Abstract
RNA editing revises the genetic code at precise locations, creating single base changes in mRNA. These changes can result in altered coding potential and modifications to protein function. Sequence analysis of the Shab potassium channel of Drosophila melanogaster revealed five such RNA editing sites. Four are constitutively edited (I583V, T643A, Y660C and I681V) and one undergoes developmentally regulated editing (T671A). These sites are located in the S4, S5–S6 loop and the S6 segments of the channel. We examined the biophysical consequences of editing at these sites by creating point mutations, each containing the genomic (unedited) base at one of the five sites in the background of a channel in which all other sites are edited. We also created a completely unedited construct. The function of these constructs was characterized using two-microelectrode voltage clamp in Xenopus oocytes. Each individual ‘unediting’ mutation slowed the time course of deactivation and the rise time during channel activation. Two of the mutants exhibited significant hyperpolarized shifts in their midpoints of activation. Constructs that deactivated slowly also inactivated slowly, supporting a mechanism of closed-state inactivation. One of the editing sites, position 660, aligns with the Shaker 449 residue, which is known to be important in tetraethylammonium (TEA) block. The aromatic, genomically-encoded residue tyrosine at this position in Shab enhances TEA block 14 fold compared to the edited residue, cysteine. These results show that both the position of the RNA editing site and the identity of the substituted amino acid are important for channel function.
Keywords: RNA editing, activation, inactivation, pore block, voltage-gated potassium channel, Shab
Introduction
Genetic information is systematically decoded according to the central dogma of genetics: DNA-to-RNA-to-protein. This orderly process is perturbed, however, by an enzyme called adenosine deaminase acting on RNA (ADAR). ADAR recognizes specific double-stranded structures in pre-mRNA after transcription, and deaminates specific adenosine (A) nucleotides to inosines(I).1 Inosines are then read as guanosines by all of the molecular machinery of a cell. This A → I change can therefore effectuate a variety of downstream consequences, for example, creating an alternative splice site or, most importantly for this study, altering the codon for an amino acid, thus resulting in a point mutation in the resultant protein. This type of RNA editing primarily affects genes that encode for neuronal signaling proteins such as voltage-gated potassium (KV) channels.2 KV channels are unique in that they are edited in diverse animal phyla: mollusks (sqKV2), arthropods (Shab and Shaker) and mammals (KV1.1).3–5 In this study, we discovered five RNA editing sites in the Drosophila Shab channel, a member of the KV2 super-family, using RT-PCR. The point mutation I583V occurs in the S4 voltage sensor; T643A in the pore helix; Y660C in the extracellular turret; T671A and I681V in the S6 segment. All five sites are highly edited in flies. We therefore characterized each site by systematically mutating each site back to the genomically encoded (i.e., ‘unedited’) amino acid on the background of a fully edited channel, and examined the biophysical consequences on voltage-clamped potassium currents. Our results showed myriad effects on the kinetics and voltage dependence of channel function, suggesting an important role of RNA editing in regulation of ion channel activity.
Results
We discovered that the Shab potassium channel of Drosophila melanogaster contains five RNA editing sites where particular amino acids can be recoded to alternate residues. All five sites are located on exon two of the Shab locus (Fig. 1A). Figure 2 shows these sites as colored symbols in a topological cartoon of the Shab channel. The five sites and editing mutations are (unedited/edited): Ile583Val in the S4 voltage sensor, RIMRILR(I/V)LKLARHST; Thr643Ala in the pore helix, VSIPE(T/A)FWWAG; Tyr660Cys in the extracellular turret, TVGYGDI(Y/C)PTTALGK; Thr671Ala ALGKVIG(T/A)VCCICGV and Ile681Val CCICGVLV(I/V)ALPIPIIV in the S6 segment
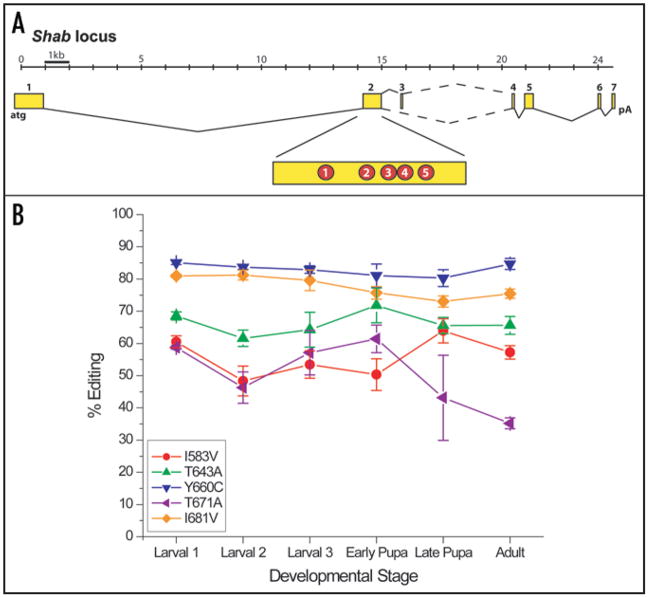
Figure 1.
Editing of the Shab locus. (A) The Shab locus transcription unit is shown with numbered exons (yellow), constitutive splice sites (solid lines) and alternative splice sites (dashed lines). The five RNA editing sites are indicated in red circles in exon 2. (B) The level of RNA editing for each site is shown as determined by whole-fly RNA preparation for three larval and two pupal stages and adult flies.
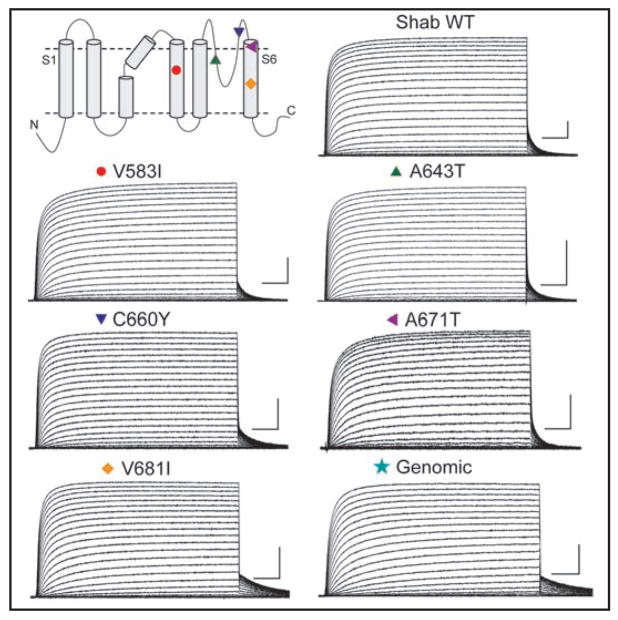
Figure 2.
Topology of the Shab channel indicating the approximate positions of the five edit sites. Also shown are representative currents for each channel. Symbols correspond to positions found on the topology model. Scale bar on each trace: 25 ms and 5 μA.
Editing sites can be developmentally regulated in both vertebrate and invertebrate systems.6–9 We determined the temporal pattern of regulation for these five sites in Shab (Fig. 1B). Four of the five sites are constitutively edited at high levels throughout development, and one site (residue 671) shows a moderate (<2-fold) decrease in editing in adult flies. These results contrast with a general increase in editing observed at several other sites in Drosophila.10,11
Because of the high level of editing in adult flies (>55% at four of the five sites), we denote the wild-type (WT) Shab channel as the fully edited variant. Consequently, each point mutant created for functional study contains the genetically encoded amino acid (the unedited form) at one of the five positions. We also created a completely unedited construct, identified as Genomic, containing the genomically encoded amino acid at each of the five sites.
To examine the functional consequences of these unediting mutations, we characterized the biophysical properties of each in channels expressed in Xenopus oocytes. Families of currents were obtained using voltage pulses from −70 mV to +70 mV in 5-mV increments from a holding potential of −100 mV (Fig. 2). None of these variants inactivates over the time and voltage range shown. Superficially the currents are comparable, although the V681I mutant and the Genomic construct deactivate more slowly than the others at the end of a depolarization (see below).
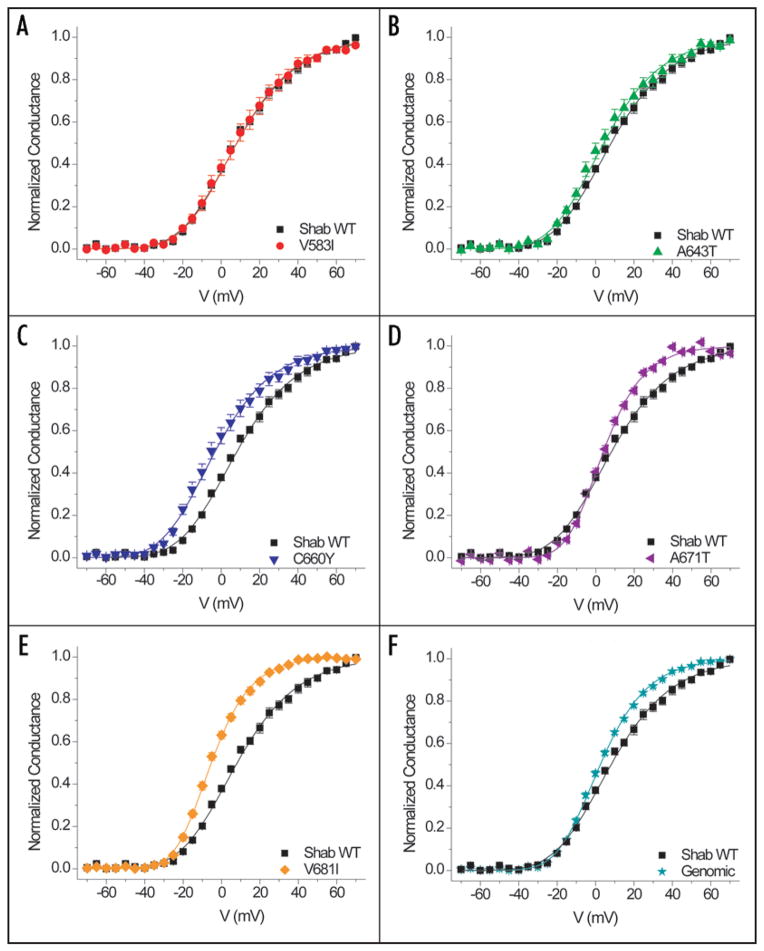
Figure 3 shows the voltage dependence of activation of each mutant compared to WT Shab. The normalized conductances were obtained from tail currents and fitted with a fourth-power Boltzmann function. Although residue 583 is housed in the critical S4 segment of the voltage sensor, V583I is the most similar to WT with indistinguishable midpoints and slopes underlying the voltage dependence of activation (Table 1 and Fig. 3A). A643T and C660Y have activation curves with shapes similar to that of the WT; however both are left shifted at their midpoints by 3.9 mV and 11.4 mV, respectively (Table 1 and Fig. 3B and C). A671T and the Genomic constructs have midpoint shifts in the same range as A643T, 4.2 mV and 5.6 mV respectively; however the activation curve is steeper at more depolarized voltages (Table 1 and Fig. 3D and F). The most prominent difference is exhibited by V681I, which has a 13.5-mV hyperpolarizing shift and a steeper voltage dependence of activation (Table 1 and Fig. 3E).
Figure 3.
Comparison of conductance-voltage relationships between WT Shab and each editing site mutant. Data fit by a Boltzmann function raised to the fourth power: Normalized conductance = [1/(1 + exp(q(V − V1/2) F/RT)]4, where q is the slope, V1/2 is the midpoint, and RT/F = 25 mV. Parameters of fits in Table 1.
Table 1.
Midpoints of activation and equivalent charge from conductance-voltage relationships
| Shab WT (n = 8) | V583I (n = 9) | A643T (n = 8) | C660Y (n = 6) | V681I (n = 8) | A671T (n = 14) | Shab Genomic (n = 9) | |
|---|---|---|---|---|---|---|---|
| V1/2 (mV) | 8.2 ± 1.2 | 8.1 ± 2.4 | 4.3 ± 2.3 | −3.2* ± 2.2 | −5.3* ± 0.8 | 4.0 ± 0.8 | 2.6 ± 0.5 |
| q (e0) | 1.3 ± 0.07 | 1.4 ± 0.1 | 1.4 ± 0.1 | 1.4 ± 0.1 | 2.0* ± 0.1 | 1.9* ± 0.06 | 1.6 ± 0.03 |
Indicates data are statistically significant compared to WT (p < 0.05, 1-way ANOVA with Scheffé correction).
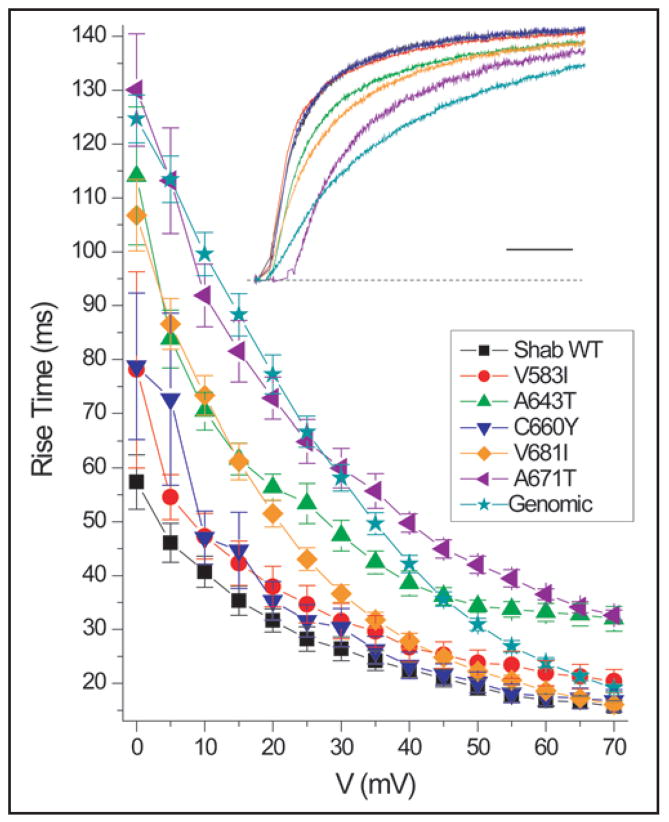
Further examination of the activation of these channels revealed differences in kinetics among the mutants. The inset of Figure 4 illustrates this comparison with color-coded sample traces showing the response to a depolarization to +10 mV. At this voltage WT (black), V583I (red) and C660Y (blue) activate most rapidly, V681I (orange) and A643T (green) activate at an intermediate rate, and the slowest activation is exhibited by A671T (violet) and Genomic (teal) constructs. These trends are also apparent in the plot of 10–90% activation rise times (Fig. 4). At more depolarized voltages, differences in rise times became less distinct for all the channel types except A643T and A671T, which remained slower than the others (Fig. 4).
Figure 4.
Activation kinetics of Shab and editing site mutants. Rise time is a measurement of the time required for channel to pass from 10% to 90% of its maximum current during a depolarization. Differences are more pronounced at more hyperpolarized voltages where the A643T (n = 7), V681I (n = 8), A671T (n = 10) and Genomic (n = 10) constructs have slower rise times. A643T and A671T continue to have slower rise times through +70 mV. For WT, V583I and C660Y (n = 7). Inset: Representative traces of each editing mutant upon activation at +10 mV. Currents were normalized to the current at 200 ms after the depolarization. Scale bar 20 ms.
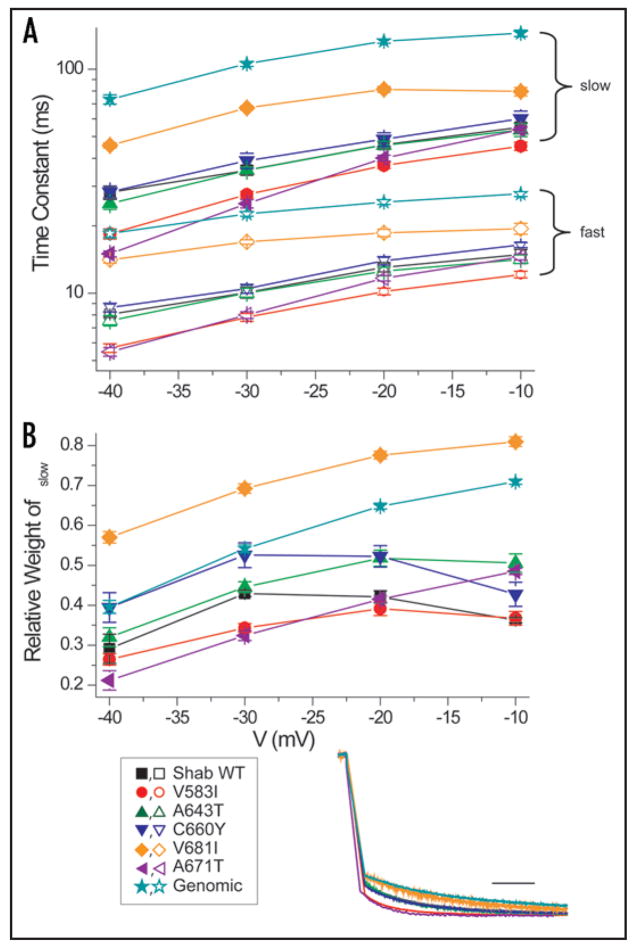
The kinetics of deactivation also differed significantly among the six variants we examined. The inset of Figure 5 shows normalized tail currents at −40 mV following a depolarization to +50 mV. As observed for activation kinetics, V683I and the Genomic construct deactivate most slowly. A671T and V583I deactivate most rapidly, and the remaining three constructs are intermediate. The time course of deactivation was fitted by a double exponential relaxation composed of a fast and slow component with time constants and weights shown in Figure 5. The slow overall kinetics of deactivation for V681I is due to both larger time constants and a preponderance of the slow component. The predominance of the slow component in the Genomic construct is especially evident at more depolarized voltages.
Figure 5.
Deactivation kinetics of WT (n = 6) and editing site mutants. (A) Deactivation kinetics of each channel have a fast (open symbol) and slow (closed symbol) component. Deactivation of V681I (n = 8) and Genomic (n = 8) are consistently slower, and V583I (n = 10) is faster, over the voltage range. The number of recorded cells was: A643T (n = 9), C660Y (n = 7) and A671T (n = 7). (B) Relative weight of the slow component of the deactivation time constant for each channel. V681I has the greatest relative weight for the slow component and V583I has the smallest. Inset: Representative traces of deactivation at −40 mV from a depolarization to +50 mV. Currents were normalized to the outward current 50 ms after the depolarization. Scale bar 10 ms.
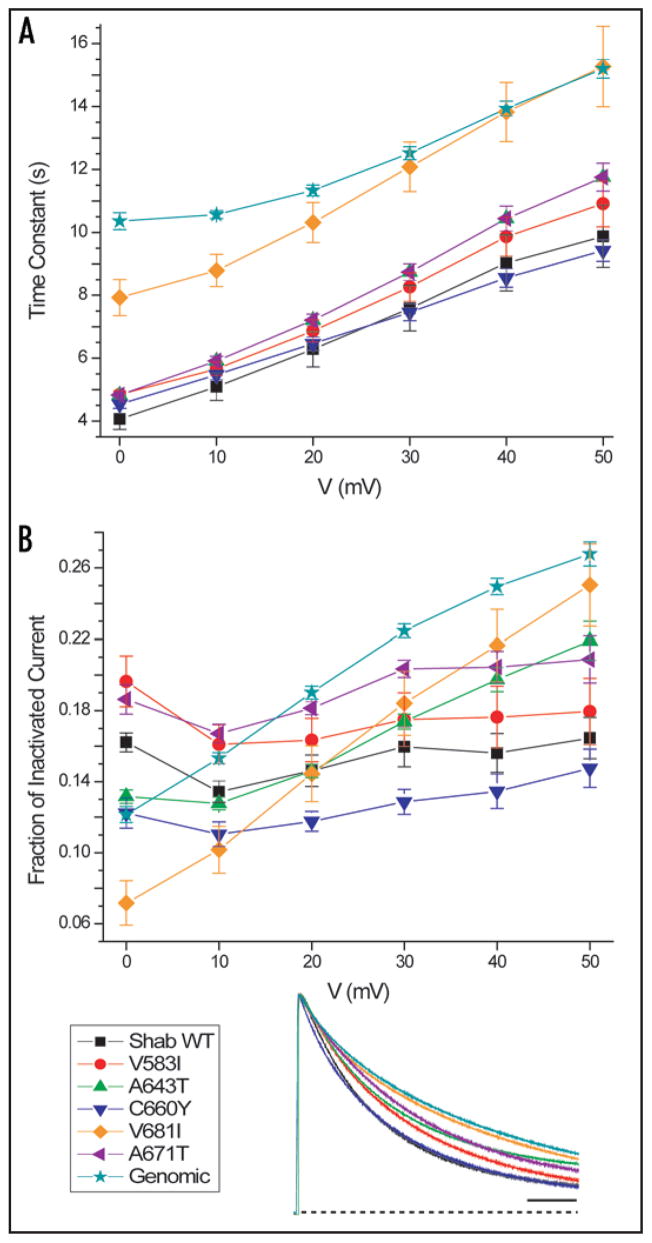
To measure inactivation kinetics we used 30-s depolarizations to six test voltages. Figure 6A shows the time constants of inactivation estimated from single exponential relaxations. The V681I and Genomic constructs inactivate most slowly. These two constructs also have the slowest kinetics of deactivation (Fig. 5A). In general, all Shab potassium currents tend to inactivate more completely at more hyperpolarized voltages (Fig. 6B). V681I and Genomic constructs exhibit both the slowest and the least complete inactivation at +50 mV (Fig. 6 inset).
Figure 6.
Inactivation kinetics of Shab RNA edit sites. (A) Inactivation time constant of editing mutant sites. V681I (n = 8) and Genomic (n = 9) have uniformly slower inactivation time constants over the voltage range. Number of recorded cells: WT (n = 8) and all other constructs (n = 7). (B) The fraction of inactivated current compares the peak current with the current at the end of the 30-s depolarization. All constructs inactivate more completely at more hyperpolarized voltages. V681I and Genomic do not inactivate as completely as the other constructs. Inset: Representative traces of inactivation at +50 mV. Currents were normalized to the peak current of the trace. Scale bar 5 s.
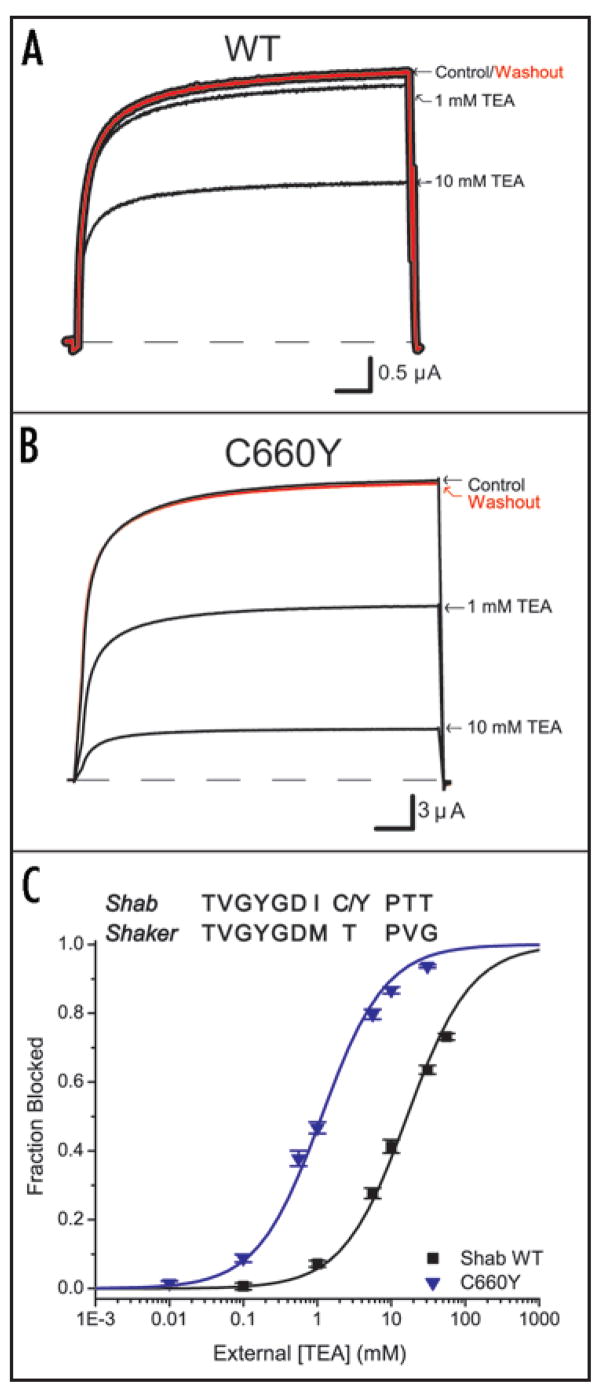
The third editing site (residue 660) aligns with residue 449 of Shaker (Fig. 7C inset). This residue at the extracellular mouth of the pore is known to contribute to the docking site for extracellular blockers like tetraethylammonium (TEA) and toxins in Shaker. Specifically, high-affinity TEA block requires either a tyrosine or phenylalanine at this position.12–14 Although the genomic sequence for this residue encodes for tyrosine, the edited (WT) residue is cysteine. To test whether this residue plays a similar role in Shab as in Shaker, we examined TEA block of WT Shab and the C660Y mutant. In accordance with previous studies, Figure 7 shows that a tyrosine at this position enhances TEA block. Figures 7A and B show reversible block at two TEA concentrations, and Figure 7C plots the fraction of blocked channels against extracellular TEA concentration. These data show that the tyrosine variant (Ki = 1.14 mM ± 0.09 mM) is 14-fold more sensitive to TEA block than the cysteine variant (Ki = 16.36 mM ± 1.04 mM), thus confirming the importance for block of an aromatic residue at this position.
Figure 7.
Influence of extracellular pore residue on TEA block. (A) and (B) Both WT and C660Y exhibit reversible TEA block at +50 mV. C660Y is more sensitive as shown at 1 mM and 10 mM TEA. Scale bars are 10 ms. (C) TEA sensitivity was measured for WT (n = 5) and C660Y (n = 6) and fit using a standard binding equation. Inset above: Amino acid sequence alignment of the pore region comparing the TEA binding site in Shab (position 660) to the corresponding site in Shaker (position 449).
Discussion
RNA editing is a strategy that allows a cell to make point mutations enzymatically without altering the archival genomic information that encodes proteins. The action of ADAR enzymes depends on a variety of factors, perhaps most critically on the formation of recognizable double-stranded RNA (dsRNA) targets.15 Further specific diversification of amino acid side-chains occurs through regulation of the levels—and activity—of different isoforms of ADAR, each with its own specificity for dsRNA structures.1 Changes in the levels of editing observed in Shab during fly development (Fig. 1B) may reflect gross changes in nervous system structure; however, they may also indicate an additional level of regulation governing ADAR enzymatic efficiency or recognition of editing sites involving yet unknown factors (e.g., chaperone proteins). The editing of mRNA encoding ion channels typically causes rather subtle changes of biophysical properties due to point mutations. However, some coding changes can produce dramatic alterations in function. The canonical example is editing of the Gln/Arg site in neuronal glutamate receptor channels.16 In this case, the unedited form of the protein is lethal, presumably because unedited channels are excessively permeant to calcium ions, which are toxic when overabundant inside neurons. By contrast, our data show that editing produces rather modest effects on the gating of Shab channels. These results are in accord with studies of the orthologous potassium channel (sqKV2) in squid. We will discuss the differences between RNA editing of these two channels below.
The predominant functional consequences of RNA editing on Shab are alterations of the voltage dependence and kinetics of channel gating. We also observed an order-of-magnitude alteration of the affinity of an extracellular blocker in response to a single point mutation. Effects on gating are not surprising in that the point mutations occur in highly conserved locations of the channel protein, either in the voltage-sensing S4 segment or in the pore domain that houses the activation, and perhaps the inactivation, gates. Nevertheless, the effects would have been difficult to predict, because except for one case (Y660C), the editing mutations are all relatively conservative, and all of them preserve the charge of the side-chain. The tyrosine-cysteine switch, although involving two uncharged residues, is accompanied by a change of aromaticity. Because Shab channels are edited at levels >50% in four of the five sites (Fig. 1B), we used the fully edited channel as our wild-type (WT) and examined the effects of single ‘unediting’ mutations at the five sites. We also created a completely unedited (Genomic) channel.
Two out of the six ‘unediting’ constructs produced significant hyperpolarizing shifts in the activation G-V curve, compared to the fully edited WT. The most prominent effect, a 13.5-mV shift, is found in V681I in the S6 segment, the construct with the steepest voltage dependence of activation. The main difference between valine and isoleucine is that the latter is ~18% larger in volume due to an extra methyl group. This residue apparently orients towards the aqueous cavity of the permeation pathway,17–19 where it can interact with intracellular pore blockers like the inactivation gate of KV1 channels.5,17 The effect of this point mutation on activation gating may be due to its proximity to a downstream Pro-Ile-Pro motif (Fig. 8) that is somehow involved in opening and closing of the activation gate.20–24 Unexpectedly, mutation of the editing site in the voltage sensing domain, residue 583 in the middle of the S4 segment, had no effect on the voltage dependence of activation, reflected in the G-V relationship. Summarizing, RNA editing of the three most downstream editing sites each caused small depolarizing shifts of activation. Therefore, editing of these three sites tends to make the Shab channel less prone to open, which would enhance the excitability of a neuron containing the edited channels.
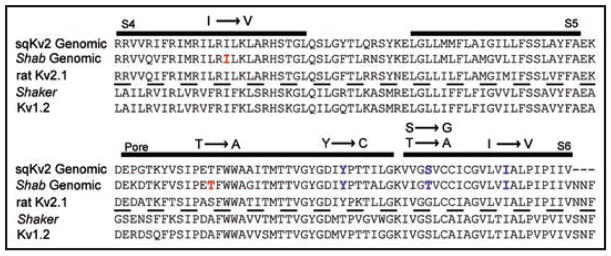
Figure 8.
Aligned sequences of squid, Drosophila Shab (fully genomic), rat KV2.1, Drosophila Shaker and rat KV1.2. In red are two RNA editing sites found only in Shab. Shown in blue are three RNA editing sites found in both Shab and sqKV2. Note the second site has the same location but a different resultant amino acid change for squid (S → G) vs. Drosophila (T → A). Figure adapted from Patton et al.3
Each mutagenic departure from the fully edited WT construct slows activation kinetics. The slowest activation is exhibited by A671T, located just above the middle of the pore helix.18 The side-chain of this residue is oriented towards the S5 segment of its own subunit. It could therefore affect gating movements that involve relative displacements of these two helices. Perhaps the larger and more hydrophilic threonine side-chain interferes mildly with such a tertiary conformational change. Notably, this site is the least edited in adult flies (<50%; Fig. 1B). Therefore, the genomic residue, threonine, is most prevalent. The slower activation of Shab channels with Thr671 would tend to make neurons with this residue more excitable.
Deactivation, the closing of the activation gate, differs for each construct. It has complicated kinetics, in that the time course requires a double-exponential fit. Deactivation is slowest in the Genomic construct, indicating that, in general, editing speeds deactivation. V681I, near the bottom of the S6 segment, also has significantly slower deactivation than WT due both to larger time constants and to the largest proportion of the slow component among the mutants. Inactivation time constants for both Genomic and V681I are also significantly slower than in the other constructs. V583I in the S4 segment deactivates faster than completely edited Shab. The reason for this effect is not obvious. In the recent crystal structure of an open KV channel,18 the homologous residue is an isoleucine and is oriented towards the space, perhaps an aqueous vestibule, between the voltage sensing and pore domains. This putative aqueous vestibule may have a dynamic, voltage-sensitive contour that contributes to voltage-dependent gating.25 Residues lining aqueous vestibules are likely to play a role in shaping the electric field in the vicinity of the S4 segment.
The aromatic, genomic residue tyrosine at the 660 position in Shab, which aligns with the well-known Shaker 449 position, enhances TEA block 14 fold compared to the edited residue, cysteine. This result verifies the importance of the aromatic residue at this position for TEA block of potassium channels. There are two reported orientations of tyrosines observed in crystal structures of KcsA potassium channels,26,27 one of which may correspond to the en face orientation of an aromatic side-chain at this position in Shaker potassium channels.28 A clue to the orientation of this tyrosine in Shab, and the peptide backbone in this region, could be provided by its inhibition constant for TEA, ~1 mM. This is however roughly midway between the affinities for TEA block of Shaker and wild-type KcsA,14,29 leaving the aromatic’s orientation undefined for the moment. Because this residue is situated at the outer mouth of the pore where it can interact with extracellular pore blockers, it raises the possibility that an editing mutation of this residue might affect the binding of some unidentified toxin or endogenous molecule that plugs the pore. Thus, mutation of this residue might play a role in the co-evolution of Drosophila with a poisonous predator or food source.
Our initial examination of Shab currents failed to reveal an inactivation process. This was surprising because of a previous study demonstrating so-called U-type inactivation in mammalian KV2.1 channels.30 This type of inactivation is evident even for short-duration depolarizations, given repetitively. It is manifest as a form of cumulative inactivation, indicating that inactivation proceeds more rapidly from the holding potential than from a depolarized potential that activates channel opening.30 We tested for cumulative inactivation, without success, using voltage protocols that clearly revealed inactivation in mammalian KV2.1 channels (data not shown). However, KV2 channels from squid have a very slow inactivation process that can be observed during activating depolarizations.3 We observed a similar inactivation process in response to 30-s depolarizations (Fig. 6). One shared feature between these orthologs is that in both cases depolarization decreased the rate of inactivation, in contrast to the KV1 phenotype observed in Shaker channels.31 The voltage dependence of KV2 channel inactivation, as well as the cumulative inactivation observed for mammalian KV2.1 channels, suggests that all of these channels tend to inactivate from pre-open closed states along the activation pathway, a feature that appears to be shared with KV3 and KV4 channels32,33
There is some overlap in the RNA editing sites and amino acid changes in Drosophila and squid potassium channels from the KV2 superfamily. Two of the five sites we describe in Drosophila are not edited in the sqKV2 gene (note the red sites in Fig. 8), and one of the three remaining editing sites, found in both Drosophila and squid, produces a different point mutation in sqKV2. Whereas editing of Thr671 in Shab changes this residue to alanine, the aligned mutation in sqKV2 converts a serine to a glycine. Moreover, the consequences of these changes differ in each channel type. For example, the Y → C editing site in sqKV2 (Fig. 8) does not significantly influence the conductance-voltage relationship, and it slows deactivation by a factor of two compared to the genomic construct.3 The opposite change in Shab (C → Y) in a fully edited background produces a significant hyperpolarizing shift in the midpoint of activation and has very little impact on the deactivation time constant relative to the all-edited construct (Table 1 and Fig. 5A). Another difference between KV2 channels in squid and Drosophila concerns inactivation. In squid, the most pronounced effect on the inactivation rate is a ~1.5-fold slowing of inactivation when cysteine is substituted for the genomic tyrosine at the residue that aligns with Shab’s 660. Our results reveal almost no effect on inactivation rate when mutating this residue in Shab. The largest effect we observed for a point mutation was at residue 681 in the S6 segment (Fig. 6). The inactivation rate is faster for the edited residue, valine, than for the genomic isoleucine. This agrees with the results for sqKV2, although the magnitude of the effect is larger in Shab. The conservation of editing sites between these two orthologs suggests that their role is preserved by evolutionary selection. The challenge for us is to learn whether this conservation is due to the biophysical consequences of the editing mutations, or perhaps due to effects of the mutations on the biogenesis or trafficking of the channels in the nervous systems of these animals. A further question concerns the need for enzymatic regulation of these sites and the role of that regulation in development and adaptation to changes in the environment. In the sqKV1.1 channel, for example, twenty-two of twenty-six RNA editing sites substitute a larger amino acid for a smaller one.34 This same pattern is seen in intracellular proteins from organisms that have adapted to cold environments; the smaller, edited amino acid may allow the proteins more flexibility at lower temperatures.34
Materials and Methods
RNA editing analysis
All RNA extractions were performed using TRIzol (Invitrogen) on whole flies. Shab transcripts were amplified by RT-PCR using gene-specific primers at all steps. Levels of editing for individual editing sites were obtained by direct sequencing of RT-PCR products from at least three independent reactions per sample. Areas under the curves were determined from electropherogram traces and editing level expressed as {% editing} = (area G/total area A + G) * 100.
Molecular biology for expression studies
All Shab constructs were inserted into the pBSTA vector for expression in oocytes. Editing mutations at the four sites were generated using the QuikChange Site-Directed Mutagenesis Kit (Stratagene) and verified by direct sequencing. Each of the five ‘unediting’ point mutations converted the edited amino acid to the genomically encoded one, in the background of the fully edited channel. We also created a completely unedited construct, identified as Genomic, containing the genomically encoded amino acid at each of the five sites. cRNA was prepared by in vitro transcription (mMessage mMachine Ultra, Ambion).
Electrophysiology
Stage V to VI Xenopus oocytes were injected with cRNA. The oocytes were kept at 18°C for 1–2 d until they were used for recording. Potassium current was recorded with the two-microelectrode voltage clamp technique using an OC-725C voltage clamp (Warner Instruments, Hamden, CT) in a standard Ringers solution (in mM): 116 NaCl, 2 KCl, 1.8 MgCl2, 2 CaCl2, 5 HEPES, pH 7.6. Recording electrodes were filled with 3 M KCl. Perfusion experiments were performed with an ALA VM8 (ALA Scientific) gravity-flow delivery system. Summarized data are presented as mean ± SEM.
Acknowledgments
The Shab11 channel was a gift from Dr. Lawrence Salkoff.
Abbreviations
- ADAR
adenosine deaminase acting on RNA
- KV
voltage-gated potassium channel
- TEA
tetraethylammonium
References
- 1.Bass BL. RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem. 2002;71:817–46. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seeburg PH, Hartner J. Regulation of ion channel/neurotransmitter receptor function by RNA editing. Curr Opin Neurobiol. 2003;13:279–83. doi: 10.1016/s0959-4388(03)00062-x. [DOI] [PubMed] [Google Scholar]
- 3.Patton DE, Silva T, Bezanilla F. RNA editing generates a diverse array of transcripts encoding squid KV2 K + channels with altered functional properties. Neuron. 1997;19:711–22. doi: 10.1016/s0896-6273(00)80383-9. [DOI] [PubMed] [Google Scholar]
- 4.Hoopengardner B, Bhalla T, Staber C, Reenan R. Nervous system targets of RNA editing identified by comparative genomics. Science. 2003;301:832–6. doi: 10.1126/science.1086763. [DOI] [PubMed] [Google Scholar]
- 5.Bhalla T, Rosenthal JJC, Holmgren M, Reenan R. Control of human potassium channel inactivation by editing of a small mRNA hairpin. Nature Struct Biol. 2004;11:950–6. doi: 10.1038/nsmb825. [DOI] [PubMed] [Google Scholar]
- 6.Lomeli H, Mosbacher J, Melcher T, Hoger T, Geiger JR, Kuner T, Monyer H, Higuchi M, Bach A, Seeburg PH. Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science. 1994;266:1709–13. doi: 10.1126/science.7992055. [DOI] [PubMed] [Google Scholar]
- 7.Bernard A, Khrestchatisky M. Assessing the extent of RNA editing in the TMII regions of GluR5 and GluR6 kainate receptors during rat brain development. J Neurochem. 1994;62:2057–60. doi: 10.1046/j.1471-4159.1994.62052057.x. [DOI] [PubMed] [Google Scholar]
- 8.Keegan LP, Brindle J, Gallo A, Leroy A, Reenan RA, O’Connell MA. Tuning of RNA editing by ADAR is required in Drosophila. Embo J. 2005;24:2183–93. doi: 10.1038/sj.emboj.7600691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohlson J, Pedersen JS, Haussler D, Ohman M. Editing modifies the GABA(A) receptor subunit alpha3. RNA. 2007;13:698–703. doi: 10.1261/rna.349107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanrahan CJ, Palladino MJ, Ganetzky B, Reenan RA. RNA editing of the Drosophila para Na+ channel transcript. Evolutionary conservation and developmental regulation. Genetics. 2000;155:1149–60. doi: 10.1093/genetics/155.3.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Palladino MJ, Keegan LP, O’Connell MA, Reenan RA. dADAR, a Drosophila double-stranded RNA-specific adenosine deaminase is highly developmentally regulated and is itself a target for RNA editing. RNA. 2000;6:1004–18. doi: 10.1017/s1355838200000248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.MacKinnon R, Yellen G. Mutations affecting TEA blockade and ion permeation in voltage-activated K+ channels. Science. 1990;250:276–9. doi: 10.1126/science.2218530. [DOI] [PubMed] [Google Scholar]
- 13.Kavanaugh MP, Varnum MD, Osborne PB, Christie MJ, Busch AE, Adelman JP, North RA. Interaction between tetraethylammonium and amino acid residues in the pore of cloned voltage-dependent potassium channels. J Biol Chem. 1991;266:7583–7. [PubMed] [Google Scholar]
- 14.Heginbotham L, MacKinnon R. The aromatic binding site for tetraethylammonium ion on potassium channels. Neuron. 1992;8:483–91. doi: 10.1016/0896-6273(92)90276-j. [DOI] [PubMed] [Google Scholar]
- 15.Higuchi M, Single FN, Kohler M, Sommer B, Sprengel R, Seeburg PH. RNA editing of AMPA receptor subunit GluR-B: a base-paired intron-exon structure determines position and efficiency. Cell. 1993;75:1361–70. doi: 10.1016/0092-8674(93)90622-w. [DOI] [PubMed] [Google Scholar]
- 16.Seeburg PH. A-to-I editing: new and old sites, functions and speculations. Neuron. 2002;35:17–20. doi: 10.1016/s0896-6273(02)00760-2. [DOI] [PubMed] [Google Scholar]
- 17.Zhou M, Morais-Cabral JH, Mann S, MacKinnon R. Potassium channel receptor site for the inactivation gate and quaternary amine inhibitors. Nature. 2001;411:657–61. doi: 10.1038/35079500. [DOI] [PubMed] [Google Scholar]
- 18.Long SB, Tao X, Campbell EB, MacKinnon R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007;450:376–82. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- 19.Panyi G, Deutsch C. Probing the cavity of the slow inactivated conformation of Shaker potassium channels. J Gen Physiol. 2007;129:403–18. doi: 10.1085/jgp.200709758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holmgren M, Shin KS, Yellen G. The activation gate of a voltage-gated K+ channel can be trapped in the open state by an intersubunit metal bridge. Neuron. 1998;21:617–21. doi: 10.1016/s0896-6273(00)80571-1. [DOI] [PubMed] [Google Scholar]
- 21.Del Camino D, Holmgren M, Liu Y, Yellen G. Blocker protection in the pore of a voltage-gated K+ channel and its structural implications. Nature. 2000;403:321–5. doi: 10.1038/35002099. [DOI] [PubMed] [Google Scholar]
- 22.Del Camino D, Yellen G. Tight steric closure at the intracellular activation gate of a voltage-gated K+ channel. Neuron. 2001;32:649–56. doi: 10.1016/s0896-6273(01)00487-1. [DOI] [PubMed] [Google Scholar]
- 23.Labro AJ, Raes AL, Bellens I, Ottschytsch N, Snyders DJ. Gating of Shaker-type channels requires the flexibility of S6 caused by prolines. J Biol Chem. 2003;278:50724–31. doi: 10.1074/jbc.M306097200. [DOI] [PubMed] [Google Scholar]
- 24.Webster SM, Del Camino D, Dekker JP, Yellen G. Intracellular gate opening in Shaker K+ channels defined by high-affinity metal bridges. Nature. 2004;428:864–8. doi: 10.1038/nature02468. [DOI] [PubMed] [Google Scholar]
- 25.Nguyen TP, Horn R. Movement and crevices around a sodium channel S3 segment. J Gen Physiol. 2002;120:419–36. doi: 10.1085/jgp.20028636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doyle DA, Cabral JM, Pfuetzner RA, Kuo AL, Gulbis JM, Cohen SL, Chait BT, MacKinnon R. The structure of the potassium channel: Molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 27.Lenaeus MJ, Vamvouka M, Focia PJ, Gross A. Structural basis of TEA blockade in a model potassium channel. Nature Struct Biol. 2005;12:454–9. doi: 10.1038/nsmb929. [DOI] [PubMed] [Google Scholar]
- 28.Ahern CA, Eastwood AL, Lester HA, Dougherty DA, Horn R. A cation-π interaction between extracellular TEA and an aromatic residue in potassium channels. J Gen Physiol. 2006;128:649–57. doi: 10.1085/jgp.200609654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heginbotham L, LeMasurier M, Kolmakova-Partensky L, Miller C. Single Streptomyces lividans K+ channels: Functional asymmetries and sidedness of proton activation. J Gen Physiol. 1999;114:551–9. doi: 10.1085/jgp.114.4.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klemic KG, Shieh CC, Kirsch GE, Jones SW. Inactivation of KV2.1 potassium channels Biophys. J. 1998;74:1779–89. doi: 10.1016/S0006-3495(98)77888-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoshi T, Zagotta WN, Aldrich RW. Two types of inactivation in Shaker K+ channels: effects of alterations in the carboxy-terminal region. Neuron. 1991;7:547–56. doi: 10.1016/0896-6273(91)90367-9. [DOI] [PubMed] [Google Scholar]
- 32.Klemic KG, Kirsch GE, Jones SW. U-type inactivation of KV3.1 and Shaker potassium channels. Biophys J. 2001;81:814–26. doi: 10.1016/S0006-3495(01)75743-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaulin YA, Santiago-Castillo JA, Rocha CA, Covarrubias M. Mechanism of the modulation of KV4:KChIP-1 channels by external K+ Biophys J. 2008;94:1241–51. doi: 10.1529/biophysj.107.117796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosenthal JJC, Bezanilla F. Extensive editing of mRNAs for the squid delayed rectifier K+ channel regulates subunit tetramerization. Neuron. 2002;34:743–57. doi: 10.1016/s0896-6273(02)00701-8. [DOI] [PubMed] [Google Scholar]