Summary
Mitochondrial inheritance, the transfer of mitochondria from mother to daughter cell during cell division, is essential for daughter cell viability. The mitochore, a mitochondrial protein complex containing Mdm10p, Mdm12p and Mmm1p, is required for mitochondrial motility leading to inheritance in budding yeast. We observe a defect in cytokinesis in mitochore mutants and another mutant (mmr1Δ gem1Δ) with impaired mitochondrial inheritance. This defect is not observed in yeast that have no mitochondrial DNA or defects in mitochondrial protein import or assembly of β-barrel proteins in the mitochondrial outer membrane. Deletion of MDM10 inhibits contractile ring closure, but does not inhibit contractile ring assembly, localization of a chromosomal passenger protein to the spindle during early anaphase, spindle alignment, nucleolar segregation or nuclear migration during anaphase. Release of the mitotic exit network (MEN) component, Cdc14p, from the nucleolus during anaphase is delayed in mdm10Δ cells. Finally, hyperactivation of the MEN by deletion of BUB2 restores defects in cytokinesis in mdm10Δ and mmr1Δ gem1Δ cells, and reduces the fidelity of mitochondrial segregation between mother and daughter cells in wild-type and mdm10Δ cells. Our studies identify a novel MEN-linked regulatory system that inhibits cytokinesis in response to defects in mitochondrial inheritance in budding yeast.
Results and Discussion
Mutations that inhibit mitochondrial inheritance produce multibudded cells in budding yeast
Equal segregation of mitochondria between mother and daughter cells during yeast cell division occurs as a result of bidirectional movement of mitochondria to the bud tip and mother cell tip and anchorage of the organelle at those sites (1). The mitochore, a mitochondrial membrane protein complex containing the proteins Mmm1p, Mdm10p and Mdm12p, is required for binding of mitochondria to actin filaments in vitro, actin cable-dependent bidirectional mitochondrial movement, and mitochondrial inheritance (1-3). In early characterizations of mitochondrial morphology and distribution mutants, Sogo and Yaffe (4) noted the presence of a multibudded phenotype in mdm10Δ cells. We find that multibudded clusters consisting of 3-5 buds are present during mid-log phase and accumulate with growth time in mdm10Δ cells. This multibudded phenotype is observed in mdm10Δ cells in three different genetic backgrounds: S288C, W303 and A264A (data not shown).
In wild-type yeast, mitochondria constitute a dynamic and tubular reticulum (Fig. 1A-B) (1). In mdm10Δ cells, mitochondria are large spherical structures that fail to move from mother cells to buds and undergo rapid loss of mitochondrial DNA (mtDNA) (2-3). The large spherical mitochondria typical of mdm10Δ cells are usually present in only one cell within a multibudded clump (Fig. 1E-F). Visualization of DNA confirmed that mdm10Δ cells have no mtDNA and revealed that each cell body in mdm10Δ clumps contains a nucleus (Fig. 1G-H). The viability of wild-type and mdm10Δ cells during mid-log phase growth, assessed using FUN-1 staining, is 93.5% and 76.5%, respectively. Thus, a mutation in MDM10 that results in severe defects in mitochondrial morphology and inheritance also produces defects in mother-daughter cell separation but does not inhibit nuclear inheritance or compromise cell viability.
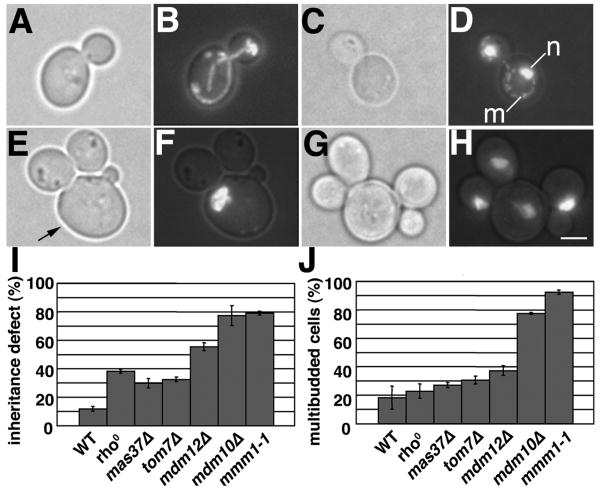
Figure 1. Cell separation defects in mitochondrial inheritance mutants.
Wild-type (BY4741) or mdm10Δ (Open Biosystems 398) cells were grown in SC medium at 30°C to mid-log phase. Cells were either stained for mitochondria using MitoTracker Red, or fixed with formaldehyde and stained using the DNA-binding dye DAPI. Images of MitoTracker Red or DAPI stained cells are 2-D projections of the reconstructed 3-D volume that are superimposed on the corresponding phase image. A-B and E-F) Phase images and MitoTracker Red staining of wild-type and mdm10Δ cells, respectively. The arrow points to the original mother cell in a multibudded mdm10Δ cluster that contains a large spherical mitochondrion. Bar: 1 μm. C-D and G-H) Phase images and DAPI staining of wild-type and mdm10Δ cells, respectively. n: nuclear DNA. m: mtDNA. I-J) Quantification of mitochondria-free buds in cell bearing small buds (I) and multibudded cells (J) in wild-type (ISY001), rho0 (ISY001-rho0), mas37Δ (ISY005), tom7Δ (ISY006), mdm12Δ (ISY003), mmm1-1 (ISY065) and mdm10Δ (ISY002) cells (n>800). Cells were grown in SC medium at 30°C for 12-16 hrs to late-log phase. The optical density (OD600) of cultures at the time of analysis was 1.2 – 1.4. Error bars are standard deviations.
Deletion of MDM10, MDM12 or MMM1 also results in defects in maintenance of mtDNA, mitochondrial morphology and assembly of β-barrel proteins in the mitochondrial outer membrane (OM) (2, 4-6). Therefore, we tested whether the multibudded phenotype of mdm10Δ cells is due to defects in these mitochondrial inheritance-independent processes by analysis of yeast bearing deletions in mtDNA, MAS37 or TOM7. rho0 cells have no mtDNA and severe defects in mitochondrial respiration (7). Mas37p is a subunit of the SAM/TOB complex, which mediates assembly of β-barrel proteins into the mitochondrial OM (8). Tom7p is a subunit of the protein-translocating pore in the mitochondrial OM (9). Deletion of TOM7 produces defects in mitochondrial morphology that are similar to those observed in mdm10Δ cells as well as defects in mitochondrial protein import (6). Tom7p also promotes the segregation of Mdm10p from the SAM/TOB complex (10).
rho0, mas37Δ, and tom7Δ cells exhibit significantly lower defects in mitochondrial inheritance and lower levels of multibudded cells compared to mitochore mutants (Fig. 1I-J). Thus, the multibudded phenotype observed in mdm10Δ cells is not a consequence of loss of mtDNA, or of defects in mitochondrial respiratory activity, protein import, or OM β-barrel protein assembly. Moreover, we observed a link between the extent of multibudded cells in late-log phase cultures and the severity of the mitochondrial inheritance defect in yeast carrying mutations in mitochore subunits: mdm10Δ = mmm1–1 ≫ mdm12Δ (Fig. 1I-J). Mdm12p coordinates mitochondrial inheritance and biogenesis through its direct interactions with the PUF family protein Puf3p (11). Thus, mdm12Δ cells may have less severe multibudded and inheritance phenotypes compared to mdm10Δ or mmm1-1 mutants because Mdm12p has regulatory effects on mitochondrial motility, while Mdm10p and Mmm1p have predominant roles in mediating mitochondrial motility. Overall, the multibudded phenotype observed in all mutants analyzed correlates with defects in mitochondrial inheritance.
mdm10Δ cells exhibit defects in contractile ring closure
mdm10Δ cells that enter the cell cycle cycle are in G2 phase 20 min later than wild-type cells (SFig. 1). Spindle assembly and disassembly as well as the appearance and disappearance of mitotic cyclin are delayed to a similar extent in mdm10Δ compared to wild-type cells (SFig. 2). Formation of the second bud (d2) in multibudded mdm10Δ cells occurs 150 min after release from pheromone-induced G1 arrest, 25 min after the first bud (d1) undergoes Clb2p degradation and spindle disassembly (SFig. 2).
rho0 cells undergo a delay in cell cycle progression similar to that observed in mdm10Δ, the decrease in cell cycle progression in mdm10Δ may be due to loss of mtDNA. However, the multibudded phenotype in mdm10Δ cells is not due to loss of mtDNA (Fig. 1J), or to defects in septation (degradation of the cell wall between mother and daughter cells), spindle alignment or nucleolar segregation (SFig. 3-4). Rather, it is due to defects in contractile ring closure. Actomyosin ring contraction was visualized in wild-type and mdm10Δ cells using a fully-functional fusion protein consisting of the type II myosin (Myo1p) fused to GFP (12), mitochondria-targeted DsRed, and 4-D imaging (time lapse imaging combined with 3-D reconstruction). Deletion of MDM10 has no effect on contractile ring assembly: Myo1p-GFP localizes to a ring at the mother-bud junction in both wild-type and mdm10Δ cells (Fig. 2A-D). Moreover, mdm10Δ cells have the capacity to undergo contractile ring closure (Fig. 2B), and to do so with kinetics (14.2 ± 3.5 min, n = 48) similar to that of wild-type cells (10.4 ± 2.1 min, n = 43). There is some loss of synchrony in mdm10Δ cells at the time of contractile ring closure. Nonetheless, mdm10Δ cells that undergo contractile ring closure do so 20-40 min later in the cell cycle compared to wild-type cells (n = 48).
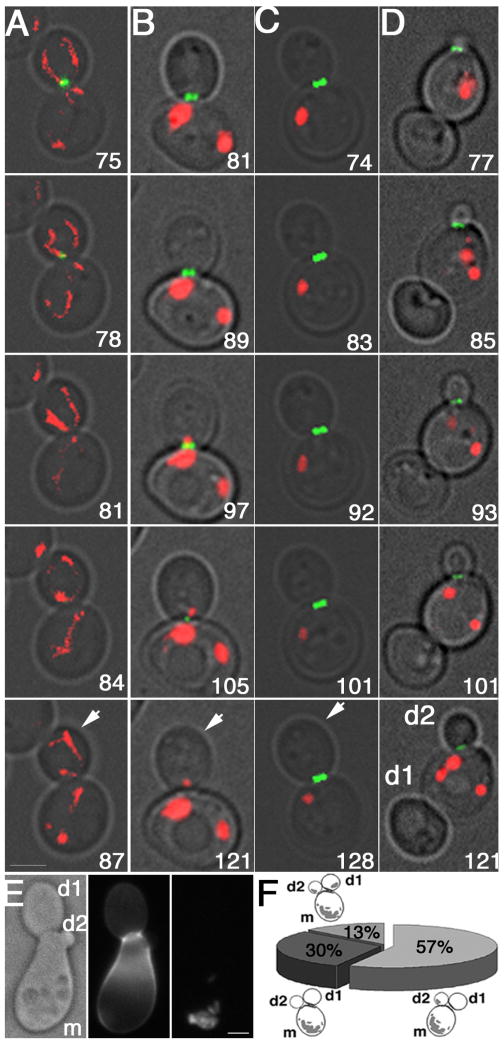
Figure 2. Multibudded clusters of mdm10Δ cells are due to defects in contractile ring closure.
A-D) Still frames from time-lapse imaging of Myo1p-GFP (green) and DsRed-labeled mitochondria (red) in synchronized wild-type (ISY008) (A) and mdm10Δ (ISY009) (B-D) cells. Unbudded cells were isolated from mid-log phase cultures by centrifugation through a 10-35% sorbitol gradient for 12 min at 56 × g and visualized by 4D time lapse imaging. Imaging was performed 1 hr after bud formation for a total of 1 hr. Images were acquired at 3 and 4 min intervals for wild-type and mdm10Δ cells, respectively. Images shown are 2D projections of 3D reconstructions. Arrows points to buds. Numbers shown indicate time of image acquisition from the onset of bud formation. Bar, 1 μm. A) Wild-type cell undergoing contractile ring closure. B) mdm10Δ cell that has mitochondria in the bud and undergoes contractile ring closure. C) mdm10Δ cells that does not undergo contractile ring closure and has no detectable mitochondria in the bud. D) Multibudded mdm10Δ cell in which the first bud (d1) has no detectable mitochondria and remains associated to the mother cell, and a contractile ring has assembled at the site of growth of the second daughter cell (d2). E) Mitochondrial morphology and distribution in multibudded cells from a synchronized mdm10Δ cells. Cells were grown in SC medium at 30°C to mid-log phase (OD600 = 0.5 – 0.8) and incubated with α-factor (10 μM) for 2.5 hrs. Cells were washed and resuspended in medium, fixed at various times after release from pheromone-induced G1 arrest and stained with Calcufluor white to stain bud scars on the mother cell (m) but not on the first or second daughter cell (d1 and d2, respectively) produced from that mother cell (middle panel). DsRed labeled mitochondria are present in the mother cell but not in daughter cells (left panel). Bar, 1 μm. F) Quantification of mitochondrial content in mother cells (m), their first (d1) and second (d2) daughter cells in multibudded mdm10Δ cells from synchronized cell cultures. n = 100 clumps with 3 cell bodies.
However, mdm10Δ cells exhibit defects in contractile ring closure, which correlates with defects in mitochondrial inheritance (Fig. 2C). To quantitate the frequency of contractile ring closure, Myo1p-GFP and DsRed-labeled mitochondria were visualized in cells that bore large buds at the onset of imaging for 2 hrs. During this time, contractile ring closure occurred in 100% of the wild-type cells examined (n=19) and in only 29% of the mdm10Δ cell examined (n=38). To evaluate mitochondrial inheritance as a function of contractile ring closure, we measured the mitochondrial content in buds of mdm10Δ cells that undergo contractile ring closure (Fig. 2B) and in the first buds (d1) of multibudded mdm10Δ that failed to undergo contractile ring closure at the mother cell:d1 junction (Fig. 2E). In wild-type and mdm10Δ cells that undergo contractile ring closure 43±2% (n = 32), and 36.7±3.12% (n=37) of mitochondria are in the bud, respectively. In contrast, there are no detectable mitochondria in 87% of d1 cells within multibudded mdm10Δ cells (n = 100).
Role for the MEN in regulation of cell cycle progression in mdm10Δ cells
The MEN regulates cell cycle progression in response to spindle alignment and elongation, and to the transfer of the nucleus from mother to daughter cell during the anaphase-to-telophase transition. Cdc14p activation and localization of the active protein to its sites of action are essential for degradation of a mitotic cyclin (Clb2p), inactivation of a mitotic cyclin-dependent kinase (CDK; Cdc28p/Clb2p), dephosphorylation of CDK substrates, and exit from mitosis (13). However, several studies indicate that the MEN also has a direct role in regulating contractile ring closure during cytokinesis in budding yeast (14-19).
mdm10Δ cells undergoes mitotic exit, as assessed by degradation of Clb2p and spindle disassembly (SFig. 2). To evaluate the role of the MEN in the observed cytokinesis defect, we studied the localization of Cdc14p-GFP in mdm10Δ and wild-type cells. Cdc14p is released from its inhibitor Cfi1p/Net1p in the nucleolus during two stages in the cell division cycle. In early anaphase, separase, as part of the Cdc fourteen early-anaphase release (FEAR) pathway, promotes a transient and partial release of Cdc14p from the nucleolus. In a second phase, signal transduction through the MEN releases the remaining Cdc14p, which facilitates mitotic exit and cytokinesis (20).
We confirmed that Cdc14p-GFP in wild-type cells localizes to the nucleolus through early stages of the cell division cycle, and is released from the nucleolus and localizes to the spindle pole bodies and bud neck as the spindle apparatus elongates (Fig. 3A). When the spindle is at its maximum length (6-8 μm), 100% of the Cdc14p-GFP is released from the nucleolus (Fig. 3C). In mdm10Δ cells, some cytosolic Cdc14p localizes to the spindle pole body in mdm10Δ cells bearing fully elongated spindles. However, release of Cdc14p-GFP from the nucleolus is inhibited by 50% in mdm10Δ cells bearing 4-6 μm spindles, and to a lesser extent in cells with 6-8 μm spindles compared to wild type cells (Fig. 3B-C). Thus, deletion of MDM10 results in a delay in release of Cdc14p from the nucleolus.
Figure 3. Cdc14p is mislocalized in mdm10Δ cells.
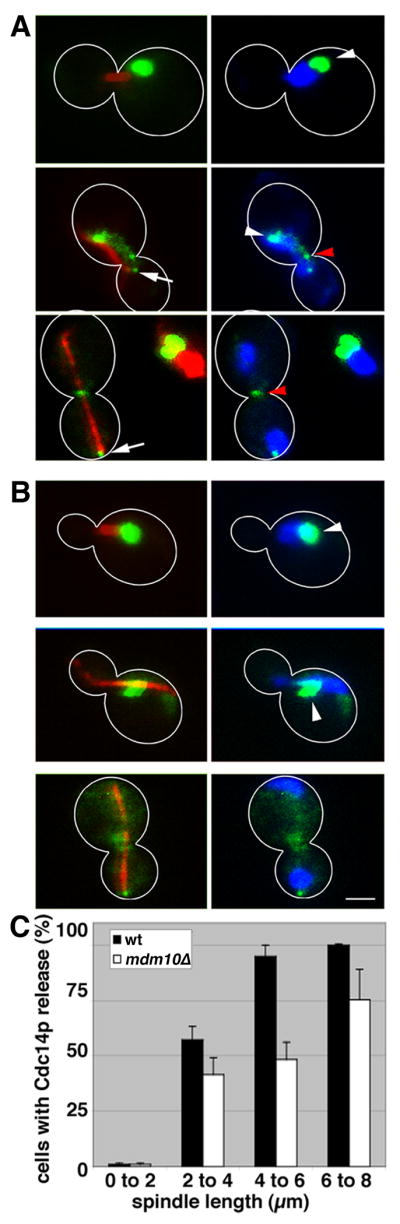
Wild-type (LGY020) and mdm10Δ (LG0Y21) cells expressing Cdc14p-GFP and mCherry-tagged tubulin were grown to mid-log phase, fixed and stained with DAPI as for Fig. 1. The images shown are 2-D projections from reconstructed 3-D volumes. An overlay of Cdc14p-GFP (green) and tubulin in the mitotic spindle (red) are shown (left). An overlay of Cdc14p-GFP (green) and DAPI (blue) are shown (right). Cell outlines are shown in white. White arrow: spindle pole body. White arrowhead: nucleus. Red arrowhead: mother-bud neck. Bar, 1 μm. A) Cdc14p-GFP localization in wild-type cells. Cdc14p-GFP localizes to the nucleolus in cells bearing short, but detectable spindles (upper panels), to the nucleus and spindle pole bodies in early anaphase when spindles are 4-6 μm in length (middle panels) and to spindle pole bodies and the mother-bud neck during telophase when spindles have elongated and reached their maximum length of 8-10 μm (lower panels). B). Defects in localization of Cdc14p-GFP in mdm10Δ cells. C) Quantitation of the release of Cdc14p from the nucleolus in wild-type and mdm10Δ cells as a function of spindle length. Error bars show standard error of the mean (n>200).
Sli15p, a chromosomal passenger protein and substrate for Cdc14p that is released from the nucleolus during early anaphase (21), localizes to the spindle apparatus to the same extent in mdm10Δ and in wild-type cells (SFig. 5). Thus, mislocalization of Cdc14p in mdm10Δ cells is due to an alteration in MEN-mediated control of Cdc14p and not the FEAR pathway. In light of these findings and our observation that release of Cdc14p from the nucleolus is partially inhibited in mdm10Δ cells, it is possible that the level of MEN-mediated Cdc14p activation in mdm10Δ cells is sufficient to support mitotic exit but insufficient to support cytokinesis.
Consistent with this, conditions that hyperactivate the MEN promote cytokinesis in mdm10Δ cells. Deletion of BUB2 suppresses the subtle mitotic exit defect observed in mdm10Δ cells, but has no effect on the time of entry of mdm10Δ cells into anaphase (SFig. 6). Deletion of BUB2 or overexpression of CDC5 in mdm10Δ cells results in a 67% decrease in the number of multibudded cells in late-log phase cell cultures compared to mdm10Δ cells (Fig. 4A-B). Thus, conditions that bypass MEN regulation bypass the cytokinesis defects observed in mdm10Δ cells.
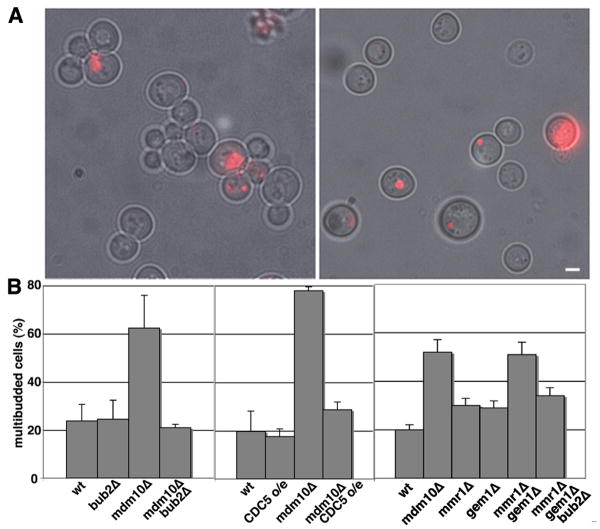
Figure 4. Hyperactivation of the MEN suppresses the defect in cytokinesis defect observed in mdm10Δ cells.
A) mdm10Δ cells that expressed mitochondria-targeted DsRed and contained either no plasmid (ISY002) or plasmid-borne CDC5 under control of the GAL promoter (ISY048) incubated in galactose-based media for 5.5 hrs. Images shown are phase-contrast images of cells superimposed upon fluorescence images of mitochondria labeled with DsRed. Bar, 3 μm. B) Quantitation of multibudded cells in wild-type cells and mdm10Δ cells that either overexpress CDC5 or carry a deletion of BUB2. Strains used for wild-type, CDC5 overexpression, mdm10Δ and mdm10Δ overexpressing CDC5 are ISY001, ISY048, ISY002 and ISY013, respectively (dark grey bars). Strains used for wild-type, bub2Δ, mdm10Δ, and bub2Δ mdm10Δ are BY4741, Open Biosystems 6189, Open Biosystems 398, and LGY025, respectively (light grey bars). Cell culture and quantitation were carried out as for Fig. 1D. Error bars show standard deviations for n>800 measurements. C) Quantitation of multibudded cells in wild-type (BY4741), mmr1Δ (Open Biosystems 4139), gem1Δ (Open Biosystems 357), mmr1Δ gem1Δ (DCY001) and mmr1Δ gem1Δ bub2Δ cells (DCY002) that were analyzed after treatment with zymolyase 20T (0.1 mg/ml for 10 min at RT). Error bars show standard deviations for n>100 measurements.
To determine whether other mutations that inhibit mitochondrial inheritance also affect cytokinesis, we studied GEM1, a member of the rho (Miro) family of GTPases and MMR1, a protein that localizes to mitochondria, binds to the type V myosin Myo2p and is required for anchorage of mitochondria in the bud tip (22-23). mmr1Δ or gem1Δ mutants exhibit subtle defects in mitochondrial inheritance, and low but detectable defects in cytokinesis. However, gem1Δ mmr1Δ double mutants exhibit mitochondrial distribution and inheritance defects that are significantly greater than those observed in either single mutant (23) and a cytokinesis defect that is more severe than that observed in either single mutants and similar to that observed in the mdm10Δ mutant. In addition, deletion of BUB2 suppresses the cytokinesis defect observed in the gem1Δ mmr1Δ double mutant (Fig. 4B). These findings provide additional evidence for the existence of a mechanism to inhibit cell cycle progression at cytokinesis when there are severe defects in mitochondrial inheritance.
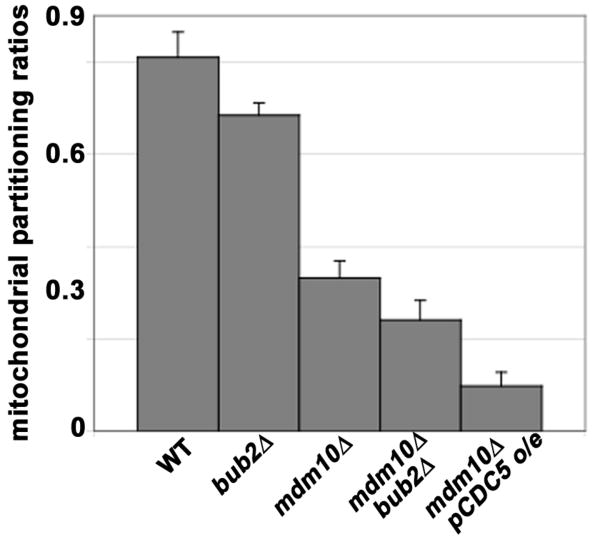
Finally, the primary function of a checkpoint is to insure that critical cell division processes occur with high fidelity and at the correct time as cells divide. Thus, if the MEN regulates cell cycle progression in response to mitochondrial inheritance, then hyperactivation of the MEN should reduce the fidelity of mitochondrial inheritance. Indeed, we find that conditions that bypass MEN regulation, deletion of BUB2 or overexpression of CDC5, result in defects in partitioning of mitochondria between mother cells and buds (Fig. 5). Deletion of BUB2 reduces the amount of mitochondria in daughter cells. Deletion of MDM10 produces more severe defects in the fidelity of mitochondrial inheritance. Finally, mdm10Δ mutants bearing a deletion in BUB2 or overexpressing CDC5 exhibit defects in mitochondrial partitioning that are more severe than that in mdm10Δ mutants.
Figure 5. Hyperactivation of the mitotic exit network results in mitochondrial partitioning defects.
Wild-type, bub2Δ, mdm10Δ, and mdm10Δ bub2Δ cells (ISY001, ISY028, ISY002, ISY029), as well as a cell bearing plasmid-borne CDC5 under control of the Gal1 promoter (pCDC5) (ISY013), were grown to mid-log phase in SC-ura. All cells contain mitochondria-targeted Ds Red. For overexpression of CDC5, ISY013 was incubated in galactose for induction (6 hrs). Cells were fixed, and images of yeast bearing large buds (buds ≥2/3 the length of their mother cells) were collected at 1-μm z-intervals through the whole cell. The mitochondrial area was calculated in each z-section using a user-defined threshold, which was the same for a given mother and its bud, and these areas were summed over the mother cell and the bud to determine mitochondrial volume. Mitochondrial partitioning ratios were obtained by dividing mitochondrial areas of buds by mitochondrial areas of mother cells. Error bars show standard error of the mean for n>250 measurements.
Overall, there are numerous cell cycle checkpoints to monitor events associated with nuclear inheritance, including replication of nuclear DNA and segregation of chromosomes and nuclei. Here, we provide evidence for a mitochondrial inheritance checkpoint that inhibits cytokinesis when there are defects in mitochondrial inheritance in budding yeast, and for a role for the MEN in this process. In Drosophila melanogaster, mitochondrial second messengers, either ROS or ATP, can function as two independent signals to enforce checkpoints at G1/S that are not due to metabolic restriction (23). Our findings indicate that a checkpoint for mitochondrial inheritance, that is also independent to metabolic restriction, exist in budding yeast. Finally, since there are mechanisms to insure the inheritance of many organelles and the MEN is a conserved pathway, our findings also raise the possibility that there are similar checkpoints for organelle inheritance in yeast and other cell types.
Experimental Procedures
A summary of the materials and methods used for this study is included. Please refer to Supplemental Information for more detailed description.
Yeast strains, plasmids, and growth conditions
Yeast strains used in this work are listed in Table S1. rho0 derivatives were generated from wild-type cells expressing plasmid-borne mitochondria-targeted DsRed (ISY001), as described by Goldring et al. (7). Other yeast methods were performed according to Sherman (24). Yeast cell viability was measured using FUN-1 (25).
The carboxy terminus of Myo1p and Cdc14p were tagged with GFP using PCR-based insertion into the chromosomal copies of the MYO1 or CDC14 loci (26). Table S2 lists primers used to tag these genes. Standard molecular techniques for cloning procedures were used (27).
Fluorescence microscopy, image analysis and cytology
Mitochondria, tubulin and Sli15p were visualized using plasmid borne GFP fusion proteins. Chitin in bud scars and DNA were visualized using Calcofluor White and DAPI. Acquisition, manipulation and analysis of fluorescence images was carried out as described previously (4).
Supplementary Material
Acknowledgments
We are grateful to Drs. A. Amon, J. Shaw, K. Bloom and E. Schiebel of for plasmids; to Drs. J. Aris, A. Amon, T. Davis, D. Kellogg, J. Kitajewski and G. Schatz for antibodies; to the members of the Pon laboratory for support and critical evaluation; to Jessica Lui for data analysis and interpretation, to K. Gordon in the Flow Cytometry Shared Resource of the Herbert Irving Comprehensive Cancer Center (Columbia U.) for assistance with flow cytometry experiments, and to Drs. F. Luca and J. Gautier for enlightening discussions. This work was supported by research grants to L. A. Pon from the NIH (GM45735), and to L.J.G-R. from the Ramón Areces Foundation (Spain).
Abbreviations List
- MEN
mitotic exit network
- SPB
spindle pole body
- mtDNA
mitochondrial DNA
- FEAR
Cdc fourteen early anaphase release
- CDK
cyclin-dependent kinase
- OM
outer membrane
- DAPI
4′,6-diamidino-2-phenylindole
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fehrenbacher KL, Yang HC, Gay AC, Huckaba TM, Pon LA. Live cell imaging of mitochondrial movement along actin cables in budding yeast. Curr Biol. 2004;14:1996–2004. doi: 10.1016/j.cub.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 2.Boldogh I, Vojtov N, Karmon S, Pon LA. Interaction between mitochondria and the actin cytoskeleton in budding yeast requires two integral mitochondrial outer membrane proteins, Mmm1p and Mdm10p. J Cell Biol. 1998;141:1371–1381. doi: 10.1083/jcb.141.6.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boldogh I, Nowakowski DW, Yang HC, Chung H, Karmon S, Royes P, Pon LA. A protein complex containing Mdm10p, Mdm12p and Mmm1p links mitochondrial membranes and DNA to the cytoskeleton-based segregation machinery. Mol Biol Cell. 2003;14:4618–4627. doi: 10.1091/mbc.E03-04-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sogo LF, Yaffe MP. Regulation of mitochondrial morphology and inheritance by Mdm10p, a protein of the mitochondrial outer membrane. J Cell Biol. 1994;126:1361–1373. doi: 10.1083/jcb.126.6.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hobbs AE, Srinivasan M, McCaffery JM, Jensen RE. Mmm1p, a mitochondrial outer membrane protein, is connected to mitochondrial DNA (mtDNA) nucleoids and required for mtDNA stability. J Cell Biol. 2001;152:401–410. doi: 10.1083/jcb.152.2.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meisinger C, Rissler M, Chacinska A, Szklarz LK, Milenkovic D, Kozjak V, Schonfisch B, Lohaus C, Meyer HE, Yaffe MP, Guiard B, Wiedemann N, Pfanner N. The mitochondrial morphology protein Mdm10 functions in assembly of the preprotein translocase of the outer membrane. Dev Cell. 2004;7:61–71. doi: 10.1016/j.devcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 7.Goldring ES, Grossman LI, Krupnick D, Cryer DR, Marmur J. The petite mutation in yeast. Loss of mitochondrial deoxyribonucleic acid during induction of petites with ethidium bromide. J Mol Biol. 1970;52:323–335. doi: 10.1016/0022-2836(70)90033-1. [DOI] [PubMed] [Google Scholar]
- 8.Wiedemann N, Kozjak V, Chacinska A, Schonfisch B, Rospert S, Ryan MT, Pfanner N, Meisinger C. Machinery for protein sorting and assembly in the mitochondrial outer membrane. Nature. 2003;424:565–571. doi: 10.1038/nature01753. [DOI] [PubMed] [Google Scholar]
- 9.Honlinger A, Bomer U, Alconada A, Eckerskorn C, Lottspeich F, Dietmeier K, Pfanner N. Tom7 modulates the dynamics of the mitochondrial outer membrane translocase and plays a pathway-related role in protein import. EMBO J. 1996;15:2125–2137. [PMC free article] [PubMed] [Google Scholar]
- 10.Meisinger C, Wiedmann N, Rissler M, Strub A, Milenkovic D, Schoenfisch B, Mueller H, Kozjak V, Pfanner N. Mitochondrial protein sorting: Differentiation of β-barrel assembly to Tom7-mediated segregation of Mdm10. J Biol Chem. 2006;281:22819–22826. doi: 10.1074/jbc.M602679200. [DOI] [PubMed] [Google Scholar]
- 11.García-Rodríguez LJ, Gay AC, Pon LA. Puf3p, a Pumilio family RNA binding protein, localizes to mitochondria and regulates mitochondrial biogenesis and motility in budding yeast. J Cell Biol. 2007;176:197–207. doi: 10.1083/jcb.200606054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lippincott J, Li R. Dual function of Cyk2, a cdc15/PSTPIP family protein, in regulating actomyosin ring dynamics and septin distribution. J Cell Biol. 1998;143:1947–1960. doi: 10.1083/jcb.143.7.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stegmeier F, Amon A. Closing mitosis: the functions of Cdc14 phosphatase and its regulation. Annu Rev Genet. 2004;38:203–232. doi: 10.1146/annurev.genet.38.072902.093051. [DOI] [PubMed] [Google Scholar]
- 14.Song S, Lee KS. A novel function for Saccharomyces cerevisiae CDC5 in cytokinesis. J Cell Biol. 2001;152:451–469. doi: 10.1083/jcb.152.3.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luca FC, Mody M, Kurischko C, Roof DM, Giddings TH, Winey M. Sacharomyces cerevisiae Mob1p is required for cytokinesis and mitotic exit. Mol Cell Biol. 2001;21:6972–6983. doi: 10.1128/MCB.21.20.6972-6983.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bembenek J, Kang J, Kurischko C, Li B, Raab JR, Belanger KD, Luca FC, Yu J. Crm1-mediated nuclear export of Cdc14 is required for completion of cytokinesis in budding yeast. Cell Cycle. 2005;4:961–971. doi: 10.4161/cc.4.7.1798. [DOI] [PubMed] [Google Scholar]
- 17.Blondel M, Bach S, Bamps S, Dobbelaere J, Wiget P, Longaretti C, Barral Y, Meijer L, Peter M. Degradation of Hof1 by SCFGrr1 is important for actomyosin ring contraction during cytokinesis in yeast. EMBO J. 2005;24:1440–1452. doi: 10.1038/sj.emboj.7600627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Corbett M, Xiong Y, Boyne JR, Wright DJ, Munro E, Price C. IQGAP and mitotic exit network (MEN) proteins are required for cytokinesis and re-polarization of the actin cytoskeleton in the budding yeast, Saccharomyces cerevisiae. Eur J Cell Biol. 2006;85:1201–1215. doi: 10.1016/j.ejcb.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 19.Clifford DM, Wolfe BA, Roberts-Galbraith RH, McDonald WH, Yates JR, 3rd, Gould KL. The Clp1/Cdc14 phosphatase contributes to the robustness of cytokinesis by association with anillin-related Mid1. J Cell Biol. 2008;181:79–88. doi: 10.1083/jcb.200709060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.D'Amours D, Amon A. At the interface between signaling and executing anaphase-Cdc14 and the FEAR network. Genes Dev. 2004;18:2581–2595. doi: 10.1101/gad.1247304. [DOI] [PubMed] [Google Scholar]
- 21.Itoh T, Watabe A, Toh-E A, Matsui Y. Complex formation with Ypt11p, a rab-type small GTPase, is essential to facilitate the function of Myo2p, a class V myosin, in mitochondrial distribution in Saccharomyces cerevisiae. Mol Cell Biol. 2002;22:7744–7757. doi: 10.1128/MCB.22.22.7744-7757.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frederick RL, Okamoto K, Shaw JM. Multiple pathways influence mitochondrial inheritance in budding yeast. Genetics. 2008;178:825–837. doi: 10.1534/genetics.107.083055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Owusu-Ansah E, Yavari A, Mandal S, Banerjee U. Distinct mitochondrial retrograde signals control the G1-S cell cycle checkpoint. Nat Genet. 2008;40:356–361. doi: 10.1038/ng.2007.50. [DOI] [PubMed] [Google Scholar]
- 24.Sherman F. Getting started with yeast. Methods Enzymol. 2002;350:3–41. doi: 10.1016/s0076-6879(02)50954-x. [DOI] [PubMed] [Google Scholar]
- 25.Millard PJ, Roth BL, Thi HP, Yue ST, Haugland RP. Development of the FUN-1 family of fluorescent probes for vacuole labeling and viability testing of yeasts. Appl Environ Microbiol. 1997;63:2897–2905. doi: 10.1128/aem.63.7.2897-2905.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook J, Russell DW. Molecular Cloning : A Laboratory Manual. 2nd. Cold Spring Harbor Laboratory Press; 1998. [Google Scholar]
- 28.Paulovich AG, Hartwell LH. A checkpoint regulates the rate of progression through S phase in S. cerevisiae in response to DNA damage. Cell. 1995;82:841–847. doi: 10.1016/0092-8674(95)90481-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.