the genetic code. J Mol Biol. 1994;235:1377–80. doi: 10.1006/jmbi.1994.1094. [DOI] [PubMed] [Google Scholar]
- 22.Paushkin SV, Patel M, Furia BS, Peltz SW, Trotta CR. Identification of a human endonuclease complex reveals a link between tRNA splicing and pre-mRNA 3' end formation. Cell. 2004;117:311–21. doi: 10.1016/s0092-8674(04)00342-3. [DOI] [PubMed] [Google Scholar]
- 23.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karolchik D, Baertsch R, Diekhans M, Furey TS, Hinrichs A, Lu YT, et al. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–4. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–15. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hughes TR, Hiley SL, Saltzman AL, Babak T, Blencowe BJ. Microarray analysis of RNA processing and modification. Methods in enzymology. 2006;410:300–16. doi: 10.1016/S0076-6879(06)10014-2. [DOI] [PubMed] [Google Scholar]
- 27.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–10. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wiegant JC, van Gijlswijk RP, Heetebrij RJ, Bezrookove V, Raap AK, Tanke HJ. ULS: a versatile method of labeling nucleic acids for FISH based on a monofunctional reaction of cisplatin derivatives with guanine moieties. Cytogenet Cell Genet. 1999;87:47–52. doi: 10.1159/000015390. [DOI] [PubMed] [Google Scholar]
- 29.Huber W, von Heydebreck A, Sultmann H, Poustka A, Vingron M. Bioinformatics. Vol. 18. Oxford; England: 2002. Variance stabilization applied to microarray data calibration and to the quantification of differential expression; pp. 96–104. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.