Abstract
In this study, our high throughput microRNA (miRNA) expression analysis revealed that the expression of miR-140 was associated with chemosensitivity in osteosarcoma tumor xenografts. Tumor cells ectopically transfected with miR-140 were more resistant to methotrexate (MTX) and 5-fluorouracil (5-FU). Overexpression of miR-140 inhibited cell proliferation in both osteosarcoma U-2 OS (wt-p53) and colon cancer HCT 116 (wt-p53) cell lines, but less so in osteosarcoma MG63 (mut-p53) and colon cancer HCT 116 (null-p53) cell lines. miR-140 induced p53 and p21 expression accompanied with G1 and G2 phase arrest only in cell lines containing wild type of p53. Histone deacetylase 4 (HDAC4) was confirmed to be one of the important targets of miR-140. The expression of endogenous miR-140 was significantly elevated in CD133+hiCD44+hi colon cancer stem-like cells which exhibit slow proliferating rate and chemoresistance. Blocking endogenous miR-140 by locked nucleic acid (LNA) modified anti-miR partially sensitized resistant colon cancer stem-like cells to 5-FU treatment. Taken together, our findings indicate that miR-140 is involved in the chemoresistance by reduced cell proliferation via G1 and G2 phase arrest mediated in part, through the suppression of HDAC4. miR-140 might be a candidate target to develop novel therapeutic strategy to overcome drug resistance.
Keywords: miR-140, chemosensitivity, histone deacetylase 4, cancer stem cells
Introduction
miRNAs are a class of small non-coding regulatory RNA molecules that recently have been shown to play important roles in a wide array of biological processes (Ambros and Lee, 2004; Bartel, 2004; Pillai et al., 2007; Plasterk, 2006; Zhang et al., 2007). miRNAs regulate mRNA translation mainly by binding to the 3′-UTR of their target mRNA transcripts to inhibit translation by sequence-specific mRNA cleavage or translational repression, although the detailed molecular mechanism of miRNAs in regulating target mRNA translation is still not being fully elucidated (Kozak, 2008). Precise chronological and topological regulation of posttranscriptional gene silencing by miRNAs is essential for animal development and tissue differentiation (Alvarez-Garcia and Miska, 2005). Abnormal expression of miRNAs is suggested to be associated with various human disorders, including cancer (Calin and Croce, 2006; Lu et al., 2005; Volinia et al., 2006).
In this study, we utilized high throughput miRNA expression analysis to reveal miRNAs associated with chemosensitivity using osteosarcoma tumor xenografts treated with chemotherapeutic agents doxorubicin, cisplatin, and or ifosfamide. Clustering and Venn diagram analysis revealed three miRNAs (miR-140, miR-217 and miR-431) overlapped with these compounds. Among these, miR-140 showed consistent high expression levels across all the three drugs, suggesting that miR-140 may contribute to a broad spectrum of chemoresistance mechanism. miR-140 was recently reported to target histone deacetylase 4 (HDAC4) based on a luciferase reporter assay in mouse cells (Tuddenham et al., 2006). Histone deacetylases (HDACs) mediate changes in nucleosome conformation and are important in the regulation of gene expression (Finnin et al., 1999). HDACs are also involved in cell-cycle progression and differentiation, and their deregulation is associated with several cancers (Yang and Grégoire, 2005). Histone acetylation is important for regulating DNA chromatin structure and transcriptional control (Eberharter and Becker, 2002; Grozinger and Schreiber, 2002; Sengupta and Seto, 2004). HDAC isozyme can be categorized into three classes and HDAC4 belongs to class II, which can be regulated and shuttled between the cytoplasm and the nucleus in response to various signal transduction stimuli. In addition, class II HDACs exert their transcriptional co-repressor functions by interaction with other co-repressors or direct binding to (and sequestering) sequence-specific transcriptional factors such as MEF2, Runx3, and nuclear factor κB (NF-κB) (Grozinger and Schreiber, 2002; Yang and Grégoire, 2005). Recent studies have shown that HDAC4 promotes growth of colon cancer cells via repression of p21 (Wilson et al., 2008). We reasoned that miR-140 have a broad impact on cell proliferation and chemosensitivity through the regulation of HDAC4 expression.
We directly investigated the functional significance of miR-140 in cancer biology and its potential role in chemoresistance in both human osteosarcoma and colon cancer cell lines. HCT 116 (wt-p53) colon cancer cells transfected with miR-140 were more resistant to chemotherapeutic agents such as methotrexate (MTX) and 5-fluorouracil (5-FU). Overexpression of miR-140 significantly inhibited cell proliferation in cancer cell lines containing wild type of p53, and this maybe the major contributing factor to the chemoresistance phenotype. It was achieved, at least in part, by the induction of both G1 and G2 cell cycle arrest along with induction of p21. This effect, however, was largely absent in cell lines with either mutant or null p53. These results indicated that the impact of miR-140 on cell proliferation and cell cycle control was dependant on the presence of functional wild type of p53. We confirmed that one of the important targets of miR-140 was HDAC4. The expression of endogenous miR-140 was highly elevated in CD133+hiCD44+hi colon cancer stem-like cells compared to control colon cancer cells, further suggesting that tumor stem-like cells may be avoiding cellular and DNA damage caused by chemotherapy with a reduced proliferating phenotype mediated, at least in part, by miR-140. The resistance can be partially reversed by treating cells with Locked Nucleic Acid (LNA) modified anti-miR140. Furthermore, miR-140 expression level was decreased in clinical colon cancer specimens compared to adjacent normal tissues of the same patients, suggesting the lowered levels of miR-140 in tumors may be contributing to proliferating phenotype in differentiated colon cancer cells. miR-140 might be a candidate target to develop novel therapeutic strategy to overcome drug resistance.
Results
miR-140 expression is associated with chemosensitivity
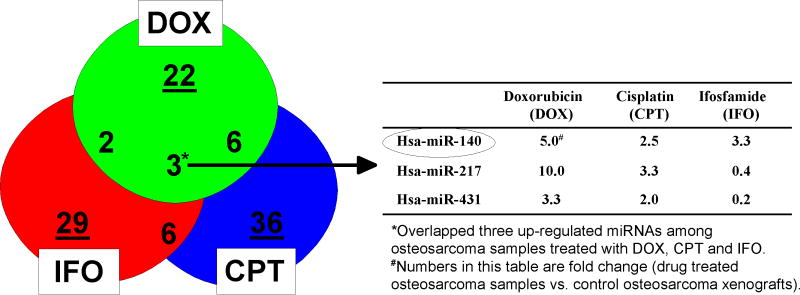
Based on the important roles of miRNAs in cancer, we reasoned that certain miRNAs may contribute to the mechanisms of cancer chemoresistance. To identify miRNAs associated with chemosensitivity, miRNA expression was screened using a panel of osteosarcoma tumor xenografts treated with doxorubicin, cisplatin and ifosfamide. Supplementary Table 1 showed the classification of human osteosarcoma xenografts according to their response (++++/+++/++ good, or -/+ poor) to doxorubicin, cisplatin, and ifosfamide. Using Luminex bead based high throughput miRNA expression analysis, we analyzed miRNA expression from control osteosarcoma xenografts and samples treated with doxorubicin, cisplatin, and ifosfamide. Normalized miRNA expression results were analyzed by ANOVA using GeneSpring expression analysis software. Differentially expressed miRNAs (average fold change) affected by these drugs were listed in Supplementary Table 2. Venn diagram analysis revealed that miR-140, miR-217 and miR-431 were overlapped among all three agents (Figure 1). Among these, miR-140 showed consistent high expression levels across all three drugs (5.0-, 2.5- and 3.3-fold respectively), suggesting miR-140 may play important roles in a broad spectrum of chemoresistance, and this has led us to focus on miR-140 in this study.
Figure 1.
Venn diagram analysis of overlapping miRNAs based on chemosensitivity to doxorubicin, cisplatin and ifosfamide in human osteosarcoma xenografts.
Overexpression of miR-140 causes chemoresistance to MTX and 5-FU
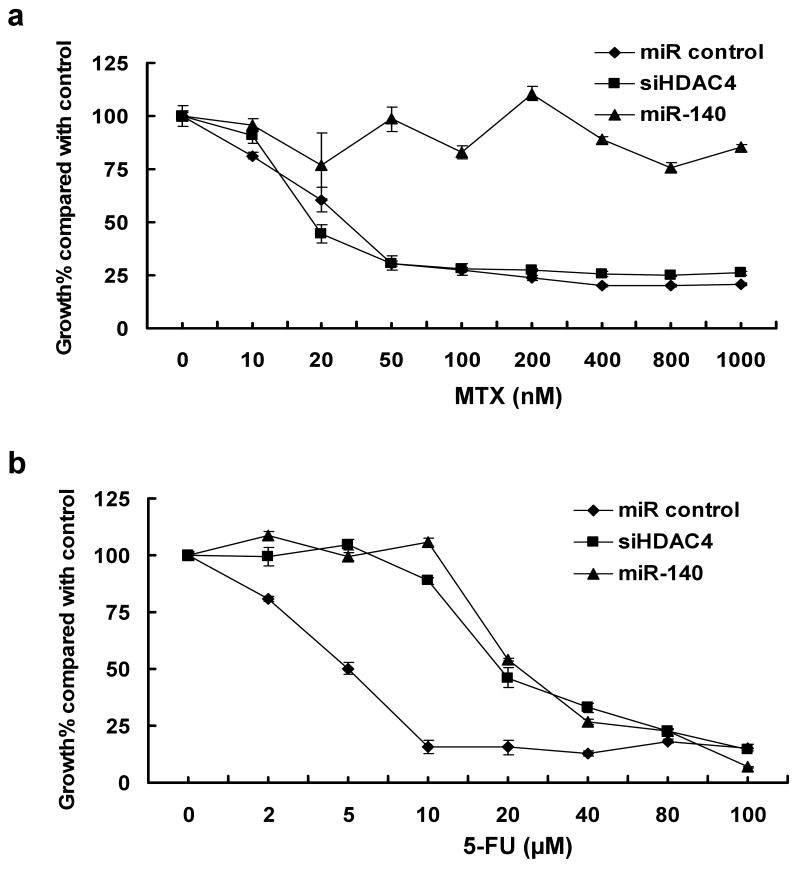
To directly test the relationship of miR-140 with the chemoresistance, HCT 116 (wt-p53) cells were transfected with miR-140, non-specific miR (miR control), and siRNA against HDAC4 to evaluate the impact of miR-140 on chemosensitivity to MTX and 5-FU treatment. We found cells with elevated miR-140 were more resistant to MTX (Figure 2a) and 5-FU (Figure 2b) compared to miR control cells.
Figure 2.
Chemosensitivity analysis in HCT 116 (wt-p53) cells. Cells were transfected with 100nM miR-140, miR control, or siHDAC4, then treated with MTX (a) or 5-FU (b) for 72 h, and cell viability was determined by the WST-1 assay. miR control was used as the negative control. Numbers are indicated as mean ± SD.
Effect of miR-140 on cell proliferation and cell cycle control
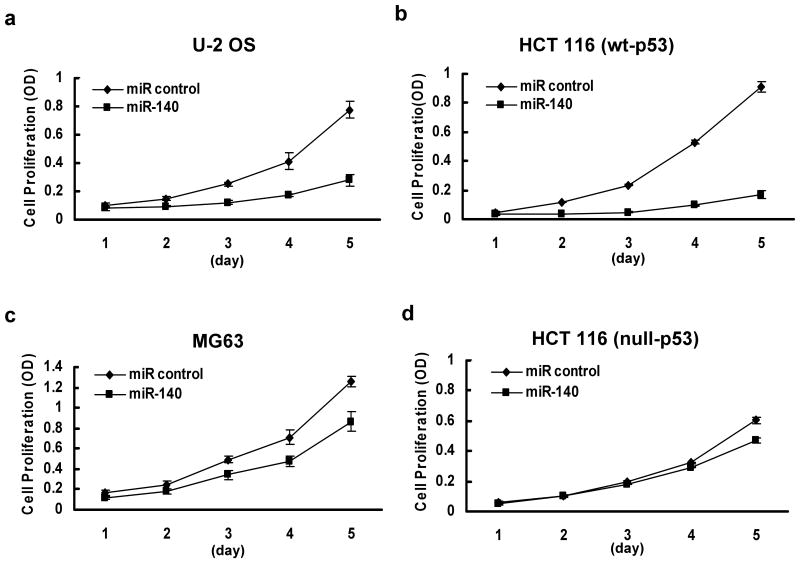
To investigate the possible mechanisms of miR-140 in chemoresistance, we first evaluated the impact of miR-140 on cell proliferation using osteosarcoma cell lines U-2 OS (wt-p53) and MG63 (mut-p53), and colon cancer cell lines HCT 116 (wt-p53) and HCT 116 (null-p53). Non-specific miR was used as the negative control. Although previous report indicated that miR-140 may be cartilage specific based on in situ hybridization (Tuddenham et al., 2006), we did find miR-140 expressed in some of the colon cancer cell lines using real time qRT-PCR analysis. This was consistent with other reports that miR-140 was expressed in a number of different tumor types (Iorio et al., 2007; Izzotti et al., 2009). Our results showed that overexpression of miR-140 can suppress cell proliferation in U-2 OS cells by 64.05±4.01% (Figure 3a), in HCT 116 (wt-p53) by 81.4±3.75% (Figure 3b), with less impact on MG63 cells (31.3±4.96%) (Figure 3c) and HCT 116 (null-p53) cells (22.42±1.88%) (Figure 3d) on day 5. By contrast, the miR control had no effect on cell proliferation, indicating that this effect caused by miR-140 is highly specific.
Figure 3.
Impact of miR-140 on cell proliferation in U-2 OS cells (wt-p53) (a), HCT 116 (wt-p53) cells (b), MG63 cells (mut-p53) (c) and HCT 116 (null-p53) cells (d). Each cell group was transfected with 100 nM miR control or miR-140, cell numbers were determined by the WST-1 assay. All the data represent mean ± SD.
To determine whether the impact of miR-140 on cell proliferation is related to cell cycle control, we analyzed the effect of miR-140 on cell cycle by flow cytometry using U-2 OS, MG63, HCT 116 (wt-p53) and HCT 116 (null-p53) cells transfected with miR control or miR-140. Our results showed that miR-140 induced both G1 (>3-fold) and G2 arrest (>2-fold) in HCT 116 (wt-p53) cells (Figure 4a). By contrast, this effect has not been observed in HCT 116 (null-p53) cells (Figure 4a). The results in U-2 OS and MG63 cells were consistent with HCT 116 (wt-p53) and HCT 116 (null-p53) cells respectively (data not shown). Thus, miR-140 inhibits cell proliferation and triggers cell cycle arrest possibly via p53 dependent manner.
Figure 4.
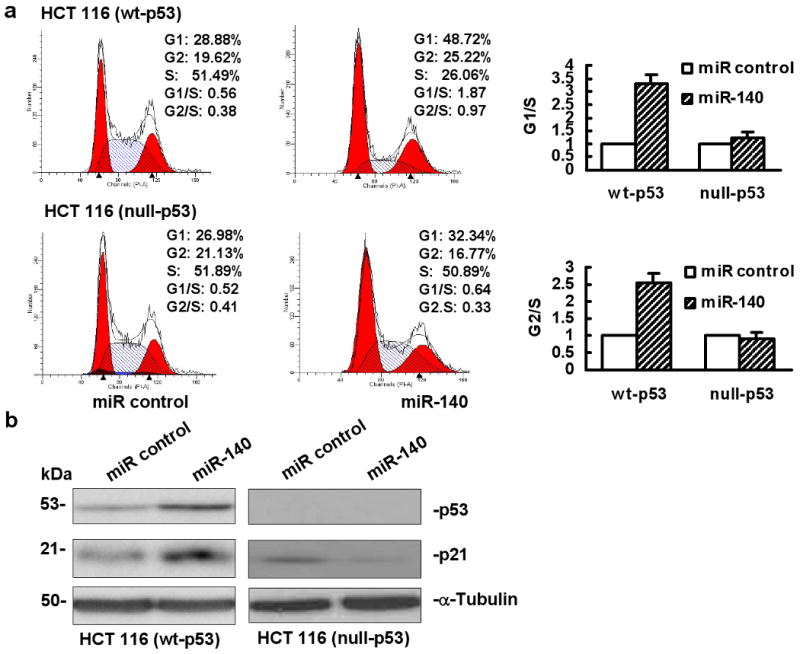
Impact of miR-140 on cell cycle. (a) Cell cycle analysis by flow cytometry in U-2 OS and MG63 cells or HCT 116 (wt-p53) and HCT 116 (null-p53) cells transfected with 100 nM miR control or miR-140. The values of G1/S and G2/S ratio in the miR control were set as 1, the bar graphs showed the relative quantity of G1/S and G2/S ratio in the miR-140 transfected cells compared to the miR control as mean ± SD. This experiment was repeated two separate times, and similar results were obtained. The representative flow cytometry pattern was shown. (b) Western immunoblot analysis of p53 and p21 expression in HCT 116 (wt-p53) (left panel) and HCT 116 (null-p53) (right panel) cells, α-tubulin was used as a protein loading control.
To further determine whether the cell cycle regulating genes are related with miR-140 overexpression, we analyzed the expression of cell cycle regulating genes p53 and p21. Figure 4b showed the results of p53 and p21 expression by Western immunoblot in HCT 116 (wt-p53) and HCT 116 (null-p53) cells. Ectopic overexpression of miR-140 increased the expression of both p53 and p21 proteins (Figure 4b, left panel, lane 2) in HCT 116 (wt-p53) cells. There was no increase of p21 expression in HCT 116 (null-p53) cells (Figure 4b, right panel, lane 2). The expression of p53 and p21 results from osteosarcoma cell lines U2-OS and MG63 were consistent with the data from colon cancer cell lines (Supplementary Figure 1).
Induction of endogenous miR-140 expression by MTX treatment in wild type p53 cell lines
To investigate whether endogenous miR-140 level may be associated with DNA damaging agent such as MTX treatment, the expression of miR-140 was quantified using real time qRT-PCR analysis from RNAs isolated in control and miR-140 transfected colon cancer cells. Our results showed that the induction of p53 protein by MTX treatment in HCT 116 (wt-p53) cells caused a significant increase of miR-140, but not in HCT 116 (null-p53) cells (Supplementary Figure 2). These results indicate that endogenous miR-140 level is associated with functional p53 after genotoxic stress by MTX treatment.
Translational regulation of HDAC4 expression by miR-140
A recent report demonstrated that miR-140 targeted HDAC4 at the 3′-UTR in mouse cells to regulate translation (Tuddenham et al., 2006). We then analyzed the human miR-140 sequence and confirmed that mouse mmu-miR-140 has the same sequence as human hsa-miR-140 and it is highly conserved (Figure 5a). HDAC4 was reported to promote the growth of colon cancer cells (Wilson et al., 2008). To experimentally confirm that the expression of HDAC4 is indeed regulated by miR-140 in human cell lines, we overexpressed miR-140 by transient transfection in U-2 OS and HCT 116 (wt-p53) cells. First we accessed the expression level of HDAC4 mRNA using real time qRT-PCR. Oligofectamine alone (vehicle control) and non-specific miR were used as the negative controls, siRNA against HDAC4 was the positive control. There was only a slight reduction in HDAC4 mRNA expression by miR-140 treatment (Figure 5b, column 4), by contrast, siHDAC4 clearly caused mRNA degradation (Figure 5b, column 3). Next, we analyzed the protein level of HDAC4 using Western immunoblot. Overexpression of miR-140 significantly decreased the expression of HDAC4 protein without considerable amount of mRNA degradation (Figure 5c, lane 3), which was consistent with the results of Tuddenham et al. and Nicolas et al. (Nicolas et al., 2008; Tuddenham et al., 2006). To further confirm that the expression of HDAC4 is regulated by miR-140, we performed the loss-of-function analysis by knocking down the endogenous miR-140 with locked nucleic acid (LNA) modified anti-miR140 in HCT 116 (wt-p53) and HCT 116 (null-p53) cells, scramble-miR (LNA-control) was used as the negative control. Our results showed that knocking down endogenous miR-140 by LNA anti-miR140 can restore the expression of HDAC4 (Figure 5d, lane 2). We also confirmed the knock down of HDAC4 protein expression by siRNA in HCT 116 (wt-p53) cells (Supplementary Figure 3a, lane 3). However, compared with the remarkable suppressing effect on cell proliferation by miR-140, the HDAC4 specific siRNA treated cells showed only a slight reduction in cell proliferation and the inhibitory effort was diminished after 4 days (Supplementary Figure 3b). Moreover, HCT 116 (wt-p53) cells transfected with siRNA specific for HDAC4 showed sensitivity to MTX treatment compared to miR-140 (Figure 2a), however, it did increase resistance to 5-FU (Figure 2b).
Figure 5.
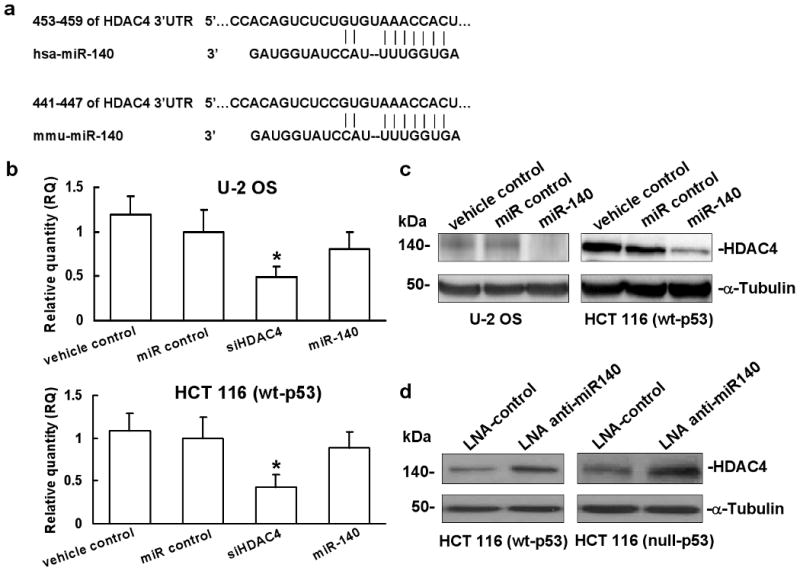
HDAC 4 is the target of miR-140. (a) Sequence comparison analysis of 3′-UTRs of mouse and human HDAC4 mRNAs with miR-140 interaction site. (b) Expression of HDAC4 mRNA in U-2 OS and HCT 116 (wt-p53) cells analyzed by real time qRT-PCR. (*P<0.05, Student's t test, n=3). (c) Protein expression analysis of HDAC4 in HCT 116 (wt-p53) and HCT 116 (null-p53) cells transfected with miR-140, oligofectamine alone (vehicle control) and miR control were used as the negative controls. (d) HCT 116 (wt-p53) and HCT 116 (null-p53) cells were transfected with LNA anti-miR140 and scramble-miR (LNA-control), HDAC4 protein was quantified by Western immunoblot.
Elevated expression of miR-140 in human colon cancer stem-like cells contributes to chemoresistance
To determine that colon cancer stem-like cells may have higher levels of miR-140 expression to process slow proliferating phenotype thereby avoiding damage caused by chemotherapeutic agents, the colon cancer stem-like cells were isolated using both CD133 and CD44 as selection markers from HCT 116 (wt-p53) cells (Figure 6a). Both CD133 and CD44 have been used by several groups to isolate colon cancer stem-like cells (Dalerba et al., 2007; Du et al., 2008; O'Brien et al., 2007; Ricci-Vitiani et al., 2007). The characterization of CD133+hiCD44+hi cancer stem-like cells has been described in details in our previous study (Botchkina et al., 2009). The expression of miR-140 in colon cancer stem-like cells was found to be nearly 4-fold higher than that in the control bulk cancer cells (Figure 6b). The results suggest that colon cancer stem-like cells may utilize miR-140 to slow down cell proliferation and avoid damage caused by chemotherapy until receiving a proliferation and differentiation signal, further verifying the impact of miR-140 on cell proliferation and chemotherapy resistance. CD133+hiCD44+hi cells were far more resistant (about 20% cell death) to high dose 5-FU treatment than non-sorted control HCT 116 (wt-p53) cells (>80% cell death) (Figure 6c, top panel). To directly demonstrate that we can reverse the chemoresistance to 5-FU treatment in CD133+hiCD44+hi cells, the expression of miR-140 was knocked down by LNA modified anti-miR140. Our results showed that CD133+hiCD44+hi cells were more sensitive to 5-FU treatment compared to LNA-control treated cells (Figure 6c, lower panel).
Figure 6.
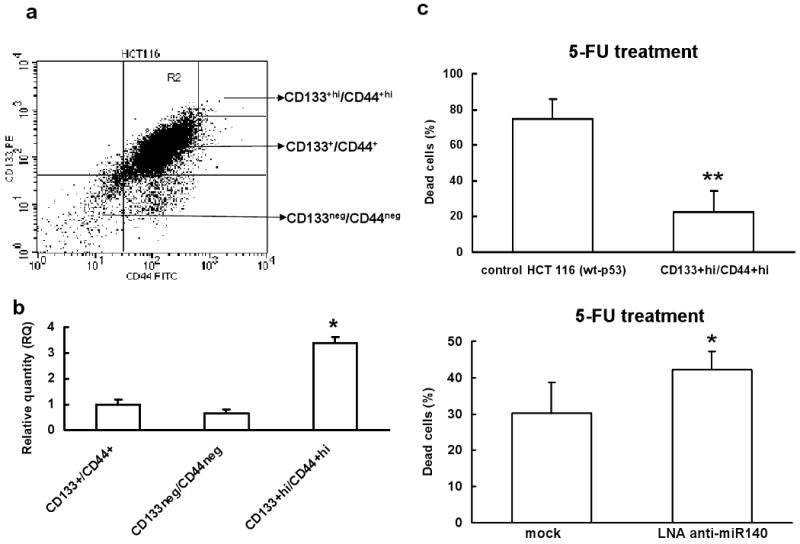
Expression of miR-140 in human colon cancer stem-like cells is elevated. (a) Flow cytometry analysis of sorted CD133+hi/CD44+hi HCT 116 (wt-p53) colon cancer stem-like cells using labeled CD133-PE and CD44-FITC antibodies. (b) Expression levels of miR-140 in colon cancer stem-like cells and colon cancer cells were analyzed by real time qRT-PCR (*P<0.05, Student's t test, n=3). (c) CD133+hi/CD44+hi colon cancer stem-like cells were more resistant to 5-FU treatment. CD133+hi/CD44+hi and control HCT 116 (wt-p53) cells were incubated with lethal dose 5-FU (100 μM) for 48 h, the dead cells were determined by the FITC Annexin V and PI detection kit (top panel, **P<0.01, Student's t test, n=3). CD133+hi/CD44+hi HCT 116 (wt-p53) colon cancer stem-like cells transfected with LNA anti-miR140 became sensitive to 5-FU treatment. CD133+hi/CD44+hi cells were transfected with 100 nM of LNA anti-miR140, 24 h later, cells were incubated with 100 μM of 5-FU for 48 h. The dead cells were determined by the FITC Annexin V and PI detection kit (lower panel, *P<0.05, Student's t test, n=3).
Expression of miR-140 is decreased in colorectal cancer specimens
Due to the impact of miR-140 on cell proliferation, we reasoned that differentiated proliferating tumor cells may have reduced miR-140 expression. To evaluate miR-140 expression level in colon cancer patients, we compared the miR-140 levels in 24 fresh frozen colon cancer specimens and their paired adjacent normal mucosa using real time qRT-PCR analysis (Supplementary Table 3). Our results showed that the expression levels of miR-140 were significantly reduced compared to adjacent normal mucosa (P<0.05) (Supplementary Figure 4).
Discussion
Our laboratory was focusing on elucidating the impact of non-coding miRNAs in chemoresistance, in particular, the resistance mechanism to antifolates (e.g. MTX) and fluoropyrimidine based compounds (e.g. 5-FU). Our interest on miR-140 was based on our high throughput miRNA expression profiling results using a panel of osteosarcoma tumor xenografts treated with high dose doxorubicin, cisplatin, and ifosfamide. The expression analysis showed that miR-140 was associated with chemoresistance in osteosarcoma (Figure 1). We reasoned that miR-140 may influence chemosensitivity to a broad spectrum of anti-cancer agents. We directly demonstrated that overexpression of miR-140 rendered colon cancer cells more resistant to both MTX and 5-FU treatment (Figure 2). This could be due to several possible reasons.
One major reason of the resistant phenotype is that miR-140 reduced the cell proliferation rate (Figure 3a and 3b) through decreased S phase and increased cell cycle arrest (Figure 4a). This was achieved in part, by the induction of p21 and p53 (Figure 4b). In general, slow proliferating or quiescent cells are more resistant to DNA damaging agent treatment as both MTX and 5-FU acting in S phase of the cell cycle to cause DNA damage. Elevated level of miR-140 in the colon cancer stem-like cells further supported this notion (Figure 6b). Cancer stem-like cells, exhibit low rate of division and proliferation in their niche that help them to avoid chemotherapy and radiation (Zou, 2008). This is the major difference between cancer stem-like cells and relative rapid proliferating differentiated cancer cells which can be eliminated effectively by chemotherapy treatment. Our results showed that colon cancer stem-like cells had higher miR-140 level than control bulk cancer cells (Figure 6b), suggesting that colon cancer stem-like cells may utilize miR-140 to slow down cell proliferation and avoid damage caused by chemotherapy. This may be an important novel mechanism in that tumor stem-like cells acquire slow proliferative or quiescent phenotype by certain miRNAs such as miR-140 to avoid damage caused by chemotherapy such as MTX and 5-FU. Results obtained from LNA modified anti-miRNA experiments demonstrated that we can partially sensitize colon cancer stem-like cells to 5-FU treatment (Figure 6c, lower panel). We also realized that it was difficult to achieve high transfection efficiency in floating colon cancer stem-like cell spheres. It is quite conceivable that this chemo-sensitization effect can be further improved by optimizing transfection efficiency. Colon cancer specimens had reduced miR-140 expression levels (Supplementary Figure 4), which supports our hypothesis that only the small fraction of tumor stem-like cells with a slow proliferating phenotype are mediated at least in part, by elevated miR-140, while differentiated tumor cells acquire relative rapid proliferation phenotype by reducing some of these miRNAs (e.g. miR-140) expression.
The other possibility is miR-140 causes chemoresistance through suppressing some key targets such as HDAC4. HDAC4 was highly expressed in the proliferative compartment in normal colonic and small intestinal epithelium (Wilson et al., 2008). We experimentally confirmed that one of the important targets of miR-140 was HDAC4, miR-140 reduced the expression level of HDAC4 protein without degradation of the target mRNA (Figure 5). We realized that the role of HDAC4 in the chemoresistance to MTX and 5-FU was quite different in our study. Although HDAC4 contributed to the slight reduction of proliferation of HCT 116 (wt-p53) cells, no direct relationship between HDAC4 and chemoresistance to MTX treatment was found (Figure 2a). Most likely that the overall impact of miR-140 on pathways is the important factor. Other efforts for discovering miR-140 mediated targets have been reported by Nicolas et al. recently that miR-140 mediated 21 potential targets using mouse cell lines such as NIH-3T3 (Nicolas et al., 2008). However, HDAC4 was not on their reported target list, this is partly due to the fact that miR-140 reduced the protein expression level of HDAC4 without altering the level of mRNA. Therefore, total mRNA based expression profiling will likely miss these targets. It's also worth noticing that one of their confirmed targets of miR-140 was Cxcl12 (Nicolas et al., 2008). Cxcl12/CXCR4 interaction is important for chemotaxis, adhesion, tumor invasion, and T cell response (Koishi et al., 2006). In fact, it has been reported that p53 attenuates cancer cell migration and invasion through suppression of Cxcl12 expression in stromal fibroblasts (Moskovits et al., 2006). Lowered expression levels of miR-140 in colorectal cancer specimens (Supplementary Figure 4) might result an elevated level of Cxcl12, which likely contributes to tumor progression and metastasis.
In contrast to MTX, there was a direct relationship between 5-FU resistance and HDAC4 suppression by miR-140. Cells transfected with siRNA against HDAC4 were resistant to 5-FU (Figure 2b), elevated p21 protein may contribute to such resistance (Figure 4b, left panel). This was consistent with a previous report that p21 induction was associated with 5-FU resistance (Bunz et al., 1999). The induction of p21 expression by miR-140 may be caused by the down-regulation of HDAC4. Wilson et al. investigated the detailed molecular mechanism of HDAC4 in regulating p21 expression (Wilson et al., 2008). HDAC4 directly interacted with Sp1 to the proximal p21 promoter. Although HDAC4 is just one of the many targets of miR-140, we reasoned that reduced expression of HDAC4 by miR-140 does release the suppressive control for p21 expression to allow cell cycle control. The induction of p21 was absent in cells lacking wild type p53 suggests that miR-140 is dependent on the presence of p53 to exert its function. The exact mechanism of regulatory relationship between p53 and miR-140 is important and remains to be determined in our future study. These findings also raise the notion that miR-140, like miR-192/215 and miR-34s, is one of the miRNAs that either directly or indirectly mediated by p53 to control cell cycle and cell proliferation (Braun et al., 2008; Georges et al., 2008; He et al., 2007; Raver-Shapira et al., 2007; Song et al., 2008). We are just at the beginning of understanding these complicated controls. Future studies are clearly needed to fully understand the underline molecular regulatory mechanisms.
In conclusion, in this study, we provide direct evidence that miR-140 influences chemosensitivity to MTX and 5-FU treatment. The function of miR-140 in cell proliferation and cell cycle control is clearly dependent on the presence of wild type p53 gene. Further studies are needed to further define additional molecular pathways mediated by miR-140.
Materials and methods
Cell culture and reagents
Human osteosarcoma cell lines U-2 OS (wt-p53) and MG63 (mut-p53) were purchased from the American Type Culture Collection (ATCC). The human colon cancer cell lines HCT 116 (wt-p53) and HCT 116 (null-p53) were gifts from Professor Bert Vogelstein (The Johns Hopkins University). U-2 OS, HCT 116 (wt-p53) and HCT 116 (null-p53) cells were maintained in McCoy's 5A medium (Invitrogen), and MG63 cells were maintained in Eagle's Minimum Essential Medium (ATCC), supplemented with 10% fetal bovine serum. MG63 cells have a rearrangement mutation of the p53 gene, which results in disruption of p53 protein function (Chandar et al., 1992).
Osteosarcoma tumor xenografts
Ten human osteosarcoma xenografts were used, eight of which have been described previously (Bruheim et al., 2004) and two newly established xenografts were included. The details of drug treatment and anti-tumor activity evaluation were also described (details see Results). The chemosensitivity results of each tumor xenograft to doxorubicin, cisplatin and ifosfamide were summarized in Supplementary Table 1.
Clinical samples
A total of 48 snap frozen colorectal patient specimens were selected (24 paired colon normal mucosa and tumor samples). These patients had undergone surgical resection of primary colorectal adenocarcinoma at the Department of Visceral and Transplantation Surgery, University of Ulm, Germany. Patient consent forms were obtained from every patient according to the institutional regulations. The characteristics of these patients were shown in Supplementary Table 3.
Transfection of miRNA and siRNA specific to HDAC4
Cells were plated in six-well plates at a density of 2×105 cells/well. The next day, cells were transfected with 100 nM of miR-140 precursor or non-specific miR control (Ambion) with Oligofectamine (Invitrogen) based on the manufacturer's instructions. Positive control siRNA specific against HDAC4 (ON-TARGET plus SMARTpool L-008799-00-0010, human HDAC4, NM_000791) was purchased from Dharmacon and transfected with Oligofectamine according to the manufacturer's protocols at a final concentration of 100 nM.
RNA isolation and real time qRT-PCR analysis of miRNA
Total RNA, including miRNAs, was isolated from cell lines, tumor xenografts or clinical specimens using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. cDNA synthesis was carried out with the High Capacity cDNA synthesis kit (Applied Biosystems) using 5 ng of total RNA as template. The miRNA sequence-specific RT-PCR primers for miR-140 and endogenous control RNU6B were purchased from Ambion. Real-time qRT-PCR analysis was carried out using Applied Biosystems 7500 Real-Time PCR System (details see ref. Song et al., 2008). The gene expression threshold cycle (CT) values of miRNAs from each sample were calculated by normalizing with internal control RNU6B and relative quantitation values were plotted.
Real time qRT-PCR analysis of mRNA expression
cDNA was synthesized with the High Capacity cDNA synthesis kit (Applied Biosystems) using 2 μg of total RNA as template. The PCR primers and probes for HDAC4 and the internal control gene GAPDH were purchased from Applied Biosystems. Real-time qRT-PCR analysis was performed on an ABI 7500HT instrument (details see ref. Song et al., 2008).
Cell proliferation analysis
Cells were plated in 96-well plates at 2×103 cells/well after transfection with miR-140 or miR control. Cells were cultured for 24, 48, 72, 96 and 120 h. The absorbance at 450 and 630 nm was measured after incubation with 10 μl of WST-1 (Roche Applied Science) for 1 h.
Cell cycle analysis
Cells were transfected with miR-140 and miR control described as above. At 36 h after transfection, cells were harvested and resuspended at 0.5-1×105 cells/ml in modified Krishan buffer (Dressler et al., 1988; Krishan, 1975). Before being analyzed by flow cytometry, cells were treated with 0.02mg/ml RNase H and stained with 0.05mg/ml propidium iodide (Sigma).
Western immunoblot analysis
Cells were lysed in 1×RIPA buffer (Sigma) supplied with 100 μM PMSF (Sigma) and proteinase inhibitor cocktail (Sigma) at 48 h after transfection. Equal amount of proteins were resolved by 8% SDS-PAGE gels (Laemmli, 1970). The primary antibodies used for the analysis included goat anti-HDAC4 polyclonal Ab (1:1000, N-18), mouse anti-p53 mAb (1:1000, DO-1), mouse anti-p21 mAb (1:1000, F-5), and mouse anti-α-tubulin mAb (1:1000, TU-02), all from Santa Cruz Biotechnology.
miR-140 knock down
HCT 116 (wt-p53) and HCT 116 (null-p53) cells were transfected with 100 nM of scramble-miR or LNA anti-miR140 oligonucleotides (Exiqon) in the six-well plates (2×105 cells/well) by Lipofectamine 2000 (Invitrogen). Cells were harvested at 72 h after transfection and cellular proteins were extracted. HDAC4 protein was detected by Western immunoblot analysis.
MTX treatment
HCT 116 (wt-p53) and HCT 116 (null-p53) cells were seeded in 6-well plates at 2×105 per well and incubated with or without MTX (100 nM) for 24 h. Then total RNA and proteins were extracted, respectively. The subsequent real time qRT-PCR for miR-140 and Western immunoblot analysis were performed as described above.
Isolation of colon cancer stem-like cells by FACS
HCT 116 (wt-p53) cells were sorted with multiparametric flow cytometry with BD FACS Aria cell sorter (Becton Dickinson) at sterile conditions. Cells were prepared and labeled with conjugated anti-human CD133-PE (clone 105902, R&D Systems) and CD44-FITC (clone F10-44-2, R&D Systems). Antibodies were diluted in buffer containing 5% BSA, 1mM EDTA and 15-20% blocking reagent (Miltenyi Biotec) to inhibit non-specific binding to non-target cells. After 15 min incubation at 4°C, stained cells were washed, resuspended in 500 μl of MACS buffer and sorted.
MTX and 5-FU cytotoxicity
HCT 116 (wt-p53) cells were replated in 96-well plates at 2×103 cells/well in triplicate after transfected with miR-140, miR control, or siRNA against HDAC4 in 100 μl of medium. Twenty-four hours later, MTX (ranged from 10-1000 nM) or 5-FU (ranged from 2 to 100 μM) was added and incubated for 72 h. Ten μl of WST-1 was added to each well. After 1 h incubation, absorbance was measured at 450 and 630 nm respectively. Non-specific miR was used as the negative control.
Colon cancer stem-like cells were transfected with 100 nM of LNA anti-miR140 using Lipofectamine 2000 described as above. Twenty-four hours later, cells were washed by FBS and then incubated with lethal dose 5-FU (100 μM) for 48 h. The dead cells were determined by the FITC Annexin V and PI detection kit (BD Biosciences, Pharmingen). Briefly, cells were harvested and re-suspended in 1×Annexin V binding and stained with Annexin V (5 μl) and PI (5 μl) for 15 min at room temperature in the dark. After additional 400 μl of binding buffer, cells were analyzed by flow cytometry. For the sensitivity of 5-FU in the colon cancer stem-like cells and control bulk cancer cells, cells were incubated with 100 μM of 5-FU for 48 h before flow cytometery analysis.
Statistic analysis
All experiments were repeated at least twice. Statistical significance was evaluated by Student's t test (two-tailed) comparison between two groups of data using GraphPad Prism software 5 (GraphPad). Expression levels of miR-140 in each clinical sample were normalized by the internal control RNU6B using SDS software v1.2 (Applied Biosystems). Sample with the lowest expression levels of miR-140 was set as 1 to generate relative expression values. Statistical difference in the expression level between tumor and normal tissues was calculated by two-tailed paired Wilcoxon test using MedCalc® 10.0.2 (MedCalc software, Belgium). Statistical significance was set as a P<0.05.
Supplementary Material
Acknowledgments
We appreciate the critical reading of our manuscript by Stephanie Burke (Stony Brook University). This work was supported by Stony-Brook Translational Research Laboratory Start-up fund and NIH CA114043 (J. Ju) and MH075020 (J. Ju).
Footnotes
Conflict of interest: The authors declare no conflict of interest.
References
- Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development. 2005;132:4653–4662. doi: 10.1242/dev.02073. [DOI] [PubMed] [Google Scholar]
- Ambros V, Lee RC. Identification of microRNAs and other tiny noncoding RNAs by cDNA cloning. Methods Mol Biol. 2004;265:131–158. doi: 10.1385/1-59259-775-0:131. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Botchkina IL, Rowehl RA, Rivadeneira DE, Karpeh MS, Jr, Crawford H, Dufour A, et al. Phenotypic subpopulations of metastatic colon cancer stem cells: genomic analysis. Cancer Genomics Proteomics. 2009;6:19–29. [PubMed] [Google Scholar]
- Braun CJ, Zhang X, Savelyeva I, Wolff S, Moll UM, Schepeler T, et al. p53-responsive microRNAs 192 and 215 are capable of inducing cell cycle arrest. Cancer Res. 2008;68:10094–10104. doi: 10.1158/0008-5472.CAN-08-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruheim S, Bruland OS, Breistol K, Maelandsmo GM, Fodstad O. Human osteosarcoma xenografts and their sensitivity to chemotherapy. Pathol Oncol Res. 2004;10:133–141. doi: 10.1007/BF03033741. [DOI] [PubMed] [Google Scholar]
- Bunz F, Hwang PM, Torrance C, Waldman T, Zhang Y, Dillehay L, et al. Disruption of p53 in human cancer cells alters the responses to therapeutic agents. J Clin Invest. 1999;104:263–269. doi: 10.1172/JCI6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin G, Croce CM. MicroRNA signatures in human cancers. Nature Reviews Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Chandar N, Billig B, McMaster J, Novak J. Inactivation of p53 gene in human and murine osteosarcoma cells. Br J Cancer. 1992;65:208–214. doi: 10.1038/bjc.1992.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalerba P, Dylla SJ, Park IK, Liu R, Wang X, Cho RW, et al. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci U S A. 2007;104:10158–10163. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dressler LG, Seamer LC, Owens MA, Clark GM, McGuire WL. DNA flow cytometry and prognostic factors in 1331 frozen breast cancer specimens. Cancer. 1988;61:420–427. doi: 10.1002/1097-0142(19880201)61:3<420::aid-cncr2820610303>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- Du L, Wang H, He L, Zhang J, Ni B, Wang X, et al. CD44 is of functional importance for colorectal cancer stem cells. Clin Cancer Res. 2008;14:6751–6760. doi: 10.1158/1078-0432.CCR-08-1034. [DOI] [PubMed] [Google Scholar]
- Eberharter A, Becker PB. Histone acetylation: a switch between repressive and permissive chromatin. Second in review series on chromatin dynamics. EMBO Rep. 2002;3:224–229. doi: 10.1093/embo-reports/kvf053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnin MS, Donigian JR, Cohen A, Richon VM, Rifkind RA, Marks PA, et al. Structures of a histone deacetylase homologue bound to the TSA and SAHA inhibitors. Nature. 1999;401:188–193. doi: 10.1038/43710. [DOI] [PubMed] [Google Scholar]
- Georges SA, Biery MC, Kim SY, Schelter JM, Guo J, Chang AN, et al. Coordinated regulation of cell cycle transcripts by p53-Inducible microRNAs, miR-192 and miR-215. Cancer Res. 2008;68:10105–10112. doi: 10.1158/0008-5472.CAN-08-1846. [DOI] [PubMed] [Google Scholar]
- Grozinger C, Schreiber SL. Deacetylase enzymes: biological functions and the use of small-molecule inhibitors. Chem Biol. 2002;9:3–16. doi: 10.1016/s1074-5521(02)00092-3. [DOI] [PubMed] [Google Scholar]
- He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–8707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- Izzotti A, Calin GA, Arrigo P, Steele VE, Croce CM, De Flora S. Downregulation of microRNA expression in the lungs of rats exposed to cigarette smoke. FASEB J. 2009;23:806–812. doi: 10.1096/fj.08-121384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koishi K, Yoshikawa R, Tsujimura T, Hashimoto-Tamaoki T, Kojima S, Yanagi H, et al. Persistent CXCR4 expression after preoperative chemoradiotherapy predicts early recurrence and poor prognosis in esophageal cancer. World J Gastroenterol. 2006;12:7585–7590. doi: 10.3748/wjg.v12.i47.7585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Faulty old ideas about translational regulation paved the way for current confusion about how microRNAs function. Gene. 2008;423:108–115. doi: 10.1016/j.gene.2008.07.013. [DOI] [PubMed] [Google Scholar]
- Krishan A. Rapid flow cytofluorometric analysis of mammalian cell cycle by propidium iodide staining. J Cell Biol. 1975;66:188–193. doi: 10.1083/jcb.66.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Moskovits N, Kalinkovich A, Bar J, Lapidot T, Oren M. p53 Attenuates cancer cell migration and invasion through repression of SDF-1/CXCL12 expression in stromal fibroblasts. Cancer Res. 2006;66:10671–10676. doi: 10.1158/0008-5472.CAN-06-2323. [DOI] [PubMed] [Google Scholar]
- Nicolas FE, Pais H, Schwach F, Lindow M, Kauppinen S, Moulton V, et al. Experimental identification of microRNA-140 targets by silencing and overexpressing miR-140. RNA. 2008;14:2513–2520. doi: 10.1261/rna.1221108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- Pillai RS, Bhattacharyya SN, Fillipowicz W. Repression of protein synthesis by miRNAs: how many mechanisms? Trends cell Biol. 2007;17:118–126. doi: 10.1016/j.tcb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- Plasterk RH. MicroRNAs in animal development. Cell. 2006;124:877–881. doi: 10.1016/j.cell.2006.02.030. [DOI] [PubMed] [Google Scholar]
- Raver-Shapira N, Meiri E, Spector Y, Rosenfeld N, Moskovits N, Bentwich Z, et al. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26:731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, et al. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- Sengupta N, Seto E. Regulation of histone deacetylase activities. J Cell Biochem. 2004;93:57–67. doi: 10.1002/jcb.20179. [DOI] [PubMed] [Google Scholar]
- Song B, Wang Y, Kudo K, Gavin EJ, Xi Y, Ju J. miR-192 Regulates dihydrofolate reductase and cellular proliferation through the p53-microRNA circuit. Clin Cancer Res. 2008;14:8080–8086. doi: 10.1158/1078-0432.CCR-08-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarasov V, Jung P, Verdoodt B, Lodygin D, Epanchintsev A, Menssen A, et al. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle. 2007;6:1586–1593. doi: 10.4161/cc.6.13.4436. [DOI] [PubMed] [Google Scholar]
- Tuddenham L, Wheeler G, Ntounia-Fousara S, Waters J, Hajihosseini MK, Clark I, et al. The cartilage specific microRNA-140 targets histone deacetylase 4 in mouse cells. FEBS Lett. 2006;580:4214–4217. doi: 10.1016/j.febslet.2006.06.080. [DOI] [PubMed] [Google Scholar]
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson AJ, Byun DS, Nasser S, Murray LB, Ayyanar K, Arango D, et al. HDAC4 promotes growth of colon cancer cells via repression of p21. Mol Biol Cell. 2008;19:4062–4075. doi: 10.1091/mbc.E08-02-0139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XJ, Grégoire S. Class II histone deacetylases: from sequence to function, regulation, and clinical implication. Mol Cell Biol. 2005;25:2873–2874. doi: 10.1128/MCB.25.8.2873-2884.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Wang Q, Pan X. MicroRNAs and their regulatory roles in animals and plants. J Cell Physiol. 2007;210:279–289. doi: 10.1002/jcp.20869. [DOI] [PubMed] [Google Scholar]
- Zou GM. Cancer initiating cells or cancer stem cells in the gastrointestinal tract and liver. J Cell Physiol. 2008;217:598–604. doi: 10.1002/jcp.21541. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.