Abstract
The Epidermal Growth Factor Receptor (EGFR) activation of downstream Signal Transducers and Activators of Transcription 3 (STAT3) plays a crucial role in the pathogenesis of lung cancer. STAT3 transcriptional activity can be negatively regulated by Protein Inhibitor of Activated STAT3 (PIAS3). We investigated PIAS3 time-dependent shuffling and binding to STAT3 in an EGF-dependent model in lung cancer by using confocal microscopy, immunoprecipitation, luciferase reporter assay and protein analysis of segregated cellular components. We also explored the role of phosphorylation at Tyr705 of STAT3 in the formation of PIAS3/STAT3 complex and intracellular shuffling. In a growth factor-free state, PIAS3 was localized to the cytoplasm and unbound to STAT3 in both H520 and A549 cells. Upon exposure to EGF, we observed STAT3 phosphorylation and rapid formation of the PIAS3/STAT3 complex. Within 5 minutes there was a progressive translocation of the complex to the nucleus and by 10 minutes PIAS3 was uniquely localized to the nuclear compartment. Thirty minutes after, PIAS3 returned to the cytoplasm. Using site directed mutagenesis, we substituted Tyr705 of STAT3 with a phenylalanine. Despite EGF stimulation, we observed a significant decrease in PIAS3 and STAT3 binding and a significant reduction in nuclear translocation of PIAS3. Furthermore, there was a significant reduction in PIAS3 capacity to reduce STAT3 mediated gene transcription. In wild type STAT3 cells, increasing concentrations of PIAS3 resulted in a proportional decrease in STAT3 phosphorylation. These data suggest an important role for the negative regulatory effect of PIAS3 on STAT3 in epidermal growth factor driven tumors.
Keywords: PIAS3, STAT3, non small cell lung cancer
INTRODUCTION
Signal transducers and activators of transcription (STAT) are latent cytoplasmic transcription factors, which become activated and phosphorylated by upstream receptor and non-receptor tyrosine kinases. Upon activation, dimer formation occurs with subsequent nuclear translocation and modulation of gene transcription of its target genes. This pathway is of importance because of its functions in hematopoiesis, immune response, and oncogenesis (1). Out of these seven STATs (1, 2, 3, 4, 5a, 5b and 6), STAT3 is of particularly importance due to its involvement in a wide spectrum of biological functions. It is activated and phosphorylated by a variety of cytokines and growth factors, such as platelet derived growth factor (PGDF) and epidermal growth factor (EGF) in large number of human malignancies (2). STAT proteins have three families of natural inhibitors, the protein inhibitors of activated STAT (PIAS) (3), the suppressors of cytokine signaling (SOCS) (4–6), and the Src homology 2 containing phosphatase (SHP) (7). All three are known to participate in the negative regulation of the STAT signal transduction pathway (8).
PIAS3 belongs to a multi-gene family, which was first identified as a transcriptional repressor of activated STAT3 that inhibits transactivation of a STAT3-responsive reporter gene and inhibition of the DNA-binding activity of STAT3 (3, 9). Limited information exists on its relevance and function in solid tumors. Foremost, the intracellular kinetics and trafficking of PIAS3 in the setting receptor tyrosine kinase activation of solid tumors is unknown. PIAS3 is expressed in prostate cancer cells and functions as a transcriptional cofactor for the androgen receptor and its over-expression can induce apoptosis in prostate cancer cells (10). Most recently a loss of PIAS3 protein expression (but not mRNA) has been shown in glioblastoma multiform with an increase in STAT3 phosphorylation and activity (11). We have previously demonstrated in lung cancer that Epidermal Growth Factor Receptor (EGFR) activation leads to PIAS3-STAT3 binding (12). We have also demonstrated an absence of PIAS3 expression in 50% of lung cancer specimens with epigenetic changes as a potential mechanism for this loss (13).
The hypotheses underlying this current work is to determine the kinetics of EGFR induced PIAS3 and STAT3 interaction in a lung cancer model as well as the importance of STAT3 tyrosine phosphorylation site for this association. In the present study we have shown the time dependent cellular localization of PIAS3 under the influence of EGF, importance of the phosphorylation on the tyrosine residue at position 705 of STAT3 in forming the complex with the PIAS3 in order to inhibit the STAT3 mediated transcription. We also demonstrate that PIAS3 intracellular concentration effects STAT3 tyr705 phosphorylation and transcriptional activity.
RESULTS
Time and ligand-dependent PIAS3 localization
We investigated the time and ligand-dependent sub-cellular localization of PIAS3 protein in two different NSCLC cell lines using confocal laser-scanning microscopy. For this purpose, we initially analyzed our cells in the serum-free state without the presence of ligand. Subsequently cells were stimulated with EGF at a concentration of 20 ng/mL, which activates the EGFR-STAT3 signaling pathway in lung cancer cells. This leads to the formation of a STAT3-PIAS3 complex. We found that, in the unstimulated cells, in both cell lines (Figures 1A and 1B), PIAS3 resides mainly in the cytoplasm. Within 5 minutes of stimulation with EGF there is evidence of PIAS3 migration toward the nucleus. This translocation remains evident for the 10 and 20 minutes time point but by 30 minutes PIAS3 returns to the cytoplasmic compartment despite the presence of EGF.
Figure 1. Subcellular time dependent localization of PIAS3 upon EGF stimulation.
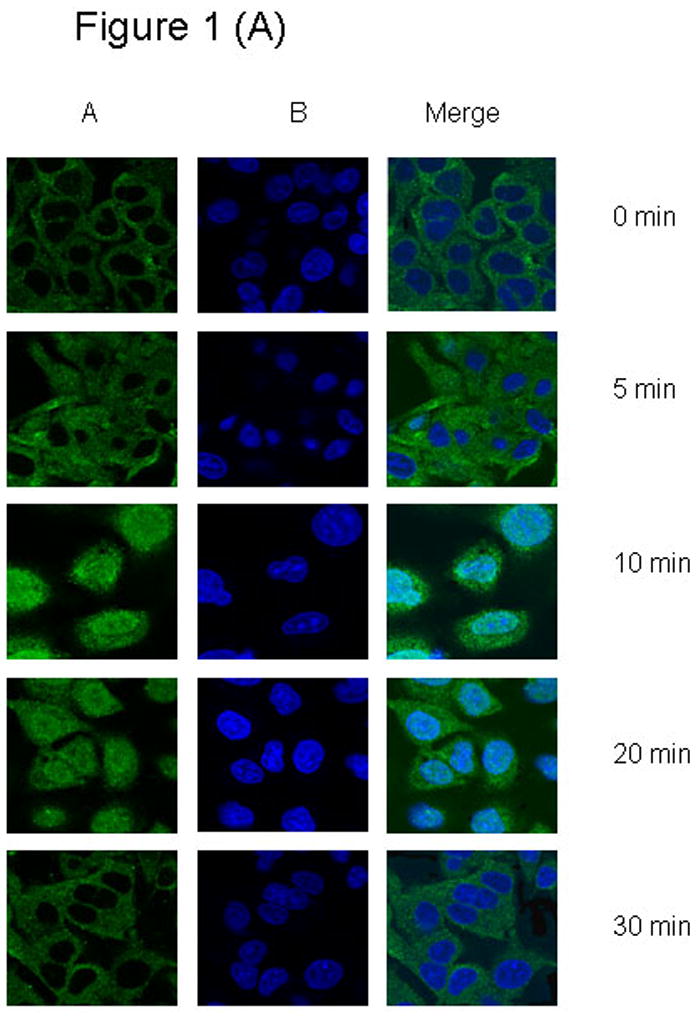
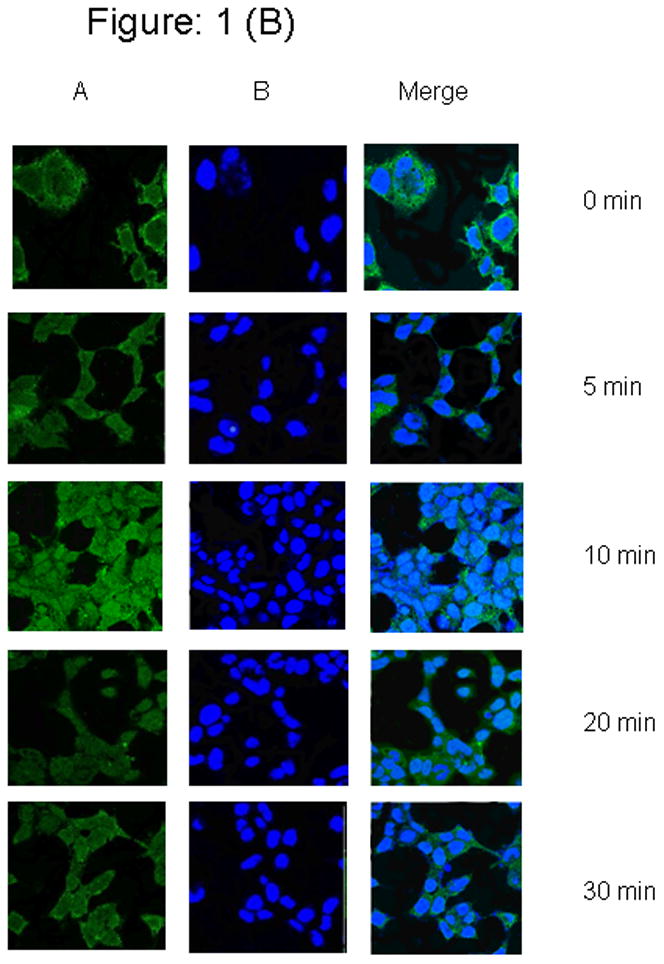
(1 A) A549 cells were seeded on poly-l-lysine treated glass bottom culture dish Starved for at least 12 h in serum-free medium and cells were unstimulated or stimulated with 20 ug/ml of EGF for 0 to 30 min. At every 5 min interval, cells were fixed, permeablized, and stained as described in the Methods., (1 B) H520 cells were seeded on poly-l-lysine treated glass bottom culture dish Starved for at least 24 h in serum-free medium and cells were unstimulated or stimulated with 20 ng/ml of EGF for 0 to 30 min. At every 5 min interval, cells were fixed, permeablized, and stained as described above. Panel (A), DRAQ5 counterstain (nuclei, blue); (B) Anti-rabbit IgG conjugated with Alexa flour 488 counterstain (Pias3, green)
Tyrosine 705 of STAT3 is critical for ligand-dependent PIAS3-STAT3 association
Given the fact that Tyr705 phosphorylation of STAT3 is an important process for STAT3 homodimer formation and nuclear translocation, we further investigated the importance Tyrosine 705 of STAT3 in the PIAS3-STAT3 complex formation and PIAS3 nuclear translocation. We transiently co-transfected A549 and H520 cell lines with either the wild type or mutant Y705 – Y705F STAT3 gene construct. After exposure to the EGF ligand, the cells were lysed and subjected to immunoprecipitation with an anti-PIAS3 antibody. The recovered immune complexes were subjected to immuno-blotting analysis with an anti-STAT3 antibody. This showed that PIAS3 interacted significantly more with wild type STAT3 (WT-STAT3) as compared to mutant STAT3 transfected cells (Figure 2A). This result indicates that the presence of Y705 residue is important for the complete association of STAT3 with PIAS3. However as seen in figure 2A a small band still appears for the mutant Y705 transfected cells. This most likely reflects the endogenous WT-STAT3 in these cells.
Figure 2.
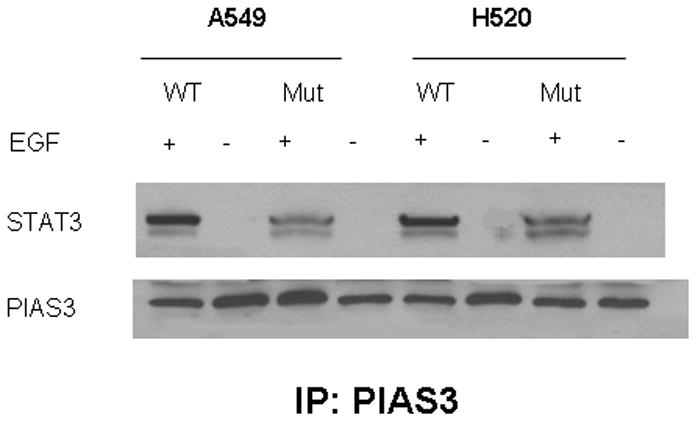
Immunoprecipitation (IP) of Pias3 with whole protein extract of NSCLC A549 and H520 cell lines transfected with wild type and 705 mutant of STAT3 expression construct with FLAG tagged pcDNA, EGF stimulated or unstimulated after 48 hrs transfection. Immunoprecipitates were resolved by Western blotting (WB) and membranes were probed with an anti, STAT3 and PIAS3 antibody. High expression of pSTAT3 is observed in wild type (lanes 1–3) whereas pSTAT3 levels are lower in mutant (lanes 2–4). In both situations, Stat3 co-precipitated with Pias3 suggesting increased Pias3/Stat3 binding.
Tyrosine 705 of STAT3 also effects PIAS3 nuclear translocation
To further demonstrate the importance of Y705 of STAT3 on EGF-induced PIAS3 nuclear translocation, we obtained nuclear and cytoplasmic extracts of A549 cells that were stimulated with EGF for 10 minutes. As had been seen in Figure 1 we would expect that maximum PIAS3 nuclear translocation to occur at the 10-minute time point. Figure 3A demonstrates that at the 10-minute time point in mutant STATs transfected cells very little PIAS3 exists in the nucleus as compared to the cytoplasmic compartment (nuclear/cytoplasmic ratio of 0.62 based on densitometry analysis of bands). This demonstrates the absence of significant STAT3 translocation to the nucleus when Y705 of STAT3 is mutated. On the other hand, in the WT-transfected cells the inverse can be seen with significantly more PIAS3 in the nucleus (nuclear/cytoplasmic ratio = 2.74). To further corroborate these findings we used confocal laser-scanning microscopy in our WT- or mutant-transfected cells. Once again cells were visualized after 10 minute of exposure to EGF. The WT cells demonstrate significant nuclear translocation of PIAS3, while much less was observed in the mutant cells (Figure 3B).
Figure 3.
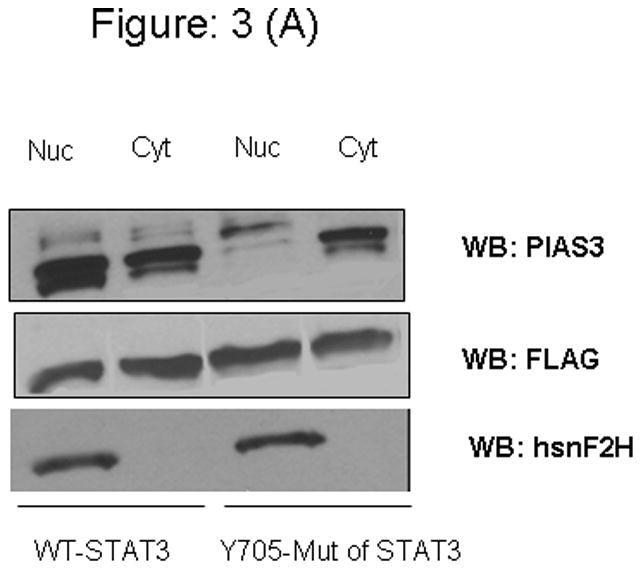
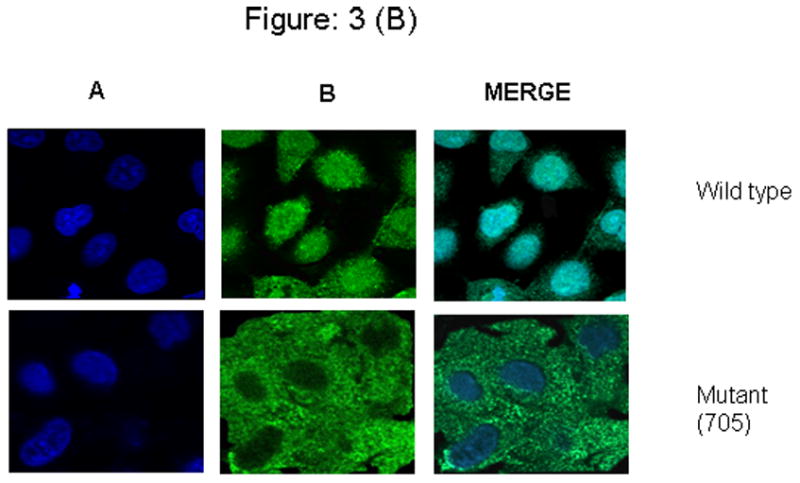
Figure 3A: Western blot analysis. A549 cells were transient transfected with a plasmid containing wild type Y705 and mutant (Y705F) STAT3 expression vector. After 48 hours of transfection, cells were starved for 12 h in serum-free medium and cells were stimulated with 20 μg/ml of EGF for 10 min, nuclear cell lysate (Nuc) and cytoplasmic cell lysate (Cyt) of wild type and Tyr-705 mutant plasmid transfected cell line subjected to SDS-PAGE and blotted with anti-PIAS3.
Figure 3B: A549 cells were incubated on the poly-l-lysine treated glass bottom culture dish, were transiently transfected with a plasmid containing wild type Y705 and mutant (Y705F) STAT3 expression vector. Both the transfected cell lines were starved for at least for 12 h in serum-free medium and were stimulated with 20 μg/ml of EGF for 10 min. Cells were fixed, permeablized, and immunostained. Wild type cells shows complete shuttling of Pias3 into nucleus, whereas mutant (Y705F) cells could not accumulate completely into the nucleus. Suggesting that the presence of tyr 705 is essential for the efficient translocation. Panel (A), DRAQ5 counterstain (nuclei, blue) and, (B) Anti-rabbit IgG conjugated with Alexa flour 488 counterstain (Pias3, green).
Negative functional role of PIAS3 on STAT3 transcriptional activity: crucial role for Y705 of STAT3
To analyze the functional aspects of Y705 on the negative regulatory effects of PIAS3 on STAT3 transcriptional activity we co-transfected the A549 cell line with wild type Y705 or mutant Y705F STAT3 along in a pCMV5 vector containing PIAS3 gene. To determine the transcriptional capacity of STAT3, a pTA-luc vector containing the luciferase reporter gene under the transcriptional control of STAT3 was used. Without the presence of EGF the luciferase activity is minimal. However with EGF stimulation and WT-STAT3 a significant increase in luciferase activity is seen (Figure 4). Co-transfection with PIAS3 expression construct results in a substantial decrease in luciferase expression when WT-STAT3 is present (p < 0.0001) and no significant effect when PIAS3 is co-transfected with mutant STAT3. This data indicates that Y705 is critical for the negative regulatory effects of PIAS3 on STAT3 transcriptional activity.
Figure 4. Effect of mutation of STAT3 at Y705 on PIAS3-STAT3 association.
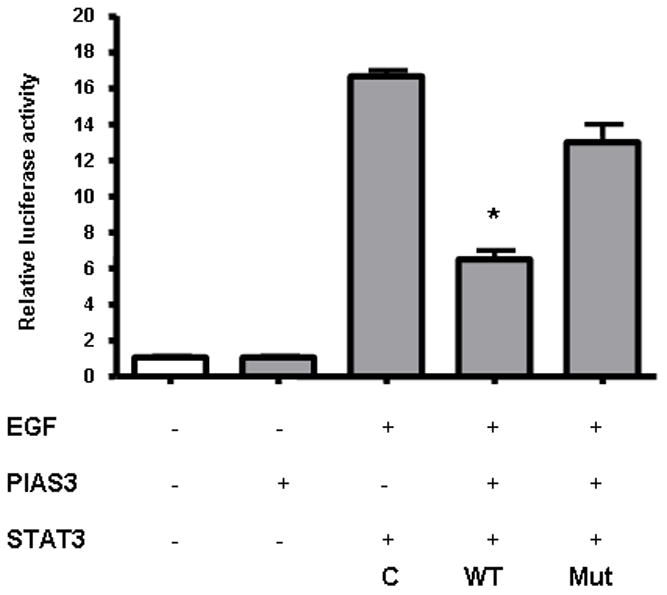
PIAS3 significantly inhibits STAT3 regulated gene expression in wild type STAT3 but fails to do so in cell lines transfected with Y705F mutant of STAT3. After 48 hrs of co-transfection with Wild type or Y705F mutant with luciferase reporter pTA-LUC vector or empty pTA-LUC vector with PIAS3 or without PIAS3, cells were then triggered with EGF for 10 min. Luciferase activity of lysed cells was measured and normalized against the protein concentration. The luciferase activity of lysed cells was measured and normalized against the protein concentration. The mean ± the standard error of the mean (SEM) of four experiments is shown. The luciferase activities of wild type and mutant were compared with the control (C) which is without PIAS3 transfection.
The negative regulation of STAT3 transcriptional activity is PIAS3 dose dependent
We then hypothesized that the intracellular amount of PIAS3, would determine the negative regulatory effect on STAT3 ability to modulate transcription of its target genes. Using our luciferase expression vector containing the STAT3 binding sequence we progressively increased the amount of intracellular PIAS3 by increasing the amount of transfected PIAS3-containing pCMV vector. EGF results in a significant increase in luciferase activity. In both the cell lines, A549 and H520, with increasing concentrations of PIAS3 a clear dose-dependent decrease in STAT3 transcriptional activity can be seen (Figure 5A and 5B).
Figure 5.
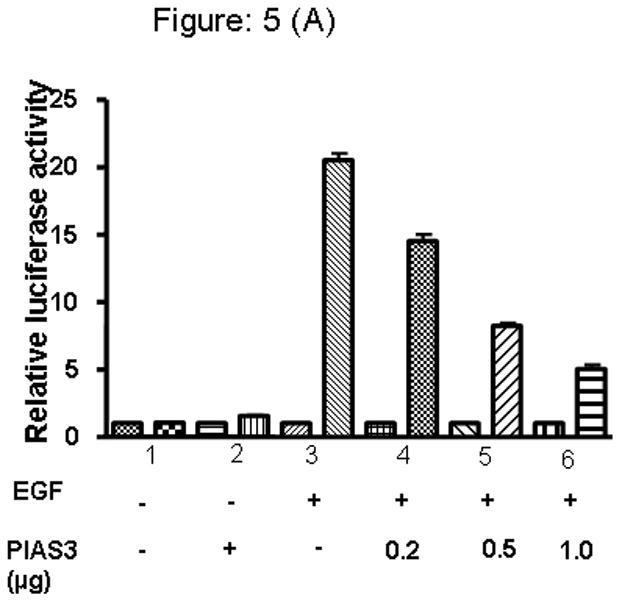
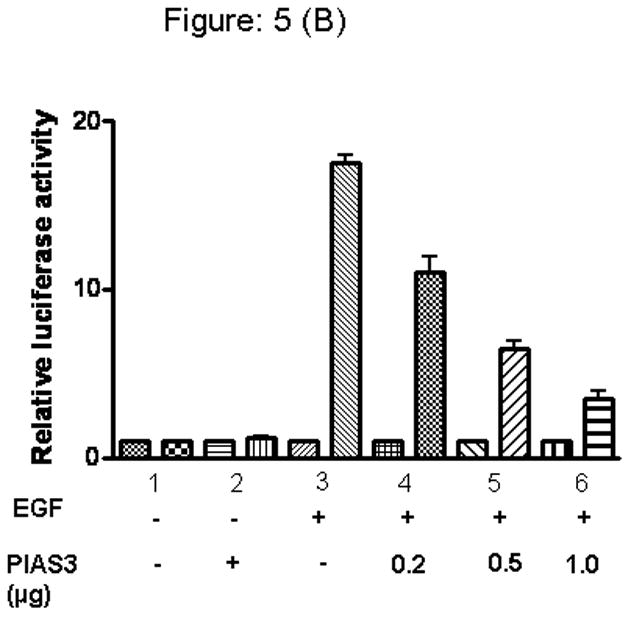
PIAS3 regulates EGF dependent transcriptional activation of STAT3 in A) A549 and, B) H520 cell lines. Cells were transfected with luciferase reporter pSTAT3-LUC vector (1μg) together with either PIAS3 expression constructs or the empty pCMV vector in increasing concentration, and then stimulated or unstimulated with EGF after 48 hrs of transfection cells were harvested and relative luciferase activity was measured. The results are presented as the relative luciferase activity from triplicate experiments.
PIAS3 dose-dependent effect on STAT3 transcriptional activity may result from the effect of STAT3 Y705 phosphorylation status
In order to elucidate the potential mechanism by which a dose dependent effect of PIAS3 would result in decreased STAT3 transcriptional activity, we looked at the relation between STAT3 phosphorylation status of the Y705 site in the presence of EGF and increasing doses of transfected PIAS3. Transfection with the same vector not containing the PIAS3 gene was used as control (Figure 6A and 6B). In this experiment both A549 and H520 cell lines were transfected with increasing amounts of PIAS3 or empty vectors. After stimulation with 20 ng/mL of EGF, cells were harvested and nuclear cell lysate were prepared. By immunoblotting with anti-phosphospecific pSTAT3 antibody (against Y705 site) we found that the p-STAT3 protein levels were significantly reduced in the nucleus, and this reduction was dose dependent on the amount of PIAS3 available. Similar results were seen in both the cell lines. Anti hsnf2H antibody was used as a loading control for nuclear extracts. This result suggests that PIAS3 inhibits STAT3 by potentially accelerating dephosphorylation process as we had previously shown that PIAS3 does not effect the initial phosphorylation of STAT3.
Figure 6.
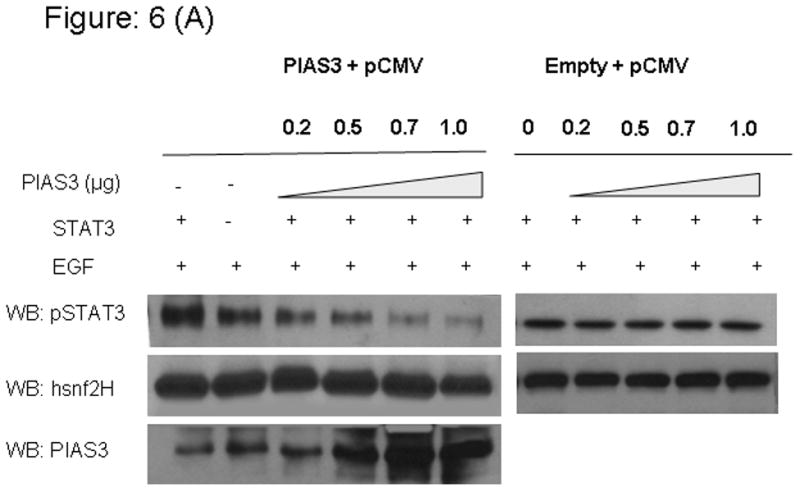
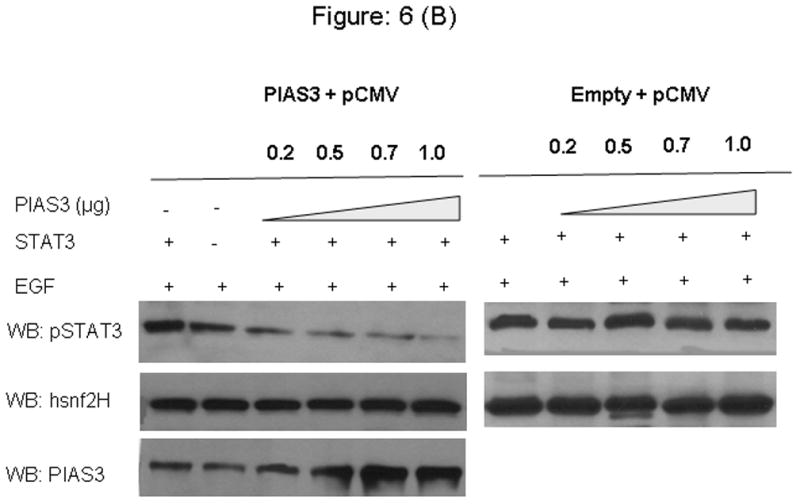
Transfection of PIAS3 expression constructs or the empty pCMV5 vector in increased concentration results in downregulation of pSTAT3 in, increasing concentration of PIAS3 and then stimulated or unstimulated with EGF after 48 hrs of transfection cells were harvested and nuclear extracts were prepared and immunoblotted for pSTAT3 and hsnF2H as a nuclear extract loading control A) A549 cell line and B) H520 cell line.
DISCUSSION
STAT3 activation has been observed to occur in many solid tumors and hematologic malignancies and is correlated with proliferative and anti-apoptotic effects in malignancies (14–16). There are several physiological STAT inhibitors of which one is the protein inhibitors of activated STAT (PIAS). PIAS proteins represents a group of five proteins PIAS1, PIAS3, PIASxα, PIASxβ and PIASy which are known to block the STAT-DNA binding activity (17). These proteins contain several conserved domains including the N-terminal SAP (Saf-A/B, Acinus, and Pias) box with the LXXLL signature; the Zn finger/RING domain, which is essential for SUMO ligase activity; and a C-terminal acidic domain, which is involved in binding to other nuclear co-activators (18) and (19). Subsequent studies in many cell systems determined that PIAS proteins interact with and modulate a broad array of nuclear proteins. The suggested mechanism of action of PIAS3 is substrate modification through the ability of PIAS proteins to function as E3 ligases and mediate small ubiquitin-related modifier-1 (SUMO-1) modification of transcription factors (2, 20–23). PIAS3 is known to inhibit gene induction through activated STAT3 by blocking the DNA binding activity of the transcription factor (3).
In this report we have demonstrate that EGF stimulation in NSCLC cells leads to the activation of STAT3 and its complex formation with PIAS3. This occurs within minutes of exposure to EGF suggesting a negative feedback loop to control STAT3 signaling. PIAS3 subsequently returns to the cytoplasm by 30 minutes of EGF stimulation. This corresponds to the same time frame in which STAT3 homodimers form with EGF stimulation as described by Wang et al. (24). Previous data in human and murine cell lines have determined that PIAS3 localizes to nuclear and cytoplasmic compartments, however the kinetics of trafficking among the two compartments was unknown (3). PIAS3’s function as a transcription factor modulator suggests that the nucleus is its site of activity. Here, we determined the presence of subcellular localization of PIAS3 protein by in situ immunofluorescence analysis and immunoblot analysis of nuclear and cytoplasmic extracts. Our data suggests that its localization is time and ligand dependent. Both, PIAS3 binding to STAT3 and its nuclear translocation are also strongly dependent on the presence of tyrosine 705 of STAT3. The importance of Tyr705 of STAT3 for PIAS3/STAT3 interaction had not been previously described.
Phosphorylation is known as an important process in the formation of protein complexes. To evaluate the importance of phosphorylation at tyrosine 705 of STAT3 in the formation of PIAS3-STAT3 complex, we constructed tyrosine 705 mutants in which tyrosine residue is substituted with phenylalanine, which mimics the charge of hydroxyl group of tyrosine. The wild type human STAT3 and Y705F mutant of STAT3 cloned in to pcDNA3 vector were transfected to A549 and H520 cell lines and stimulated or unstimulated with EGF and these cell lysate was immunoprecipitated with anti PIAS3 antibody. Substitution of tyrosine with phenylalanine resulted into decreased binding with PIAS3. These results shows that phosphorylation at tyrosine residue on the position 705 of STAT3 is critical in the PIAS3 and STAT3 association. In addition PIAS3 trafficking to the nucleus is hampered when in Y705F mutant STAT3 cells.
To understand the functional aspects of Y705 on the negative regulatory effects of PIAS3 on the STAT3 transcriptional activity we co-transfected the A549 cell line with wild type Y705 or mutant Y705F STAT3 cloned into luciferase reporter vector under the control of TA promoter, along with the pCMV5 vector containing PIAS3 expression construct. We found that PIAS3 inhibited wild type STAT3 transcriptional activity but failed to inhibit mutant STAT3 (Fig. 4). From the PIAS3 portion of the interaction, Levy et al showed that short stretch of 50 amino acids in the PINIT domain of PIAS3 is directly responsible for binding and down-regulation of STAT3 (25). We now demonstrate that the tryrosine 705 of STAT3 is critical from the STAT3 portion of the interaction.
We also demonstrate the negative inhibitory effect of PIAS3 is concentration dependent. Increasing intracellular levels of PIAS3 clearly proportionally increases the negative inhibitory role on STAT3 signal transduction. A549 and H520 cells were cotransfected with the luciferase reporter construct containing STAT3 encoding gene under the TA promoter in the presence of increasing amount of PIAS3 expression vector. EGF-induced STAT3 activation was inhibited by the expression of PIAS3 in a dose dependent manner. Recently we demonstrated that PIAS3 protein is not expressed in roughly 50% of human NSCLC specimens (13), with a similar down regulation shown in glioblastoma multiforme specimens (11). This thus sets the stage to consider upregulation of PIAS3 as potential antitumor strategy in lung cancer. Our data suggests that this concentration dependent effect occurs by two mechanisms which may be related. First we show that there is PIAS3 concentration dependent decrease in STAT3 transcriptional activity. This is consistent with the previous described effect of PIAS3 on the DNA binding activity of STAT3 (3). However, we also show a potential novel mechanism by which increasing PIAS3 concentration may determine its anti-STAT3 activity. We thus show that increasing PIAS3 concentrations decreases phosphorylation of tyr705 of STAT3. This suggests that PIAS3 may have a role in the dephosphorylation process of STAT3. As PIAS3 dose not alter the capacity of EGF stimulation to initially induce STAT3 phosphorylation, our data suggests that PIAS3 may accelerate the dephosphorylation of phospho-STAT3.
In this study, we have presented the experimental evidence that supports the hypothesis of the functional and biochemical interaction between the STAT3 and PIAS3 in NSCLC. Understanding the mechanism of interactions between the STAT3 and PIAS3 in more detailed would be important in providing the basis for drug development in NSCLC.
MATERIALS AND METHODS
Cell Lines
Human pulmonary epithelial cell line A549 cells and H520 were purchased from American Type Culture Collection (Manassas, VA). A549 and H520 were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% (v/v) heat-inactivated fetal bovine serum (FBS, Hyclone, South Logan, UT), 50 unit/ml penicillin, and 50 μg/ml streptomycin in a 5% CO2 humidified incubator at 37 °C. Cell number and viability were assessed by Trypan Blue (Sigma) dye exclusion using a hemacytometer.
Confocal microscopy
A549 and H520 cells incubated on the glass bottom culture dish (MatTek Corp., Ashland, MA) were starved for at least 12 h in serum-free medium. Cells were unstimulated or stimulated with 20 ng/ml of EGF for 5–30 min, were fixed with 4% formaldehyde diluted in phosphate-buffered saline. Fixed cells were permeabilized with 0.1% Triton X in phosphate-buffered saline for 10 min. After 1 h blocking with 1% BSA in PBS, cells were incubated with the rabbit polyclonal anti-PIAS3 (Santa Cruz Biotechnology, 1:100) antibody at room temperature for 2 h and washed 3–4 times with phosphate-buffered saline. Incubation of the Alexa Fluor® 488 goat anti-rabbit IgG (Molecular Probes, 1:500) was done for 1 h at room temperature followed by repeated washes using phosphate-buffered saline. After the subsequent staining with 1umol DRAQ-5 in PBS for 30 min, confocal images were obtained on Zeiss LSM510 NLO laser scanning microscope using single line (488 nm) or multitrack sequential excitation (488 and 633 nm). Images were acquired and processed with Zeiss LSM Image Browser software.
Plasmid construction
The STAT3 wild type and mutant (Y705F) constructs were generated as follows. The cDNA encoding the open reading frame of human STAT3 and STAT3 mutant, in which tyrosine is replaced with phenylalanine at position 705 and each were subcloned into the NheI and XhoI site at the multiple cloning site of the luciferase reporter pTA-luc vector (CLONTECH, Palo Alto, CA). The FLAG tagged pcDNA3 vector containing the coding region for wild-type (wt) STAT3 and tyrosine 705 mutant STAT3 was generously provided by Dr. J. Stark at Learner Research Institute, Cleveland. The cDNA encoding PIAS3 in a pCMV5 expression vector was generously provided by Dr. Ken Shuai from the David Geffen School of Medicine, UCLA. The fidelity of all the constructs was verified by direct sequencing.
Transient transfection and Luciferase assay
A549 cells were seeded at 1X105 cells per well in 6-well plates. The HD FuGENE (Roche) was used as a transfection reagent to cotransfect the cells with luciferase reporter construct pSTAT3 TA-Luc (wild type or mutant (Y705F)), or pTA-Luc alone as a non specific control and pcMV5 PIAS3 expression construct or pcMV5 alone as a contrrol. The cells were incubated in DMEM/HF12 media for 48 hrs treated or not with 20ng/mL EGF for 15 minutes, cells were washed with cold PBS, lysed with passive lyses buffer (Promega) and then centrifuged at 12,000 × g for 4 min. The supernatant was collected and stored at −80 °C until assessment of luciferase activity. Luminescence was read in a Berthold luminometer (Lumat LB9501) after briefly mixing the supernatant (20 μl) with 100 μl firefly luciferase assay substrate solution. The activity of luciferase was normalized to protein concentrations in lysate. Transfections were repeated at least three times, and the relative changes are presented as means ± standard errors.
Co-immunoprecipitation
A549 cells were transfected with the wild type or mutant Y705F STAT3 with a luciferase reporter vector. Transfection was carried out as described above with the transfection reagent HD Fugene (Roche). After 24 hr of transfection, media was replaced with serum free media and incubated for overnight at 37°C. Next day, cells were stimulated for 10 min with the 20 ng/ml of EGF. Nuclear and cytoplasmic cell lysate were prepared separately using Panomics kit under the manufacturer’s instructions. Hundred micrograms of protein lysate was complexed with 2μg antibody, anti-PIAS3 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) for 2 hours at 4°C, on rocking platform. Protein A-Sepharose beads (50μl) (Zymed Laboratories, Inc., San Francisco, CA) were added, the solution was incubated for one hr, washed three times with lysis buffer, centrifuged, and the supernatant removed. The bead, protein-antibody complex was then resuspended in 1X Laemmli buffer, boiled at 95°C for 5 minutes, and ten microliters of the supernatant containing the protein-antibody complex was resolved on 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), electrotransferred to nitrocellulose membranes, and detected by immunoblotting with the appropriate antibodies using the enhanced chemiluminescence (ECL) substrate from Amersham Pharmacia Corp.
Western Blotting
To obtain protein lysate, cells that were in log-phase growth (50–70% confluence) were lysed in buffer containing 1% Triton X-100, 0.15M sodium chloride, 50mM Tris, pH 7.4, and protease inhibitors (aprotinin 50ug/mL, pepstatin 50ug/mL, leupeptin 10ug/mL, EDTA 0.4mM, sodium orthovanadate 0.4mM, sodium fluoride 10mM, sodium pyrophosphate 10mM, phenylmethylsulfonyl fluoride 5mM). Protein concentrations were determined by the Coomassie method (BioRad Protein Assay, Invitrogen, Carlsbad, CA). Fifteen micrograms of protein from each sample was separated on 10% SDS-PAGE, transferred to PVDF membrane (Immobilon, Millipore, Bedford, MA), and blotted with specific antibodies (1:200 dilution for all primary antibodies and 1:5000 for all secondary antibodies Santa Cruz Biotechnology, Inc., Santa Cruz, CA).
Statistical analysis
All the described experiments were performed more than 3 times and the data are presented as mean values ± S.E.M. P values were determined by T-test using Prism software. P values < 0.05 were considered statistically significant.
Acknowledgments
Supported by grant K23 CA109348 from the National Institutes of Health
References
- 1.Buettner R, Mora LB, Jove R. Activated STAT signaling in human tumors provides novel molecular targets for therapeutic intervention. Clin Cancer Res. 2002;8:945–54. [PubMed] [Google Scholar]
- 2.Levy DE, Darnell JE., Jr Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651–62. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 3.Chung CD, Liao J, Liu B, et al. Specific inhibition of Stat3 signal transduction by PIAS3. Science. 1997;278:1803–5. doi: 10.1126/science.278.5344.1803. [DOI] [PubMed] [Google Scholar]
- 4.Endo TA, Masuhara M, Yokouchi M, et al. A new protein containing an SH2 domain that inhibits JAK kinases. Nature. 1997;387:921–4. doi: 10.1038/43213. [DOI] [PubMed] [Google Scholar]
- 5.Naka T, Narazaki M, Hirata M, et al. Structure and function of a new STAT-induced STAT inhibitor. Nature. 1997;387:924–9. doi: 10.1038/43219. [DOI] [PubMed] [Google Scholar]
- 6.Starr R, Willson TA, Viney EM, et al. A family of cytokine-inducible inhibitors of signalling. Nature. 1997;387:917–21. doi: 10.1038/43206. [DOI] [PubMed] [Google Scholar]
- 7.Shen SH, Bastien L, Posner BI, Chretien P. A protein-tyrosine phosphatase with sequence similarity to the SH2 domain of the protein-tyrosine kinases. Nature. 1991;352:736–9. doi: 10.1038/352736a0. [DOI] [PubMed] [Google Scholar]
- 8.Rakesh K, Agrawal DK. Controlling cytokine signaling by constitutive inhibitors. Biochem Pharmacol. 2005;70:649–57. doi: 10.1016/j.bcp.2005.04.042. [DOI] [PubMed] [Google Scholar]
- 9.Liu B, Liao J, Rao X, et al. Inhibition of Stat1-mediated gene activation by PIAS1. Proc Natl Acad Sci U S A. 1998;95:10626–31. doi: 10.1073/pnas.95.18.10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ogata Y, Osaki T, Naka T, et al. Overexpression of PIAS3 suppresses cell growth and restores the drug sensitivity of human lung cancer cells in association with PI3-K/Akt inactivation. Neoplasia. 2006;8:817–25. doi: 10.1593/neo.06409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brantley EC, Nabors LB, Gillespie GY, et al. Loss of protein inhibitors of activated STAT-3 expression in glioblastoma multiforme tumors: implications for STAT-3 activation and gene expression. Clin Cancer Res. 2008;14:4694–704. doi: 10.1158/1078-0432.CCR-08-0618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kluge A, Dabir S, Kern J, et al. Cooperative interaction between protein inhibitor of activated signal transducer and activator of transcription-3 with epidermal growth factor receptor blockade in lung cancer. Int J Cancer. 2009 doi: 10.1002/ijc.24553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kluge AND, Kern J, Eisenberg R, Halmos B, Ma P, Dowlati A. Protein Inhibitor of Activated STAT3 (PIAS3) is expressed in human lung cancer and is under epigenetic control. Proc Am Assoc Cancer Res. 2008 [Google Scholar]
- 14.Darnell JE., Jr Transcription factors as targets for cancer therapy. Nat Rev Cancer. 2002;2:740–9. doi: 10.1038/nrc906. [DOI] [PubMed] [Google Scholar]
- 15.Levy DE, Lee CK. What does Stat3 do? J Clin Invest. 2002;109:1143–8. doi: 10.1172/JCI15650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu H, Jove R. The STATs of cancer--new molecular targets come of age. Nat Rev Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 17.Sharrocks AD. PIAS proteins and transcriptional regulation--more than just SUMO E3 ligases? Genes Dev. 2006;20:754–8. doi: 10.1101/gad.1421006. [DOI] [PubMed] [Google Scholar]
- 18.Duval D, Duval G, Kedinger C, Poch O, Boeuf H. The ‘PINIT’ motif, of a newly identified conserved domain of the PIAS protein family, is essential for nuclear retention of PIAS3L. FEBS Lett. 2003;554:111–8. doi: 10.1016/s0014-5793(03)01116-5. [DOI] [PubMed] [Google Scholar]
- 19.Jimenez-Lara AM, Heine MJ, Gronemeyer H. PIAS3 (protein inhibitor of activated STAT-3) modulates the transcriptional activation mediated by the nuclear receptor coactivator TIF2. FEBS Lett. 2002;526:142–6. doi: 10.1016/s0014-5793(02)03154-x. [DOI] [PubMed] [Google Scholar]
- 20.Kotaja N, Karvonen U, Janne OA, Palvimo JJ. PIAS proteins modulate transcription factors by functioning as SUMO-1 ligases. Mol Cell Biol. 2002;22:5222–34. doi: 10.1128/MCB.22.14.5222-5234.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakagawa K, Yokosawa H. PIAS3 induces SUMO-1 modification and transcriptional repression of IRF-1. FEBS Lett. 2002;530:204–8. doi: 10.1016/s0014-5793(02)03486-5. [DOI] [PubMed] [Google Scholar]
- 22.Tallec LP, Kirsh O, Lecomte MC, et al. Protein inhibitor of activated signal transducer and activator of transcription 1 interacts with the N-terminal domain of mineralocorticoid receptor and represses its transcriptional activity: implication of small ubiquitin-related modifier 1 modification. Mol Endocrinol. 2003;17:2529–42. doi: 10.1210/me.2003-0299. [DOI] [PubMed] [Google Scholar]
- 23.Ungureanu D, Vanhatupa S, Kotaja N, et al. PIAS proteins promote SUMO-1 conjugation to STAT1. Blood. 2003;102:3311–3. doi: 10.1182/blood-2002-12-3816. [DOI] [PubMed] [Google Scholar]
- 24.Wang L, Banerjee S. Differential PIAS3 expression in human malignancy. Oncol Rep. 2004;11:1319–24. [PubMed] [Google Scholar]
- 25.Levy C, Lee YN, Nechushtan H, et al. Identifying a common molecular mechanism for inhibition of MITF and STAT3 by PIAS3. Blood. 2006;107:2839–45. doi: 10.1182/blood-2005-08-3325. [DOI] [PubMed] [Google Scholar]


