Abstract
The S. cerevisiae α-factor pheromone receptor, Ste2p, has been studied as a model for G protein-coupled receptor (GPCR) structure and function. Dimerization has been demonstrated for many GPCRs, although the role(s) of dimerization in receptor function is disputed. Transmembrane domains one (TM1) and four (TM4) of Ste2p were shown previously to play a role in dimerization. In this study, single cysteine substitutions were introduced into a Cys-less Ste2p, and disulfide-mediated dimerization was assessed. Six residues in TM1 (L64 to M69) that had not been previously investigated and nineteen residues in TM7 (T278 to A296) of which fifteen were not previously investigated were mutated to create 25 single Cys-containing Ste2p molecules. Ste2p mutants V68C in TM1 and nine mutants in TM7 (cysteine substituted into residues 278, 285, 289, and 291 to 296) showed increased dimerization upon addition of an oxidizing agent in comparison to the background dimers formed by the Cys-less receptor. The formation of dimers was decreased for TM7 mutant receptors in the presence of α-factor indicating that ligand binding resulted in a conformational change that influenced dimerization. The effect of ligand on dimer formation suggests that dimers are formed in the resting state and the activated state of the receptor by different TM interactions.
G protein-coupled receptors (GPCRs) are membrane proteins that form one of the largest and most diverse families of proteins in eukaryotes ranging from yeast to human. Though the primary sequences are different among the GPCRs, all GPCRs share common structural features: seven transmembrane helical domains (TMs) across the lipid bilayer, with the TMs connected by intracellular and extracellular loops, an extracellular N-terminus and an intracellular C-terminus (1). GPCRs mediate responses to various stimuli such as hormones, odors, peptides and neurotransmitters. Binding of ligand to a GPCR triggers receptor-specific signals through a heterotrimeric G protein. Since it has been reported that genetic variation of GPCRs often alters receptor functions such as ligand binding, G protein coupling, and receptor life cycle, GPCR mutation is considered a causative agent of many of human diseases (2). GPCRs have been the most successful molecular drug targets in clinical medicine (3).
Ste2p is the α-factor pheromone receptor in Saccharomyces cerevisiae and has been used as a model for the study of the molecular basis of GPCR function (4-6). Ste2p can be replaced in yeast cells with mammalian receptors with functionality conserved (7), and Ste2p can be expressed and trigger signal transduction upon ligand binding in HEK293 cells (8). Also, Ste2p can serve as an established model for fungal GPCRs. Recently, many more GPCRs in fungi have been identified and classified into six different categories based on sequence homology and ligand sensing [for reviews see (9)]. Ste2p is the most well studied receptor among fungal GPCRs, some of which are suggested to be related to fungal pathogenesis [for reviews see (9)].
Recently, evidence has been growing that many GPCRs form homo- and/or hetero- dimeric or oligomeric complexes [for reviews see (9-11)]. Oligomerization has been discovered by techniques such as crosslinking, bioluminescence resonance energy transfer, fluorescence resonance energy transfer, and immunoprecipitation (10). Dimerization is thought to be important for various aspects of GPCR function such as receptor biogenesis, formation of ligand-binding sites, signal transduction, and down-regulation (11, 12). However, the view that dimers are involved in the rhodopsin-like (Class 1A) receptor-activated signaling has been challenged (13-16).
It has been demonstrated that Ste2p is internalized as a dimer/oligomer complex (17, 18), and oligomerization-defective mutants can bind α-factor but signaling is impaired (19). It has also been shown that the dominant/negative effect on wild-type signaling of a signaling-defective mutation in Ste2p (Ste2p-Y266C) can be partially reversed by mutations in the G56XXXG60 dimerization motif, indicating that signal transduction by oligomeric receptors requires an interaction between functional monomers (20). Recently, dimer interfaces were identified in Ste2p near the extracellular end of TM1 and TM4 (21). In that study it was found that dimerization was symmetric, occurring between receptors at the TM1-TM1 interface or the TM4-TM4 interface. In our current study, using the disulfide cross-linking methodology, we studied the participation of specific residues at the intracellular boundary between TM1 and intracellular loop one and the entire TM7 in Ste2p dimerization.
Experimental Procedures
Strains, Media, and Plasmids
Saccharomyces cerevisiae strain LM102 described by Sen and Marsh (22) was used in the growth arrest and LacZ assays. The genotype for the LM102 strain is: MATa, bar1, his4, leu2, trp1, met1, ura3, FUS1-lacZ∷URA3, ste2-dl (deleted for the α-factor receptor). The protease-deficient strain BJS21 (MATa, prc1-407 prb1-1122 pep4-3 leu2 trp1 ura3-52 ste2∷KanR) was used in disulfide cross-linking and western blot assays to decrease receptor degradation during analyses (23). The parental plasmid, pHY4 expressing the template construct used for mutagenesis, FT-HT-Xa (cys-less Ste2p with the FLAG and His epitope tags with Factor Xa cleavage cite, see Table 1 for description of the various receptor constructs used in this study) was generated by introducing a tandem Factor Xa cleavage site between Val192 and Thr193 into pBec2 expressing FT-HT (FLAG and His tagged Cys-less Ste2p) under a consititutive GPD promoter (24). Twenty-five single Cys mutations ranging from Leu64 through Met69 on TM1 and Thr278 through Ala296 on TM7 were generated in the pHY4 background by PCR based site-directed mutagenesis (25). For co-expression experiments, plasmid pHY6 was constructed from p426GPD, a 2-μm based shuttle vector with a GPD promoter, CYC1 terminator, and URA marker for selection in yeast (26). STE2 containing C-terminal FLAG and His epitope tags and a tandem Factor Xa digestion site in EL2 was PCR-amplified from plasmid pHY4 using primers that introduced BamHI and EcoRI restriction sites. The resulting PCR product was subcloned into the complementary sites of p426GPD. Also, to create pHY6 an epitope tag comprising codons encoding a 9-amino acid sequence of rhodopsin (rho-tag) was substituted for the FLAG and His epitope tags by ligation of a C-terminus part of STE2 product amplified from pBKY1(27). Cys mutants in this template were generated as described above. The sequences of constructs were verified by DNA sequence analysis completed by the Molecular Biology Resource Facility located on the campus of the University of Tennessee. Primers were purchased from Sigma Genosys or IDT (Coralville, IA). After sequence confirmation, constructs were transformed into the ste2-deletion strains LM102 and BJS21 by the method of Gietz et al (28). Transformants bearing the pHY4 or pHY6 constructs were selected by their growth in the absence of tryptophan for pHY4 on MLT medium (Medium lacking tryptophan) or in the absence of uracil for pHY6 on MLU medium (Medium lacking uracil) (29). All media components were obtained from Difco.
Table 1.
Receptors used in this study
| Receptor name | Description |
|---|---|
| Wild-type | Native Ste2p |
| Ste2p-FT-HT | Wild-type with FLAG /His epitope tags |
| FT-HT | Cys-less Ste2p-FT-HT |
| FT-HT-Xa | FT-HT with tandem Factor Xa cleavage sites |
| Template for all Cys mutants generated in this study | |
Growth Arrest (Halo) Assay
Growth arrest was measured as described previously (30). Briefly, filter disks were impregnated with 10-μl portions of peptide solutions at various concentrations and placed onto the overlay containing S. cerevisiae LM102 cells. The plates were incubated at 30 °C for 24-36 h and then observed for clear zones (halos) around the disks. The halo measured included the diameter of the disc. The normalized activity of each mutant was determined by comparing halo size for the FT-HT-Xa receptor at 2μg of α-factor. All assays were carried out at least three times with no more than a 2-mm variation in halo size at a particular amount applied for each peptide.
FUS1-lacZ Gene Induction Assay
S. cerevisiae LM102 contains a FUS1-lacZ gene that is inducible by mating pheromone. Cells were grown overnight in SD (synthetic defined) medium (Yeast nitrogen base medium (Difco) without aminoacid) supplemented with the required amino acids at 30 °C to 5 × 106 cells/ml, washed by centrifugation, and grown for one doubling (hemocytometer count) at 30 °C. Induction was performed by adding 10-6 M of α-factor to 1 ml of concentrated cells (1 × 107 cells/ml). The mixtures were vortexed and, after incubation at 30 °C with shaking for 2 h, cells were harvested by centrifugation, and each pellet was resuspended and assayed for β-galactosidase activity (expressed as Miller units) in duplicate by using a β-galactosidase assay kit (Pierce) according to the manufacturer's instructions. The activity of each mutant was normalized by comparing β-galactosidase activity for the wild-type strain. The standard deviation was determined from three independent experiments.
Preparation of membranes
Membrane preparation of Ste2p was carried out essentially as described previously (30). Cells were grown to log phase, and then 1 × 108 cells were harvested by centrifugation and lysed by agitation with glass beads in a lysis buffer containing 50 mM Hepes, pH 7.5, 1 mM EDTA, 10 μg/ml phenylmethylsulfonyl fluoride, 2 μg/ml leupeptin, and 2 μg/ml pepstatin. The lysate was cleared by centrifugation at 2,000 × g for 5 min, and then membranes were harvested by centrifugation at 15,000 × g for 45 min. The membrane pellet was washed and then resuspended in 100 μl of a buffer (pH 7.4) containing 10% glycerol, 50mM Hepes, 0.15mM NaCl, 2mM CaCl2, 5mM KCl, 5mM MgCl2, 4mM EDTA (30). The protein concentration was determined by the Lowry assay (Pierce), and the membrane preparation was stored at -20 °C overnight and used for further assay the next day.
Disulfide Cross-linking with Cu-Phenanthroline
The 100 μg of membrane protein preparation were treated with a fresh preparation (pH 7.4) of Cu(II)-1,10-phenanthroline (Cu-P; final concentration, 2.5 μM CuSO4 and 7.5μM phenanthroline). The reaction was carried out at room temperature for 30 min, terminated with 50 mM EDTA and kept on ice for 20 min followed by adding Laemmli sample buffer. For study of the time course of cross-linking, Cu-P treated samples were aliquotted at 45 sec, 2 min, 5 min, 10 min, 20 min, and 30 min before termination. In all cases where a high level of crosslinking was observed the membranes were treated with Cu-P and the reaction terminated by EDTA only and by EDTA and NEM to prevent crosslinking that might occur upon subsequent manipulation of the sample. No differences were found in the extent of crosslinking with the additional NEM treatment. In experiments designed to prevent disulfide bond formation, the membranes were treated with 5mM of NEM for 20min prior to incubation with Cu-P reagent. Alpha-factor or antagonist ([desW1,desH2]α-factor) (10 μM final concentration) were added to the membrane preparation and incubation allowed to proceed for 30 min prior to Cu-P treatment in experiments performed to examine the influence of ligand on dimerization.
Factor Xa digestion
The membrane protein preparation (40 μg) was incubated with 0.4 unit of Factor Xa (Novagen) in Factor Xa cleavage buffer (0.1M NaCl, 50 mM Tris-HCl, 5 mM CaCl2, pH 8.0) containing 0.1% Triton X-100 for 30min. Each sample was divided into two aliquots. The reactions were terminated by adding one-third the volume of Laemmli sample buffer (30% glycerol, 3% SDS, 0.01% bromphenol blue, 0.1875 M Tris, pH 6.8). To one aliquot β-mercaptoethanol (final 1%, v/v) was added for reducing conditions. Samples were analyzed by SDS-PAGE and Western blotting.
Western blot
Immunoblot analysis of Ste2p was carried out as described previously (30). Each sample was incubated at room temperature and then separated on NuPAGE 10% Bis-Tris SDS-polyacrylamide gel (Invitrogen) using either non-reducing or reducing conditions and electrophoretically transferred to Immobilon™-P membrane (Millipore Corp., Bedford, MA). The blot was probed with anti-FLAG M2 antibody (Eastman Kodak Co.) or 1D4 antibody (a monoclonal antibody for rho-tag, purchased from Flintbox, BC, Canada), and the bands were visualized with the West Pico chemiluminescent detection system (Pierce). Blots were imaged, and the total density of all Ste2p bands in each lane was determined using a ChemiDoc XRS photodocumentation system with Quantity One one-dimensional analysis software (Bio-Rad). The intensity of the monomer and dimer signals was measured by densitometry, and the percentage of dimer was calculated as [Dimer/(Dimer+Monomer)×100]. The averages of the ratio were measured from at least three independent experiments and standard deviations are presented in Table 3.
Table 3.
Effect of ligand binding on Cu-P(Cu (II)-1, 10-phenanthroline) stimulated disulfide bond formation1
| Receptor | No treatment | Cu -P2 | α, Cu-P3 | A, Cu-P4 | |
|---|---|---|---|---|---|
| FT-HT-Xa | 15.0 ± 4.4 | 12.0 ± 0.1 | 13.1 ± 0.2 | 12.2 ± 0.1 | |
| TM1 | L64C | 15.8 ± 0.1 | 11.3 ± 0.4 | 14.6 ± 1.0 | 13.6 ± 0.8 |
| T65C | 16.2 ± 0.1 | 14.4 ± 0.1 | 15.9 ± 0.3 | 12.8 ± 0.3 | |
| L66C | 12.3 ± 0.4 | 11.9 ± 1.0 | 13.2 ± 0.4 | 11.9 ± 0.8 | |
| I67C | 19.3 ± 4.5 | 17.0 ± 0.1 | 17.4 ± 4.1 | 18.0 ± 2.0 | |
| V68C | 28.5 ± 3.2 | 99.2 ± 0.1 | 98.3 ± 0.8 | 98.8 ± 0.1 | |
| M69C | 17.7 ± 6.9 | 14.2 ± 5.0 | 17.0 ± 6.4 | 28.9 ± 10.2 | |
| TM7 | T278C | 15.1 ± 0.8 | 65.8 ± 8.8 | 12.5 ± 0.1 | 12.4 ± 0.1 |
| T279C | 17.3 ± 0.3 | 10.1 ± 2.3 | 15.8 ± 0.2 | 10.9 ± 2.7 | |
| V280C | 15.3 ± 2.1 | 14.7 ± 1.5 | 16.3 ± 0.6 | 12.4 ± 0.1 | |
| A281C | 16.8 ± 0.5 | 11.9 ± 0.6 | 14.3 ± 1.5 | 11.3 ± 0.1 | |
| T282C | 11.7 ± 0.3 | 15.4 ± 0.1 | 12.0 ± 0.8 | 11.7 ± 0.1 | |
| L283C | 16.6 ± 0.4 | 15.7 ± 0.2 | 13.3 ± 0.1 | 10.7 ± 3.5 | |
| L284C | 15.2 ± 0.1 | 12.0 ± 0.2 | 17.7 ± 3.0 | 15.3 ± 0.1 | |
| A285C | 16.5 ± 0.8 | 53.5 ± 8.3 | 16.5 ± 2.1 | 17.7 ± 0.5 | |
| V286C | 15.6 ± 0.4 | 17.0 ± 0.3 | 13.2 ± 2.0 | 15.9 ± 0.4 | |
| L287C | 16.0 ± 0.1 | 14.3 ± 1.3 | 11.3 ± 1.7 | 16.0 ± 1.8 | |
| S288C | 14.7 ± 1.0 | 14.7 ± 1.5 | 11.5 ± 0.6 | 12.6 ± 0.2 | |
| L289C | 26.8 ± 2.0 | 61.0 ± 9.8 | 50.2 ± 12.9 | 40.9 ± 8.2 | |
| L291C | 31.8 ± 0.4 | 66.4 ± 0.1 | 52.0 ± 8.8 | 44.5 ± 5.6 | |
| S292C | 30.2 ± 8.3 | 82.3 ± 0.4 | 75.5 ± 11.4 | 65.9 ± 0.2 | |
| S293C | 21.1 ± 0.7 | 70.2 ± 5.3 | 38.5 ± 8.3 | 44.0 ± 0.1 | |
| M294C | 27.0 ± 7.6 | 75.6 ± 1.0 | 57.8 ± 0.7 | 50.8 ± 1.0 | |
| W295C | 15.7 ± 0.2 | 95.2 ± 0.7 | 76.2 ± 0.6 | 86.3 ± 0.1 | |
| A296C | 14.8 ± 2.1 | 91.1 ± 1.0 | 73.6 ± 1.5 | 82.1 ± 0.8 | |
The intensity of the monomer and dimer signal was measured by densitometry and the ratio of the signals was determined. The percentage of dimer was calculated as [Dimer/(Dimer+Monomer) ×100].
Cu-P = Cu-P(Cu (II)-1, 10-phenanthroline) treatment.
α, Cu-P = membranes treated with alpha-factor and Cu-P(Cu (II)-1, 10-phenanthroline).
A,Cu-P = membranes treated with antagonist and Cu-P(Cu (II)-1, 10-phenanthroline).
Saturation Binding assay with [3H]α-Factor
Tritiated α-factor (10.2 Ci/mmol) (31) was used in saturation binding assays on total membrane preparations as described previously (29) Specific binding data were analyzed by nonlinear regression analysis for single-site binding using Prism software (GraphPad Software, San Diego, CA) to determine the Bmax value (receptors/cell) for each mutant receptor. Each experiment was carried out at least 3 times. The close similarity among three replicates is indicated by the standard deviations shown in Table 2.
Table 2.
Biological activities of Cys mutant receptors
| Receptor | β-galactosidase Activity | Growth Arrest activity3 | Binding Assay4 | |||
|---|---|---|---|---|---|---|
| Basal Activity1 | Induced Activity2 | B-Max | Kd(nM) | |||
| FT-HT-Xa | 1.0 ± 0.1 | 1.0 ± 0.1 | 1.00 | 1.0 ± 0.3 | 1.0 ± 0.3 | |
| TM1 | L64C | 1.0 ± 0.1 | 0.9 ± 0.2 | 1.00 | 1.7 ± 0.2 | 2.5 ± 0.7 |
| T65C | 0.9 ± 0.1 | 0.9 ± 0.1 | 1.08 | 1.2 ± 0.2 | 1.5 ± 0.7 | |
| L66C | 1.0 ± 0.1 | 1.3 ± 0.5 | 1.00 | 0.5 ± 0.1 | 0.5 ± 0.2 | |
| I67C | 1.0 ± 0.1 | 1.1 ± 0.3 | 1.04 | 1.5 ± 0.1 | 2.2 ± 0.5 | |
| V68C | 1.0 ± 0.1 | 0.9 ± 0.3 | 1.00 | 0.9 ± 0.2 | 1.1 ± 0.4 | |
| M69C | 1.1 ± 0.1 | 0.7 ± 0.2 | 1.00 | 0.7 ± 0.1 | 1.0 ± 0.4 | |
| TM7 | T278C | 0.7 ± 0.1 | 0.9 ± 0.2 | 0.88 | 0.7 ± 0.1 | 0.7 ± 0.3 |
| T279C | 1.1 ± 0.2 | 0.9 ± 0.1 | 1.08 | 0.4 ± 0.1 | 0.4 ± 0.1 | |
| V280C | 0.8 ± 0.1 | 1.3 ± 0.5 | 1.00 | 0.3 ± 0.1 | 0.5 ± 0.1 | |
| A281C | 0.5 ± 0.1 | 1.1 ± 0.3 | n.d | 0.4 ± 0.1 | 0.8 ± 0.2 | |
| T282C | 1.8 ± 0.5 | 0.9 ± 0.3 | 1.00 | 0.4 ± 0.1 | 0.9 ± 0.2 | |
| L283C | 1.6 ± 0.3 | 0.7 ± 0.2 | 0.96 | 1.7 ± 0.7 | 4.7 ± 1.3 | |
| L284C | 1.0 ± 0.1 | 0.8 ± 0.2 | 0.92 | 0.6 ± 0.1 | 1.1 ± 0.3 | |
| A285C | 1.4 ± 0.1 | 1.0 ± 0.1 | 0.88 | 0.7 ± 0.2 | 0.9 ± 0.2 | |
| V286C | 1.3 ± 0.1 | 1.0 ± 0.3 | 1.12 | 0.7 ± 0.1 | 1.9 ± 0.7 | |
| L287C | 1.7 ± 0.1 | 1.0 ± 0.3 | 1.01 | 0.8 ± 0.1 | 1.1 ± 0.1 | |
| S288C | 1.1 ± 0.1 | 1.0 ± 0.3 | 1.00 | 0.7 ± 0.1 | 4.1 ± 1.5 | |
| L289C | 1.4 ± 0.1 | 0.8 ± 0.2 | 0.92 | 1.3 ± 0.2 | 2.1 ± 0.6 | |
| P290C | 1.4 ± 0.1 | 1.0 ± 0.1 | 0.88 | n.d. | n.d. | |
| L291C | 1.3 ± 0.1 | 1.0 ± 0.3 | 0.88 | 0.6 ± 0.1 | 1.7 ± 0.8 | |
| S292C | 1.4 ± 0.1 | 1.0 ± 0.3 | 1.04 | 0.6 ± 0.1 | 0.2 ± 0.1 | |
| S293C | 1.5 ± 0.1 | 1.0 ± 0.3 | 1.00 | 1.2 ± 0.1 | 2.5 ± 0.4 | |
| M294C | 1.2 ± 0.1 | 1.2 ± 0.4 | 0.94 | 0.8 ± 0.1 | 0.9 ± 0.2 | |
| W295C | 1.7 ± 0.3 | 1.5 ± 0.3 | 1.01 | 1.3 ± 0.1 | 1.3 ± 0.1 | |
| A296C | 1.7 ± 0.4 | 2.5 ± 0.8 | 1.00 | 0.7 ± 0.1 | 0.4 ± 1.5 | |
Relative activity (±standard deviation) compared with basal activity of FT-HT-Xa (Miller unit of FT-HT-Xa was 3.9)
Relative activity (±standard deviation) compared with induced activity of FT-HT-Xa at 1μM of α-factor (Miller unit of FT-HT-Xa was 31.7)
The relative halo size (growth arrest) was compared to the halo size of the FT-HT-Xa receptor at 2μg of α-factor applied to a disc (the halo size of FT-HT-Xa was 24 mm). The standard deviation of the halo activity for all receptors was within ± 0.2 (three replicates). n.d. = not detected
The Bmax and Kd values are presented relative to those of the FT-HT-Xa receptor. The Bmax and the Kd were determined by saturation binding of radioactive α-factor according to the protocol in Methods. n.d. = not detected, as the binding was not greater than background (B-Max of FT-HT-Xa was 3491 and Kd was 10.7nM).
HIS-select HC Nickel affinity
Membrane preparations (500 μg protein) from each sample were resuspended in a solubilization buffer (50 mM Tris HCl, 150 mM NaCl, 1% Triton X-100, 5 mM imidazole, pH 8.0) overnight. The solubilized proteins were then mixed with HIS-select HC Nickel affinity gel (Sigma) and incubated at 4°C for 30 minutes with end-over-end mixing. The Nickel gel was separated from the mixture by centrifugation and then washed five times with buffer (50 mM sodium phosphate, pH 8.0, 0.3 M NaCl, 5 mM imidazole). Ste2p was eluted from the Nickel gel two times using elution buffer (50 mM sodium phosphate, pH 8.0, 0.3 M NaCl, 250 mM imidazole). The eluted samples were analyzed by SDS-PAGE silver staining and western blotting using antibodies against FLAG, a 9-amino acid sequence of rhodopsin (rho-tag), or an antibody to the N-terminus of Ste2p (32).
Results
Expression and Biological Activities of Cys Mutant Receptors
To begin the analysis of Ste2p dimerization we chose twelve residues, six in TM1 and six in TM7 (L289 – M294) proximal to the cytoplasmic face of Ste2p (Fig. 1A) for mutation to Cys. To extend the initial cross-linking results, thirteen additional residues in TM7 were mutated (T278C – S288C, W295C, and A296C) (Fig. 1A). These targeted residues were chosen for several reasons: (i) The TM1 region chosen was one helix turn away from the sequence G56XXXG60 previously established as being involved in Ste2p dimerization (19), (ii) TM7 contains the AXXXA motif also suggested to play a role in dimerization of other proteins (33) and 20 known growth factor receptors with tyrosine kinase activity (34), (iii) eight of the TM7 residues mutated (S288-S293,W295 and A296) are conserved among fungal pheromone receptors (35), and (iv) TM1 and TM7 are in close proximity to one another in crystal structures of GPCRs (36, 37) and a model of Ste2p (35). To eliminate non-specific cross-linking the template for these mutations was a Cys-less receptor. This template (FT-HT-Xa) also contained two C-terminal epitope tags (FLAG and 6XHis) and tandem Factor Xa cleavage sites (IEGRIEGR) in the second extracellular domain in order to facilitate detection of inter-domain cross-linking (Fig. 1A). Wild-type, FT-HT (Cys-less Ste2p with the FLAG and His epitope tags) and FT-HT-Xa (the template construct) receptors used in this study demonstrated almost identical biological activities in a growth arrest assay indicating that incorporating the protease site and the epitope tags did not alter receptor function (Fig. 1B). In addition, FT-HT-Xa and Ste2p-FT-HT showed identical expression (data not shown). The expression level of each single-Cys mutant receptor was determined by western blot analysis. All mutants except P290C showed several bands between 44kDa and 55kDa and expression levels similar to that of FT-HT-Xa (Fig. 2A and Fig 4A). The multiple bands are typical of Ste2p expression and are due to differences in glycosylation state, which does not influence receptor function (38). Although two intrinsic Cys residues have been substituted, a weak band at 110 kDa, corresponding to a dimerized form of Ste2p, was observed for the FT-HT receptor (Fig. 2B). This band is likely a native, non-covalent dimer which was not disrupted by membrane protein preparation or SDS-PAGE. Such dimers have been observed on SDS-PAGE gels by other investigators working with Ste2p (20, 39, 40) and are also seen with the FT-HT-Xa receptor (Fig. 5A; Lane 1). This weak dimer observed in the Cys-less receptor is consistent with immunoprecipition results showing that the two intrinsic Cys are not involved in dimerization (18). Membrane expression of P290C receptor was very low when judged by western blot (data not shown). It is known that one of the roles of the conserved proline in TM7 in GPCRs is correct folding (41). The reduced expression of P290C was observed previously when it was shown that this receptor was primarily defective in plasma membrane localization using a P290C-GFP receptor, but the P290C-GFP construct did not demonstrate a defect in signaling (42).
Figure 1.
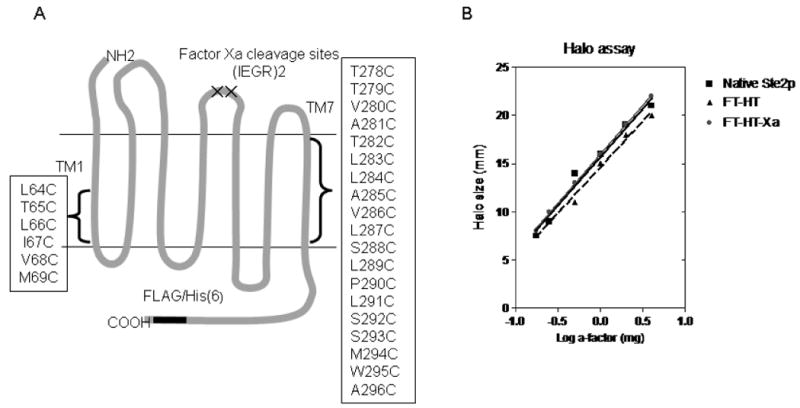
A. The two-dimensional topology of Saccharomyces cerevisiae Ste2p. The cartoon indicates the location of the genetically engineered Factor Xa digestion site in EL2 and the epitope tags in the C-terminus. The two endogenous Cys residues (C59 and C252) were mutated to Ser to generate FT-HT-Xa which was the parental GPCR used to generate the TM1 and TM7 cysteine mutations in positions 64 to 69 in TM1 and 278 to 296 in TM7 as indicated in the boxes. B. Dose-response analysis of growth arrest assay. The zone of growth inhibition of strains carrying the indicated receptors was measured at various concentrations of α-factor. FT-HT-Xa is the Cys-less receptor containing C-terminal FLAG and His epitope tags and a tandem Factor Xa digestion site in EL2. FT-HT is the Cys-less receptor without the Xa digestion site containing the FLAG and His epitopes, and Native Ste2p is the wild-type receptor that has no epitope tags and has two Cys residues (C59 and C252).
Figure 2.
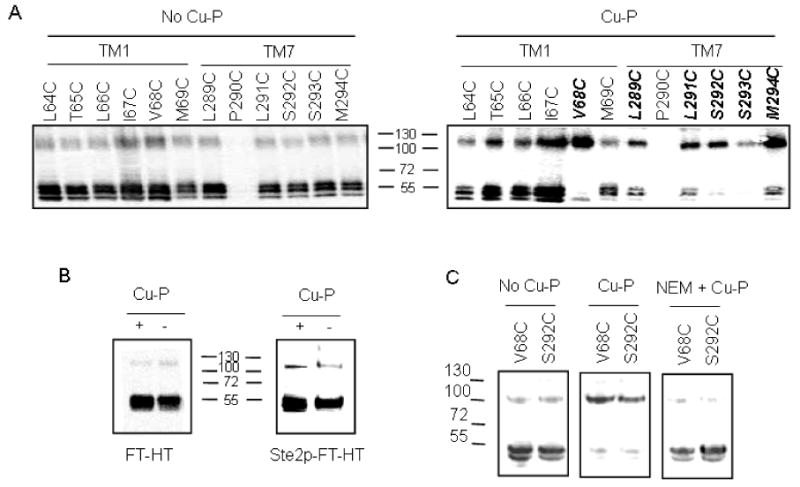
Effect of Cu-P (Cu (II)-1, 10-phenanthroline) treatment on the dimerization of Ste2p containing single cysteine mutations. Membrane proteins were treated with Cu-P or left untreated, followed by SDS-PAGE, then immunoblotted and probed with anti-FLAG antibody. The upper band (∼110 kDa) represents dimerized receptor and the lower band (∼55 kDa) represents monomer. A. Receptors with Cys in TM1 or TM7 were treated with Cu-P (Right panel) or were untreated (Left Panel). Receptors indicated in bold exhibited a shift in signal from monomeric to dimeric form. B. Controls with Cys-less Ste2p-FT-HT and Ste2p-FT-HT. C. V68C and S292C receptors were untreated or treated with Cu-P in the presence or absence of NEM(N-ethylmaleimide) added prior to Cu-P treatment.
Figure 4.
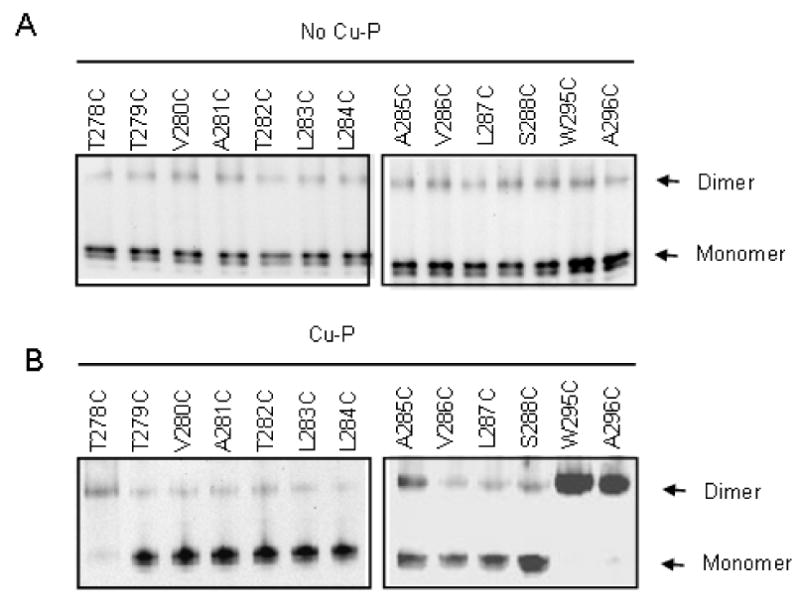
Effect of Cu-P (Cu (II)-1, 10-phenanthroline) treatment on FT-HT-Xa containing cysteine replacements in TM7. Membranes were prepared, treated without (A) or with (B) Cu-P reagent, solubilized and separated on SDS-PAGE. Each sample was immunoblotted and probed with anti-FLAG antibody.
Figure 5.
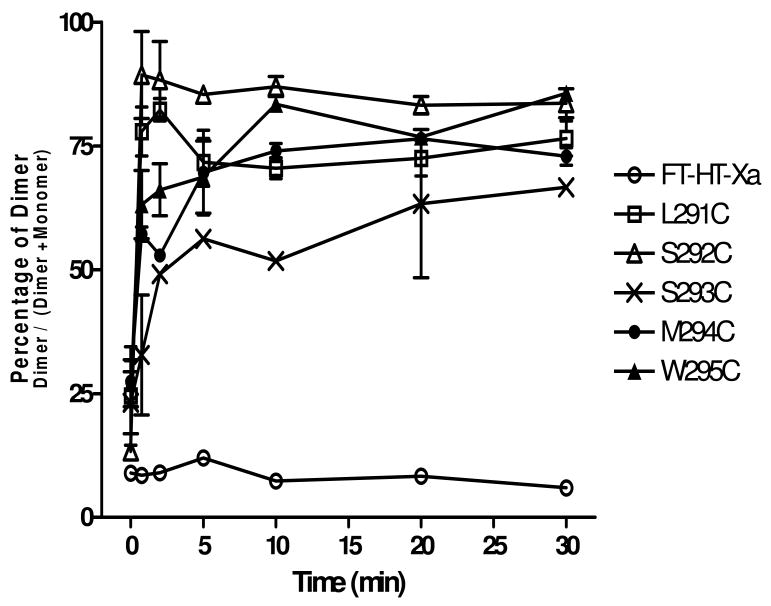
Time course analysis of dimer formation at residues C-terminal to P290 of TM7. Total membranes were prepared from cells expressing each receptors and treated with Cu-P (Cu (II)-1, 10-phenanthroline) for 45 sec, 2 min, 5 min, 10 min, 20 min, and 30 min. The samples were analyzed by SDS-PAGE, following by immunoblotting with anti-FLAG antibody. The average percentage of dimer population from three independent experiments was calculated and plotted.
Biological activity of each mutant was measured by growth arrest, β-galactosidase activity and binding assays and is normalized relative to the parent receptor (Table 2). Most mutant receptors showed similar β-galactosidase activity and growth arrest activity as compared to FT-HT-Xa. Two mutants (M69C and L283C) showed slightly lower induced β-galactosidase activities and A281C did not show growth arrest activity which is consistent with a previous study (43). The difference between β-galactosidase activity and cell division arrest has been observed in many mutagenic studies of Ste2p (44-46). It has been proposed that a short term signaling effect is measured by the β-galactosidase assay, whereas the long term effect is measured by the halo assay. The α-factor binding affinities of most Cys mutants were within the experimental error range (± 2.5 fold), with the exceptions of the S292C, receptor which showed an increase in binding affinity (5 fold lower Kd value), and the L283C and S288C which showed a decrease in binding affinity (up to 4.7 fold higher Kd values) as compared to that of FT-HT-Xa. B-Max values determined by a saturation binding assay indicated that the numbers of binding sites were similar to FT-HT-Xa for most mutant receptors, with the exceptions of T279C V280C, A281C and T282C, which showed expression about 50% lower than that of FT-HT-Xa. The P290C receptor showed no detectable alpha-factor binding, consistent with lack of expression in the western blot analysis. Nonetheless, this receptor exhibited comparable growth arrest and β-galactosidase activities, suggesting that there were sufficient receptors expressed on the cell surface to elicit these biological responses upon ligand treatment (Table 2). Previous studies have shown that remarkably low levels of Ste2p at the cell surface are sufficient to manifest the biological responses (47). Taken together, we conclude that Cys mutation of the targeted residues did not severely interfere with receptor expression and function. We did not use the P290C receptor further in this study as it was expressed at a very low level.
Dimerization of Some Cys Mutants in TM1 and TM7 Is Markedly Enhanced by the Oxidizing Reagent Cu (II)-1, 10-Phenanthroline
Initially we tested the involvement of twelve TM residues, six on TM1 and six on TM7 (L289 – M294) proximal to the cytoplasmic face of Ste2p (Fig. 1A), in dimerization. As stated above, FT-HT-Xa and virtually all of the single Cys containing Ste2p mutants exhibited weak bands at about 110 kDa consistent with a small amount of dimerized Ste2p. However, in all cases the predominant band was near 55 kDa, the molecular mass of monomeric Ste2p. When these same receptors were treated with Cu (II)-1, 10-phenanthroline (Cu-P), a reagent that has been used as an oxidizing reagent to drive disulfide bond formation in membrane proteins (48-51), many of the Cys mutated receptors showed marked increases in the 110 kDa band and a concomitant decrease in the monomer band (Fig 2A, Table 3). Notably, V68C, a TM1 mutant, and five mutants on TM7 (L289C, L291C, S292C, S293C and M294C) showed strong (>60%) dimerization upon Cu-P treatment (Fig. 2A, Table 3). When FT-HT (epitope tagged Cys-less receptor) and Ste2p-FT-HT (epitope tagged receptor with intrinsic cysteine residues at C59 in TM1 and C252 in TM5) receptors were tested, there was no difference in dimerization in the presence and absence of Cu-P treatment (Fig. 2B) indicating that increased dimerization of mutant receptors was due to the specific cysteine residues engineered into Ste2p. To further test whether dimerization was mediated by disulfide bond formation involving cysteine residues, NEM (N-ethylmaleimide) pre-treatment was used. NEM alkylates the free –SH group of cysteine irreversibly, so that disulfide bond formation cannot occur after NEM treatment. As shown in Figure 2C, pretreatment of V68C or S292C with NEM completely eliminated the Cu-P mediated dimerization. Similar results were found with the other TM7 residues tested (data not shown). In contrast to these six residues, placement of cysteine at positions 64, 65, 66, 67 and 69 followed by Cu-P treatment did not result in a dimer population that exceeded 20% (Fig. 2A, right panel). Taken together, the results indicate that insertion of cysteine residues at position 68 on TM1 and positions 289, 291, 292, 293 or 294 on TM7 led to high levels of Ste2p dimerization through disulfide formation.
Although the studies described above provided evidence for Cu-P catalyzed Ste2p dimerization, it was possible that the single Cys mutant Ste2p is cross-linking with other proteins with similar molecular weight to yield the observed high molecular weight bands. To confirm that the observed dimer band in the western blot was a Ste2p-Ste2p homo-dimer, we took advantage of the protease (Factor Xa) digestion site engineered into EL2 of Ste2p. Cross-linking between two Cys residues in TM7 followed by Factor Xa digestion would yield a homodimeric complex of 66 kDa (Fig. 3A). Indeed TM7 mutants (L289C, L291C, S292C, S293C and M294C) digested with Factor Xa led to the appearance of a band near 66 kDa when SDS-PAGE was run under non-reducing condition (Fig. 3B; left panel). This band was markedly reduced when the gel was run in the presence of β-mercaptoethanol, a potent disulfide reducing agent. The 66 kDa band was specific for dimerized receptors with a disulfide connection between monomers as the band was not observed for FT-HT-Xa (Cys-less) and a TM1 mutant (M69C) in which there was no disulfide-mediated dimer formation (Fig. 3B). The monomer bands (∼55 kDa) are due to an incomplete Factor Xa digestion. We performed partial digestion because a longer incubation led to the degradation of all bands. Bands at ∼33 kD are Factor Xa digestion products of the monomeric receptor species (remaining in the samples). The generation of this band from the S292C and S293C mutant receptors, in which the majority of receptors formed dimers (Fig. 2A, right panel), is likely the result of detergent induced (Triton X-100 in Factor Xa digestion buffer) dissociation of background dimers. Indeed, the presence of background dimers from all the receptors tested including FT-HT-Xa (Cys-less) and M69C were reduced in the presence of Triton X-100. Further support for the disulfide bond induced dimer formation was provided when a Rho tagged M294C receptor co-expressed with a HIS tagged M294C receptor was retrieved as a dimer after pull-down using anti-HIS antibody both with and without Cu-P (data not shown). These results and the cross-linking results demonstrate the proximal location of the residues (L289-M294) on the intracellular parts of the TM7 domains of two Ste2p molecules, and provide evidence for homo-dimerization of Ste2p involving TM7 as well as TM1.
Figure 3.
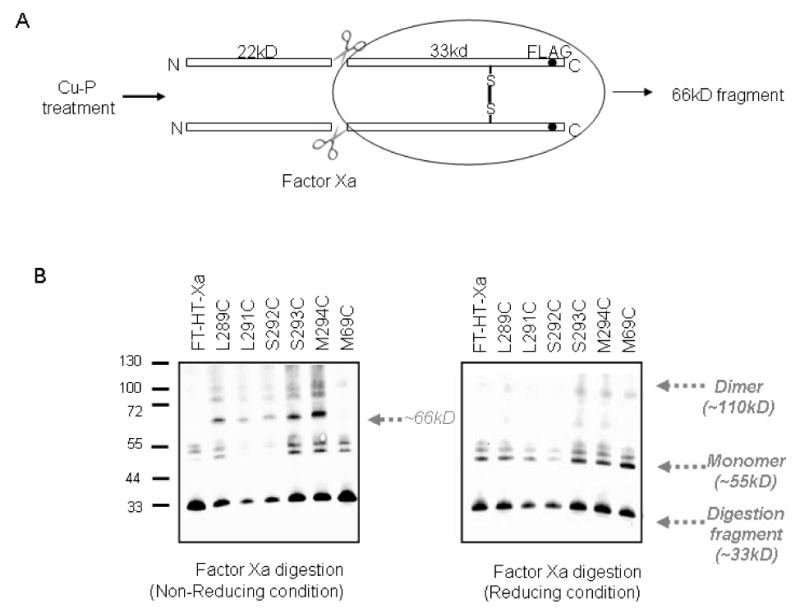
Protease Factor Xa digestions of single cysteine containing Ste2p receptors. A. Diagram showing possible explanation for ∼66 kDa band from TM7-TM7 interaction. B. Total membrane proteins derived from cells expressing indicated receptors. Cu-P treated membranes with Ste2p containing single Cys mutations at various positions as indicated were treated with Factor Xa as described in the Methods. The digests were subjected to SDS-PAGE under reducing or non-reducing conditions, immunoblotted and probed by anti-FLAG antibody.
The finding that six consecutive residues in TM7, with the exception of P290C, showed an increase in dimerization was unexpected since this region is believed to be α-helical. To see whether dimerization would occur throughout TM7, we expanded Cys replacement to include the full TM7 by generating eleven additional cysteine mutants N-terminal to the L289 residue (T278C, T279C, V280C, A281C, T282C, L283C, L284C, A285C, V286C, L287C, and S288C) and two cysteine mutants C-terminal to the M294 residue (W295C, A296C). Membranes from each mutant were prepared and processed with or without Cu-P treatment. Compared to the Cys-less FT-HT-Xa receptor, the T278C and A285C receptors exhibited at least a 3-fold increase in dimerization (Fig. 4). Though dimer formation on the T278C mutant was not observed in a recent study (21), this mutant showed strong dimer formation with Cu-P treatment in our study. The difference observed for this one residue is likely the result of variations in experimental design. In our current study Cu-P was used at a final concentration of 2.5 μM, while in the previous study the concentration was 500 μM. In addition, in our study we used membrane preparations whereas whole cells were used by Wang et al. Receptors W295C and A296C showed a large increase in dimerization upon treatment with Cu-P compared to FT-HT-Xa, (Fig 4A; Table 3). In contrast, nine mutants (T279C-L284C, V286C, L287C and S288C) gave virtually identical dimerization results in the presence and absence of the oxidizing agent (Fig. 4B; Table 3). These results indicate that unlike the residues C-terminal to Pro290, residues located N-terminal to Pro290 exhibit a periodicity with respect to dimer formation of Cys mutants which is consistent with the α-helical structure predicted for typical transmembrane domains of GPCRs. Because of a concern that disulfide crosslinking might trap and thereby favor the accumulation of dimer during the 30 min oxidization reaction, we analyzed the efficiency of the dimer formation at different times (Fig. 5). The results indicated that L291C, S292C, S293C, M294C, and W295C started to form dimers after only 45 seconds of Cu-P incubation and reached half maximum levels within 5 minutes. Previously, the relative distance between TM residues in rhodopsin was estimated by the reaction time for disulfide formation between to Cys residues on different TMs (52). Thus, our data indicate that TM7 C-terminal to P290 is highly flexible allowing all of the residues in this part of TM7 to form disulfide bonds with more or less the same rapidity.
Effect of Ligand Addition on Dimerization
It is generally believed that activation of GPCRs upon ligand binding results in a conformational change involving rearrangement of transmembrane domains (53-56). To ascertain whether this conformational change would affect Ste2p dimerization, we investigated the changes in the dimerization pattern of Ste2p receptors in the presence and absence of ligand. The membranes expressing each mutant receptor were incubated with agonist (α-factor) or an antagonist ([desW1, desH2]α-factor) and then treated with Cu-P. Dimer and monomer formation were monitored by western blots (Fig. 6A and 6B) and the percent dimer formation was determined (see Methods, Table 3). Dimer formation of the Cys-less FT-HT-Xa receptor in the presence or absence of Cu-P was not affected by agonist or antagonist (Fig. 6A and Table 3). Also, the ratio of dimer to monomer formed by Cu-P mediated cross-linking of the TM1 cysteine mutants did not change significantly after incubation with α-factor or an α-factor antagonist (representative results for V68C are shown in Fig. 6B and Table 3). A time course of crosslinking at 0.75, 2, 5, 10, 20, and 30 min was also performed with the V68C receptor incubated with either α-factor or antagonist. The reaction was complete at 45 seconds (0.75 min) and no change in crosslinking relative to that observed in the absence of ligand was observed (data not shown). TM7 mutants which did not exhibit increased levels of dimerization upon Cu-P treatment (T279C-L284C, V286C, L287C and S288C), also showed no difference in dimer formation in the presence of α-factor or the α-factor antagonist (Table 3; raw data for S288C is shown in Fig. 6B). However, for those TM7 receptors which exhibited a significant increase in dimer formation upon Cu-P treatment (T278C, A285C, L289C, L291C, S292C, S293C, M294C, W295C, and A296C), ligand treatment decreased the relative amount of dimer formation (Table 3; raw data for T278C and W295C are shown in Fig. 6B). Taken together, the results suggest that the dimer interface of TM7 of Ste2p is changed in response to either agonist or antagonist binding.
Figure 6.
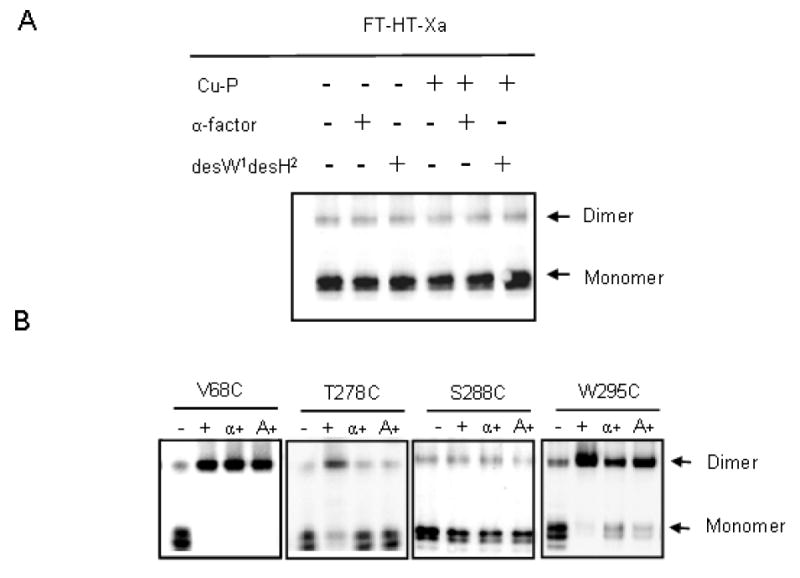
Effect of ligand binding on Cu-P (Cu (II)-1, 10-phenanthroline) stimulated disulfide bond formation. A, B. Total membrane protein was derived from cells expressing indicated receptors. Each sample was immunoblotted and probed with anti-FLAG antibody. FT-HT-Xa receptor was treated as indicated B ‘-’ indicates no treatment, ‘+’ indicates Cu-P treated samples, ‘α +’ indicates samples incubated with α-factor followed by Cu-P treatment and ‘A+’ indicates samples incubated with the antagonist [desW1, desH2]α-factor followed by Cu-P treatment.
Discussion
In this study, using a disulfide cross-linking methodology, we identified a specific residue in TM1 that interacts in Ste2p dimers, and we present the first evidence that residues in TM7 of this receptor participate in its dimerization. All of our studies were conducted with Ste2p in its membrane-bound state. Previously, it was shown that the maximum distance between α-carbons linked by disulfide bonds is about 7 Å (57). Thus, cross-linking experiments should identify amino acid side chains that are in close proximity when using the Cu-P oxidation reagent, which facilitates oxidation of sulfhydryl groups in cysteine residues. In analyzing our data it is important to note that disulfide crosslinking might trap transient intermediates. Depending on the time of cross-linking relative to the rates of interconversion of the monomeric and dimeric states of the receptor, covalent cross-linking might affect the equilibrium and bias the sample to yield more dimeric species than are present in the native population of the receptor. However, by comparing the cross-linked population of a receptor mutated in a specific region we believe we can learn about the relative tendencies of individual residues to participate in receptor-receptor contacts. Cross-linking between cysteine residues engineered into GPCRs and the use of Cu-P as an oxidizing reagent to facilitate disulfide bond formation between TMs in GPCRs has been used extensively by the Wess and Oprian laboratories (48, 51, 55, 58, 59) and was recently applied to Ste2p dimerization (21).
It had been suggested that TM1 formed a component of the interface between the two receptors in the Ste2p dimer (19, 21, 60). Our results support these findings and furthermore provide direct evidence for the involvement of V68 in TM1 in Ste2p dimerization. The mutant receptor V68C showed markedly increased dimerization over that of the FT-HT-Xa after Cu-P treatment, while under identical conditions all of the remaining TM1 Cys receptors showed at most a minor increase in dimer formation (Fig. 2A; Table 3). The ability of V68C to form dimers is in good agreement with the recently published finding of a V45C-V45C cross-linking (21) since V45 and V68 are both located on the same face of TM1 and on opposite ends of TM1 (Fig. 7A). It was previously proposed that G56XXXG60 residues in TM1 of one GPCR monomer interacted with a hydrophobic-rich surface of TM1 (perhaps involving residues I53, V57, A61, and L64), on the second monomer by a kind of “groove-in-ridge” association with Gly being the “groove” residues and the hydrophobic residues acting as the “ridges” (19). In this study, we found that V68C formed a disulfide bond. The fact that only one residue in the sequence (L64T66L67I67V68M69) formed this linkage suggests that the Ste2p-Ste2p interactions involving this region of the TM1 helix have significant spatial restrictions and that the TM1 helix at the carboxyl side of the G56XXXG60 sequence may be relatively rigid. We note that a biophysical analysis of TM1 in micelles indicates that the G56XXXG60 motif itself is flexible [(61) and unpublished results] and may facilitate TM-TM interactions between proximal helical elements. In any event our mutational analysis of the TM1 domain defines a specific residue (V68) that appears to be involved in Ste2p dimerization.
Figure 7.
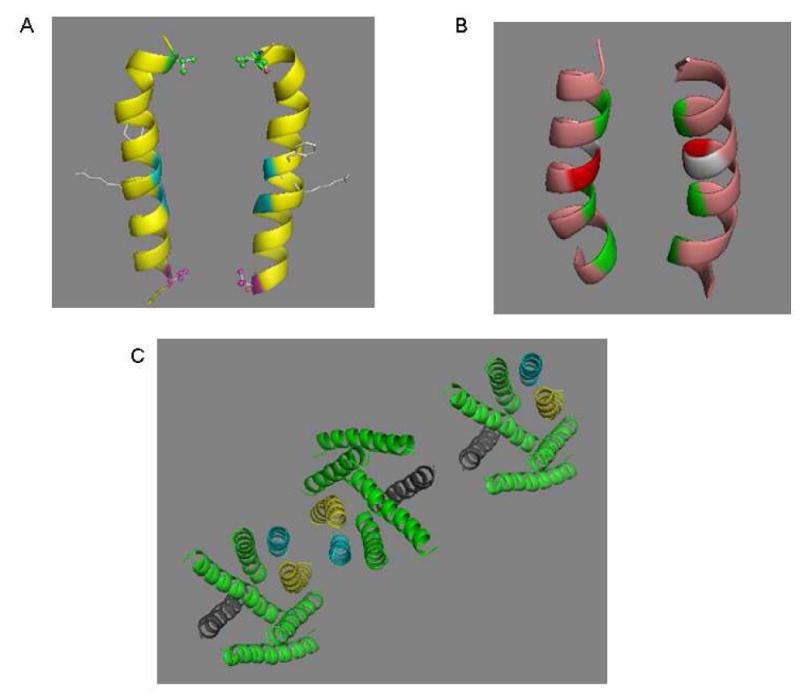
Dimerization interfaces of Ste2p. A. V68 (red), a dimer contact found in the current study, is located on the same plane with V45 (green) a dimer contact at the extracellar end of TM1 (21). The backbones of two glycine residues (blue) known to play a role in Ste2p dimerization are shown. Two resdues (F55, R58) involved in ligand binding are presented as a stick model located on the opposite side of the dimer interface. B. TM7/TM7 dimer interface. The backbone of three dimer contacts identified from the current study (T278, A285 and L289, from top to bottom) are colored green. A281 (light gray) and T282 (red) would appear to be candidates for dimer contact based on a sideview of TM7 (Fig. 8A), but the model here shows that alignments of these two residues are not favorable for disulfide bond formation. C. Proposed oligomerization interfaces of Ste2p. Dimerization of Ste2p involving TM1/TM1 (magenta) and/or TM7/TM7 (yellow) interface allows TM4/TM4 interface (dark grey) to mediate a new dimerization contact enabling oligomerization. An extracellular view of a representative trimer is shown.
In a previous FRET analysis, Overton and Blumer (60) analyzed the transmembrane domains of Ste2p that are involved in dimerization by expressing various combinations of Ste2p fragments. They concluded that TM1 is necessary and probably sufficient for dimerization, although they speculated that the N-terminus and TM2 may contribute to stabilize the dimers. However, our disulfide cross-linking data demonstrated that TM7 is also involved in the interface for dimerization of Ste2p. In the present study we have analyzed full-length receptors and it is possible that the fusion of fluorescent protein at the C-terminal of truncated receptors in the previous FRET studies hampered the interactions between TM7 domains. The involvement of TM7 in dimer formation is consistent with our previous observation that a TM6-TM7 fragment of Ste2p runs as a dimer in SDS-PAGE gel (62).
The finding that TM7 residues are involved in Ste2p dimerization leads us to propose that at least three dimerization interfaces can exist in Ste2p. In addition to the TM1 and TM4 interfaces previously found (21, 60), our data suggests that TM7-TM7 interactions are also involved in direct contacts in the Ste2p dimer. Since TM1-TM1 and TM4-TM4 contacts have been previously suggested to be involved in higher order oligomers (21) and a Ste2p trimer was demonstrated by crosslinking in a gel and by atomic force microscopy (40) it is possible that a single receptor can interact simultaneously with two additional receptors with different interfaces between the monomers: TM7 and/or TM1 can interact with TM7 and/or TM1 of a second receptor, while TM4 can interact with TM4 of a third receptor to form oligomers. A representation of a trimer is shown in Fig. 7C with higher oligomers envisaged to contain additional interactions. Based on the existing experimental data it is not possible to conclude whether TM1-TM1 and TM7-TM7 dimers can be formed simultaneously in the cell.
The results described in this study show that cysteine residues introduced in positions T278, A285 and L289 on the extracellular half of TM7 form a disulfide bond with their counterpart in another Ste2p monomer (Fig. 8A). This finding provides valuable information relating to the arrangement of the TM bundles of Ste2p and allow us to present a helical wheel projection of TM7 (Fig. 8B) in which these three residues are oriented outward instead of facing inside the TM bundles as proposed previously in the Ste2p model based on rhodopsin (35). A281 and T282, which in a two dimensional representation (Fig. 8A and B) appear to lie on the same helix face, do not form dimers. However, our 3D model suggests that A281 and T282 are not close enough to form disulfide bonds when replaced by Cys as illustrated in the modified model (Fig. 7B). Therefore, the dimerization mediated by T278, A285, and L289 is consistent with an alpha-helical periodicity in TM7 that continues up to Pro 290. In the absence of a crystal structure for Ste2p, the disulfide crosslinking results contributes to understanding structural features of the functional receptor such as inter-helical interactions that may be involved in oligomerization.
Figure 8.
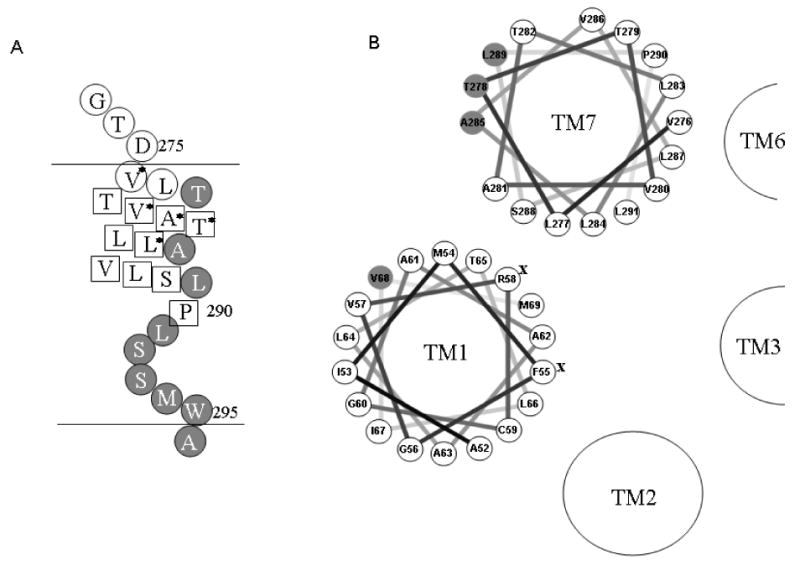
Representation of cross-linking results of TM7 and helical wheel presentation of TM1 and TM7 in Ste2p. A. Summary of TM7 cysteine cross-linking results. The horizontal lines represent the boundary of the cell membrane. Residues indicated by a circle filled in gray when mutated to Cys are involved in dimer formation, whereas those residues shown in a box do not form disulfide bonds. Residues marked by an * compensate for the non-functional Y266C mutation. B. Helical wheel presentation of TM1 and TM7. Residues indicated by a circle filled in gray are involved in dimer formation as found in this study. Residues marked by an X are important for ligand binding.
In contrast to the specific pattern of residues which could participate in cross-linking in the portion of TM7 most adjacent to the extracellular surface, when the engineered cysteine residues were located at the intracellular end of the TM7, C-terminal to Pro290 which creates a kink in TM7, all residues were able to form disulfide bonds (Fig. 8A) implying an absence of an ordered structure. We do not believe that the experimental protocol, specifically the use of the oxidizing agent, leads to a global denaturation of Ste2p because native cysteine residues in TM1 (C59) and TM5 (C252) did not form an inter-GPCR disulfide linkage under the same experimental conditions (Fig. 2B). Additionally, only one of the TM1 cysteine mutants formed Ste2p dimers and only three residues out of twelve in the extracellular part of the TM7 showed increased dimerization. Rather we propose that the L291-W295 region of Ste2p may be flexible enough to allow these residues to be exposed for disulfide bond formation with their counterpart in a second Ste2p monomer. Proline residues disrupt hydrogen bonding and have been shown to twist the standard structure of helices by introducing a kink between the segments contiguous with each other and to form molecular hinges (41, 63). The dynamic nature of the residues at the carboxyl side of Pro290 is consistent with the results of a high resolution analysis of a 73 residue fragment of Ste2p in DPC micelles which indicated that the Pro290 resulted in a kinked helix in this membrane mimetic environment (64). An irregular helical structure in part of a TM has been documented in the crystal structures of other GPCRs. In rhodopsin, TM7 contains a helical segment near the extracellular face of the receptor, followed by a non-helical segment in the vicinity of P303 (36). The crystal structure of the beta-adrenergic receptor also indicates the presence of a short extended area in TM7 (65). In addition, it has been proposed, using computational simulations of the NPXXY motif, that this region was not an ideal alpha helical structure (66). Thus, there is precedent to propose that the part of TM7 in Ste2p that is near the cytoplasmic membrane interface is not helical in structure. Although there is a possibility that the residues C-terminal to Pro290 are not in the transmembrane domain, this is unlikely because a previous study (42) and our cysteine accessibility results (data not shown) for these residues, strongly suggested that W295 and A296 are the boundary between TM7 and the intracellular space. Obviously, non-hydrogen bonded elements of a peptide in a membrane represent a high energy state and it is possible that critical water molecules satisfy some of the required interactions for Ste2p. Water molecules that bridge various receptor regions have been implicated in the crystal structure of other GPCRs (36, 65). Their involvement in the structure of Ste2p must await a high resolution structure of this GPCR.
It has been observed that the homo-dimer interface of the dopamine D2 receptor can be altered upon ligand binding (50). To monitor possible ligand-induced conformational changes in Ste2p dimerization, membrane samples were incubated with either agonist or antagonist followed by Cu-P treatment. TM1 mutants showed similar degrees of dimer formation in the presence or absence of ligand, which is consistent with a recent study showing that dimer formation of V45C at TM1 was insensitive to α-factor treatment (21). In contrast, both agonist and antagonist treatment led to a decrease in the level of dimer formed by TM7-TM7 interaction indicating that ligand binding caused a conformational change in this region.
The effect of ligand binding on TM7-mediated dimer formation suggests that resting state dimers and activated state dimers involve different TM arrangements. Previously it was shown that the ratio of monomer to dimer did not change in the presence of ligand in Ste2p as tested by FRET (17). Thus, it is possible that the equilibrium distribution between monomer-dimer is maintained during receptor activation, even while the dimer interface may differ. In addition, a recent study of the serotonin 5HT2c receptor demonstrated that ligand binding induces a differential effect on the dimer interfaces. While the TM1/TM1 interface was insensitive to ligand treatment, the TM4/5 interface changed during receptor activation (67).
The similar effects of agonist and antagonist binding on disulfide formation involving residues in the TM7 dimer interface can be understood in terms of distinct binding and signaling domains for α-factor (68). Extensive structure-function analysis of α-factor and Ste2p has revealed that the C-terminus of α-factor is critical for receptor binding. Since agonist (WHWLQLKPGQPNleY) and antagonist (WLQLKPGQPNleY) are identical except for the antagonist's N-terminal truncation, it is reasonable to suppose that the binding sites of both ligands in the receptor are highly overlapping especially at the C-terminal end of the ligands. Moreover, the carboxyl terminus has been shown to interact with residues in TM1 (23, 69). Based on this understanding it is not unreasonable that both agonists and antagonists first bind to TM1 promoting similar changes in the conformation of both TM1 and of TM7.
Interestingly, a number mutations of residues at the extracellular regions of TM1 and TM7 were found to compensate for the activity of the signaling defective mutant receptor Y266C (Fig. 7A, TM7 residues labeled as *)(70) indicating these two domains are important for receptor activation. The decrease in dimer formation at TM7 after ligand binding that we showed for T278C and A285C, both located in the extracellular part of TM7, also suggest a conformational change in this portion of Ste2p. Therefore, the extracellular ends of TM1 and TM7 may play a key role in initiating receptor activation after ligand binding to TM1 and the subsequent conformational change of TM7 may facilitate the receptor to adopt a fully activated state. Many GPCRs assume different conformations in their active and inactive states, and it is widely accepted that GPCRs sample many states even when occupied by ligand [for review see (71)]. Under our experimental conditions Cu-P mediated disulfide formation appears to distinguish these different conformations, possibly implicating a change of the dimer interface as part of the receptor activation mechanism. Nonetheless, we cannot rule out the possibility that the changes in the TM7 dimer interface upon ligand binding reflect structural changes elsewhere in Ste2p
Much remains to be understood concerning the significance of GPCR dimerization in the signaling process. The residues in TM7 examined in this study, S288LPLSSMWA296, are highly conserved in Class D receptors and are considered to have a similar function to the NPXXY residues in Class A receptors (35). In Class A receptors, the NPXXY motif is regarded to be important for receptor activation. (72-75). It is possible that TM7 plays an important role in both receptor activation and dimerization in Ste2p and in other GPCRs as well, and that dimerization is linked to signaling by these ubiquitous proteins. The results presented herein provide residue level information concerning the proximity of different sites on two Ste2p monomers in their native membrane environment. The physiological relevance of this information and of dimerization in GPCRs continues to be an intriguing area of GPCR biology.
Acknowledgments
We thank George Umanah and LiYin Huang for help in the binding assays and Badri Krishnan for carrying out the cysteine accessibility assays. This work was supported by grant GM-22087 from the National Institute of General Medical Sciences of the National Institutes of Health.
†This work was supported by grants GM-22086 and GM-22087 from the National Institute of General Medical Sciences of the National Institutes of Health.
Abbreviations
- Cu-P
copperII-1,10, phenanthroline
- EL
extracellular loop
- G protein
heterotrimeric GTP-binding protein
- GPCR
G protein-coupled receptor
- FT-HT
Cys-less Ste2p with the FLAG and His epitope tags
- FT-HT-Xa
the template construct or Cys-less Ste2p with FLAG Tag, His tag, and Factor Xa cleavage site
- MLT
medium lacking tryptophan
- MLU
medium lacking uracil
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- Ste2p-FT-HT
Ste2p with FLAG and His epitope tags
- SD medium
Synthetic defined medium
- TM
transmembrane domain
References
- 1.Lefkowitz RJ. The superfamily of heptahelical receptors. Nat Cell Biol. 2000;2:E133–136. doi: 10.1038/35017152. [DOI] [PubMed] [Google Scholar]
- 2.Sadee W, Hoeg E, Lucas J, Wang D. Genetic variations in human G protein-coupled receptors: implications for drug therapy. AAPS PharmSci. 2001;3:E22. doi: 10.1208/ps030322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Insel PA, Tang CM, Hahntow I, Michel MC. Impact of GPCRs in clinical medicine: monogenic diseases, genetic variants and drug targets. Biochim Biophys Acta. 2007;1768:994–1005. doi: 10.1016/j.bbamem.2006.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dohlman HG, Thorner J, Caron MG, Lefkowitz RJ. Model systems for the study of seven-transmembrane-segment receptors. Annu Rev Biochem. 1991;60:653–688. doi: 10.1146/annurev.bi.60.070191.003253. [DOI] [PubMed] [Google Scholar]
- 5.Dohlman HG, Thorner JW. Regulation of G protein-initiated signal transduction in yeast: paradigms and principles. Annu Rev Biochem. 2001;70:703–754. doi: 10.1146/annurev.biochem.70.1.703. [DOI] [PubMed] [Google Scholar]
- 6.Naider F, Becker JM. The alpha-factor mating pheromone of Saccharomyces cerevisiae: a model for studying the interaction of peptide hormones and G protein-coupled receptors. Peptides. 2004;25:1441–1463. doi: 10.1016/j.peptides.2003.11.028. [DOI] [PubMed] [Google Scholar]
- 7.Dirnberger D, Seuwen K. Signaling of human frizzled receptors to the mating pathway in yeast. PLoS ONE. 2007;2:e954. doi: 10.1371/journal.pone.0000954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yin D, Gavi S, Shumay E, Duell K, Konopka JB, Malbon CC, Wang HY. Successful expression of a functional yeast G-protein-coupled receptor (Ste2) in mammalian cells. Biochem Biophys Res Commun. 2005;329:281–287. doi: 10.1016/j.bbrc.2005.01.130. [DOI] [PubMed] [Google Scholar]
- 9.Xue C, Hsueh YP, Heitman J. Magnificent seven: roles of G protein-coupled receptors in extracellular sensing in fungi. FEMS Microbiol Rev. 2008;32:1010–1032. doi: 10.1111/j.1574-6976.2008.00131.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rios CD, Jordan BA, Gomes I, Devi LA. G-protein-coupled receptor dimerization: modulation of receptor function. Pharmacol Ther. 2001;92:71–87. doi: 10.1016/s0163-7258(01)00160-7. [DOI] [PubMed] [Google Scholar]
- 11.Milligan G. G protein-coupled receptor dimerisation: molecular basis and relevance to function. Biochim Biophys Acta. 2007;1768:825–835. doi: 10.1016/j.bbamem.2006.09.021. [DOI] [PubMed] [Google Scholar]
- 12.Milligan G. G protein-coupled receptor dimerization: function and ligand pharmacology. Mol Pharmacol. 2004;66:1–7. doi: 10.1124/mol.104.000497.. [DOI] [PubMed] [Google Scholar]
- 13.Bayburt TH, Leitz AJ, Xie G, Oprian DD, Sligar SG. Transducin activation by nanoscale lipid bilayers containing one and two rhodopsins. J Biol Chem. 2007;282:14875–14881. doi: 10.1074/jbc.M701433200. [DOI] [PubMed] [Google Scholar]
- 14.Chabre M, le Maire M. Monomeric G-protein-coupled receptor as a functional unit. Biochemistry. 2005;44:9395–9403. doi: 10.1021/bi050720o. [DOI] [PubMed] [Google Scholar]
- 15.White JF, Grodnitzky J, Louis JM, Trinh LB, Shiloach J, Gutierrez J, Northup JK, Grisshammer R. Dimerization of the class A G protein-coupled neurotensin receptor NTS1 alters G protein interaction. Proc Natl Acad Sci U S A. 2007;104:12199–12204. doi: 10.1073/pnas.0705312104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whorton MR, Bokoch MP, Rasmussen SG, Huang B, Zare RN, Kobilka B, Sunahara RK. A monomeric G protein-coupled receptor isolated in a high-density lipoprotein particle efficiently activates its G protein. Proc Natl Acad Sci U S A. 2007;104:7682–7687. doi: 10.1073/pnas.0611448104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Overton MC, Blumer KJ. G-protein-coupled receptors function as oligomers in vivo. Curr Biol. 2000;10:341–344. doi: 10.1016/s0960-9822(00)00386-9. [DOI] [PubMed] [Google Scholar]
- 18.Yesilaltay A, Jenness DD. Homo-oligomeric complexes of the yeast alpha-factor pheromone receptor are functional units of endocytosis. Mol Biol Cell. 2000;11:2873–2884. doi: 10.1091/mbc.11.9.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Overton MC, Chinault SL, Blumer KJ. Oligomerization, biogenesis, and signaling is promoted by a glycophorin A-like dimerization motif in transmembrane domain 1 of a yeast G protein-coupled receptor. J Biol Chem. 2003;278:49369–49377. doi: 10.1074/jbc.M308654200. [DOI] [PubMed] [Google Scholar]
- 20.Gehret AU, Bajaj A, Naider F, Dumont ME. Oligomerization of the yeast alpha-factor receptor: implications for dominant negative effects of mutant receptors. J Biol Chem. 2006;281:20698–20714. doi: 10.1074/jbc.M513642200. [DOI] [PubMed] [Google Scholar]
- 21.Wang HX, Konopka JB. Identification of Amino Acids at Two Dimer Interface Regions of the alpha-Factor Receptor (Ste2) Biochemistry. 2009 doi: 10.1021/bi900424h. [DOI] [PubMed] [Google Scholar]
- 22.Sen M, Marsh L. Noncontiguous domains of the alpha-factor receptor of yeasts confer ligand specificity. J Biol Chem. 1994;269:968–973. [PubMed] [Google Scholar]
- 23.Son CD, Sargsyan H, Naider F, Becker JM. Identification of ligand binding regions of the Saccharomyces cerevisiae alpha-factor pheromone receptor by photoaffinity cross-linking. Biochemistry. 2004;43:13193–13203. doi: 10.1021/bi0496889. [DOI] [PubMed] [Google Scholar]
- 24.Hauser M, Kauffman S, Lee BK, Naider F, Becker JM. The first extracellular loop of the Saccharomyces cerevisiae G protein-coupled receptor Ste2p undergoes a conformational change upon ligand binding. J Biol Chem. 2007;282:10387–10397. doi: 10.1074/jbc.M608903200. [DOI] [PubMed] [Google Scholar]
- 25.Akal-Strader A, Khare S, Xu D, Naider F, Becker JM. Residues in the first extracellular loop of a G protein-coupled receptor play a role in signal transduction. J Biol Chem. 2002;277:30581–30590. doi: 10.1074/jbc.M204089200. [DOI] [PubMed] [Google Scholar]
- 26.Mumberg D, Muller R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- 27.Lee BK, Jung KS, Son C, Kim H, VerBerkmoes NC, Arshava B, Naider F, Becker JM. Affinity purification and characterization of a G-protein coupled receptor, Saccharomyces cerevisiae Ste2p. Protein Expr Purif. 2007;56:62–71. doi: 10.1016/j.pep.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gietz D, St Jean A, Woods RA, Schiestl RH. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.David NE, Gee M, Andersen B, Naider F, Thorner J, Stevens RC. Expression and purification of the Saccharomyces cerevisiae alpha-factor receptor (Ste2p), a 7-transmembrane-segment G protein-coupled receptor. J Biol Chem. 1997;272:15553–15561. doi: 10.1074/jbc.272.24.15553. [DOI] [PubMed] [Google Scholar]
- 30.Lee YH, Naider F, Becker JM. Interacting residues in an activated state of a G protein-coupled receptor. J Biol Chem. 2006;281:2263–2272. doi: 10.1074/jbc.M509987200. [DOI] [PubMed] [Google Scholar]
- 31.Raths SK, Naider F, Becker JM. Peptide analogues compete with the binding of alpha-factor to its receptor in Saccharomyces cerevisiae. J Biol Chem. 1988;263:17333–17341. [PubMed] [Google Scholar]
- 32.Konopka JB, Jenness DD, Hartwell LH. The C-terminus of the S. cerevisiae alpha-pheromone receptor mediates an adaptive response to pheromone. Cell. 1988;54:609–620. doi: 10.1016/s0092-8674(88)80005-9. [DOI] [PubMed] [Google Scholar]
- 33.Kleiger G, Grothe R, Mallick P, Eisenberg D. GXXXG and AXXXA: common alpha-helical interaction motifs in proteins, particularly in extremophiles. Biochemistry. 2002;41:5990–5997. doi: 10.1021/bi0200763. [DOI] [PubMed] [Google Scholar]
- 34.Sternberg MJ, Gullick WJ. A sequence motif in the transmembrane region of growth factor receptors with tyrosine kinase activity mediates dimerization. Protein Eng. 1990;3:245–248. doi: 10.1093/protein/3.4.245. [DOI] [PubMed] [Google Scholar]
- 35.Eilers M, Hornak V, Smith SO, Konopka JB. Comparison of class A and D G protein-coupled receptors: common features in structure and activation. Biochemistry. 2005;44:8959–8975. doi: 10.1021/bi047316u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 37.Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, Stevens RC. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mentesana PE, Konopka JB. Mutational analysis of the role of N-glycosylation in alpha-factor receptor function. Biochemistry. 2001;40:9685–9694. doi: 10.1021/bi0108507. [DOI] [PubMed] [Google Scholar]
- 39.Blumer KJ, Reneke JE, Thorner J. The STE2 gene product is the ligand-binding component of the alpha-factor receptor of Saccharomyces cerevisiae. J Biol Chem. 1988;263:10836–10842. [PubMed] [Google Scholar]
- 40.Shi C, Paige MF, Maley J, Loewen MC. In vitro characterization of ligand-induced oligomerization of the S. cerevisiae G-protein coupled receptor, Ste2p. Biochim Biophys Acta. 2009;1790:1–7. doi: 10.1016/j.bbagen.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 41.Sansom MS, Weinstein H. Hinges, swivels and switches: the role of prolines in signalling via transmembrane alpha-helices. Trends Pharmacol Sci. 2000;21:445–451. doi: 10.1016/s0165-6147(00)01553-4. [DOI] [PubMed] [Google Scholar]
- 42.Choi Y, Konopka JB. Accessibility of cysteine residues substituted into the cytoplasmic regions of the alpha-factor receptor identifies the intracellular residues that are available for G protein interaction. Biochemistry. 2006;45:15310–15317. doi: 10.1021/bi0614939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin JC, Duell K, Konopka JB. A microdomain formed by the extracellular ends of the transmembrane domains promotes activation of the G protein-coupled alpha-factor receptor. Mol Cell Biol. 2004;24:2041–2051. doi: 10.1128/MCB.24.5.2041-2051.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCaffrey G, Clay FJ, Kelsay K, Sprague GF., Jr Identification and regulation of a gene required for cell fusion during mating of the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:2680–2690. doi: 10.1128/mcb.7.8.2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dube P, DeCostanzo A, Konopka JB. Interaction between transmembrane domains five and six of the alpha -factor receptor. J Biol Chem. 2000;275:26492–26499. doi: 10.1074/jbc.M002767200. [DOI] [PubMed] [Google Scholar]
- 46.Parrish W, Eilers M, Ying W, Konopka JB. The cytoplasmic end of transmembrane domain 3 regulates the activity of the Saccharomyces cerevisiae G-protein-coupled alpha-factor receptor. Genetics. 2002;160:429–443. doi: 10.1093/genetics/160.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shah A, Marsh L. Role of Sst2 in modulating G protein-coupled receptor signaling. Biochem Biophys Res Commun. 1996;226:242–246. doi: 10.1006/bbrc.1996.1340. [DOI] [PubMed] [Google Scholar]
- 48.Li JH, Han SJ, Hamdan FF, Kim SK, Jacobson KA, Bloodworth LM, Zhang X, Wess J. Distinct structural changes in a G protein-coupled receptor caused by different classes of agonist ligands. J Biol Chem. 2007;282:26284–26293. doi: 10.1074/jbc.M704875200. [DOI] [PubMed] [Google Scholar]
- 49.Han SJ, Hamdan FF, Kim SK, Jacobson KA, Brichta L, Bloodworth LM, Li JH, Wess J. Pronounced conformational changes following agonist activation of the M(3) muscarinic acetylcholine receptor. J Biol Chem. 2005;280:24870–24879. doi: 10.1074/jbc.M500379200. [DOI] [PubMed] [Google Scholar]
- 50.Guo W, Shi L, Filizola M, Weinstein H, Javitch JA. Crosstalk in G protein-coupled receptors: changes at the transmembrane homodimer interface determine activation. Proc Natl Acad Sci U S A. 2005;102:17495–17500. doi: 10.1073/pnas.0508950102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu H, Kono M, McKee TD, Oprian DD. A general method for mapping tertiary contacts between amino acid residues in membrane-embedded proteins. Biochemistry. 1995;34:14963–14969. doi: 10.1021/bi00046a002. [DOI] [PubMed] [Google Scholar]
- 52.Struthers M, Yu H, Kono M, Oprian DD. Tertiary interactions between the fifth and sixth transmembrane segments of rhodopsin. Biochemistry. 1999;38:6597–6603. doi: 10.1021/bi9902384. [DOI] [PubMed] [Google Scholar]
- 53.Bukusoglu G, Jenness DD. Agonist-specific conformational changes in the yeast alpha-factor pheromone receptor. Mol Cell Biol. 1996;16:4818–4823. doi: 10.1128/mcb.16.9.4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Farrens DL, Altenbach C, Yang K, Hubbell WL, Khorana HG. Requirement of rigid-body motion of transmembrane helices for light activation of rhodopsin. Science. 1996;274:768–770. doi: 10.1126/science.274.5288.768. [DOI] [PubMed] [Google Scholar]
- 55.Zeng FY, Hopp A, Soldner A, Wess J. Use of a disulfide cross-linking strategy to study muscarinic receptor structure and mechanisms of activation. J Biol Chem. 1999;274:16629–16640. doi: 10.1074/jbc.274.23.16629. [DOI] [PubMed] [Google Scholar]
- 56.Luo X, Zhang D, Weinstein H. Ligand-induced domain motion in the activation mechanism of a G-protein-coupled receptor. Protein Eng. 1994;7:1441–1448. doi: 10.1093/protein/7.12.1441. [DOI] [PubMed] [Google Scholar]
- 57.Srinivasan N, Sowdhamini R, Ramakrishnan C, Balaram P. Conformations of disulfide bridges in proteins. Int J Pept Protein Res. 1990;36:147–155. doi: 10.1111/j.1399-3011.1990.tb00958.x. [DOI] [PubMed] [Google Scholar]
- 58.Li JH, Hamdan FF, Kim SK, Jacobson KA, Zhang X, Han SJ, Wess J. Ligand-specific changes in M3 muscarinic acetylcholine receptor structure detected by a disulfide scanning strategy. Biochemistry. 2008;47:2776–2788. doi: 10.1021/bi7019113. [DOI] [PubMed] [Google Scholar]
- 59.Ward SD, Hamdan FF, Bloodworth LM, Siddiqui NA, Li JH, Wess J. Use of an in situ disulfide cross-linking strategy to study the dynamic properties of the cytoplasmic end of transmembrane domain VI of the M3 muscarinic acetylcholine receptor. Biochemistry. 2006;45:676–685. doi: 10.1021/bi051503q. [DOI] [PubMed] [Google Scholar]
- 60.Overton MC, Blumer KJ. The extracellular N-terminal domain and transmembrane domains 1 and 2 mediate oligomerization of a yeast G protein-coupled receptor. J Biol Chem. 2002;277:41463–41472. doi: 10.1074/jbc.M205368200. [DOI] [PubMed] [Google Scholar]
- 61.Arshava B, Taran I, Xie H, Becker JM, Naider F. High resolution NMR analysis of the seven transmembrane domains of a heptahelical receptor in organic-aqueous medium. Biopolymers. 2002;64:161–176. doi: 10.1002/bip.10157. [DOI] [PubMed] [Google Scholar]
- 62.Cohen LS, Arshava B, Estephan R, Englander J, Kim H, Hauser M, Zerbe O, Ceruso M, Becker JM, Naider F. Expression and biophysical analysis of two double-transmembrane domain-containing fragments from a yeast G protein-coupled receptor. Biopolymers. 2008;90:117–130. doi: 10.1002/bip.20950. [DOI] [PubMed] [Google Scholar]
- 63.Cordes FS, Bright JN, Sansom MS. Proline-induced distortions of transmembrane helices. J Mol Biol. 2002;323:951–960. doi: 10.1016/s0022-2836(02)01006-9. [DOI] [PubMed] [Google Scholar]
- 64.Neumoin A, Arshava B, Becker J, Zerbe O, Naider F. NMR studies in dodecylphosphocholine of a fragment containing the seventh transmembrane helix of a G-protein-coupled receptor from Saccharomyces cerevisiae. Biophys J. 2007;93:467–482. doi: 10.1529/biophysj.106.103770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rosenbaum DM, Cherezov V, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Yao XJ, Weis WI, Stevens RC, Kobilka BK. GPCR engineering yields high-resolution structural insights into beta2-adrenergic receptor function. Science. 2007;318:1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- 66.Konvicka K, Guarnieri F, Ballesteros JA, Weinstein H. A proposed structure for transmembrane segment 7 of G protein-coupled receptors incorporating an asn-Pro/Asp-Pro motif. Biophys J. 1998;75:601–611. doi: 10.1016/S0006-3495(98)77551-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mancia F, Assur Z, Herman AG, Siegel R, Hendrickson WA. Ligand sensitivity in dimeric associations of the serotonin 5HT2c receptor. EMBO Rep. 2008;9:363–369. doi: 10.1038/embor.2008.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Abel MG, Zhang YL, Lu HF, Naider F, Becker JM. Structure-function analysis of the Saccharomyces cerevisiae tridecapeptide pheromone using alanine-scanned analogs. J Pept Res. 1998;52:95–106. doi: 10.1111/j.1399-3011.1998.tb01363.x. [DOI] [PubMed] [Google Scholar]
- 69.Lee BK, Khare S, Naider F, Becker JM. Identification of residues of the Saccharomyces cerevisiae G protein-coupled receptor contributing to alpha-factor pheromone binding. J Biol Chem. 2001;276:37950–37961. doi: 10.1074/jbc.M103579200. [DOI] [PubMed] [Google Scholar]
- 70.Lin JC, Duell K, Saracino M, Konopka JB. Identification of residues that contribute to receptor activation through the analysis of compensatory mutations in the G protein-coupled alpha-factor receptor. Biochemistry. 2005;44:1278–1287. doi: 10.1021/bi048050u. [DOI] [PubMed] [Google Scholar]
- 71.Park PS, Lodowski DT, Palczewski K. Activation of G protein-coupled receptors: beyond two-state models and tertiary conformational changes. Annu Rev Pharmacol Toxicol. 2008;48:107–141. doi: 10.1146/annurev.pharmtox.48.113006.094630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Barak LS, Menard L, Ferguson SS, Colapietro AM, Caron MG. The conserved seven-transmembrane sequence NP(X)2,3Y of the G-protein-coupled receptor superfamily regulates multiple properties of the beta 2-adrenergic receptor. Biochemistry. 1995;34:15407–15414. doi: 10.1021/bi00047a003. [DOI] [PubMed] [Google Scholar]
- 73.Fritze O, Filipek S, Kuksa V, Palczewski K, Hofmann KP, Ernst OP. Role of the conserved NPxxY(x)5,6F motif in the rhodopsin ground state and during activation. Proc Natl Acad Sci U S A. 2003;100:2290–2295. doi: 10.1073/pnas.0435715100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Prioleau C, Visiers I, Ebersole BJ, Weinstein H, Sealfon SC. Conserved helix 7 tyrosine acts as a multistate conformational switch in the 5HT2C receptor. Identification of a novel “locked-on” phenotype and double revertant mutations. J Biol Chem. 2002;277:36577–36584. doi: 10.1074/jbc.M206223200. [DOI] [PubMed] [Google Scholar]
- 75.Wess J, Nanavati S, Vogel Z, Maggio R. Functional role of proline and tryptophan residues highly conserved among G protein-coupled receptors studied by mutational analysis of the m3 muscarinic receptor. Embo J. 1993;12:331–338. doi: 10.1002/j.1460-2075.1993.tb05661.x. [DOI] [PMC free article] [PubMed] [Google Scholar]