Abstract
The alphaviruses and flaviviruses include many important human pathogens, such as the dengue, West Nile, and Chikungunya viruses. These enveloped viruses infect cells by a membrane fusion reaction triggered by the low pH in endosomes. Fusion is mediated by viral membrane proteins through their acid-dependent conversion from a dimer on the virus surface to a homotrimer inserted in the host cell membrane. Here we review recent studies on the regulatory mechanisms that silence these fusion proteins during virus exit, and that sense low pH and mediate protein refolding during virus entry. We discuss results using truncated proteins to dissect the fusion reaction, and future research directions including the development of antiviral therapies against these medically important viruses.
Virus entry: insights from the alphaviruses and flaviviruses
Enveloped viruses are surrounded by a membrane bilayer that protects the virus genome from the extracellular milieu and fuses with a host cell membrane to release the genome into the cytoplasm. This critical membrane fusion reaction is mediated by specialized transmembrane (TM) proteins known as virus membrane fusion proteins. Recent work has detailed the pre-fusion and post-fusion structures of a number of virus fusion proteins (reviewed in [1,2]). While the structural features of such fusion proteins can differ markedly, the evidence indicates that they mediate fusion through a common mechanism. A triggering event such as receptor/co-receptor binding or the low pH of the endocytic pathway first initiates the process. The fusion protein inserts a hydrophobic region (fusion peptide or fusion loop) into the host cell target membrane, thus forming a bridge between the virus and cell membranes. The protein then refolds to a hairpin-like structure in which the fusion peptide and the transmembrane domain are at the same end of the molecule. This post-fusion form is more stable than the pre-fusion conformation, and the energy released by refolding to the final hairpin drives the fusion reaction. The final conformation of all virus membrane fusion proteins whose structure has been determined is a trimer of such hairpins. There has been considerable interest in these fusion proteins as experimental systems to study membrane fusion and as possible targets for antiviral therapy.
Alphaviruses and flaviviruses are small enveloped viruses containing plus-sense RNA genomes (reviewed in [3,4]). These viruses include many medically important species such as the alphaviruses Chikungunya virus and Venezuelan equine encephalitis virus and the flaviviruses dengue virus (DV), West Nile virus, Japanese encephalitis virus, yellow fever virus (YFV) and tick-borne encephalitis virus (TBEV). The structure, entry, and membrane fusion properties of the alphaviruses and flaviviruses have been intensively studied and will be summarized in this review (see also [5-7]). These viruses have structurally-related fusion proteins that drive the fusion reaction through their low pH-triggered conversion from a dimer form on the surface of the virus to a target-membrane-inserted homotrimer. Recent studies have defined the regulation of the alphavirus and flavivirus fusion proteins during virus assembly and fusion, and will be the major focus of this review. These advances have greatly contributed to our understanding of the mechanisms of low-pH triggering during entry, the means by which viruses protect themselves from low pH during exit, and the process of formation of trimeric hairpins from pre-fusion dimers. We finish the article by discussing possible avenues for future research, including those that might lead to development of new antiviral therapies.
Alphavirus and flavivirus organization and fusion protein structure
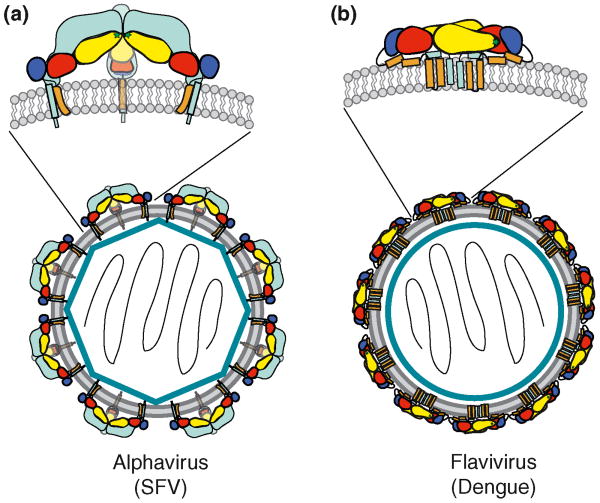
The alphavirus particle contains 240 copies of the capsid protein associated with the RNA genome to form a nucleocapsid with icosahedral symmetry (Figure 1a) [3]. This core is surrounded by a membrane containing 240 copies each of the transmembrane E2 and E1 glycoproteins, also organized with icosahedral symmetry. E2 and E1 associate as trimers of heterodimers (E2-E1)3 on the particle surface. E1 is the membrane fusion protein. It lies tangential to the virus membrane and forms an icosahedral lattice on the virus surface. The “companion protein” E2 interacts with E1, covers the E1 fusion loop and forms the spiky center of the trimer.
Figure 1.
Schematic diagrams of mature alphavirus and flavivirus particles. The plus-sense viral RNAs are represented inside the capsids in black. The capsids are shown as either a blue polygon to represent the icosahedral organization in the alphaviruses, or as a blue circle for the flaviviruses. The viral membrane bilayers are shown as a grey double layer containing the inserted viral glycoproteins. Viral particles are not to scale. (a) Organization of the Semliki Forest virus particle. The inset shows the trimer of heterodimers (two in side view, one end-on) formed by E1 and E2 inserted in the membrane. E1 is colored by domains as in Figure 2, with the transmembrane (TM) domain in light brown. The E2 protein is shown in cyan and demonstrates the interaction of E2 with E1 that protects the fusion loop (green star). (b) Organization of the dengue virus particle. The inset shows the E protein homodimer, with the fusion loops (green stars) protected by the dimeric interaction. The E domains are colored as in Figure 2, and the two alpha helices of the stem and the two TM domains are shown in light brown. The mature M protein is shown in cyan.
Flaviviruses are also highly organized particles in which the glycoproteins are arranged with icosahedral symmetry [4,6]. They contain a core of the RNA genome associated with 180 capsid proteins (Figure 1b). The membrane envelope contains 180 copies of the fusion protein E associated as homodimers and organized in a herringbone or “raft” pattern on the virus surface. The fusion loop of each E protein is hidden by its association with its dimeric partner. The membrane also contains 180 copies of the companion protein M, which is synthesized as a precursor protein, prM, whose role is discussed below.
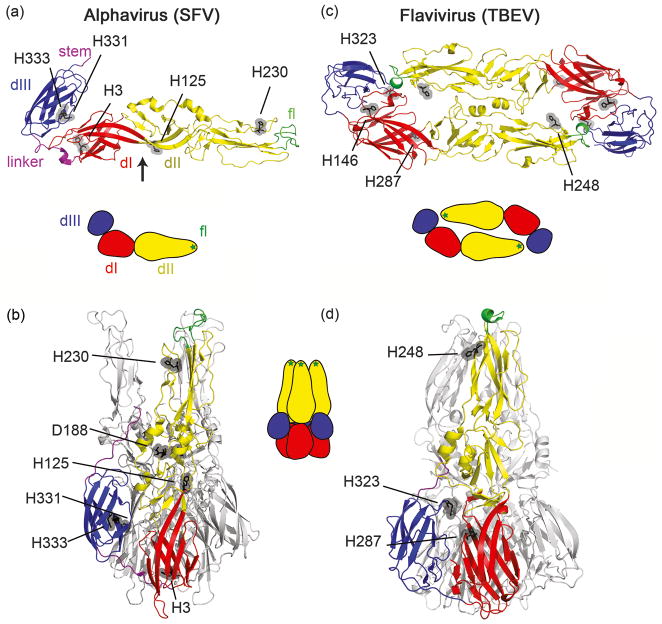
The pre-fusion structures of the E1 protein of the alphavirus Semliki Forest virus (SFV) [8,9] and of several flavivirus E proteins [10-15] have been solved. The overall architecture of these virus fusion proteins is composed primarily of β-sheets and is remarkably similar between the two groups of viruses (Figures 2a, 2c). Each protein contains an amino terminal domain I (DI) from which two long extensions form the finger-like domain II (DII) with the hydrophobic fusion loop at the tip. DI connects to domain III (DIII) via a linker region. DIII has an immunoglobulin-like fold, and connects to the C-terminal stem region and TM domain.
Figure 2.
Pre- and post-fusion structures of the alphavirus and flavivirus fusion proteins. (a) Structure of the prefusion SFV E1 protein ectodomain (PDB 2ALA [8,9]). The three domains (D) of E1 are shown in the standard color scheme: DI in red, DII in yellow with the fusion loop (fl) shown in green, and DIII in blue. The linker between DI and DIII is shown in purple. A stem region connects DIII with the TM domain, and the portion of the stem present in the structure is shown, also colored purple. Histidine residues discussed in the text are highlighted in gray, and the hinge region between DI and DII is indicated by the black arrow. (b) SFV post-fusion homotrimer (PDB 1RER [17]), with histidine residues from panel A indicated. (c) Structure of the prefusion TBEV E protein ectodomain homodimer (PDB 1SVB [10]). Each fusion loop is hidden within a “pocket” formed by DI and DIII of the dimeric partner. Conserved histidine residues that play a role in the fusion mechanism are highlighted in gray, along with H146. Domains are colored as in panel A, and the hinge, linker and stem are in analogous positions as those indicated in panel A. (d) TBEV post-fusion homotrimer (PDB 1URZ [18]), with histidine residues from (c) indicated. This figure was prepared using the program PyMOL [62]. Cartoons of each structure colored in the same domain scheme are also shown.
Both alphaviruses and flaviviruses enter cells primarily through uptake in clathrin-coated vesicles, and fuse in the low pH environment of endosomes [7,16]. The post-fusion structures of E1 and E reveal a dramatic reorganization of the originally dimeric fusion proteins to homotrimers in which DIII has moved towards the fusion loop (Figures 2b, 2d) [17-19]. The final postfusion hairpin is thus formed by the packing of DIII and the stem region against the central trimer, thus bringing the fusion loops and TM domains to the same end of the rod-like trimer. The alphavirus and flavivirus fusion proteins are often referred to as “class II”. This is a useful (although simplified) shorthand to differentiate them from the “class I” and “class III” fusion proteins [1,2,20]. The class I fusion proteins contain a central α-helical coiled-coil domain and are exemplified in influenza virus and human immunodeficiency virus-1. The class III proteins, exemplified by the vesicular stomatitis virus G protein and the herpes simplex virus 1 glycoprotein B, have five domains that include both a central trimeric coiled-coil and considerable β-sheet structure.
Virus assembly and maturation
The structural proteins of the alphaviruses and flaviviruses are synthesized as polyproteins that are co- and post-translationally processed by viral and cellular proteases. The alphavirus envelope proteins are translocated into the endoplasmic reticulum (ER) and transported through the secretory pathway to the plasma membrane, where virus budding occurs [3]. The E2 protein is synthesized as a precursor termed p62, which associates with E1 and assists in its folding and transport. Based on the relative resistance of the p62-E1 heterodimer to low pH dissociation [21], it is believed that p62 protects E1 from the mildly acidic pH (∼pH 6.0) of the trans-Golgi network (TGN). The p62 precursor is processed by the cellular protease furin [22] to produce mature E2 protein plus E3, a small polypeptide that is released from many alphaviruses after processing. Structural studies of immature and mature alphavirus particles demonstrate that their overall organization is similar, with each containing trimeric spikes composed of three heterodimers of p62 (or E2) and E1 [23,24]. Maturation to E2 permits subsequent heterodimer dissociation and virus fusion in the endosomal pH range (∼pH 5.5-6.5), and strongly increases virus infectivity [22,25].
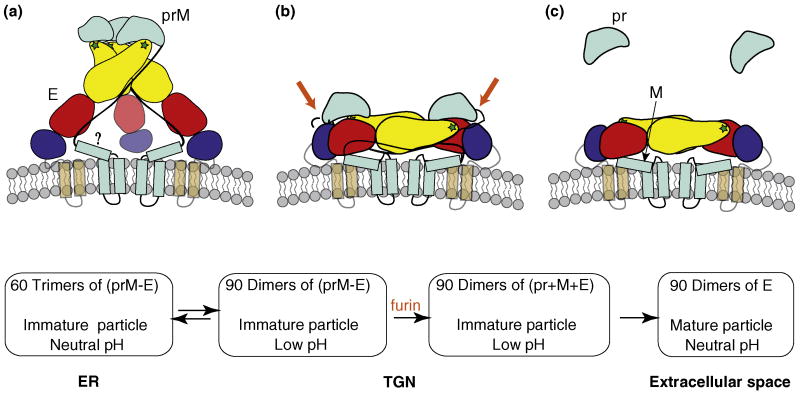
The flavivirus envelope proteins are similarly translocated into the ER where the TM prM protein dimerizes with the fusion protein E and assists in its folding [4]. PrM is processed by furin to the mature M protein plus the pr polypeptide [26]. However, flaviviruses assemble and bud into the ER and the virus particles are transported through the secretory pathway and released from the cell. Recent studies have determined the structure of the pr region of DV prM and have greatly advanced our understanding of flavivirus maturation [27,28] (Figure 3). The immature flavivirus particle contains 60 trimers of prM-E heterodimers that form spiky protrusions on the virus surface, with the pr region of prM covering the fusion loop of E [27,29,30]. The low pH of the TGN induces a dramatic but reversible reorganization of the envelope proteins to a herringbone-like, relatively flat arrangement of dimers of prM-E [28]. This rearrangement makes the prM protein accessible to cleavage by furin as the virus transits through the secretory pathway [26,28]. The pr polypeptide remains bound after cleavage and thus protects E from the mildly acidic pH of the exocytic pathway. Once the virus particle is released from the cell, the neutral pH environment allows the dissociation of pr, completing the maturation process [28]. The mature particle shows a dramatic increase in fusion activity and infectivity compared with the prM-containing immature form [26,28].
Figure 3.
Flavivirus maturation pathway. (a) Immature flavivirus particle at neutral pH. The virus buds into the neutral pH environment of the endoplasmic reticulum (ER), and the immature particle contains spiky trimers of prM-E heterodimers. One (prM-E) trimer is shown, with E colored by domains as in Figure 2; the stem and transmembrane domains are displayed brown and translucent. The prM protein is colored cyan. It is unclear whether the region of M (denoted by a question mark) that normally forms a membrane-proximal alpha-helix in the mature particle has this conformation in the immature particle. (b) Immature flavivirus particle at low pH. As the virions progress through the trans-Golgi network (TGN), the acidic environment induces the rearrangement from (prM-E) trimers to (prM-E) dimers (one of which is shown) with similar orientation and organization as in the mature virus. This low pH-induced transition is reversible in DV [28], but not in TBEV [26]. The transition allows access for furin to cleave prM to pr plus M, as indicated by the red arrows. At low pH, pr remains bound to E. (c) Mature flavivirus particle at neutral pH. The particle is secreted into the extracellular space. The pr polypeptide dissociates from E at neutral pH, readying the virus for fusion during entry.
The remarkable reorganization during flavivirus maturation thus depends on the changes in pH that the virus sees during its transit through the secretory pathway. The initial low pH-dependent rearrangment of prM-E trimers to dimers may be mediated at least in part by a histidine (H) residue in the M protein region [27,31]. The structures of the pr peptide and E protein suggest that their pH-dependent interaction includes complementary charge pairing between highly conserved residues, H244 in E and two aspartic acid residues in pr, D63 and D65 (DV numbering) [27]. Together these conformational changes leave the mature virus primed to respond to low pH during virus entry.
Inhibition of alphavirus and flavivirus fusion and infection
Understanding the pre- and post-fusion conformations of class I viral fusion proteins has led to inhibitors that block protein refolding during virus fusion (reviewed in [32-34]). Here we will summarize strategies and progress to date on developing inhibitors of the class II fusion proteins.
The hinge region connecting DI and DII (Figure 2) undergoes important changes during flavivirus maturation (reviewed in [6]), and during class II protein refolding to the postfusion homotrimer [17-19]. In silico docking was used to screen libraries of small molecules for binding to a hydrophobic pocket in the hinge region of the DV or YFV E protein [35-37]. An optimized candidate was found to inhibit DV infection at an early step, suggestive of inhibition of virus fusion [35]. This strategy may prove important for structure-based design of antiviral agents that could affect virus maturation or fusion with host cell membranes.
The recent structural studies of the pr peptide indicate that it bridges two E proteins and stabilizes the E dimer interface [27,28] (Figure 3). Small molecules that bind at this site might similarly stabilize the E dimer and prevent fusion by preventing the dimer-to-trimer transition [28].
The class II stem region extends along the groove formed by two E or E1 proteins in the homotrimer, thus connecting DIII to the TM domain and forming part of the “outer layer” of the trimeric hairpin (Figures 2, 4). Peptides derived from the outer layer of the class I fusion proteins can act as potent and clinically useful inhibitors [34,38], and by analogy the class II stem was suggested as a potential inhibitor [18,19]. However, to date peptides spanning the SFV stem region or antibodies directed against the SFV stem have failed to inhibit virus fusion or infection [39]. This reflects, in part, the relative inaccessibility of the stem region in the prefusion conformation [39]. In addition, mutagenesis studies show that, while the wild type stem sequence is optimal for fusion, neither a specific sequence nor length of the SFV stem is required [40]. The flavivirus stem appears to have a more defined structure, and can promote trimerization [41] and trimer stability [42]. Peptides corresponding to the stem region of DV E protein inhibit infection by DV and West Nile Virus [43]. Thus the stem-central trimer interaction might be a feasible antiviral target for the flaviviruses but not the alphaviruses.
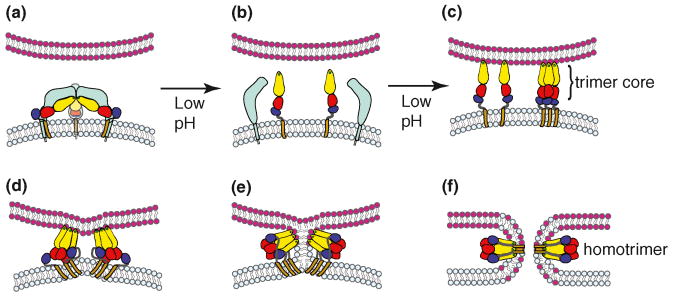
Figure 4.
Proposed model of the alphavirus membrane fusion mechanism. (a) Virus particle in the pre-fusion state. The virus membrane, depicted in light blue, contains a trimer of E2-E1 heterodimers, shown colored as in Figures 1 and 2. The target membrane is shown in pink. The fusion protein E1 is in a metastable conformation. (b) Triggering. Upon exposure to low pH, dissociation of the E2-E1 heterodimer occurs, exposing the E1 fusion loop. The disposition of E2 after heterodimer dissociation is unknown. (c) The fusion loop inserts in the target membrane through a low pH and cholesterol-dependent mechanism. A core trimer is formed by DI and DII. This step is promoted by residues H3 (DI) and D188 (DII), as discussed in the text. (d-e) In a pH-independent interaction, DIII and the stem region are folded against the core trimer in the groove formed by two E1 proteins. The distortion of the target membrane by fusion loop insertion, the fold-back of DIII and stem, and the cooperative action of several trimers (of which only two are shown) are proposed to provide the force to mediate membrane fusion. (e) Fusion proceeds through a hemifusion step in which the two outer leaflets merge. (f) E1 forms the final stable post-fusion homotrimer, in which the fusion loops and the transmembrane domains are located at the same side of the molecule. Concomitantly, this refolding drives complete fusion via formation of the fusion pore. While this model depicts alphavirus fusion, the general features of steps (c-f) appear similar for flavivirus fusion.
The outer layer of the class II hairpin is also composed of DIII, which dramatically reorients towards the fusion loop during hairpin formation (Figures 2 and 4). Recombinant SFV and DV DIII proteins specifically inhibit virus fusion and infection when present during the low pH-triggered conformational change [44]. This inhibition is due to the binding of exogenous DIII to a trimeric intermediate in which the endogenous DIII has not completely folded back against the central trimer. Although not required, inhibition by DIII is enhanced by the presence of the stem, suggesting that the stem confers increased stability. Thus, DIII inhibition demonstrates the existence of a trimeric intermediate during class II fusion and suggests that the specific and stable interaction between DIII and the core trimer could serve as a target for class II fusion inhibitors.
Control of low pH-dependent fusion
Viral fusion proteins that are triggered by the acidic environment of endosomes have evolved specific mechanisms to respond to low pH. The term “pH sensor” will be used here to denote regions or residues that, upon proton binding, destabilize the prefusion conformation to initiate fusion protein refolding. Low pH can also act by titrating residues that are involved in forming or stabilizing the post-fusion conformation, thus helping to drive the fusion reaction to completion. Important examples of both types of mechanisms have been recently described for the flaviviruses and alphaviruses and will be reviewed here.
Given the pH threshold of these viruses, residues whose side chains titrate with a pKa in the ∼ 5-7 range could be relevant players in the pH response, and thus histidines (pKa ∼6.5) have been proposed to play key roles in fusion [45]. It is also worth noting that the pKa of a titratable group can vary widely, depending on the electrostatic potential and solvent accessibility of its local environment [46]. For example, while the carboxylate of free glutamate and aspartate titrates in the 4.4 range in solution, within proteins its pKa can range from ∼2 to 10 [47]. Surface residues are directly affected by solvent pH, but even buried residues can be influenced by pH changes through interactions that network with residues on the surface of a protein [48].
The hydrophobic fusion loop of the flavivirus fusion protein is hidden in the dimer interface (Figure 2c), and the dimer must dissociate at low pH to permit fusion loop insertion in the target membrane [49]. A recent study of TBEV carefully defined the role of conserved histidine residues in this response to low pH [50, also discussed in 51] (Figures 2c, 2d, and Table 1). H323, located in a region of DIII that protects the fusion loop of its dimeric partner, acts as an initial pH sensor. Mutation of H323 to alanine inhibits dimer dissociation, fusion loop exposure and membrane insertion, and fusion. The conserved H149 on DI may also be involved in initial pH sensing, although this residue could not be tested by mutagenesis [50]. H323 and H149 are located in the interface between DI and DIII and are thus positioned to disrupt the dimer interface and fusion loop “pocket” upon protonation.
Table 1.
Histidine mutations that affect the flavivirus and alphavirus fusion mechanisms.
| Mutation(s) | Locationa | Fusion phenotype | Proposed role(s) in fusion |
|---|---|---|---|
| Flavivirus (TBEV) | |||
| H248N b + H287A | DII: ij loopc close to fusion loop; in trimer groove DI: hinge, trimer interface |
Fusion block | Trimer formation and stabilization |
| H323A | DIII: at interface with DI | Fusion block | pH sensor for dimer dissociation; trimer stabilization |
| Alphavirus (SFV) | |||
| H3A | DI: near linker region in trimer | pH shift, decreased fusion | Regulation of trimerization |
| H125A | DII: hinge, E1-E1 interface in virus glycoprotein shell | No significant phenotype | None |
| H230A | DII: ij loopc close to fusion loop; in trimer groove | Fusion block | Late step in fusion after trimer formation; no pH effect |
| H331A + H333A | DIII: at interface with DI | No significant phenotype | None |
Domain location and position in prefusion conformation, unless indicated as trimer.
Note that H244 of DV (discussed in text) is analogous to H248 of TBEV.
The ij loop is at the tip of DII, connecting β-strands i and j.
In addition to mediating the initial release of the dimer, H323 also functions to increase the stability of the post-fusion trimer. This probably occurs through formation of a salt bridge between H323 and E373 on DIII, thereby inhibiting return to the prefusion interaction of E373 with an arginine (R9) on DI [50]. The new results may also explain interesting earlier findings. Treatment of TBEV at pH 10 causes dissociation of dimers and membrane-insertion of fusion loops, but does not trigger stable trimer formation or fusion [52]. Under these conditions, H323 would not be protonated and the release of the pre-fusion DIII-DI interface interactions and the stabilization of the final trimer conformation would not occur. These data also indicate that fusion loop insertion is pH-independent.
Several other regions of the flavivirus E protein affect its pH-dependence. Mutations in the hinge region between DI and DII alter the pH threshold of flavivirus fusion [10,11]. In general, these mutations convert long hydrophobic chains to short hydrophobic chains, perhaps allowing for tighter packing of the hydrophobic pocket of the hinge, thus making dimer dissociation more difficult. A double mutation of H287A and H248N blocks TBEV fusion by decreasing the stability of the trimer, presumably through dual effects on the trimer interface (H287A) and stem packing (H248N) [50] (Figure 2, Table 1). Thus there is strong evidence for important roles of both initial pH-sensing and fusion protein refolding in the regulation of flavivirus fusion.
The first observed response of alphaviruses to low pH treatment is the dissociation of the E2-E1 heterodimer [53] (Figure 4). Based on mutants in E2 that require a lower pH [54-56], and on the relative resistance of the p62-E1 dimer to low pH dissociation [21,25], dimer dissociation appears to be mediated primarily through effects of low pH on the E2 protein. However, evidence indicates that low pH is also involved in triggering subsequent conformational changes in the E1 protein [57,58]. A recent study addressed the role of conserved histidine residues in the response of SFV E1 to low pH [59] (Figure 2a, 2b, and Table 1). Unlike the flaviviruses, histidine residues in SFV DIII (H331, H333) are not involved in pH sensing, even though they are located at the interface of DIII with DI in the prefusion form. Although E1 has a hinge region and forms the icosahedral lattice on the alphavirus surface, mutation of a conserved histidine (H125) that lies in the hinge and at an interface between E1 proteins on the virus surface does not significantly affect virus fusion or pH dependence. However, mutation of H3, a conserved histidine in DI, to alanine decreases virus infectivity, fusion, and trimer formation, and markedly shifts the pH-threshold required for fusion and E1 trimerization. H3 is located near the DI/DIII linker region that extends during the fold-back of DIII to the post-fusion conformation and is also positioned to affect intermolecular interactions within the trimer (Figure 2). It will be important to determine the interactions of H3 that mediate its effects on E1 trimerization.
Mechanistic studies of alphavirus E1 trimerization
Trimerization of the E1 or E protein is a critical step in alphavirus and flavivirus fusion. Soluble E1 or E ectodomains can interact to form trimers, demonstrating that the interactions in trimerization are inherent to the proteins, rather than requiring the specific geometry found on the virus particle. The structures of the alphavirus and flavivirus homotrimers show inter-chain interactions through contacts in DI and DII, with DIII clamping into the grooves of this central trimer region [17-19]. Analysis of the steps in trimerization, their requirements, and the stages of fusion they mediate is important to our understanding of the fusion mechanism. Recent findings on SFV E1 trimerization will be summarized here.
As discussed above, inhibition studies using exogenous DIII identified a trimeric pre-fusion intermediate in which DIII is not completely folded-back [44]. This result suggests that the central trimer may be formed prior to DIII fold-back. Recent mutagenesis studies of a conserved aspartate D188 located in the trimer interface region of E1 DII (Figure 2a, 2b) indicate that neutralization of D188 by either protonation or salt-bridge formation is important in formation of the central trimer and in fusion [60].
Truncated E1 proteins containing only DI and DII (termed E1 DI/II) were used to analyze the properties of E1 trimer formation [58]. Similar to the complete E1 ectodomain, E1 DI/II specifically interacts with cholesterol-containing membranes at low pH and forms a stable trimer. Thus the DI/II protein contains the elements required for low pH-dependent trimerization. Negative stain electron microscopy was used to compare the trimers formed by the complete E1 ectodomain and E1 DI/II. Both types of trimers insert into membranes through the fusion loop-containing tip, and interact with adjacent trimers to form rings of 5 or 6 trimers and hexagonal trimer lattices [58,61]. The E1 DI/II trimers promote deformation of rounded liposomes into membrane tubules [58], indicating that the inter-trimer fusion loop interactions can cause membrane bending in the absence of DIII and final hairpin formation.
These E1 DI/II trimers were used as targets for the binding of exogenous SFV DIII proteins [58]. Binding is specific, efficient, and stable. While the stem region is not required for DIII binding, it does increase the overall stability of the DIII-core trimer complex. Although formation of the core trimer requires low pH, once this intermediate is formed the binding of DIII is independent of pH. Thus these truncated E1 proteins reconstitute the interactions that produce the fusogenic E1 hairpin. The in vitro interaction of DIII with the core trimer may enable the identification of small molecules that both inhibit this protein-protein interaction and block virus fusion.
Concluding remarks and future directions
The alphavirus and flavivirus fusion proteins are assembled with companion proteins that protect them from acid pH during exocytic transport and that are processed by furin to permit fusion in the physiological pH range. During virus entry in the low pH environment of endosomes, the E1 and E fusion proteins dissociate from their dimeric interactions, insert into the endosome membrane, trimerize, and refold to a hairpin conformation, thus driving membrane fusion (see model in Figure 4). As summarized in this review, recent studies have defined the role of pH and processing during flavivirus maturation, identified important histidine residues involved in flavivirus dimer dissociation and in the refolding of the E and E1 proteins, and developed in vitro systems to reconstitute the protein-protein interactions during virus fusion. The combined results from structural, biochemical and molecular biology approaches have been synergistic and exciting. These results also identify outstanding questions in the field, and suggest potential experimental strategies to begin to address them.
Studies of the response of the alphaviruses and flaviviruses to low pH have primarily focused on the role of histidine residues. Computer modeling programs are becoming increasingly sophisticated at predicting the pKa of an ionizable group in a known protein structure (reviewed in [46]). Such modeling programs may be helpful in identifying other ionizable residues involved in pH regulation, and in modeling the effects of histidine substitutions on adjacent titratable groups. Results from mutagenesis studies already suggest specific regions that may play a role during protein refolding and trimerization, particularly the DI-DIII linker and the trimer interface region.
The recent findings on the structure, processing and pH sensitivity of the flavivirus prM/pr protein demonstrate the importance of this companion protein. For the alphaviruses, many fundamental questions on the companion protein p62/E2 remain, including structural characterization, the role of the E3 polypeptide, and the control of the differential pH sensitivity of the p62 vs. E2 interaction with E1. More broadly, it will be important to understand how viruses in the same families, such as hepatitis C and rubella virus, protect their fusion proteins during biosynthesis, since the putative companion proteins of these viruses do not appear to be proteolytically-processed.
Current evidence indicates that the insertion of the flavivirus fusion loop into the target membrane does not require low pH. Will this also be the case for the alphaviruses? What is the relative role of pH vs. cholesterol for the alphaviruses, and will there be differences in the requirements for initial fusion loop-membrane insertion vs. stable membrane interaction of the final refolded trimer? Studies of membrane-inserted SFV E1 and E1 DI/II trimers reveal striking lateral interactions. Further experiments will be required to determine the function of these inter-trimer interactions during fusion. One potentially useful approach will be to use two-dimensional E1 lattices in combination with cryo-electron microscopy to image the interactions of the fusion loop with the target membrane and its cooperative interactions with the fusion loops of adjacent trimers.
The studies of alphavirus and flavivirus biosynthesis, processing, and membrane fusion suggest a number of interesting targets for inhibitors. On the biosynthetic side, these include the interaction of pr with E and of p62 with E1, as disruption of this interface could leave the fusion protein unprotected against acidic pH in the TGN. Conversely, stabilization of various dimer interfaces would also prevent fusion, and the pr peptide is a paradigm for a molecule that bridges two E proteins and prevents their dissociation [27]. During virus entry, potential targets include the interactions between the three subunits that form the DI/II core trimer, and the binding of DIII to the core trimer. We have learned over the past few years that experiments to define these targets provide important information about fusion protein intermediates. In the future, this information may translate into new approaches for antiviral therapies.
Acknowledgments
We dedicate this review to the memory of our colleague and friend Dr. Dennis Shields. We thank all of the members of our lab for helpful discussions, and Gwen Taylor, Gleyder Roman-Sosa, Aihua Zheng, Kartik Chandran and Félix Rey for their comments on the manuscript. We also thank Mark Girvin for insightful discussions of amino acid titration. Work in our laboratory was supported by grants to M.K. from the National Institutes of Health (AI075647, AI067931, GM057454, U54AI057158) and by Cancer Center Core Support Grant NIH/NCI P30-CA13330. C.Y.L. was supported in part through the Medical Scientist Training Program of the Albert Einstein College of Medicine (NIH T32 GM07288). We acknowledge the important contributions of those researchers whose work was not fully cited due to space limitations.
Glossary
- Ectodomain
a portion of a membrane protein that extends into the extracellular space.
- In silico docking
a computational method to characterize the complementarity of a ligand and a protein target. It can be used to identify small molecules that bind to a defined region of a target protein structure.
- Icosahedral symmetry
an icosahedron is composed of 20 triangular faces, contains 12 vertices, and has five-fold, three-fold, and two-fold axes of rotational symmetry. The organization of many virus particles is based on such 5:3:2 symmetry principles.
- Metastable conformation
a protein conformation that exists due to an energy barrier that prevents the formation of the most stable form. The metastable conformation can be triggered to overcome the energy barrier, releasing energy as it refolds to the final stable conformation.
- Plus-sense RNA genome
viral RNA genome with message-sense polarity that serves as an mRNA for translation of viral proteins.
- Polyprotein
larger protein that is cleaved to produce functionally distinct polypeptides. A viral polyprotein can by processed by viral and/or cellular proteases.
- Trans-Golgi network (TGN)
a network of vesicles and tubules at the trans face of the Golgi apparatus that functions in processing and sorting of newly synthesized membrane proteins and other molecules.
References
- 1.Harrison SC. Viral membrane fusion. Nat Struct Mol Biol. 2008;15(7):690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.White JM, et al. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit Rev Biochem Mol Biol. 2008;43(3):189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuhn RJ. Togaviridae: The Viruses and Their Replication. In: Knipe DM, editor. Fields Virology. Vol. 1. Lippincott, Williams and Wilkins; 2007. pp. 1001–1022. [Google Scholar]
- 4.Lindenbach BD, et al. Flaviviruses: the viruses and their replication. In: Knipe DM, Howley PM, editors. Fields' Virology. Lippincott, Williams and Wilkins; 2007. pp. 1101–1152. [Google Scholar]
- 5.Kielian M. Class II virus membrane fusion proteins. Virol. 2006;344:38–47. doi: 10.1016/j.virol.2005.09.036. [DOI] [PubMed] [Google Scholar]
- 6.Mukhopadhyay S, et al. A structural perspective of the flavivirus life cycle. Nat Rev Micro. 2005;3(1):13–22. doi: 10.1038/nrmicro1067. [DOI] [PubMed] [Google Scholar]
- 7.Stiasny K, Heinz FX. Flavivirus membrane fusion. J Gen Virol. 2006;87(Pt 10):2755–2766. doi: 10.1099/vir.0.82210-0. [DOI] [PubMed] [Google Scholar]
- 8.Lescar J, et al. The fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell. 2001;105(1):137–148. doi: 10.1016/s0092-8674(01)00303-8. [DOI] [PubMed] [Google Scholar]
- 9.Roussel A, et al. Structure and interactions at the viral surface of the envelope protein E1 of Semliki Forest virus. Structure. 2006;14:75–86. doi: 10.1016/j.str.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 10.Rey FA, et al. The envelope glycoprotein from tick-borne encephalitis virus at 2A resolution. Nature. 1995;375:291–298. doi: 10.1038/375291a0. [DOI] [PubMed] [Google Scholar]
- 11.Modis Y, et al. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc Natl Acad Sci USA. 2003;100(12):6986–6991. doi: 10.1073/pnas.0832193100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Modis Y, et al. Variable surface epitopes in the crystal structure of dengue virus type 3 envelope glycoprotein. J Virol. 2005;79(2):1223–1231. doi: 10.1128/JVI.79.2.1223-1231.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y, et al. Conformational changes of the flavivirus E glycoprotein. Structure (Camb) 2004;12(9):1607–1618. doi: 10.1016/j.str.2004.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nybakken GE, et al. Crystal structure of the West Nile virus envelope glycoprotein. J Virol. 2006;80(23):11467–11474. doi: 10.1128/JVI.01125-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanai R, et al. Crystal structure of west nile virus envelope glycoprotein reveals viral surface epitopes. J Virol. 2006;80(22):11000–11008. doi: 10.1128/JVI.01735-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kielian M. Membrane fusion and the alphavirus life cycle. Adv Virus Res. 1995;45:113–151. doi: 10.1016/s0065-3527(08)60059-7. [DOI] [PubMed] [Google Scholar]
- 17.Gibbons DL, et al. Conformational change and protein-protein interactions of the fusion protein of Semliki Forest virus. Nature. 2004;427:320–325. doi: 10.1038/nature02239. [DOI] [PubMed] [Google Scholar]
- 18.Bressanelli S, et al. Structure of a flavivirus envelope glycoprotein in its low-pH-induced membrane fusion conformation. EMBO J. 2004;23(4):728–738. doi: 10.1038/sj.emboj.7600064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Modis Y, et al. Structure of the dengue virus envelope protein after membrane fusion. Nature. 2004;427(6972):313–319. doi: 10.1038/nature02165. [DOI] [PubMed] [Google Scholar]
- 20.Backovic M, Jardetzky TS. Class III viral membrane fusion proteins. Curr Opinion Struct Biol. 2009;19(2):189–196. doi: 10.1016/j.sbi.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wahlberg JM, et al. The heterodimeric association between the membrane proteins of Semliki Forest virus changes its sensitivity to low pH during virus maturation. J Virol. 1989;63:4991–4997. doi: 10.1128/jvi.63.12.4991-4997.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang X, et al. Furin processing and proteolytic activation of Semliki Forest virus. J Virol. 2003;77(5):2981–2989. doi: 10.1128/JVI.77.5.2981-2989.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang W, et al. Placement of the structural proteins in sindbis virus. J Virol. 2002;76(22):11645–11658. doi: 10.1128/JVI.76.22.11645-11658.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferlenghi I, et al. The first step:maturation of the Semliki Forest virus spike occurs through a dramatic localized conformational change. J Mol Biol. 1998;283:71–81. doi: 10.1006/jmbi.1998.2066. [DOI] [PubMed] [Google Scholar]
- 25.Salminen A, et al. Membrane fusion process of Semliki Forest virus II: Cleavage- dependent reorganization of the spike protein complex controls virus entry. J Cell Biol. 1992;116:349–357. doi: 10.1083/jcb.116.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stadler K, et al. Proteolytic activation of tick-borne encephalitis virus by furin. J Virol. 1997;71:8475–8481. doi: 10.1128/jvi.71.11.8475-8481.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li L, et al. The flavivirus precursor membrane-envelope protein complex: structure and maturation. Science. 2008;319(5871):1830–1834. doi: 10.1126/science.1153263. [DOI] [PubMed] [Google Scholar]
- 28.Yu IM, et al. Structure of the immature dengue virus at low pH primes proteolytic maturation. Science. 2008;319(5871):1834–1837. doi: 10.1126/science.1153264. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, et al. Structure of immature West Nile virus. J Virol. 2007;81(11):6141–6145. doi: 10.1128/JVI.00037-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y, et al. Structures of immature flavivirus particles. EMBO J. 2003;22(11):2604–2613. doi: 10.1093/emboj/cdg270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin YJ, Wu SC. Histidine at residue 99 and the transmembrane region of the precursor membrane prM protein are important for the prM-E heterodimeric complex formation of Japanese encephalitis virus. J Virol. 2005;79(13):8535–8544. doi: 10.1128/JVI.79.13.8535-8544.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harrison SC. Mechanism of membrane fusion by viral envelope proteins. Adv Virus Res. 2005;64:231–261. doi: 10.1016/S0065-3527(05)64007-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Earp LJ, et al. The many mechanisms of viral membrane fusion proteins. Curr Topics Microbiol Immunol. 2005;285:25–66. doi: 10.1007/3-540-26764-6_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eckert DM, Kim PS. Mechanisms of viral membrane fusion and its inhibition. Annu Rev Biochem. 2001;70:777–810. doi: 10.1146/annurev.biochem.70.1.777. [DOI] [PubMed] [Google Scholar]
- 35.Wang QY, et al. A Small Molecule Dengue Virus Entry Inhibitor. Antimicrob Agents Chemother. 2009 doi: 10.1128/AAC.01148-08. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Z, et al. Design, synthesis, and biological evaluation of antiviral agents targeting flavivirus envelope proteins. J Med Chem. 2008;51(15):4660–4671. doi: 10.1021/jm800412d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou Z, et al. Antiviral compounds discovered by virtual screening of small-molecule libraries against dengue virus E protein. ACS Chem Biol. 2008;3(12):765–775. doi: 10.1021/cb800176t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moore JP, Doms RW. The entry of entry inhibitors: A fusion of science and medicine. Proc Natl Acad Sci USA. 2003;100(19):10598–10602. doi: 10.1073/pnas.1932511100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liao M, Kielian M. Site-directed antibodies against the stem region reveal low pH-induced conformational changes of the Semliki Forest virus fusion protein. J Virol. 2006;80(19):9599–9607. doi: 10.1128/JVI.01054-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liao M, Kielian M. Functions of the stem region of the Semliki Forest virus fusion protein during virus fusion and assembly. J Virol. 2006;80(22):11362–11369. doi: 10.1128/JVI.01679-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Allison SL, et al. Mapping of functional elements in the stem-anchor region of tick-borne encephalitis virus envelope protein E. J Virol. 1999;73:5605–5612. doi: 10.1128/jvi.73.7.5605-5612.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stiasny K, et al. Differences in the postfusion conformations of full-length and truncated class II fusion protein E of tick-borne encephalitis virus. J Virol. 2005;79(10):6511–6515. doi: 10.1128/JVI.79.10.6511-6515.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hrobowski YM, et al. Peptide inhibitors of dengue virus and West Nile virus infectivity. Virol J. 2005;2(1):49. doi: 10.1186/1743-422X-2-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liao M, Kielian M. Domain III from class II fusion proteins functions as a dominant-negative inhibitor of virus-membrane fusion. J Cell Biol. 2005;171(1):111–120. doi: 10.1083/jcb.200507075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kampmann T, et al. The Role of Histidine Residues in Low-pH-Mediated Viral Membrane Fusion. Structure. 2006;14(10):1481–1487. doi: 10.1016/j.str.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 46.Srivastava J, et al. Intracellular pH sensors: design principles and functional significance. Physiology (Bethesda) 2007;22:30–39. doi: 10.1152/physiol.00035.2006. [DOI] [PubMed] [Google Scholar]
- 47.Li H, et al. Very fast empirical prediction and rationalization of protein pKa values. Proteins. 2005;61(4):704–721. doi: 10.1002/prot.20660. [DOI] [PubMed] [Google Scholar]
- 48.Qin J, et al. Ionization equilibria for side-chain carboxyl groups in oxidized and reduced human thioredoxin and in the complex with its target peptide from the transcription factor NF kappa B. Biochem. 1996;35(1):7–13. doi: 10.1021/bi952299h. [DOI] [PubMed] [Google Scholar]
- 49.Stiasny K, et al. Membrane interactions of the tick-borne encephalitis virus fusion protein E at low pH. J Virol. 2002;76(8):3784–3790. doi: 10.1128/JVI.76.8.3784-3790.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fritz R, et al. Identification of specific histidines as pH sensors in flavivirus membrane fusion. J Cell Biol. 2008;183(2):353–361. doi: 10.1083/jcb.200806081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harrison SC. The pH sensor for flavivirus membrane fusion. J Cell Biol. 2008;183(2):177–179. doi: 10.1083/jcb.200809175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stiasny K, et al. Characterization of a structural intermediate of flavivirus membrane fusion. PLoS Pathog. 2007;3(2):e20. doi: 10.1371/journal.ppat.0030020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bron R, et al. Membrane fusion of Semliki Forest virus in a model system: Correlation between fusion kinetics and structural changes in the envelope glycoprotein. EMBO J. 1993;12:693–701. doi: 10.1002/j.1460-2075.1993.tb05703.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang X, Kielian M. An interaction site of the envelope proteins of Semliki Forest virus that is preserved after proteolytic activation. J Virol. 2005;337:344–352. doi: 10.1016/j.virol.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 55.Zhang X, Kielian M. Mutations that promote furin-independent growth of Semliki Forest virus affect p62-E1 interactions and membrane fusion. Virol. 2004;327:287–296. doi: 10.1016/j.virol.2004.06.037. [DOI] [PubMed] [Google Scholar]
- 56.Glomb-Reinmund S, Kielian M. fus-1, a pH-shift mutant of Semliki Forest virus, acts by altering spike subunit interactions via a mutation in the E2 subunit. J Virol. 1998;72:4281–4287. doi: 10.1128/jvi.72.5.4281-4287.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gibbons DL, et al. Multistep regulation of membrane insertion of the fusion peptide of Semliki Forest virus. J Virol. 2004;78(7):3312–3318. doi: 10.1128/JVI.78.7.3312-3318.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sanchez San Martin C, et al. A stable prefusion intermediate of the alphavirus fusion protein reveals critical features of class II membrane fusion. Cell Host Microbe. 2008;4:600–608. doi: 10.1016/j.chom.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Qin ZL, et al. Role of conserved histidine residues in the low pH-dependence of the Semliki Forest virus fusion protein. J Virol. 2009;83(9):4670–4677. doi: 10.1128/JVI.02646-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu CY, Kielian M. E1 mutants identify a critical region in the trimer interface of the Semliki Forest virus fusion protein. J Virol. 2009 doi: 10.1128/JVI.01147-09. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gibbons DL, et al. Visualization of the target-membrane-inserted fusion protein of Semliki Forest virus by combined electron microscopy and crystallography. Cell. 2003;114(5):573–583. doi: 10.1016/s0092-8674(03)00683-4. [DOI] [PubMed] [Google Scholar]
- 62.DeLano WL. The PyMOL User's Manual. DeLano Scientific; 2002. [Google Scholar]