Abstract
Purpose
MLH1 is a key DNA mismatch repair (MMR) protein involved in maintaining genomic stability by participating in the repair of endogenous and exogenous mispairs in the daughter strands during S-phase. Exogenous mispairs can result following treatment with several classes of chemotherapeutic drugs as well as with ionizing radiation (IR). In this study, we investigated the role of the MLH1 protein in determining the cellular and molecular responses to prolonged low dose rate (LDR) IR, which is similar to the clinical use of cancer brachytherapy.
Experimental design
An isogenic pair of MMR+ (MLH1+) and MMR− (MLH1−) human colorectal cancer HCT116 cells were exposed to prolonged LDR-IR (1.3–17cGy/h × 24–96 h). The clonogenic survival and gene mutation rates were examined. Cell cycle distribution was analyzed with flow cytometry. Changes in selected DNA damage repair proteins, DNA damage response proteins and cell death marker proteins were examined with Western blotting.
Results
MLH1+ HCT116 cells showed greater radiosensitivity with enhanced expression of apoptotic and autophagic markers; a reduced HPRT gene mutation rate; and more pronounced cell cycle alterations (increased late S population and a G2/M arrest) following LDR-IR compared to MLH1− HCT116 cells. Importantly, a progressive increase in MLH1 protein levels was found in MLH1+ cells during prolonged LDR-IR, which was temporally correlated with a progressive decrease in Rad51 protein (involved in homologous recombination, HR) levels.
Conclusions
MLH1 status significantly affects cellular responses to prolonged LDR-IR. MLH1 may enhance cell radiosensitivity to prolonged LDR-IR through inhibition of HR (via inhibition of Rad51).
Keywords: mismatch repair, low dose rate IR, MLH1, Rad51, late S phase
Statement of translational relevance
DNA mismatch repair (MMR) is a highly conserved DNA repair pathway for maintaining genomic integrity. It is also involved in cellular responses to a variety of chemotherapeutic drugs and ionizing radiation. Thus, MMR-deficiency can result in the development of human cancers which are intrinsically resistant to some forms of cancer treatment.
Environmental low dose rate ionizing radiation (LDR-IR) has carcinogenic effects on humans. However, LDR-IR is also commonly employed as an effective radiotherapeutic strategy (termed brachytherapy) for some cancers. This study investigates how a key MMR protein, MLH1, status affects the cellular response to LDR-IR.
The important clinical-translational relevance of our data suggesting that MLH1 proficiency enhances radiosensitization to prolonged LDR-IR are two fold: first, MLH1 proficiency may reduce cancer susceptibility by preventing potentially mutagenic lesions from being passed on to progeny after prolonged LDR-IR; and second, MLH1 proficiency may increase the efficacy of the cancer treatment regimens which utilize prolonged LDR-IR.
Introduction
Ionizing radiation (IR) is a well known carcinogenic hazard to humans. There are many natural and man-made sources of low dose rate ionizing radiation (LDR-IR) affecting humans, including occupational, medical and environmental exposures. The carcinogenic effects of LDR-IR on humans are recently reviewed (1). However, LDR-IR is also commonly employed as an effective radiotherapeutic strategy (termed brachytherapy) for some cancers including prostate, gynecologic, lung, breast, head and neck, anal/rectal, and esophageal cancers, as well as soft tissue sarcomas. Indeed, the use of permanent LDR brachytherapy is a common treatment approach to low risk, early stage prostate cancer, where the radioactivity of the permanently implanted iodine or palladium seeds decays over several months within the gland. LDR brachytherapy is also sometimes used in the treatment of coronary artery disease to prevent restenosis after angioplasty. Hence, a better understanding of the cellular response to LDR-IR is of clinical interest.
IR induces a complex spectrum of DNA damage including double-strand breaks (DSBs), single-strand breaks (SSBs), DNA cross links, and oxidized bases. Clearly, a cell’s ability to repair IR-induced DNA damage will affect the overall cellular response to IR. All five major DNA repair pathways, including homologous recombination (HR), non-homologous end joining (NHEJ), nucleotide excision repair (NER), base excision repair (BER) and mismatch repair (MMR) are involved in the repair of the IR-induced DNA damages (2–5). These DNA repair pathways are tightly inter-related with IR-induced cell cycle checkpoint arrest pathways, which allow time to complete DNA repair, as well as apoptotic (and other) cell death pathways, which eliminate severely damaged cells (6–10).
MMR is a highly conserved DNA repair pathway for maintaining genomic integrity (11–15). MMR-deficient (MMR−) cells demonstrate an increased gene mutation rate. The link between MMR-deficiency arising from germline MMR gene mutations and hereditary non-polyposis colorectal cancer (HNPCC) is well documented (11,12,14, 15). Many other cancers can also arise from sporadic MMR gene mutations as well as gene promoter methylation (11,12,14,15). Although the primary function of MMR is to correct mismatches generated during DNA replication, it is also involved in cellular responses to a variety of chemotherapeutic drugs (16, 17). Contrary to its role in correcting replication errors, the role of MMR in response to drug-induced DNA damages is usually detrimental to cells. Consequently, MMR− cells acquire resistance (or damage tolerance) to certain types of chemotherapeutic drugs. Thus, MMR-deficiency can result in the development of human cancers which are intrinsically resistant to some forms of cancer treatment.
Several human MMR proteins have been identified based on homology to E. coli MMR proteins. (15). Human MutS and MutL homologues are heterodimers. MSH2 heterodimerizes with MSH6 or MSH3 to form MutSα or MutSβ, respectively, both of which play a critical role in mismatch recognition and initiation of repair. MLH1 forms a heterodimer with PMS2, PMS1, or MLH3 (MutLα, MutLβ, or MutLγ respectively). MutLα is required for MMR processing of chemotherapy and IR-induced DNA damage and MutLγ plays a role in meiosis while the role of MutLβ is less clear. The impact of MMR status on cell survival following ionizing radiation was first reported by Fritzell et al (18). In their study, mouse fibroblast cells deficient in MLH1, MSH2, or PMS2 were shown to be resistant to high-dose rate ionizing radiation (225 cGy/min) compared to wild type cells. How the status of MMR proteins affects the cellular responses to protracted LDR-IR has not been fully investigated. Previously, using mouse embryonic stem cells differing in MSH2 expression, DeWeese et al demonstrated that MSH2−/− cells exhibited improved survival, decreased apoptosis and increased gene mutation following prolonged LDR-IR (24 cGy/h for 24 to 72 h) (19). Subsequently, Zeng et al showed that PMS2 null mouse cells demonstrated improved survival after prolonged LDR-IR (16–27cGy/h for 12.5 to 30 h) (20). However, how MLH1, an essential component of MutL and one of the most commonly mutated MMR genes in HNPCC patients and in MMR-deficient sporadic cancers (11, 12, 15, 21, 22), affects cellular responses to LDR-IR is unclear. We hypothesized that MLH1 status may significantly affect cellular responses to prolonged LDR-IR. In this study, we compared MLH1-deficient and MLH1-proficient cells in an attempt to gain insights into the role of MLH1 in cellular responses to prolonged LDR-IR, which may be of interest to cancer prevention and to the use of brachytherapy for cancer treatment.
Here, we show that MLH1-proficiency enhances cell killing, reduces gene mutation rate and alters cell cycle distribution to a greater extent after protracted LDR-IR. We demonstrate that MLH1 protein accumulates as a result of compromised MLH1 protein degradation during prolonged LDR-IR. Importantly, we show that MLH1 protein accumulation occurs concomitantly with Rad51 protein reduction in a dose rate- and time-dependent fashion. Our findings suggest that MLH1 may be involved in the inhibition of HR during prolonged LDR-IR, which leads to enhanced cell killing.
Materials and Methods
Cell Culture
A pair of isogenically-matched HCT116 cell lines, HCT116vector (MLH1−) and HCT116MLH1 (MLH1+), are utilized as the predominant experimental model system in this study (kindly provided by Dr. Francoise Praz, Centre National de la Recherche Scientifique, Villejuif, France). The parental cell line HCT116 is a human colon carcinoma cell line deficient in the MLH1 gene (MMR−). Some experiments are also performed on HT29 (human colorectal carcinoma cell line, MMR+), U251 (human glioblastoma cell line, MMR+) and HEC59 (human endometrial carcinoma cell line, MSH2−, MMR−) cells. All cell lines are grown in DMEM (Mediatech, Herndon, VA) with 10% FBS (HyClone, Logan, UT), 2 mM glutamine and 0.1 mM non-essential amino acids (Invitrogen, Carlsbad, CA) in 10% CO2 at 37°C.
LDR-IR
The LDR-IR source used was Iridium-192 (Best Medical International, Springfield, VA) in a standard tissue culture incubator. Iridium-192 has a half life 75 days and the dose rates used for prolonged LDR-IR ranged from 17 cGy/h to 1.3 cGy/h, which are clinically applicable LDR-IR rates as used during permanent prostate cancer brachytherapy (23). For prolonged LDR-IR, the cells were irradiated for 24 h to 96 h, with the dose rates and the LDR-IR doses varying by the distance of dishes from the iridium-192 source. The control cells were seeded at the same time as the irradiated cells but incubated in a regular incubator for the same time points.
Clonogenic survival assay
Log phase cells were plated into 60-mm dishes at the appropriate cell numbers to ensure the formation of about 100 colonies per dish. Twenty hours later, the cells were placed in the LDR-IR source incubator. At the desired time points, the cells were removed to a regular incubator and allowed to continue growing for 12 days for colony formation.
Western blotting analyses
The experiments were carried out as described previously (4). The antibodies used were as follows: β-actin (Sigma-Aldrich, St. Louis, MO); MLH1, PMS2, MSH2 (BD Biosciences Pharmingen, San Diego, CA); MSH6 (BD Biosciences, San Jose, CA); APE1, Polβ, FEN1, OGG1, Mre11, NBS1, LC3 (Novus Biologicals, Littleton, CO); Ku70, Ku80, Rad51 (Abcam, Inc., Cambridge, MA); XPA (LabVision, Fremont, CA); XPC (GeneTex, Irvine, CA); γ-H2AX (Upstate, Charlottesville, VA), PARP p85 (Promega, Madison, WI); p53, p21 and secondary antibody IgG-HRP conjugates (Santa Cruz, Santa Cruz, CA).
Flow cytometry analysis
The determination of the cell cycle profile was described previously (24). For dual-parameter flow cytometry, 1 × 106 fixed cells were incubated with a phospho-histone H3 (Ser10) antibody (Cell Signaling Technology, Boston, MA) and a secondary antibody conjugated with Alexa Fluor 488 (Invitrogen/Molecular Probes, Carisbad, CA) and then propidium iodide (PI, Sigma-Aldrich). The flow cytometric data were analyzed with Modfit 3.0 and Winlist 5.0 (Verity Software, Topsham, ME).
HPRT gene mutation assay
The cells were irradiated for 96 h (4.5 cGy/h), and the irradiated cells were then re-seeded into 100-mm dishes for another 8 days. The surviving cells were then counted and reseeded at 1×106 cells/100-mm dish and assayed for colony formation after 12 days in the presence of 40 µM 6-thioguanine (6-TG). Irradiated cells were also seeded at appropriate numbers for colony formation without 6-TG treatment. The HPRT mutant frequency was expressed as the ratio of cloning efficiency in selective (6-TG) medium to that in nonselective medium.
Statistics
The data, where applicable, represent the means ± SE. Data were analyzed using the Student’s t test. The bar graph of flow cytometry data was derived from representative experiments and, therefore, is presented without error bars. The Linear Regression test was determined using Microsoft Excel statistics.
Results
MLH1− (HCT116vector) cells showed an increased resistance and an increased HPRT gene mutation rate following protracted LDR-IR compared to MLH1+ (HCT116MLH1) cells
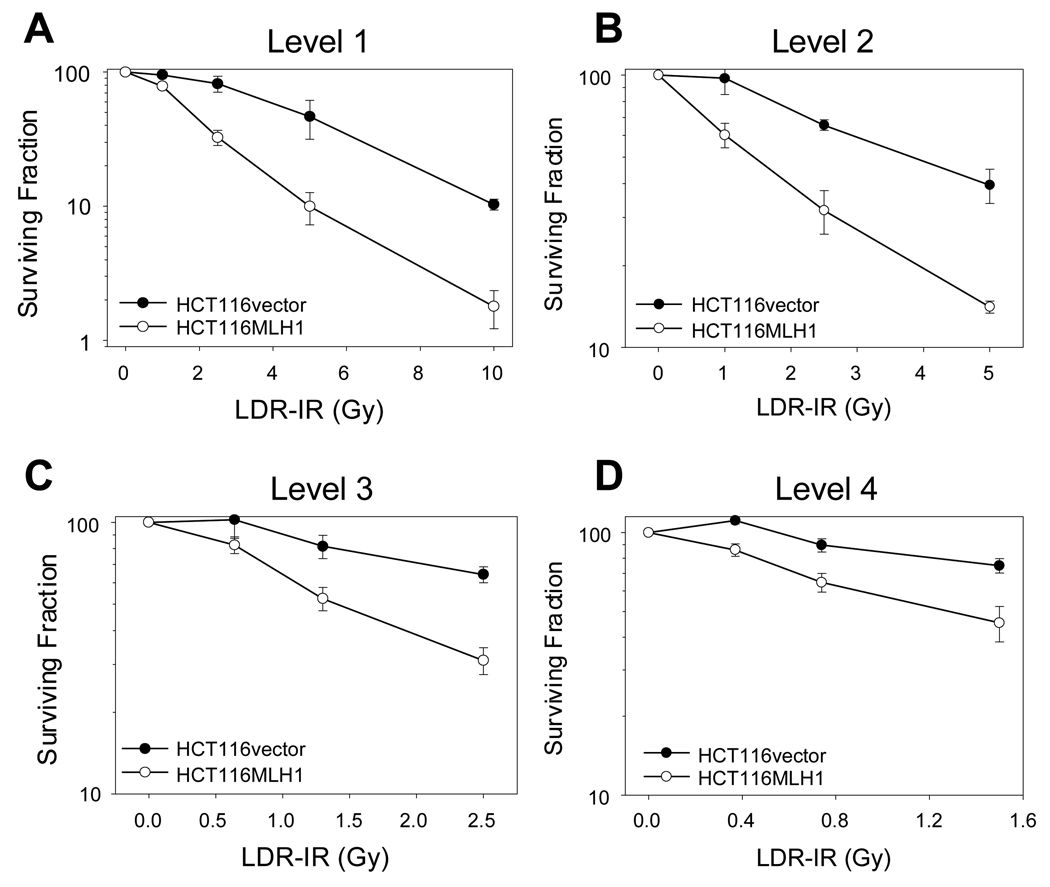
We initially performed a standard clonogenic survival assay following prolonged LDR-IR. Although the LDR-IR source normally decayed and the cells received a range of total IR doses, a valid comparison of survival was made possible by always irradiating the two HCT116 cell lines simultaneously. As shown in Fig. 1, at all 4 LDR-IR dose rate levels, the MLH1− cells demonstrated an increased resistance (survival) to LDR-IR compared to the MLH1+ cells. The ratios of the surviving fraction between the MLH1− and the MLH1+ cells were 1.2, 2.5, 4.7, and 5.9 for the total doses of 1, 2.5, 5, and 10 Gy respectively at Level 1 (Fig. 1A); 1.6, 2.1 and 2.8 for the total doses of 1, 2.5, and 5 Gy respectively at Level 2 (Fig. 1B); 1.6 and 2.1 for the total doses of 1 and 2.5 Gy respectively at Level 3 (Fig. 1C); and was 1.6 for the dose of 1 Gy at Level 4 (Fig. 1D).
Figure 1.
MLH1 status affects cell sensitivity to prolonged LDR-IR. HCT116vector and HCT116MLH1 cells were exposed to prolonged LDR-IR simultaneously. A. Level 1, 9.3–11.1cGy/h. B, Level 2, 4.2–5.2 cGy/h. C, Level 3, 2.4–2.9 cGy/h. D, Level 4, 1.3–1.7 cGy/h. The data are from three experiments with significance at the P < 0.05 level.
We next carried out a HPRT gene mutation assay (6-TG resistance assay). As expected, the MLH1− cells showed a significantly increased HPRT gene mutation rate after prolonged LDR-IR (4.5cGy/h for 96 h), while the mutation rate was only slightly increased in the MLH1+ cells (Table 1).
Table 1.
HPRT gene mutation rate (6-TG resistance)
| Cells | Treatment | Mutation rate | Induced mutation rate |
|---|---|---|---|
| HCT116vector | No radiation | 195 ± 18/1×106 | - |
| LDR-IR 4.5cGy/h | 326 ± 33/1×106 | 131/1×106 | |
| HCT116MLH1 | No radiation | 3.8 ± 0.8/1×106 | - |
| LDR-IR 4.5cGy/h | 10.2 ± 3.1/1×106 | 6.4/1×106 |
Prolonged LDR-IR led to a greater G2/M arrest and a greater late S phase population in the MLH1+ cells than in the MLH1− cells
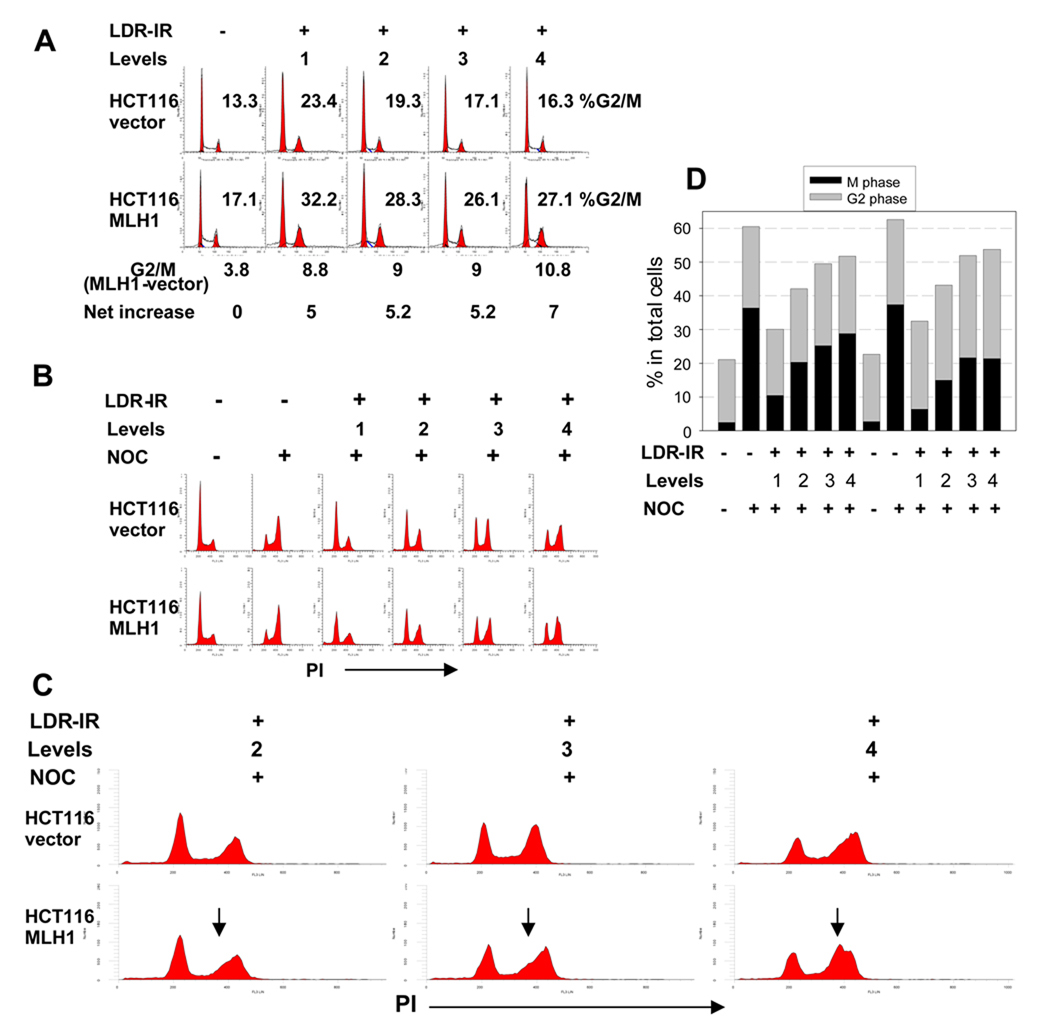
Since the MLH1 protein has been implicated in cell cycle regulation (4), we examined the cell cycle distribution during prolonged LDR-IR. Fig. 2A shows representative flow cytometry histograms after a 72 h treatment with LDR-IR. We did not observe a significant G2/M arrest peak during the 24 h to 96 h LDR-IR. Rather, a moderately increased G2/M population was found in both cell lines (Fig. 2A). Notably, the MLH1+ cells showed about 5 percentage points (equal to 20–30%) more of a G2/M population than the MLH1− cells (Fig. 2A). Although small, these differences were highly reproducible. These differences were also found at the other time points (data not shown).
Figure 2.
HCT116MLH1 (MLH1+) cells demonstrate a greater late S population and a greater G2/M checkpoint arrest during prolonged LDR-IR than HCT116vector (MLH1−) cells. A. flow cytometry histograms show moderately increased G2/M population after 72 h LDR-IR in both cell lines with about 5 percentage points more G2/M fraction in the MLH1+ cells than in the MLH1− cells. B. flow cytometry histograms show inhibition of G1/S progression in both cell lines. The cells were exposed to LDR-IR for 72 h and NOC was added for the last 8 h of LDR-IR prior to harvesting. C. the expanded portion of Fig. 2B demonstrates a late S phase population (arrows) in the MLH1+ cells. D. Bar graph derived from the representative scatter plots of dual-parameter flow cytometry for phospho-histone H3 (Ser10) and PI showing that the MLH1+ cells have less mitotic cells than the MLH1− cells. All experiments were repeated at least twice.
To better define LDR-IR effects on cell cycle progression, we next utilized nocodazole (NOC) trapping to stop the cell cycle in metaphase. Here, cells were exposed to LDR-IR for 72 h and NOC was added for the last 8 h of LDR-IR prior to harvesting. Fig. 2B shows representative flow cytometry histograms. Compared to the NOC treated controls, prolonged LDR-IR significantly inhibited cell cycle progression through the G1/S border in both cell lines. Interestingly, the MLH1+ cells also showed a greater late S phase accumulation. Fig. 2C illustrates an expanded portion of Fig. 2B, where an asymmetric 4N peak, which indicates the existence of a late S phase population, can be more easily seen in the MLH1+ cells (see arrows in Fig. 2C). This late S phase population in the MLH1+ cells is inversely correlated with the LDR-IR dose rate, i.e., the lower the dose rate (therefore, the more cells enter S phase), the more clear the late S phase. For example, this late S phase population is manifested as less steep initial slopes following Level 2 and 3 LDR-IR. But at Level 4, this late S phase population becomes a separate peak, distinguishable from the G2/M peak.
Although prolonged LDR-IR resulted in only a moderate increase in the overall G2/M population in both cell lines as defined by standard flow cytometry (Fig. 2A), we found that the MLH1+ cells had a greater G2/M checkpoint arrest than the MLH1− cells by using NOC trapping plus dual-parameter flow cytometry for the mitotic marker phospho-histone H3 (Ser10) and PI staining,. The bar graph in Fig. 2D illustrates data derived from representative dual-parameter flow cytometry scatter plots (flow data not shown). Again, cells were exposed to LDR-IR for 72 h and NOC was added for the last 8 h of LDR-IR prior to harvesting. In both un-irradiated cell populations, there were only about 2.5% mitotic cells in the total populations without NOC treatment, but the % mitotic cells increased to 36.5% with the NOC exposure. After completion of LDR-IR and NOC treatment, the % mitotic cells was reduced to 10.5%, 20.3%, 25.2% and 28.8% in the MLH1− cells for Levels 1–4, respectively, and to 6.4%, 15%, 21.6% and 21.4% in the MLH1+ cells for Levels 1–4, respectively. Compared with the NOC treated controls, the reduction in the NOC-trapped mitotic cells during protracted LDR-IR was 71%, 44%, 30% and 20% in the MLH1− cells for Levels 1–4, respectively, and was 82%, 59%, 40.8% and 41% in the MLH1+ cells for Levels 1–4, respectively. Thus, prolonged LDR-IR inhibited cell cycle progression through the G2/M checkpoint in a LDR-IR dose rate-dependent manner with a greater inhibition found in the MLH1+ cells. Furthermore, since the G2 fraction was not directly measured but calculated by subtracting the mitotic fraction from the total 4N population, it can be reasonably assumed that the greater G2 fraction in the MLH1+ cells (Fig. 2D) includes both late S phase and G2 cells.
Prolonged LDR-IR led to a progressive increase of MLH1 and PMS2 proteins in MMR+ (HCT116MLH1) cells
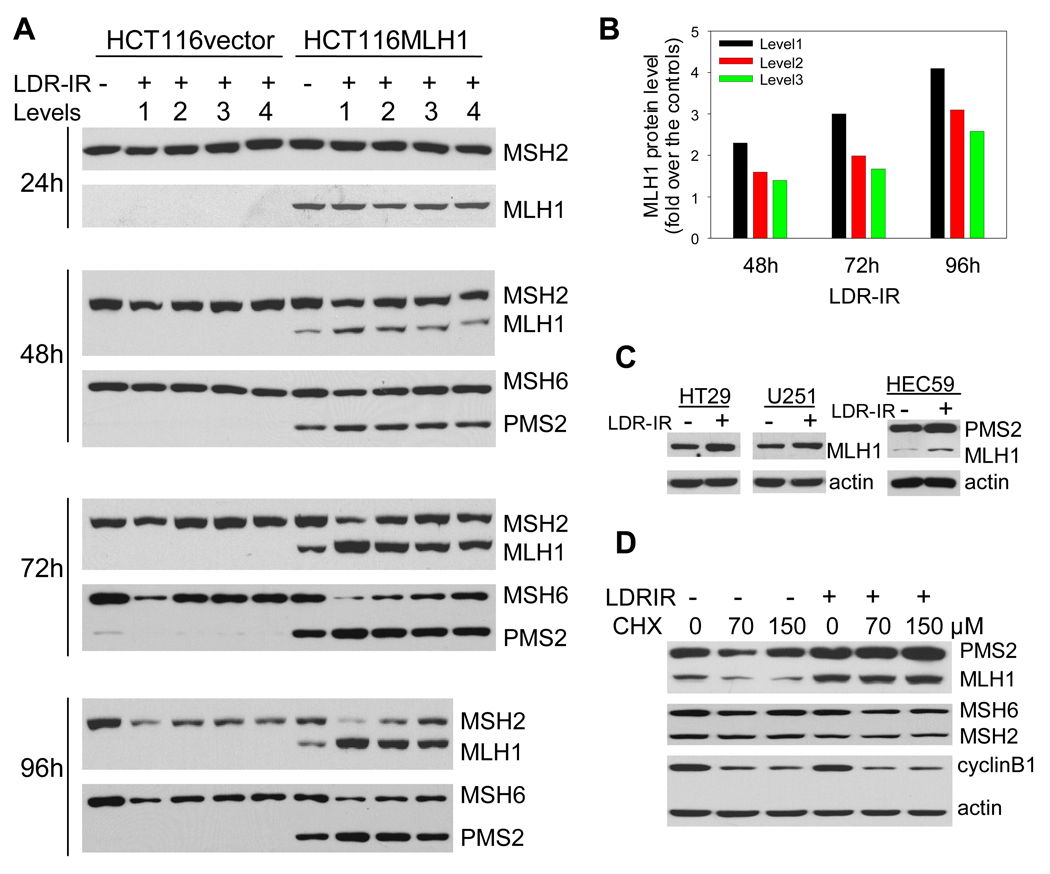
We also examined MMR protein levels by Western blotting during prolonged LDR-IR. We found that in the MLH1+ cells, the MLH1 protein level was unchanged after a 24 h exposure to LDR-IR, but progressively increased after a 48 h to 96 h exposure to LDR-IR (Fig. 3A and 3B). The level of PMS2, the dimeric partner of MLH1 in the MutLα complex, was similarly increased, albeit to a lesser degree (Fig. 3A). The increase in MLH1 and PMS2 protein levels was LDR-IR dose rate- and time-dependent. In contrast, we found that protein levels of MSH2 and MSH6 in the MutSα complex were either decreased or unchanged depending on LDR-IR dose rate and exposure time. Interestingly, the extent of the decrease in MSH2 and MSH6 proteins appears to be slightly greater in the MLH1+ cells than in the MLH1− cells.
Figure 3.
MLH1 protein accumulates in MLH1+ cells during prolonged LDR-IR as a result of compromised protein degradation. A. Western blots show that the increase in MLH1 and PMS2 proteins occurs concomitantly with decrease in MSH2/MSH6 proteins in HCT116MLH1 cells. B. MLH1 protein levels in Fig. 3A were quantified with Image J. C. Increased MLH1 protein can be seen in other MLH1+ cell lines HT29, U251 and HEC59 after LDR-IR (3.1 cGy/h for 72h). D. Protein synthesis inhibitor CHX cannot change the LDR-IR induced MLH1 accumulation (2.4 cGy/h for 96 h) in HCT116MLH1 cells. Cyclin B1 serves as a positive control, whereas PMS2, MSH2 and MSH6 are the negative controls. All Western blotting assays were repeated at least twice.
Since MLH1 protein expression in the MLH1+ cells is under the control of an exogenous CMV promoter, we questioned whether this observed increase in MLH1 protein during prolonged LDR-IR was also found in endogenous MLH1-expressing cells. To address this, we examined two other cell lines, HT29 and U251, which are both MLH1+ (MMR+). A similar increase in MLH1 protein level after prolonged LDR-IR at a very low dose rate (3.1 cGy/h for 72 h, it was the highest dose rate available at the time of the experiment) was also found in these cell lines (Fig. 3C). Furthermore, the same result was found in a MSH2− (MMR−) cell line, HEC59, which expresses MLH1 protein (Fig. 3C). Collectively, these data suggest that the protracted LDR-IR-induced increase in MLH1 and PMS2 proteins is a general phenomenon and that this increase is independent of MSH2 and MSH6, and therefore, independent of functional MMR.
Since in HCT116MLH1 cells, MLH1 protein expression resulted from a cDNA transfection, endogenous MLH1 gene regulation causing higher MLH1 protein levels in response to prolonged LDR-IR was unlikely. Thus, we hypothesized that the increased MLH1 protein might result from reduced MLH1 protein degradation. Using the protein synthesis inhibitor cycloheximide (CHX) during LDR-IR, we demonstrated that MLH1 protein degradation was indeed reduced (Fig. 3D). Here, cells were treated with LDR-IR (2.4 cGy/h) for 96 h and CHX was added 3 h prior to harvesting. While the MLH1 protein level decreased with CHX treatment without LDR-IR, the LDR-IR-induced accumulation of MLH1 protein was sustained in the presence of CHX (Fig. 3D). However, MSH2 and MSH6 protein levels did not change significantly under the same treatment conditions. These data suggest that the degradation of MLH1 protein is compromised during prolonged LDR-IR, and that the half-life of MLH1 protein is shorter than that of MSH2, MSH6 and actin proteins.
Rad51 protein expression was down-regulated to a greater extent in MLH1+ (HCT116MLH1) cells than in MLH1− (HCT116vector) cells during protracted LDR-IR
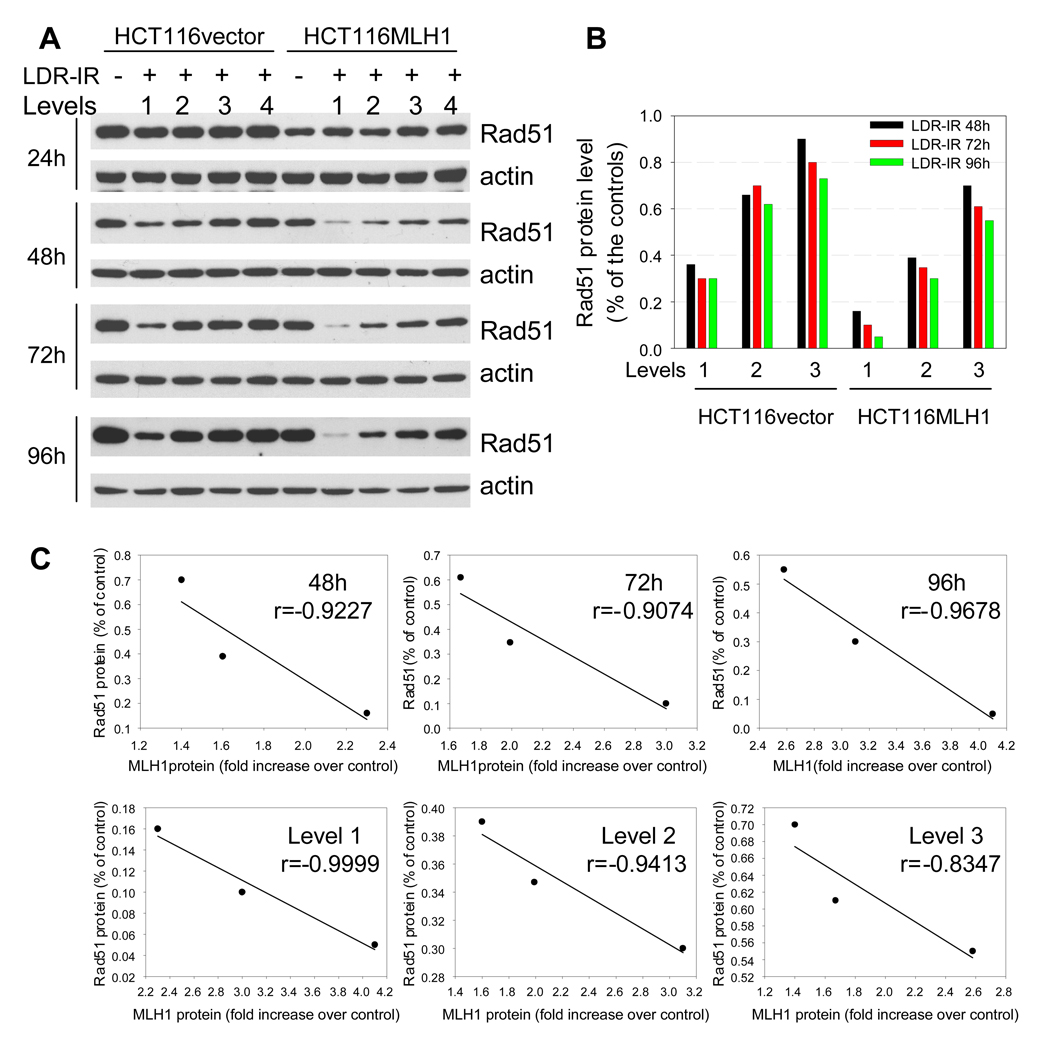
In order to verify whether MLH1 status had an effect on other DNA repair systems during prolonged LDR-IR, we examined the expression levels of four BER proteins (OGG1, APE1, Polβ and FEN1), two NER proteins (XPA and XPC) and two NHEJ proteins (Ku70 and Ku80) by Western blotting analysis. No significant changes were found in the levels of all these proteins in both cell lines in response to prolonged LDR-IR (Supplementary Fig. 1A). We also examined three HR proteins (Mre11, NBS1, Rad51). Although Mre11 and NBS1 protein levels showed no significant changes (Supplementary Fig. 1A), Rad51, which plays a pivotal role in HR, decreased during prolonged LDR-IR in both cell lines in an inverse dose rate- and time-dependent manner (Fig. 4A and 4B).
Figure 4.
Rad51 protein decreases during prolonged LDR-IR. A. Western blots demonstrate that Rad51 protein decreases progressively during prolonged LDR-IR with more significant reduction found in HCT116MLH1 (MLH1+) cells. B. Rad51 protein levels in Fig. 4A were quantified with software Image J. C. Linear regression test shows a strong correlation between MLH1 protein increase and Rad51 protein decrease in the MLH1+ cells. Correlation coefficients are calculated on data sets from Fig. 4B using Excel statistics. All Western blotting assays are repeated at least twice.
With respect to changes in Rad51 and MLH1 protein levels in response to LDR-IR, we made following interesting observations. First, the basal level of Rad51 is about 30% less in the MLH1+ cells compared to the MLH1− cells, indicating that the presence of MLH1 protein per se may affect Rad51 protein dynamics. Second, as with MLH1 protein levels, Rad51 levels do not change significantly during the first 24 h LDR-IR exposure, but decrease progressively when exposure time increases. Third, Rad51 protein level reduction in response to LDR-IR is greater in the MLH1+ cells than in the MLH1− cells. The absolute amount of Rad51 protein would be much smaller in the MLH1+ cells than in the MLH1− cells, if one considers the lower basal level of Rad51 protein in the MLH1+ cells. Fourth, the Rad51 protein decrease and the MLH1 protein increase are strongly correlated in the MLH1+ cells, as indicated by the correlation coefficients (−0.83 to −0.99) (Fig. 4C). These observations suggest that the MLH1 protein may be involved in the inhibition of HR via enhanced inhibition of Rad51 during prolonged LDR-IR.
Prolonged LDR-IR increased p53, p21, PARPp85 and LC3-II levels to greater extents in MLH1+ (HCT116MLH1) cells compared to MLH1− (HCT116vector) cells
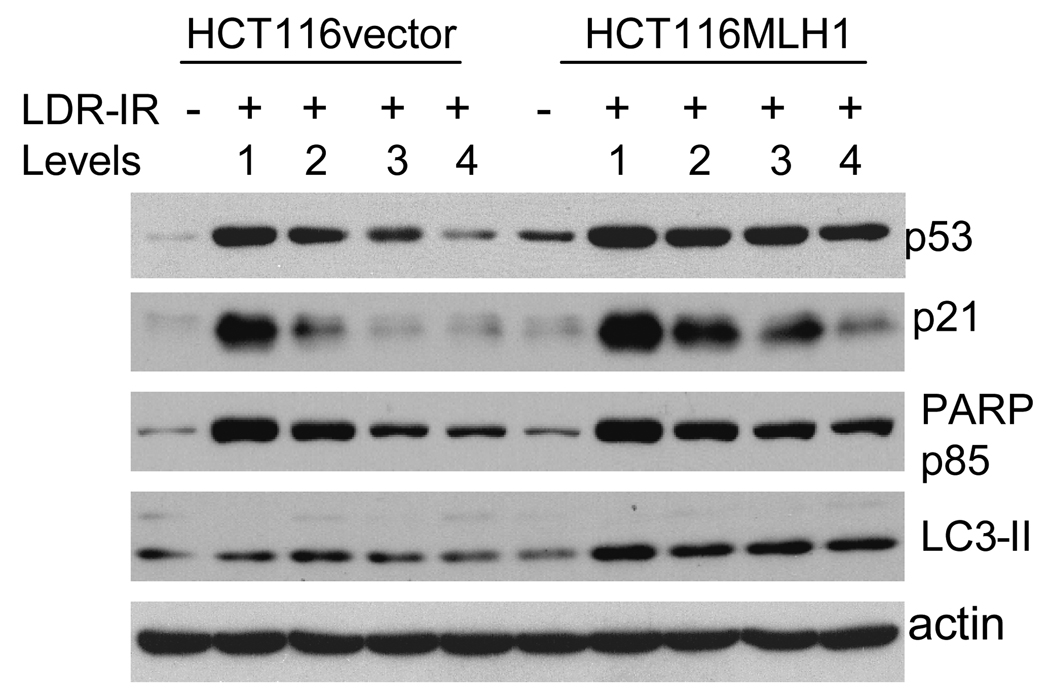
Since the MLH1 protein has been implicated in DNA damage signaling, we were interested in determining the levels of a double strand break marker, γH2AX, and the DNA damage response proteins p53 and p21 in the MLH1+ versus the MLH1− cells. γH2AX levels were only moderately altered during prolonged LDR-IR, but there was no clear difference between the MLH1+ and the MLH1− cells (Supplementary Fig. 1B). A fluorescent immunocytochemistry assay also failed to reveal γH2AX foci formation above the basal levels (data not shown). However, p53 and p21 protein levels increased in both cell lines during prolonged LDR-IR for 24 to 96 h (Fig. 5, 72 h data, other time points not shown). Notably, the increased levels of p53 and p21 proteins appear to be enhanced in the MLH1+ cells compared to that in the MLH1− cells.
Figure 5.
Western blots demonstrate that levels of p53 and p21 proteins as well as cleaved PARP p85 fragment, are all elevated in both cell lines after a 72h exposure to prolonged LDR-IR but to a greater extent in the MLH1+ cells, while LC3-II is increased in the MLH1+ cells but not in the MLH1− cells. All Western blotting assays are repeated at least twice.
Finally, since MLH1 protein is also implicated in DNA damage-induced cell death, we checked the apoptotic cell death marker, cleaved PARP p85 fragment, and the autophagic cell death marker, LC3-II. As shown in Fig. 5, PARP p85 levels are elevated in both cell lines during LDR-IR, but to a greater extent in the MLH1+ cells compared to the MLH1− cells (LDR-IR 72 h). LC3-II does not change significantly in the MLH1− cells but increases in the MLH1+ cells. These date indicate that MLH1 may increase both apoptotic and autophagic cell death pathway signaling in response to prolonged LDR-IR. However, the cell death rate at the early times was not significant enough for quantitation of different forms of cell death.
Discussion
In this LDR-IR study, we used lower dose rates (1.3–17 cGy/h for 24 to 96 h) than previous studies (19, 20) and these dose rates mimic the dose rates used during permanent prostate cancer brachytherapy (23). After prolonged LDR-IR with these low dose rates, the MLH1− cells exhibited improved clonogenic survival (i.e., relative radiation resistance) and an increased gene mutation rate (Fig. 1 and Table 1) compared to the MLH1+ cells. Additionally, MLH1-proficiency appears to augment LDR-IR induced cell killing by enhancing both apoptotic and autophagic cell death pathways (Fig. 5). The important clinical-translational relevance of our data suggesting that MLH1 proficiency enhances radiosensitization to prolonged LDR-IR are two fold: first, MLH1 proficiency may reduce cancer susceptibility by preventing potentially mutagenic lesions from being passed on to progeny after prolonged LDR-IR; and second, MLH1 proficiency may increase the efficacy of the cancer treatment regimens which utilize prolonged LDR-IR.
MMR is known to play an important role in a G2/M arrest following treatment with a variety of clinically relevant chemotherapeutic drugs (25, 26). Our group has previously shown that MMR also leads to a slower release from G2/M checkpoint arrest in MMR+ cells following high dose rate (> 1Gy/min) ionizing radiation (HDR-IR) exposures compared to MMR− cells (4). In the current study, we found that LDR-IR activates a modest G2/M checkpoint arrest in both the MLH1+ and the MLH1− cells (Fig. 2A), indicating weak activation of the ATM/ATR pathway. These data are consistent with the previous observation that LDR-IR induced DNA damage results in an inefficient activation of ATM and γH2AX (27). In agreement with these findings (27), we were also unable to detect significant increases in γH2AX levels (Supplement Fig. 1B) as well as increases in pChk2 and pChk1 (data not shown). However, by using the mitotic marker phospho-histone H3, we found that the inhibition of a G2 to M transition was greater in the MLH1+ cells than in the MLH1− cells (Fig. 2D). This observation indicates that MLH1 may be involved in the activation and/or maintenance of the G2/M checkpoint in response to prolonged LDR-IR.
Interestingly, we also found an increased late S phase population in the MLH1+ cells during protracted LDR-IR, which is not previously reported (Fig. 2C). This late S phase accumulation is not easy to recognize without nocodazole treatment. Indeed, it is only made possible when the nocodazole-trapped mitotic cell population increases significantly, resulting in a shift of the G2/M peak to the right. Previously, Brown et al have reported that MMR is required for the S phase checkpoint following HDR-IR (10). Our data also suggest a role for MLH1 in slowing late S phase progression during prolonged LDR-IR, although the mechanism underlying this late S phase accumulation in MLH1+ cells remains to be elucidated. We speculate that there may be interplay between MLH1 protein (or MMR) and HR in the late S phase during prolonged LDR-IR as the persistent LDR-IR leads to continuous generation of DNA damage and HR is believed to occur mainly in the late S and G2 phases (28).
In our study, we did not find significant changes in the protein levels of many different DNA repair proteins during prolonged LDR-IR (Supplementary Fig. 1A). However, we did find that the levels of MLH1 protein and its dimeric partner PMS2 protein were elevated (up to 4-fold increase in MLH1) after protracted LDR-IR in the MLH1+ cells, while the levels of MSH2 protein and its dimeric partner MSH6 protein were concomitantly reduced (Fig. 3A). To the best of our knowledge, these alterations in human MMR protein levels after protracted LDR-IR have not been previously reported. These results using the isogenic HCT116 cell lines were confirmed in two additional MLH1+ human cancer cell lines (Fig.3C). Additionally, a similar result is found in HEC59 endometrial cancer cells, which are MSH2− (MMR−) (Fig. 3C), suggesting that functional MMR is not a prerequisite for MLH1 protein accumulation during prolonged LDR-IR. Importantly, the increased MLH1 protein levels seem to result from reduced MLH1 protein degradation (Fig.3D). However, MLH1 mRNA levels also need to be assessed to exclude an up-regulation of MLH1 transcription. Further experiments are planned to more clearly define the mechanism and biological significance of the increased MLH1 protein (and possibly mRNA) levels during prolonged LDR-IR.
The other DNA repair protein found to be altered in this study was Rad51, which, in contrast to MLH1, was found to be progressively decreased in response to prolonged LDR-IR (Fig. 4). Rad51, a strand transferase, is a key component in HR and over-expression of Rad51 results in increased HR capability (29, 30). The Rad51 gene has previously been shown to be up-regulated after HDR-IR (31) and following acute LDR-IR (32). In contrast, Tomita et al (33) reported that the cellular response to prolonged LDR-IR is remarkably different from that to HDR-IR, and that NHEJ-defective cells are more sensitive to LDR-IR than HR-defective cells, suggesting a predominant role of NJEJ compared to HR in response to LDR-IR induced DNA damage (in presumably MLH1+ cells). In our study, we found that the basal level of Rad51 was about 30% lower in the MLH1+ cells than in the MLH− cells (Fig. 4A). Prolonged LDR-IR led to a decrease in Rad51 protein levels in both the HCT116 cell lines, but the Rad51 reduction was more significant in the MLH1+ cells than in the MLH1− cells (Fig.4A and 4B). Importantly, a linear regression defines the correlation coefficients from −0.83 to −0.99 between the MLH1 protein increase and the Rad51 protein decrease, indicating a strong inverse correlation between these two proteins (Fig. 4C).
MMR is previously reported to be involved in HR (34), as mismatches can be produced within the heteroduplex generated by strand exchange during DNA replication to allow bypass of unrepaired damage (35–37). During prolonged LDR-IR, low levels of continuous DNA damage (including low levels of DSBs) result as manifested by a sustained elevation of p53 and p21 proteins but without significant γH2AX induction. Since HR is the major repair pathway of IR-induced DSBs and is most active during late S-phase, our data demonstrating the inverse correlation of MLH1 and Rad51 protein levels during LDR-IR leads us to speculate that altered MLH1 protein degradation (with enhanced MutLα levels) affects HR via reduced expression of Rad51. A possible role of MLH1 in transcriptional and/or post-transcriptional regulation of Rad51 during prolonged LDR-IR will be assessed by our group in the near future. Our current observations are supported by a recent report demonstrating down regulation of HR and MutLα, independent of MMR activity (38). While our report provides the first link of possible inhibition of HR by increased MLH1 protein levels during prolonged LDR-IR, further studies are needed to better understand these observations. From a clinical-translational perspective, our data suggest that MLH1-proficiency may decrease cancer susceptibility following prolonged LDR-IR and may also increase the efficacy of localized prolonged LDR-IR as used in cancer brachytherapy.
Acknowledgments
This work was supported in part, by a National Cancer Institute grant (U56 CA112963) and a NASA grant (G1072) as well as the DBJ and University Radiation Medicine Foundations.
Footnotes
Disclosure of Potential Conflicts of Interest: No potential conflicts of interest are disclosed.
References
- 1.Cardis E, Vrijheid M, Blettner M, et al. The 15-Country Collaborative Study of Cancer Risk among Radiation Workers in the Nuclear Industry: estimates of radiation-related cancer risks. Radiat Res. 2007;167:396–416. doi: 10.1667/RR0553.1. [DOI] [PubMed] [Google Scholar]
- 2.Karagiannis TC, El-Osta A. Double-strand breaks: signaling pathways and repair mechanisms. Cell Mol Life Sci. 2004;61:2137–2147. doi: 10.1007/s00018-004-4174-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Georgakilas AG. Processing of DNA damage clusters in human cells: current status of knowledge. Mol Biosyst. 2008;4:30–35. doi: 10.1039/b713178j. [DOI] [PubMed] [Google Scholar]
- 4.Yan T, Schupp JE, Hwang HS, et al. Loss of DNA mismatch repair imparts defective cdc2 signaling and G(2) arrest responses without altering survival after ionizing radiation. Cancer Res. 2001;61:8290–8297. [PubMed] [Google Scholar]
- 5.Kuraoka I, Bender C, Romieu A, Cadet J, Wood RD, Lindahl T. Removal of oxygen free-radical-induced 5',8-purine cyclodeoxynucleosides from DNA by the nucleotide excision-repair pathway in human cells. Proc Natl Acad Sci U S A. 2000;97:3832–3837. doi: 10.1073/pnas.070471597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cann KL, Hicks GG. Regulation of the cellular DNA double-strand break response. Biochem Cell Biol. 2007;85:663–674. doi: 10.1139/O07-135. [DOI] [PubMed] [Google Scholar]
- 7.Jeggo PA, Lobrich M. Contribution of DNA repair and cell cycle checkpoint arrest to the maintenance of genomic stability. DNA Repair (Amst) 2006;5:1192–1198. doi: 10.1016/j.dnarep.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 8.Berry SE, Kinsella TJ. Targeting DNA mismatch repair for radiosensitization. Semin Radiat Oncol. 2001;11:300–315. doi: 10.1053/s1053-4296(01)80067-9. [DOI] [PubMed] [Google Scholar]
- 9.O'Brien V, Brown R. Signalling cell cycle arrest and cell death through the MMR System. Carcinogenesis. 2006;27:682–692. doi: 10.1093/carcin/bgi298. [DOI] [PubMed] [Google Scholar]
- 10.Brown KD, Rathi A, Kamath R, et al. The mismatch repair system is required for S-phase checkpoint activation. Nat Genet. 2003;33:80–84. doi: 10.1038/ng1052. [DOI] [PubMed] [Google Scholar]
- 11.Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 12.Kolodner RD, Marsischky GT. Eukaryotic DNA mismatch repair. Curr Opin Genet Dev. 1999;9:89–96. doi: 10.1016/s0959-437x(99)80013-6. [DOI] [PubMed] [Google Scholar]
- 13.Umar A, Kunkel TA. DNA-replication fidelity, mismatch repair and genome instability in cancer cells. Eur J Biochem. 1996;238:297–307. doi: 10.1111/j.1432-1033.1996.0297z.x. [DOI] [PubMed] [Google Scholar]
- 14.Marra G, Jiricny J. DNA mismatch repair and colon cancer. Adv Exp Med Biol. 2005;570:85–123. doi: 10.1007/1-4020-3764-3_4. [DOI] [PubMed] [Google Scholar]
- 15.Li GM. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008;18:85–98. doi: 10.1038/cr.2007.115. [DOI] [PubMed] [Google Scholar]
- 16.Pors K, Patterson LH. DNA mismatch repair deficiency, resistance to cancer chemotherapy and the development of hypersensitive agents. Curr Top Med Chem. 2005;5:1133–1149. doi: 10.2174/156802605774370883. [DOI] [PubMed] [Google Scholar]
- 17.Fedier A, Fink D. Mutations in DNA mismatch repair genes: implications for DNA damage signaling and drug sensitivity (review) Int J Oncol. 2004;24:1039–1047. [PubMed] [Google Scholar]
- 18.Fritzell JA, Narayanan L, Baker SM, et al. Role of DNA mismatch repair in the cytotoxicity of ionizing radiation. Cancer Res. 1997;57:5143–5147. [PubMed] [Google Scholar]
- 19.DeWeese TL, Shipman JM, Larrier NA, et al. Mouse embryonic stem cells carrying one or two defective Msh2 alleles respond abnormally to oxidative stress inflicted by low-level radiation. Proc Natl Acad Sci U S A. 1998;95:11915–11920. doi: 10.1073/pnas.95.20.11915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeng M, Narayanan L, Xu XS, Prolla TA, Liskay RM, Glazer PM. Ionizing radiation-induced apoptosis via separate Pms2- and p53-dependent pathways. Cancer Res. 2000;60:4889–4893. [PubMed] [Google Scholar]
- 21.Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 22.Jiricny J. The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol. 2006;7:335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- 23.Sahgal A, Roach M., 3rd Permanent prostate seed brachytherapy: a current perspective on the evolution of the technique and its application. Nat Clin Pract Urol. 2007;4:658–670. doi: 10.1038/ncpuro0971. [DOI] [PubMed] [Google Scholar]
- 24.Yan T, Berry SE, Desai AB, Kinsella TJ. DNA mismatch repair (MMR) mediates 6-thioguanine genotoxicity by introducing single-strand breaks to signal a G2-M arrest in MMR-proficient RKO cells. Clin Cancer Res. 2003;9:2327–2334. [PubMed] [Google Scholar]
- 25.Fink D, Aebi S, Howell SB. The role of DNA mismatch repair in drug resistance. Clin Cancer Res. 1998;4:1–6. [PubMed] [Google Scholar]
- 26.Irving JA, Hall AG. Mismatch repair defects as a cause of resistance to cytotoxic drugs. Expert Rev Anticancer Ther. 2001;1:149–158. doi: 10.1586/14737140.1.1.149. [DOI] [PubMed] [Google Scholar]
- 27.Collis SJ, Schwaninger JM, Ntambi AJ, et al. Evasion of early cellular response mechanisms following low level radiation-induced DNA damage. J Biol Chem. 2004;279:49624–49632. doi: 10.1074/jbc.M409600200. [DOI] [PubMed] [Google Scholar]
- 28.Tamulevicius P, Wang M, Iliakis G. Homology-directed repair is required for the development of radioresistance during S phase: interplay between double-strand break repair and checkpoint response. Radiat Res. 2007;167:1–11. doi: 10.1667/RR0751.1. [DOI] [PubMed] [Google Scholar]
- 29.Daboussi F, Dumay A, Delacote F, Lopez BS. DNA double-strand break repair signalling: the case of RAD51 post-translational regulation. Cell Signal. 2002;14:969–975. doi: 10.1016/s0898-6568(02)00052-9. [DOI] [PubMed] [Google Scholar]
- 30.Vispe S, Cazaux C, Lesca C, Defais M. Overexpression of Rad51 protein stimulates homologous recombination and increases resistance of mammalian cells to ionizing radiation. Nucleic Acids Res. 1998;26:2859–2864. doi: 10.1093/nar/26.12.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Snyder AR, Morgan WF. Gene expression profiling after irradiation: clues to understanding acute and persistent responses? Cancer Metastasis Rev. 2004;23:259–268. doi: 10.1023/B:CANC.0000031765.17886.fa. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y, Rohde LH, Emami K, et al. Suppressed expression of non-DSB repair genes inhibits gamma-radiation-induced cytogenetic repair and cell cycle arrest. DNA Repair (Amst) 2008;7:1835–1845. doi: 10.1016/j.dnarep.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 33.Tomita M, Morohoshi F, Matsumoto Y, Otsuka K, Sakai K. Role of DNA double-strand break repair genes in cell proliferation under low dose-rate irradiation conditions. J Radiat Res. 2008;49:557–564. doi: 10.1269/jrr.08036. [DOI] [PubMed] [Google Scholar]
- 34.Surtees JA, Argueso JL, Alani E. Mismatch repair proteins: key regulators of genetic recombination. Cytogenet Genome Res. 2004;107:146–159. doi: 10.1159/000080593. [DOI] [PubMed] [Google Scholar]
- 35.Dudenhoffer C, Rohaly G, Will K, Deppert W, Wiesmuller L. Specific mismatch recognition in heteroduplex intermediates by p53 suggests a role in fidelity control of homologous recombination. Mol Cell Biol. 1998;18:5332–5342. doi: 10.1128/mcb.18.9.5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saintigny Y, Rouillard D, Chaput B, Soussi T, Lopez BS. Mutant p53 proteins stimulate spontaneous and radiation-induced intrachromosomal homologous recombination independently of the alteration of the transactivation activity and of the G1 checkpoint. Oncogene. 1999;18:3553–3563. doi: 10.1038/sj.onc.1202941. [DOI] [PubMed] [Google Scholar]
- 37.Susse S, Janz C, Janus F, Deppert W, Wiesmuller L. Role of heteroduplex joints in the functional interactions between human Rad51 and wild-type p53. Oncogene. 2000;19:4500–4512. doi: 10.1038/sj.onc.1203809. [DOI] [PubMed] [Google Scholar]
- 38.Siehler SY, Schrauder M, Gerischer U, Cantor S, Marra G, Wiesmuller L. Human MutL-complexes monitor homologous recombination independently of mismatch repair. DNA Repair (Amst) 2009;8:242–252. doi: 10.1016/j.dnarep.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]