Abstract
Within the developing pancreas Hepatic Nuclear Factor 6 (HNF6) directly activates the pro-endocrine transcription factor, Ngn3. HNF6 and Ngn3 are each essential for endocrine differentiation and HNF6 is also required for embryonic duct development. Most HNF6−/− animals die as neonates, making it difficult to study later aspects of HNF6 function. Here, we describe, using conditional gene inactivation, that HNF6 has specific functions at different developmental stages in different pancreatic lineages. Loss of HNF6 from Ngn3-expressing cells (HNF6Δendo) resulted in fewer multipotent progenitor cells entering the endocrine lineage, but had no effect on β cell terminal differentiation. Early, pancreas-wide HNF6 inactivation (HNF6Δpanc) resulted in endocrine and ductal defects similar to those described for HNF6 global inactivation. However, all HNF6Δpanc animals survived to adulthood. HNF6Δpanc pancreata displayed increased ductal cell proliferation and metaplasia, as well as characteristics of pancreatitis, including up-regulation of CTGF, MMP7, and p8/Nupr1. Pancreatitis was most likely caused by defects in ductal primary cilia. In addition, expression of Prox1, a known regulator of pancreas development, was decreased in HNF6Δpanc pancreata. These data confirm that HNF6 has both early and late functions in the developing pancreas and is essential for maintenance of Ngn3 expression and proper pancreatic duct morphology.
Keywords: pancreas development, lineage tracing, multipotent progenitor, pancreatitis
Introduction
Recent advances in experimental techniques allow for more precise control over gene expression revealing that temporal regulation of specific transcription factors is crucial to allocating the correct number of pancreatic progenitor cells to specific pancreatic lineages (Hale et al., 2005; Harmon et al., 2004; Johansson et al., 2007; Schwitzgebel et al., 2000). Understanding the temporal and spatial regulation of these transcription factors is crucial to delineating the molecular mechanisms controlling pancreatic lineage allocation and organogenesis.
The exocrine pancreas, which produces and transports digestive enzymes to the rostral duodenum, is composed of acinar and ductal epithelial cells and accounts for ~98% of adult pancreatic mass. The endocrine pancreas occupies the remaining 2% of pancreatic mass, and is subdivided into five hormone-producing cell types: glucagon-secreting α cells, insulin-secreting β cells, somatostatin-secreting δ cells, pancreatic polypeptide-secreting PP cells, and ghrelin-secreting ε cells (Prado et al., 2004). Endocrine differentiation occurs in two waves: primary transition (pre-e13.5) insulin-expressing and glucagon-expressing cells are not thought to contribute to mature islets, while endocrine cells differentiating during the secondary transition (e13.5–e16.5) will give rise to mature islets (Jensen, 2004; Pictet et al., 1972).
The Hepatic Nuclear Factor 6 (HNF6) homeodomain-containing transcription factor is an important regulator of endocrine development. HNF6 was first identified in the liver, and 75% of animals with a global HNF6 deletion (HNF6−/−) die within the first ten days after birth due to liver defects (Jacquemin et al., 2000). HNF6 is expressed early in pancreatogenesis in all endodermally-derived cells, but is not detected in differentiated endocrine cells at late gestation (Landry et al., 1997; Rausa et al., 1997). HNF6 null embryos show impaired endocrine differentiation and perturbed duct morphogenesis during embryogenesis (Jacquemin et al., 2000; Pierreux et al., 2006). HNF6 acts upstream of the critical pancreatic and β cell transcription factor, pancreas/duodenum homeobox-1 (Pdx1) (Jacquemin et al., 2003) and activates expression of the pro-endocrine gene Neurogenin 3 (Ngn3) (Jacquemin et al., 2000). Ngn3 is the earliest known marker of specified endocrine cells (Gradwohl et al., 2000; Jensen et al., 2000; Schwitzgebel et al., 2000). Global deletion of HNF6 causes a dramatic down-regulation in Ngn3 expression, and a concomitant decrease in endocrine cells (Jacquemin et al., 2000), while over-expression of HNF6 specifically in the endocrine lineage results in an increase in NGN3-positive cells (Wilding Crawford et al., 2008). Mice null for Ngn3 also lack hormone-expressing cells (Gradwohl et al., 2000), and lineage tracing experiments have shown that all islet endocrine cells are derived from a Ngn3-expressing progenitor (Gu et al., 2002; Schonhoff et al., 2004). Transgenic expression of Ngn3 demonstrates that it is sufficient to direct pancreatic progenitors to all endocrine cell types (Johansson et al., 2007).
In this study, we generated mice in which HNF6 is inactivated throughout the pdx1 domain (HNF6Δpanc) to further characterize the specific roles of HNF6 in pancreas development and postnatal pancreas function. Similar endocrine defects to those described in the global HNF6 null animals (Jacquemin et al., 2000) were observed in HNF6Δpanc animals. To determine whether HNF6 plays additional roles in endocrine differentiation subsequent to Ngn3 activation, we first clarified the expression of HNF6 protein within the developing pancreas. Based on its lack of expression even in early hormone-positive cells, we hypothesized that HNF6 is required only to initiate endocrine specification. To test this hypothesis, HNF6 was inactivated specifically in cells that activated Ngn3 (HNF6Δendo). Loss of HNF6 from Ngn3-expressing cells did not affect β cell function or glucose homeostasis suggesting that HNF6 is dispensable for later events of endocrine differentiation. Lineage tracing analyses revealed, however, that a subset of Ngn3-expressing cells that lost HNF6 became incorporated into exocrine tissue thereby indicating that activation of Ngn3 does not irreversibly commit a cell to the endocrine lineage. HNF6 may then function to allow Ngn3 to reach a level within the proendocrine compartment that ensures faithful commitment of cells to the endocrine lineage.
In addition to defects in endocrine development, HNF6Δpanc embryos showed defects in duct development similar to those observed in HNF6−/− embryos (Pierreux et al., 2006). HNF6Δpanc animals all survived past weaning, allowing us to determine the consequences of HNF6 inactivation in the ductal epithelium. HNF6Δpanc adult pancreata displayed morphological changes consistent with pancreatitis, an inflammatory process in which pancreatic enzymes autodigest the pancreas. Several molecular markers of pancreatitis, including connective tissue growth factor (CTGF), metalloproteinase 7 (MMP7), and p8/Nupr1 were elevated in HNF6Δpanc mice. Moreover, these mice exhibited defects in primary cilia of pancreatic epithelial cells, a finding previously associated with increased cyst formation, pancreatitis, and ductal epithelial proliferation (Cano et al., 2006; Pugacheva et al., 2007). Pancreatitis significantly elevates the risk of pancreatic cancer in humans (Lowenfels et al., 1993), and indeed, we observed preneoplastic lesions in HNF6Δpanc pancreata including squamous cell metaplasia and acinar-to-ductal metaplasia.
Results
HNF6 is rapidly down-regulated as pancreatic progenitors are specified to the endocrine lineage
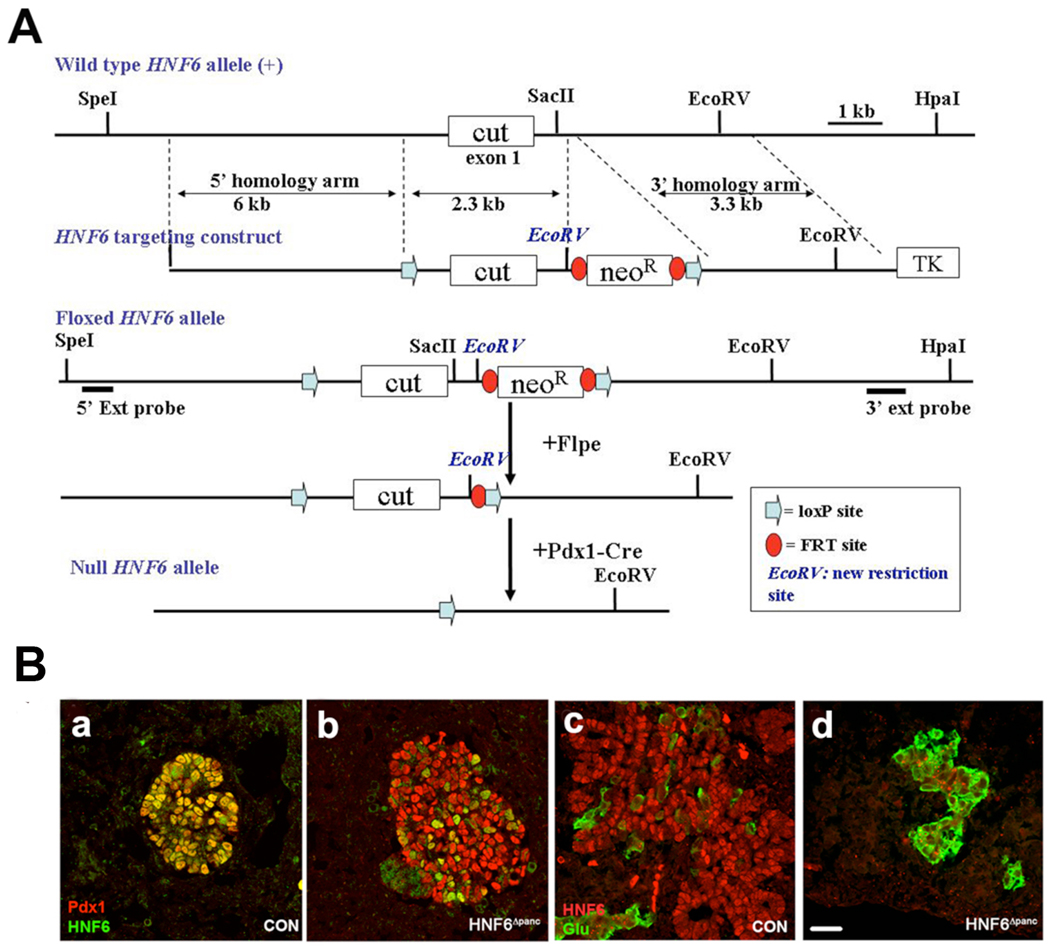
To determine the function of HNF6 in pancreas development subsequent to specification and early bud outgrowth, HNF6 was inactivated by interbreeding of HNF6flox/flox mice with pdx1-Creearly transgenic mice (Gu et al., 2002). pdx1-Creearly mediates recombination throughout the endogenous pdx1 domain: antral stomach, rostral duodenum, and entire pancreatic epithelium at e9.5 (Zhang et al., 2006). In HNF6flox/flox;pdx1-Creearly (HNF6Δpanc) mice, Cre recombinase mediates the deletion of the entire cut-domain, generating a null allele (see Experimental Procedures and Figure 1A). HNF6Δpanc mice were born at the expected frequency and survived past weaning. In control pancreatic buds at e11.5, HNF6 was detected in all Pdx1-positive cells (Figure 1Ba), but the majority of Pdx1-positive cells were HNF6 negative in HNF6Δpanc animals (Figure 1Bb). By e13.5, HNF6 protein was no longer detectable in HNF6Δpanc pancreata (Figure 1Bd), whereas expression in the control animals was as expected throughout the branching pancreatic epithelium (Figure 1Bc). No alteration in HNF6 protein expression was detected in HNF6flox/+;pdx1-Creearly animals (data not shown).
Figure 1. Schematic diagram of HNF6flox-neo targeting vector and Cre-mediated HNF6 inactivation.
(A) Representation of the mouse HNF6 locus showing exon one (the cut DNA binding domain). Also shown: HNF6flox-targeting construct, HNF6flox-targeted allele, and recombined HNF6 allele following Cre exposure. The 5' and 3' Southern blot hybridization probes are indicated as horizontal lines below the floxed allele. Restriction sites shown in italics indicate sites introduced to facilitate genotyping. (B) At e11.5, HNF6 protein (green) is co-expressed with Pdx1 (red) in nuclei of the developing pancreatic epithelium in control pancreata (a). However, HNF6 and Pdx1 are coexpressed in only few cells in HNF6Δpanc pancreata at e11.5 (b). At e13.5, HNF6 is expressed throughout the pancreatic epithelium in control pancreata (c), but not in HNF6Δpanc pancreata (d). Glucagon-positive cells (green) appear in large clusters in HNF6Δpanc at e13.5 (d), compared with controls in which glucagon-positive cells are scattered throughout the developing pancreas adjacent to the epithelium (c). Scale bar: in D, 20 µm for A–D.
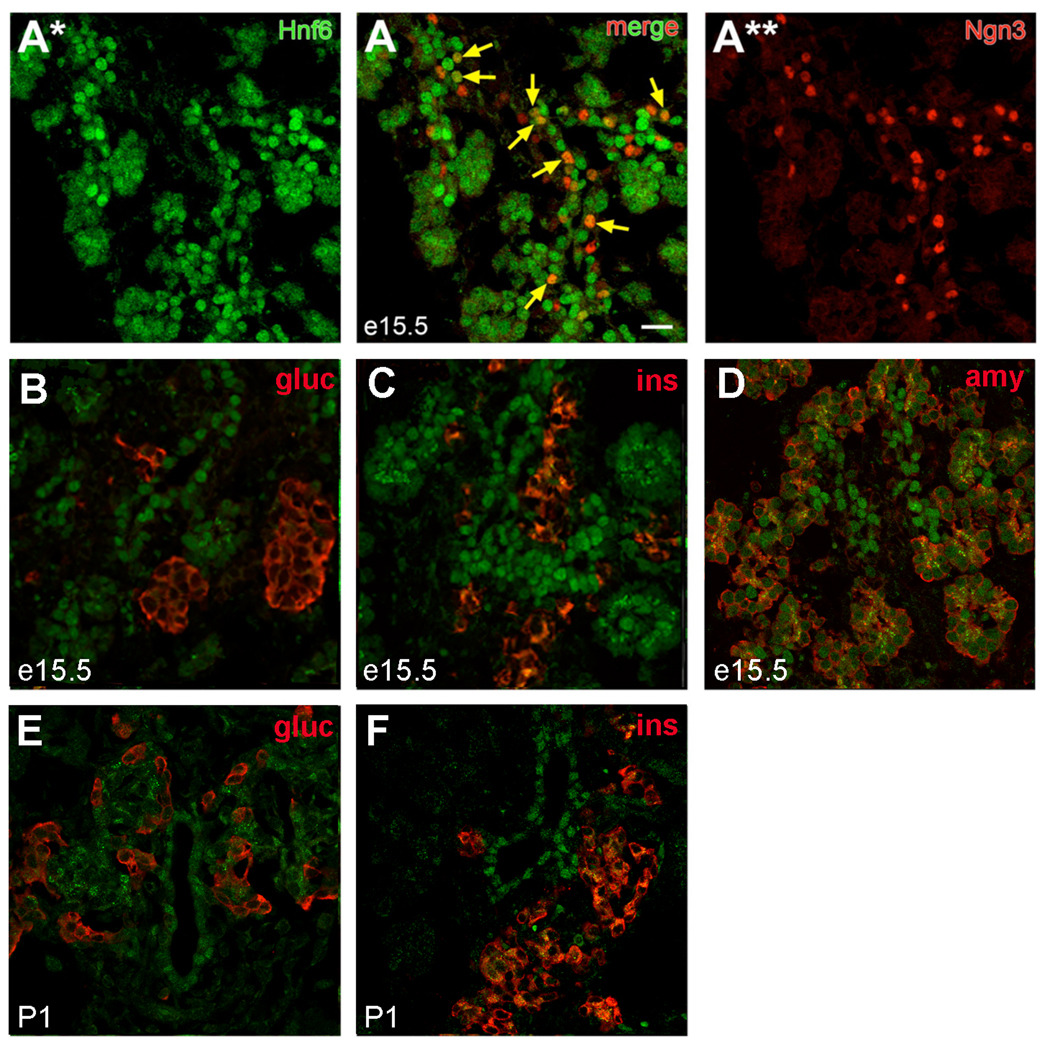
Previous in situ hybridization analysis showed that HNF6 is broadly expressed in the pancreatic epithelium during early organogenesis, but becomes restricted to ductal and acinar cells prior to birth (Landry et al., 1997; Rausa et al., 1997). However, temporal regulation of HNF6 protein has not yet been addressed in detail. Upon examining HNF6 protein localization in the developing pancreas, nuclear-localized HNF6 was detected within the pancreatic epithelium at e15.5 (Figure 2A), consistent with previous reports (Maestro et al., 2003; Pierreux et al., 2006). The central epithelium at this stage has been suggested to be the location of pancreatic multipotent progenitors, (Fujitani et al., 2006; Zhou et al., 2007), although recent results suggest that cells within the ductal trunk region give rise to endocrine and ductal cells, while those at the tip retain multipotency until late gestation at which time they give rise to acinar cells (Zhou et al., 2007). A large percentage of Ngn3-expressing cells within the trunk region co-expressed high levels of HNF6 (Figure 2A) at e13.5 (69.1 ± 2.7%; 293 Ngn3-positive cells) and e15.5 (57.1 ± 2.4%; 789 Ngn3-positive cells). HNF6 was undetectable in either glucagon- or insulin-expressing cells in embryonic pancreata at e13.5 (data not shown), e15.5 (Figure 2B,C) or postnatal day (P) 1 (Figure 2E,F). Lower levels of nuclear HNF6 were detectable at distal ductal regions and in acinar cells at e15.5 (Figure 2D); high levels were maintained in the ductal epithelium at P1 (Figure 2E,F). These latter data are consistent with previous reports demonstrating expression of HNF6 in ducts and abnormalities in duct morphogenesis in Hnf6−/− mice (Pierreux et al., 2006).
Figure 2. HNF6 protein expression in the developing pancreas.
(A) Co-localization of HNF6 (A*, green) and Ngn3 (A**, red) within the endocrine cords at e15.5. Arrows indicate co-expression of Ngn3 and HNF6. (B–D) Localization of HNF6 (green) and insulin or glucagon (B, C; red), or amylase (D; red) at e15.5. (E, F) Localization of HNF6 (green) and hormones (red) in P1 pancreatic tissue. Nuclear HNF6 was not detected in hormone-expressing cells at any time point. Scale bars represent 20 µm; visualized by confocal microscopy on 1µm optical sections.
Inactivation of HNF6 throughout the early pancreas results in defects in epithelial branching and outgrowth
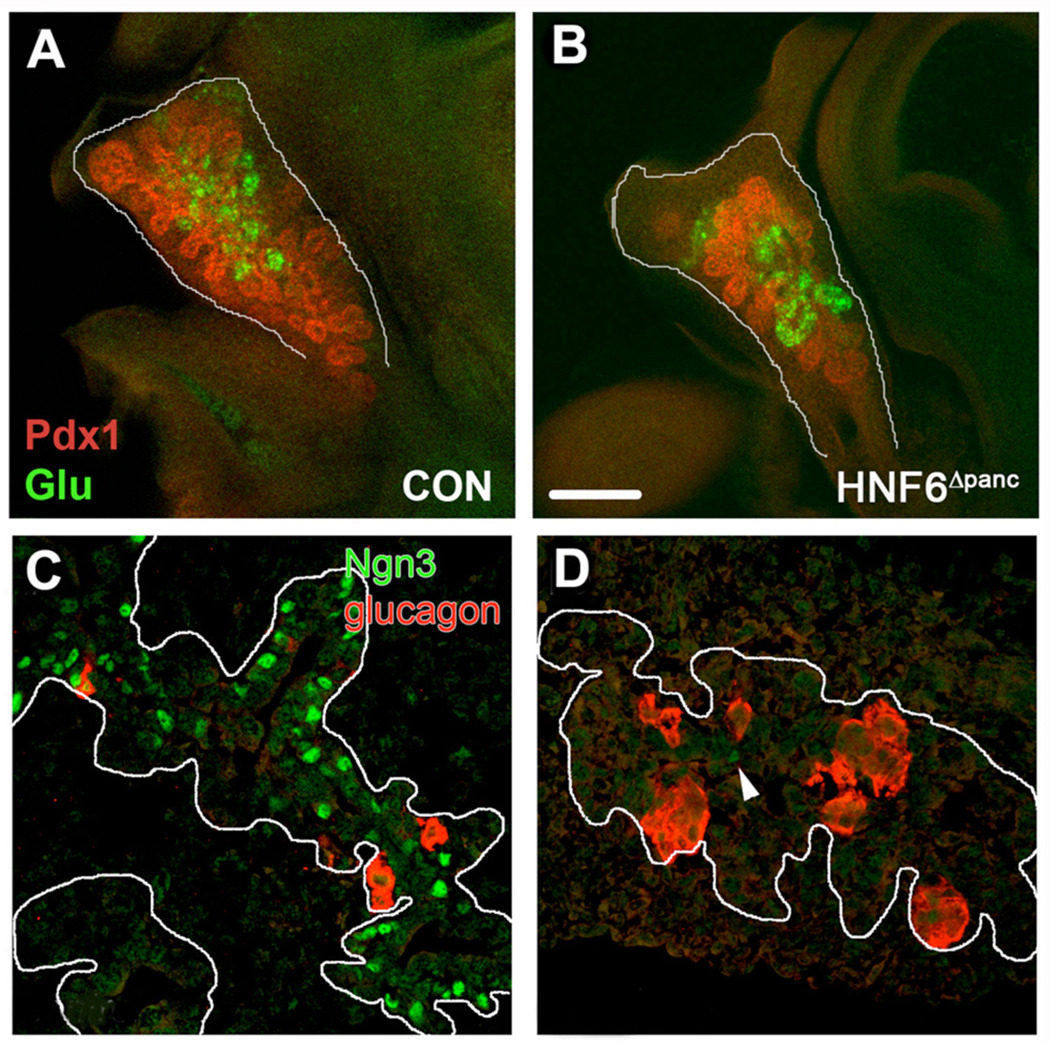
The embryonic phenotype of HNF6Δpanc pancreata, while having some similarities to global HNF6 null mice (Jacquemin et al., 2000) also exhibited distinct differences. Consistent with the global HNF6 knockout, at e14.5 the epithelium of HNF6Δpanc pancreata displayed less branching (compare Figure 3A and B) and failed to fully extend toward the distal surrounding mesenchyme compared with controls. The mutant pancreatic epithelium never fully filled the mesenchyme postnatally (data not shown), suggesting that e14.5 HNF6Δpanc pancreata were not simply developmentally delayed in their distal outgrowth. We observed a dramatic decrease in Ngn3-positive cells in HNF6Δpanc pancreata at e13.5 (Figure 3D) similar to what was reported for HNF6−/− mice (Jacquemin et al., 2000). Previous studies indicated that the number of glucagon-expressing cells was reduced by 85% in HNF6−/− embryos at e12.5 and e15.5, although the total number of α cell clusters was not different in the mutants (Jacquemin et al., 2000). We did not observe any reduction in glucagon-positive cell clusters at e13.5 or e14.5 (Figure 1B and Figure 3B,D), although the number of α and β cells was reduced at e18.5 (data not shown). The dramatic reduction in endocrine cell number resulted in elevated blood glucose levels in HNF6Δpanc mice at P2 and 3.5 weeks of age (Supplemental figure 1). No phenotypic differences were observed between males and females. At six weeks of age, random blood glucose levels in some HNF6Δpanc mice were greater than 500 mg/dl (the limit of detection of the glucometer), so fasting glucose concentrations were examined instead. HNF6Δpanc mice displayed a significantly higher fasting blood glucose level than controls (Supplemental figure 1). Thus, HNF6Δpanc mice show impaired glucose homeostasis soon after birth and this worsens with age.
Figure 3. Defective epithelial branching and endocrine specification in HNF6Δpanc pancreata.
Control (A) and HNF6Δpanc (B) pancreata were analyzed in whole mount at e14.5 for Pdx1 (red) and glucagon (green) expression. Pdx1-positive immunolabeling showed that there was less branching of the epithelium in HNF6Δpanc pancreata as compared with controls. (C,D) Localization of Ngn3 (green) and glucagon (red) in control (C) and Hnf6Δpanc (D) pancreata at e13.5. White lines delineate endocrine cords. Arrowhead in D indicates the single Ngn3+ cell detected in this field of view.
Loss of HNF6 in Ngn3-expressing cells has no effect on β cell terminal differentiation
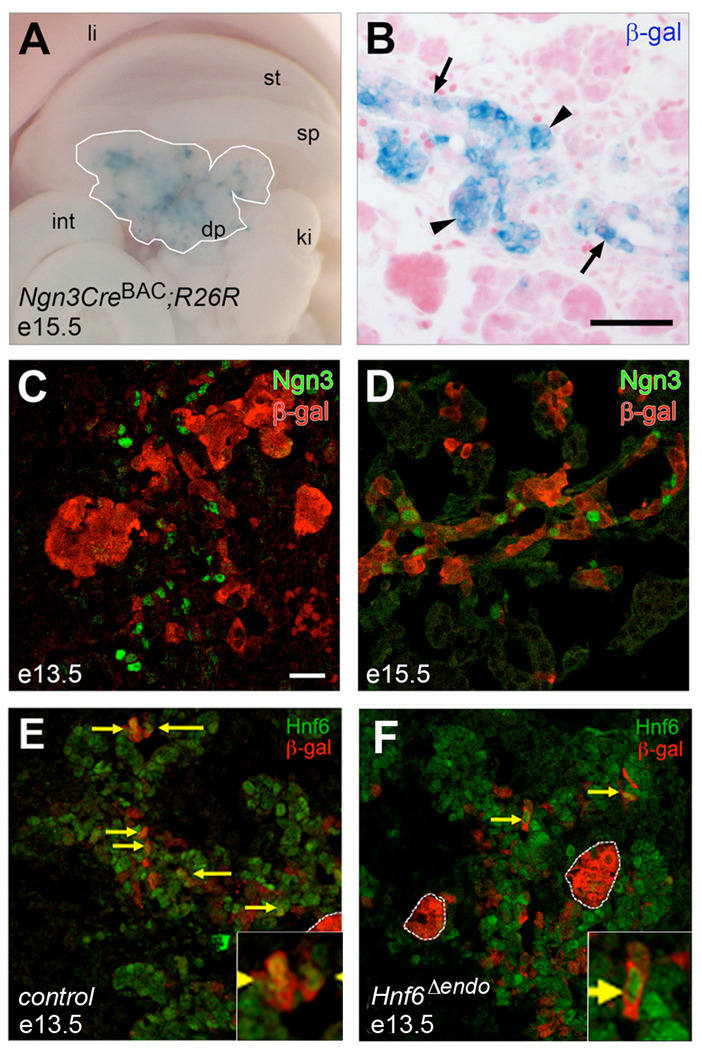
The expression data described above provides support for a model wherein HNF6 is required only early in development to initiate the endocrine program (via activation of Ngn3), but is not necessary for further steps of maturation or terminal differentiation. To directly test this hypothesis, we conditionally inactivated HNF6 in cells that had entered the proendocrine lineage using Ngn3CreBAC (Schonhoff et al., 2004). The pattern of recombination mediated by this Cre transgene recapitulates endogenous Ngn3 expression in both the endocrine pancreas and the enteroendocrine cells of the intestine, with minimal (>1%) non-endocrine recombination. Using the R26R reporter strain (Soriano, 1999), we detected recombination [as visualized by X-gal staining to detect β-galactosidase (β-gal)] in developing endocrine clusters at e15.5 and a subset of cells within the endocrine cords (Figure 4A,B). β-gal-positive cells were detected as early as e13.5 (Figure 4C) and by e15.5, and nearly every Ngn3-positive cell expressed β-gal (Figure 4D). Many β-gal-positive cells did not express Ngn3, most likely due to the fact that these cells had already begun to differentiate and were no longer expressing Ngn3, which is not expressed in hormone-positive cells (Gradwohl et al., 2000). These data indicate that the Ngn3CreBAC transgene is able to direct cell-specific recombination early and efficiently within the developing pancreas.
Figure 4. Ngn3CreBAC-mediated recombination efficiently inactivates HNF6 in multipotent pancreatic progenitors.
Ngn3CreBAC;R26R (control) e15.5 pancreas in whole mount (A) and cross-section (B) stained with X-gal to detect recombination and counterstained with eosin. White line in A delineates dorsal pancreas. X-gal staining was detected in cells in the endocrine cords (arrows) and in endocrine clusters that have migrated away from the cords (arrowheads). Recombination was detected in control pancreata as early as e13.5 (C) by co-localization of β-gal (red) with Ngn3 (green) on confocal 1µm optical sections; by e15.5 (D), recombination had occurred in nearly all Ngn3+ cells. Co-localization of β-gal (red) and HNF6 (green) at e13.5 indicated that the majority of cells had lost HNF6 protein in HNF6Δendo pancreata (F) as compared to wild type (E). Arrows indicate co-expression of β-gal and HNF6. li, liver; st, stomach; sp, spleen; int, intestine; dp, dorsal pancreas; ki, kidney. Scale bars represent 10 µm (A, B) or 20 µm (C–F).
Because Ngn3+/HNF6+ cells are a transitory population, we were concerned that the Ngn3CreBAC transgene might not mediate recombination of the HNF6flox allele early enough to produce an effect on the endocrine lineage. Ngn3 expression begins at e9.5 and reaches a peak at around e15.5 (Gradwohl et al., 2000; Jensen et al., 2000; Schwitzgebel et al., 2000). We find a significant proportion (~70%) of Ngn3-expressing cells co-express HNF6 at e13.5. Analysis of HNF6 and β-gal expression at e13.5 (Figure 4E,F) revealed that HNF6 was co-expressed in approximately 79.3 ± 5.5% of β-gal-positive cells in HNF6+/+;Ngn3CreBAC;R26R (control) pancreata (n = 266 cells). In comparison, only 27.5 ± 4.8% of β-gal-positive cells (n = 202 cells) co-expressed HNF6 in HNF6flox/flox;Ngn3CreBAC;R26R (HNF6Δendo) pancreata. These data indicate that the Ngn3CreBAC is active prior to the normal timing of HNF6 down-regulation in the endocrine lineage and successfully inactivates HNF6 in a large percentage of the transitory Ngn3+/HNF6+ cell population. The remaining β-gal-positive cells that still express HNF6 at this time point may represent a longer half-life of the HNF6 protein, or alternatively could reflect the delayed timing in recombining the two HNF6 floxed alleles compared with just a single R26R allele.
To determine the consequences of endocrine-specific HNF6 inactivation on subsequent β cell differentiation and function, we examined whether HNF6−/− cells incorporated into both β and α cell lineages, and whether or not mutant cells expressed known markers of mature β cells. The majority of X-gal-positive cells in both control and HNF6Δendo pancreata co-expressed either insulin or glucagon (Supplemental figure 2). To further assess β cell differentiation, we analyzed expression of Pdx1, MafA, and the glucose transporter GLUT2 (Supplemental figure 2). Localization of each of these proteins was comparable to controls.
β cell function was assessed in vivo by performing IP-GTT at eight weeks of age. Fasting blood glucose, plasma insulin levels, and glucose tolerance profiles were indistinguishable between control and HNF6Δendo animals (data not shown). In addition, total pancreatic insulin content at either P1 or eight weeks of age was similar between HNF6Δendo and control animals (data not shown). Taken together, these data indicate that inactivation of HNF6 specifically in cells presumed to be committed to the endocrine lineage does not prevent β cell terminal differentiation or impair β cell function.
Sustained HNF6 is necessary to allocate appropriate numbers of progenitor cells to the endocrine lineage
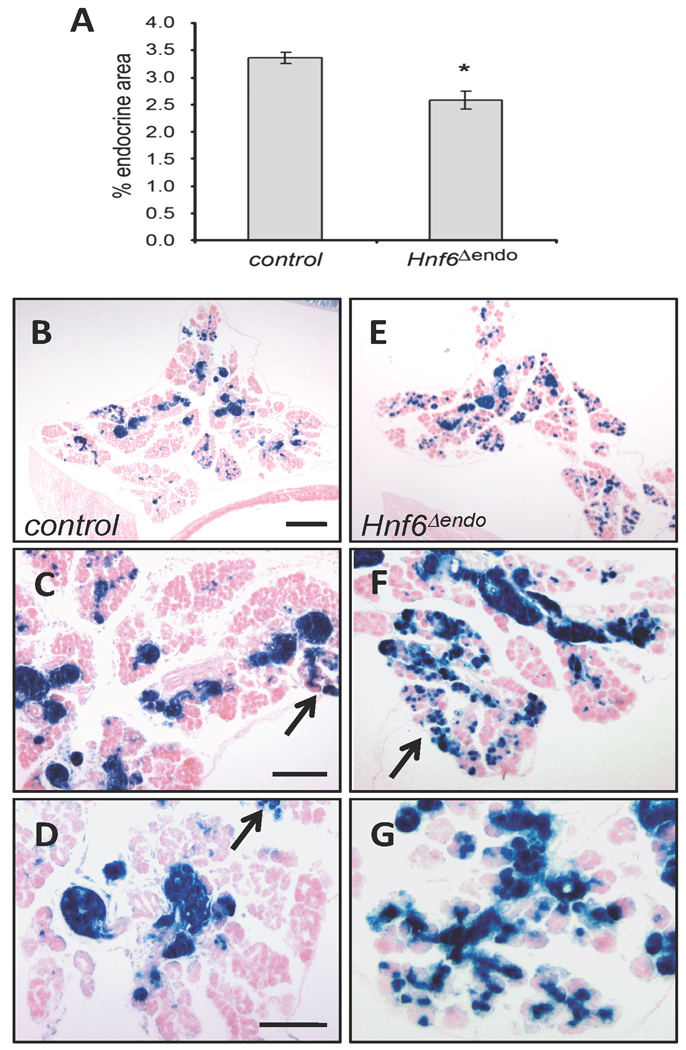
Morphometric analyses of P2 pancreata revealed a small, but statistically significant decrease in total endocrine area in HNF6Δendo pancreata as determined by synaptophysin immunohistochemistry (Figure 5) as compared to age-matched controls. This difference did not negatively impact glucose homeostasis even when mice were placed on a high fat diet to induce peripheral insulin resistance (data not shown). Morphometric examination of the lineage label (Figure 5B–E) revealed that the number of non-endocrine β-gal-expressing cells in HNF6Δendo pancreata was increased over controls. Recombination in Ngn3CreBAC;R26R mice was previously reported in a small percentage of acinar (0.5–1.8%) or ductal (1–5%) cells (Schonhoff et al., 2004). Similarly, at P2 we observed that 2.48 ± 0.6% of X-gal-expressing cells are non-endocrine in control mice (n = 5; HNF6+/+;Ngn3CreBAC;R26R or HNF6fl/+;Ngn3CreBAC;R26R). Interestingly, HNF6Δendo pancreata have an approximate six-fold increase in the percentage of X-gal-expressing cells incorporated into non-endocrine tissue (14.60 ± 3.2%; n = 3, p = 0.06). X-gal-expressing cells were detected within both acinar and ductal structures, and did not express endocrine hormones (data not shown).
Figure 5. Inactivation of HNF6 in Ngn3-expressing cells results in increased allocation of progenitor cells to the exocrine lineage.
(A) Morphometric analysis of percent endocrine area (synaptophysin+ cells) in control (n = 3) and HNF6Δendo (n = 5) pancreata at P2. *p = 0.007, as determined by Student’s t-test; error bars represent standard error of the mean. (B, C) X-gal staining of P2 pancreata indicates a greater proportion of recombined cells incorporate into the exocrine compartment in HNF6Δendo (C) pancreata as compared to age-matched controls (B). X-gal+ cells in HNF6Δendo pancreata are detected in acinar (D) and ductal (E) structures (arrows). Is, islet. Scale bars represent 200 µm (B, C) or 50 µm (D, E).
Defective duct formation in HNF6Δpanc mice
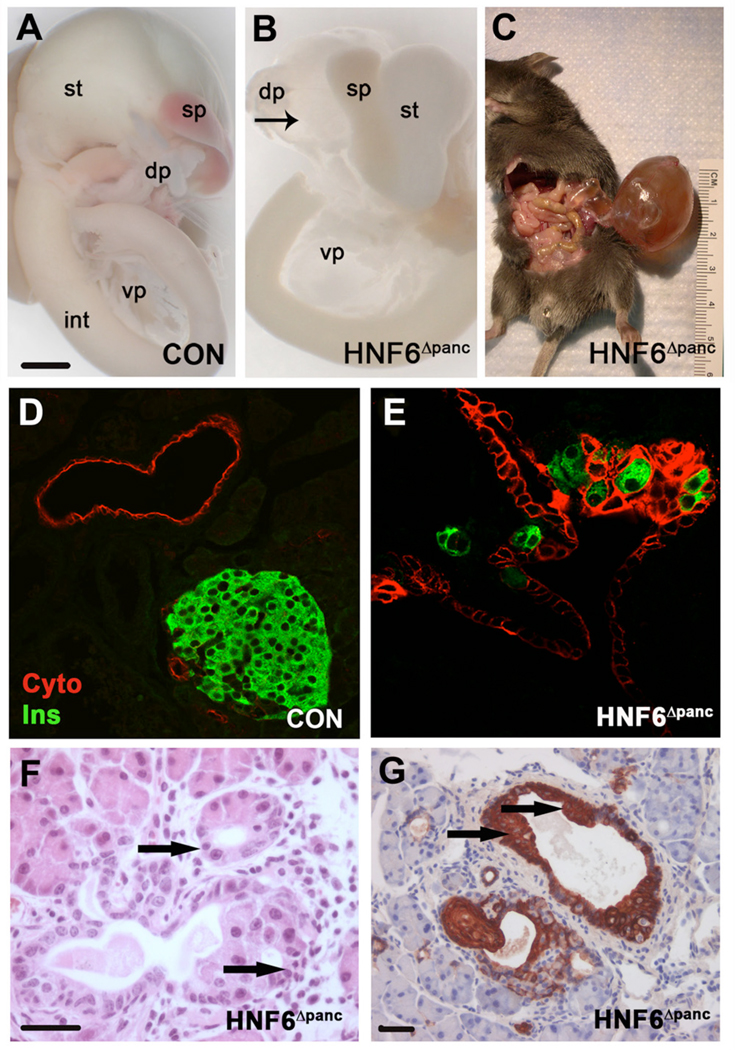
Liver defects in HNF6−/− result in the death of 75% of these animals between P1 and P10 (Jacquemin et al., 2000). Since HNF6 expression in liver is unaltered in HNF6Δpanc animals (data not shown), these animals should allow for analysis of HNF6 pancreatic function postnatally. Beginning at e18.5, HNF6Δpanc pancreata developed a single, fluid-filled cyst lined by a cuboidal epithelium in the region of the tail and body (dorsal pancreas; Figure 6B). The cysts progressively increased in size such that by six weeks of age, 62.5% of mutant mice (n = 16) were observed to have a cysts averaging 4.19 ± 2.19 cm3 (Figure 6C). Although, the large cyst was mainly located in the dorsal pancreas, dilated ductal epithelium was observed throughout the whole pancreas. Only a small portion of acinar tissue was detectable in mutant animals in close association with the cystic epithelium, whereas animals lacking cysts displayed only a minimally branched dorsal pancreas (data not shown) and/or possessed twisted irregular ducts and calcifications (data not shown). The epithelium of the dilated ducts in HNF6Δpanc pancreata was multilayered (Figure 6E,G), in contrast to the simple cuboidal to low-columnar epithelium of normal pancreatic ducts (Figure 6D). Glucagon-(data not shown), insulin- (Figure 6E), or Pdx1-positive cells (data not shown) were detected within the multilayered epithelium of HNF6Δpanc pancreata, however no definitive islets were observed.
Figure 6. Ductal abnormalities in HNF6Δpanc animals.
(A) Wild type digestive organs at e18.5. (B) Gross analysis of digestive organs from HNF6Δpanc animals showed cystic structures in pancreata by e18.5 (arrow). Pancreatic cysts progressively increased in size as the animals aged (C). (D) Antibodies against cytokeratin (red) outline the simple cuboidal epithelium in wild type pancreatic ducts. In HNF6Δpanc pancreata (E) the epithelium was multilayered, and insulin-positive cells (green) were found abnormally within the multilayered epithelium. H&E staining of HNF6Δpanc pancreata suggested acinar-to-ductal metaplasia (F, arrows). Cytokeratin labeling also revealed regions of squamous cell metaplasia in HNF6Δpanc pancreata (G, arrows). dp: dorsal pancreas, int: intestine, sp: spleen, st: stomach, vp: ventral pancreas. Scale bars: in A, 100 mm for A and B; in F, 20 µm for D–F; in G, 20 µm.
To examine whether the multilayered epithelium in HNF6Δpanc mice was actively proliferating, BrdU was injected 6 h before mice were sacrificed. Anti-pan cytokeratin antibody was used to label ductal epithelial cells. A significant increase in ductal cell proliferation was observed in HNF6Δpanc mice both at e17.5 (control, n = 4: 4.02% versus HNF6Δpanc, n = 4: 7.79%, p≤0.01) and six weeks of age (control, n = 5: 0.81% versus HNF6Δpanc, n = 4: 4.66%, p≤0.01).
Morphological and molecular characteristics of pancreatitis in HNF6Δpanc animals
Acinar-to-ductal metaplasia was observed in all HNF6Δpanc mice as early as P2 (Figure 6F). Squamous cell metaplasia was also observed in the ductal epithelium (Figure 6G). All HNF6Δpanc pancreata (from 3–6 weeks of age) displayed morphological characteristics of pancreatitis, including profound atrophy, loss of acinar tissue, and acinar inflammation that was mostly lymphocytic (Figure 7B). Pancreatic ducts were highly tortuous (Supplemental figure 3), containing secretions, hemosiderin-laden macrophages, and neutrophils in the lumen (data not shown). The dilated ducts displayed periductal inflammation (Figure 7B,C), hemorrhage (Figure 7D) and fibrosis (Figure 7F). Dystrophic calcifications were present in adjacent fat (Supplemental figure 3), indicative of previous fat necrosis. Some HNF6Δpanc mice displayed additional characteristics of pancreatitis including (data not shown): neutrophils in peripancreatic fat; neutrophils in the lumen and ductal epithelium of smaller ducts; intraparenchymal mixed inflammatory infiltrate with neutrophils and lymphocytes, with areas of acinar injury; neutrophils scattered in periductal stroma; large ducts with erosion and acute inflammation.
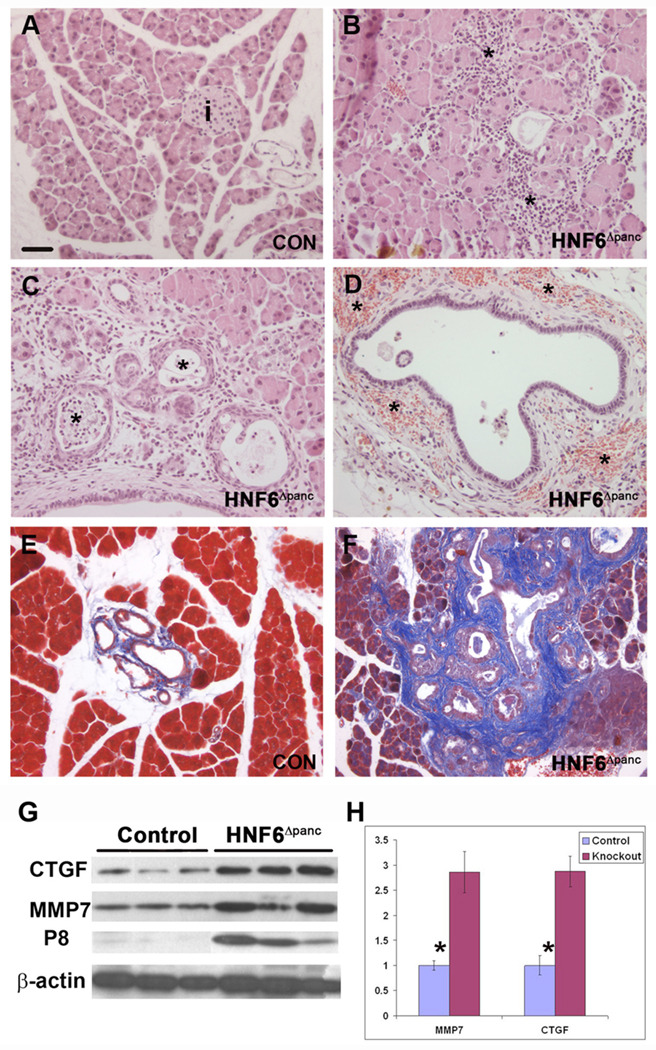
Figure 7. Pancreatitis in HNF6Δpanc animals.
(A) Wild type adult pancreas section (i: islet). (B–D) Islet structures were undetectable in adult HNF6Δpanc pancreata. Morphological changes characteristic of pancreatitis included: acinar inflammation which is mostly lymphocytic (B, asterisks), periductal inflammation (C), neutrophils in the lumen (C, asterisks), and periductal hemorrhage (D, asterisks). Trichrome staining of pancreata indicated periductal fibrosis (blue) in HNF6Δpanc animals (F) in comparison with controls (E). (G, H) Western blot analysis of total pancreatic protein from control and HNF6Δpanc animals revealed a 2.8-fold increase in both CTGF and MMP7 expression in HNF6Δpanc animals in comparison with controls. Protein levels were normalized to β-actin using densitometry. Western blot also revealed a significant increase in p8 in HNF6Δpanc pancreata, with very little p8 expression observed in control pancreata (G). Scale bar: in A, 20 µm for A–F.
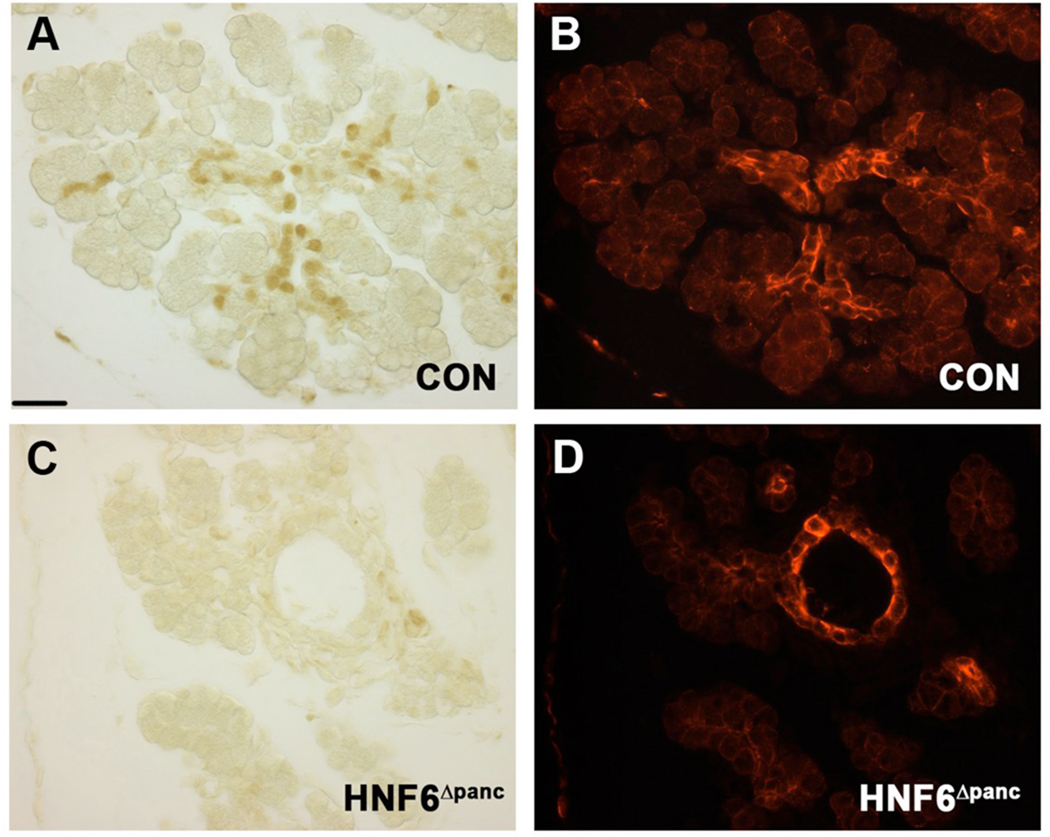
Previous studies have shown a 4.5-fold increase in expression of the secreted protein, CTGF, in human chronic pancreatitis (di Mola et al., 1999). CTGF is involved in several fibrotic diseases and plays a critical role in fibrogenesis and tissue remodeling after injury (di Mola et al., 2002). A recently identified gene, p8/(nuclear protein 1; Nupr1), is also up-regulated in human pancreatitis but is barely detectable in normal pancreata (Mallo et al., 1997). p8/Nupr1 is induced in acinar cells during the onset of chronic pancreatitis as part of the self-defense mechanism against proapoptotic insults (Motoo et al., 2001), and is also up-regulated in acute pancreatitis (Mallo et al., 1997). In addition, matrix metalloproteinase-7 (MMP7, matrilysin), a member of the MMP family of zinc-dependent extracellular proteases is associated with acinar-to-ductal metaplasia and pancreatitis (Crawford et al., 2002). Since HNF6Δpanc mice showed pancreatitis and fibrosis surrounding the ducts, we examined CTGF expression in HNF6Δpanc and control mice. Immunohistochemistry revealed very low levels of CTGF expression in the ducts of normal adult pancreata with a significant increase in CTGF expression in the ducts of HNF6Δpanc animals (Supplemental figure 4). Western blot analysis of total pancreatic protein extract confirmed that HNF6Δpanc pancreata displayed a 2.8-fold increase in both CTGF and MMP7 (Figure 7G,H). We also observed a significant increase in p8 in HNF6Δpanc pancreata, with very little p8 expression observed in controls as expected (Figure 7G).
HNF6Δpanc mice exhibited defects in primary cilia and decreased Prox1 expression
The primary cilia in the pancreatic ducts are immotile and have been proposed to function as chemo- or mechano-sensors (Davenport and Yoder, 2005). A connection between cilia defects and pancreatitis is suggested by the fact that lesions reminiscent of pancreatitis are found in approximately 10% of patients suffering from the dominant form of polycystic kidney disease (Başar et al., 2006), and the absence of pancreatic cilia in mice results in lesions that resemble those found in patients with pancreatitis or cystic fibrosis (Cano et al., 2006). Using antibodies against either β-tubulin or Arl13b (Caspary et al., 2007), primary cilia that protruded into the ductal lumen were detected in control embryonic (Supplemental figure 5) and adult pancreatic duct epithelia (data not shown). In HNF6Δpanc animals, cilia were detectable in ducts displaying a normal morphology at e18.5 (Supplemental figure 5C,D), although these were fewer in number and did not protrude into the duct lumen. However, dilated ducts in HNF6Δpanc pancreata showed a dramatic decrease in cilia number at e18.5 (Supplemental figure 5E,F) and six weeks (data not shown).
The divergent homeodomain transcription factor Prox1 is a crucial regulator of mouse pancreas organogenesis (Wang et al., 2005). Prox1 is expressed in nearly all pancreatic progenitor cells early in development but becomes restricted to islets and ducts at late-gestation (Wang et al., 2005). Pancreas-wide inactivation of Prox1 also results in dilated pancreatic ducts and acinar-to-ductal metaplasia, similar to the phenotype observed in HNF6Δpanc mice, however there is no change in HNF6 expression in embryos lacking pancreatic Prox1 (Dr. Beatriz Sosa-Pineda, personal communication). Thus, we hypothesized that HNF6 might act upstream of Prox1. At e18.5, Prox1 is highly expressed in pancreatic ducts in the control animals (Figure 8A,B). The number of Prox1-positive cells was significantly reduced in HNF6Δpanc pancreata (Figure 8C,D).
Figure 8. Decreased Prox1 in HNF6Δpanc ducts.
At e18.5, Prox1 expression (A, brown) was detected in pancreatic ducts labeled by cytokeratin (B, red) in control animals. A significant decrease in Prox1 expression in pancreatic ducts was observed in HNF6Δpanc at the same time (C,D). Scale bar: 20 µm. A and B, C and D are the same sections. Pictures were taken sequentially either under bright field or with epifluorescence.
Discussion
Precedent exists in the pancreas for transcription factors to have multiple functions at different developmental time points. For example, Pdx1 controls early steps of pancreatic bud outgrowth and proliferation (Jonsson et al., 1994; Offield et al., 1996), but it also regulates later steps of acinar cell differentiation (Hale et al., 2005), embryonic β cell proliferation (Gannon et al., 2008), and β cell function (Ahlgren et al., 1998; Holland et al., 2002) as its expression becomes gradually down-regulated in the pancreatic epithelium and enhanced in β cells.
The present study addresses the specific consequences of HNF6 inactivation in early pancreatic buds or committed endocrine cells on both pancreas development and postnatal pancreas function. The present study confirms that HNF6 plays an essential role in both endocrine and duct development, and is the first to demonstrate that loss of HNF6 activity can induce pancreatitis and cellular changes associated with pancreatic neoplasia.
Comparison of HNF6 mutant phenotypes reveals similarities and differences
Although the number of glucagon-positive cells appeared unchanged at e14.5 in HNF6Δpanc embryos, we did observe a decrease in Ngn3 expression in HNF6Δpanc at e14.5, and a subsequent reduction in the number of insulin- and glucagon-positivẹcells, at e18.5 and P2, similar to what was reported for HNF6−/− null animals at early postnatal stages. Thus, in both models, the reduction in endocrine cells is most likely due to the reduction in the number of Ngn3-expressing cells.
A difference between HNF6−/− and HNF6Δpanc animals was observed in early postnatal glucose homeostasis. Previous analysis of HNF6−/− neonates revealed decreased plasma glucose at P4 (Jacquemin et al., 2000); however, HNF6Δpanc animals showed significantly elevated glucose levels at P2. HNF6 is highly expressed in the liver, and previous studies have shown that HNF6 stimulates transcription of both glucokinase (Lannoy et al., 2002) and glucose-6-phosphatase in the liver (Streeper et al., 2001). The observed hypoglycemia in HNF6 null neonates could be caused by loss of HNF6 in the liver resulting in defects in glycogen storage, glycogenolysis or gluconeogenesis. The early hyperglycemia observed in HNF6Δpanc neonates is revealed in the absence of the confounding effects from loss of HNF6 in the liver.
In the original description of the HNF6−/− phenotype, islets were observed in the animals that survived the neonatal period and lived to adulthood, but these islets had a disrupted architecture and impaired function (Jacquemin et al., 2000). In contrast, HNF6Δpanc adult pancreata contained individual insulin-positive cells or small endocrine clusters of less than 8 cells, but no definitive islets. The cause of this discrepancy is currently unclear, but may be due to the difference in genetic background of the mice used in the two studies.
Ductal defects in HNF6Δpanc animals include pancreatitis and metaplasia
The vast majority of HNF6 global null animals die between P1 and P10 (Jacquemin et al., 2000). Restriction of HNF6 inactivation to the pdx1-expression domain using Cre-lox technology permitted an analysis of HNF6 function in all mutant animals after birth. Although cystic structures were observed in pancreata of HNF6 null animals (Jacquemin et al., 2000), this is the first study in which pancreatitis has been described due to HNF6 inactivation in the pancreas.
CTGF exhibits a diverse range of cellular functions including cell adhesion, cell migration, potentiation of growth factor–induced proliferation, inflammation, fibrosis, tumor growth, and tumor metastasis (Dornhofer et al., 2006). Its expression strongly correlates with increased pancreatic fibrosis (di Mola et al., 1999). Thus, the fibrosis observed in HNF6Δpanc pancreata may be caused by increased CTGF. Interestingly, we have found that over-expression of HNF6 in the developing pancreas leads to a decrease in CTGF expression (Wilding Crawford et al., 2008). Taken together, our results suggest that HNF6 negatively regulates CTGF expression, although it is currently unknown whether this is due to direct binding of HNF6 to the CTGF promoter.
The increase in p8 observed in HNF6Δpanc pancreata further supports the development of pancreatitis in these animals. p8/Nupr1 is a transcriptional cofactor expressed at only low levels in normal pancreata but is induced in the initial phases of pancreatitis and is associated with acinar regeneration and proliferation (Mallo et al., 1997), and more recently, β cell proliferation (Yanagita et al., 2006). Thus, induction of p8 expression in HNF6Δpanc pancreata may be indicative of mutant acinar cells mounting a protective response to cellular stress (Motoo et al., 2001), and/or stimulate proliferation of acinar cells and the few β cells present.
Although bile duct blockage is known to be a cause of pancreatitis in humans, and HNF6 is deleted from the common bile duct in our model (data not shown), we do not believe that loss of HNF6 from the bile duct is the cause of the observed pancreatitis. The bile ducts from HNF6Δpanc animals were slightly smaller, but were not distended or blocked (data not shown). Instead, we hypothesize that the defects in primary cilia are the main cause for the ductal cysts and metaplasia present in HNF6Δpanc pancreata. Primary cilia are found on most vertebrate epithelial cells and play a negative role in epithelial cell proliferation (Pugacheva et al., 2007) by acting as chemo- or mechano-sensors particularly in liver, kidney and pancreas (Davenport and Yoder, 2005). Primary cilia function is required for the maturation and maintenance of proper tissue organization in the pancreas (Cano et al., 2004): reductions in cilia in the Oak Ridge polycystic kidney (orpk) mutant mouse results in abnormal tubular structures and massive acinar cell loss, similar to what we observed in HNF6Δpanc pancreata. Cysts were also observed in HNF6−/− embryos, and were associated with abnormal development of primary cilia in duct cells and reduced expression of genes involved in polycystic disease, namely those encoding HNF-1β and cilium-associated proteins polyductin, fibrocystin, and cystin (Pierreux et al., 2006).
The homeobox-containing transcription factor Prox1, the mammalian homologue of the Drosophila gene prospero, is expressed in nearly all pancreatic progenitor cells early in development (Wang et al., 2005). Our immunohistochemical analyses suggest that HNF6 functions upstream of Prox1, and may regulate pancreatic duct morphogenesis in part through Prox1.
Both acinar-to-ductal and squamous cell metaplasia were observed in HNF6Δpanc pancreata. Patients with ductal metaplasia arising in the setting of chronic pancreatitis have a 16-fold increase in relative risk for pancreatic ductal adenocarcinoma (PDAC) (Lowenfels et al., 2000). MMP7 is expressed in the epithelium of pre-malignant lesions in multiple glandular tissues, including breast, prostate, and intestine (Powell et al., 1996) and is thought to influence the initiation and maintenance of metaplastic events in the pancreatic epithelium (Crawford et al., 2002). We detected elevated MMP7 levels in HNF6Δpanc animals. In addition, elevated CTGF levels have been detected in PDAC, and inhibition of CTGF with a human monoclonal antibody has been shown to reduce primary and metastatic tumor growth in a mouse model of PDAC (Dornhofer et al., 2006). Thus, taken together, these results suggest that loss of HNF6 in the pancreas may cause increased risk of PDAC or more rare forms of pancreatic cancer such as squamous cell carcinoma.
Maintained HNF6 is required to ensure proper allocation of progenitors to the endocrine lineage
We found that β cell terminal differentiation proceeds normally in cells that have lost HNF6 subsequent to activation of the Ngn3 promoter. These results are consistent with the protein expression pattern of HNF6, wherein HNF6 appears to be rapidly down-regulated from Ngn3-expressing cells as they are specified to the endocrine lineage. These data strongly support the hypothesis that HNF6 is not necessary at subsequent steps of endocrine differentiation, but functions specifically at the initial steps of endocrine specification. Indeed, our previous studies showing that maintenance of HNF6 in the endocrine lineage is incompatible with proper β cell differentiation and function (Gannon et al., 2000; Tweedie et al., 2006) also support this model of HNF6 activity in the pancreas.
Altered allocation of Ngn3-expressing cells to non-endocrine lineages in HNF6Δendo pancreata suggests that Ngn3-expressing cells retain some degree of plasticity during the initial stages of lineage commitment. Pro-endocrine progenitors in HNF6Δendo mice are exposed to a reduced duration of HNF6/Ngn3 co-expression and thus a reduced number of cells that have activated Ngn3 remain committed to the endocrine lineage. This suggests that a threshold level of Ngn3 must be attained to irreversibly commit cells to an endocrine fate. Our data are consistent with a recent study in which Ngn3 expression was induced during specific developmental time windows in the context of the Ngn3 null background (Johansson et al., 2007). The authors suggest that the pancreatic epithelium responds in a cell-autonomous manner to threshold levels of Ngn3 that determine the number of cells shunted into the endocrine pathway. Thus, HNF6 may be necessary to maintain sufficient levels of Ngn3 within presumptive endocrine progenitors, such that they are ensured of an endocrine fate. That HNF6 is rapidly down-regulated in committed endocrine cells also suggests that some signal, specific to the endocrine cell lineage, is involved in silencing HNF6. Further studies identifying an HNF6 repressor will be necessary to confirm or disprove this hypothesis.
Experimental Procedures
Generation of mice
To generate mice with a loxP-containing HNF6 allele (HNF6flox), we constructed a loxP-FRT HNF6flox-neo targeting vector using BAC recombineering (Liu et al., 2003). To avoid disrupting potential regulatory regions within the HNF6 locus, mouse and human HNF6 gene homology was compared (using http://genome.ucsc.edu) and loxP sites introduced flanking the cut-domain in regions of less than 50% sequence conservation (Figure 1A). A clone containing HNF6 genomic DNA was obtained from a mouse RPCI-22 BAC library (BACPAC Resources, Children’s Hospital Oakland, CA). The HNF6 targeting construct also contained a neomycin resistance cassette (neoR) located 5’ to the second loxP site and flanked by FRT sites to facilitate deletion by Flp recombinase, and a Herpes Simplex Virus-thymidine kinase (TK) cDNA located outside of the HNF6 gene homology region. The HNF6flox-neo targeting vector was electroporated into mouse TL1 ES cells (129S6), which were propagated on mouse embryo fibroblast feeder cells and selected for neoR (G418) and TK (gancyclovir) resistance (Hogan, 1994). ES cell colonies containing the HNF6flox-neo targeted allele were determined by Southern blot analysis with 5' and 3' external probes (Figure 1A, Supplemental figure 6). Using a 5' genomic probe and SpeI/EcoRV digestion, the HNF6flox allele was detected as a smaller molecular weight DNA fragment because of the EcoRV site engineered 1.7kb upstream of the endogenous EcoRV site (Supplemental figure 6A). Using a 3' genomic probe and SacII/HpaI digestion (Figure 1A), the HNF6flox allele was detected as a larger band due to the 2kb PGK-neoR positive selection cassette (Supplemental figure 6A). 450 HNF6flox-neo heterozygous ES cell clones were analyzed by Southern blot; 45 clones (10%) were correctly targeted. Two independent ES cell clones with the appropriate HNF6flox-neo-targeted locus were used to generate chimeric mice by injection into mouse C57BL/6 blastocysts. High percentage chimeras were bred with C57BL/6 wild type mice, and agouti offspring were screened for heterozygosity for the targeting construct. The neoR cassette was removed in vivo by breeding HNF6flox-neo/+ animals to human ACTB (β-actin)-driven Flpe transgenic mice (Rodriguez et al., 2000) that mediates excision of FRT-flanked DNA in somatic and germ cells. HNF6flox/+ mice were used to generate mice homozygous for the floxed allele (HNF6flox/flox). Controls for these studies included both HNF6flox/+;pdx1-Cre and HNF6flox/flox mice; no phenotypic differences were observed between the two genotypes.
Genotyping of HNF6flox/flox mice was by PCR amplification using primers that flank the first loxP site [5’-GTCGTCGACCTCTCTCCTGTCTCCCTCAGTATCC and 5’-ATAAGCGGCCGCCCTCCCTCTCTCTTTCCATC]. These primers amplify a 550 bp band in the endogenous allele and a 590 bp band in the floxed allele (Supplemental figure 6B).
Two Cre driver lines were used for these studies. The presence of each Cre transgene was determined by PCR using primers for Cre (Zhang et al., 2006). In the pdx1-Creearly line, 5.5 kb of the pdx1 5’ regulatory region drive early pancreas-wide Cre expression (Gu et al., 2002). HNF6flox/flox;pdx1-Cre (HNF6Δpanc) mice were sacrificed by three months of age, due to severe diabetes and large pancreatic cysts. The Ngn3CreBAC transgene is comprised of a bacterial artificial chromosome (BAC) spanning approximately 180 kb of the Ngn3 locus, with a nuclear-localized Cre recombinase inserted into the translational start site of Ngn3 (Schonhoff et al., 2004). Recombination was monitored using the ROSA26 (R26R) reporter strain (Soriano, 1999).
An inbred hybrid line (B6D2; Jackson Labs) was used for endogenous HNF6 protein expression studies. For embryonic analyses, the morning of the vaginal plug was considered e0.5. Mice were maintained on mouse chow 5015 (24% fat, 57% carbohydrates, 19% protein; TestDiet) ad libitum. All mouse studies were performed in accordance with the Vanderbilt Institutional Animal Care and Use Committee guidelines under the supervision of the Division of Animal Care.
Histology and immunolabeling
Dissected tissues were fixed (4% paraformaldehyde, 4°C, 2h), dehydrated in an ethanol series, embedded in paraffin, and sectioned to a thickness of 5 µm. Tissues for cryosectioning were fixed overnight 4°C, incubated in 30% sucrose overnight at 4°C, embedded in OCT compound (VWR Scientific, West Chester, PA), and 5 μm sections were cut on a Leica CM 3050 S cryostat.
Paraffin sections were deparaffinized in Citrisolv (Fisher) and rehydrated in a decreasing ethanol series to distilled water. Hematoxylin and eosin (H&E) staining was as described (Offield et al., 1996). Masson trichrome staining was performed following the manufacturer’s instruction (Sigma). Primary antibodies were used at the following dilutions: guinea pig anti-insulin (1/1000, Linco), rabbit anti-glucagon (1/1000, Linco), rabbit anti-MafA (1/500, Calbiochem), rabbit anti-GLUT2 (1/500, a gift of Bernard Thorens, Univ. of Lausanne), guinea pig anti-Pdx1 (1/1000, a gift of Chris Wright, Vanderbilt Univ.), mouse anti-synaptophysin (1/500, Chemicon), guinea-pig anti-Ngn3 (1/1000, a gift from Dr. Maike Sander, Univ. of California at Irvine), rat anti-BrdU (1/400, Accurate Chem Scientific Corp.), rabbit anti-CTGF (1/1000, a gift from Dr. David Brigstock, The Ohio State Univ.), rabbit anti-cytokeratin (1/1000, Dako), mouse anti-acetylated β-tubulin (1/5000, Sigma), rabbit anti-Arl13b/Hennin (1/500, a gift from Dr. Tamara Caspary, Emory University), rabbit anti-Prox1 (1/500, Angiobio Co.). Detection of MafA required Tris/EGTA antigen retrieval, detection of Pdx1 and GLUT2 required sodium citrate antigen retrieval as previously described (Tweedie et al., 2006); detection of cytokeratin and Arl13b/Hennin required antigen retrieval with proteinase-K diluted 1:1000 (Dako) for 5 min prior to primary antibody.
Frozen sections were thawed to room temperature and permeabilized in 0.1% Triton in PBS for 45 minutes. Primary antibodies were used at the following dilutions: rabbit anti-HNF6 (1/250, Santa Cruz Biotechnology, Inc.), guinea pig anti-Ngn3 (1/1000), rabbit anti-β-gal (1/5000, MP Biomedicals), rabbit anti-amylase (1/500, a gift of Ray MacDonald, Univ. of Texas Southwestern), and anti-insulin and anti-glucagon (as above). All primary antibodies were incubated overnight at 4°C in a humidified chamber. Primary antibodies were detected by species-specific donkey secondary antibodies conjugated with Cy2, Cy3, or Cy5 fluorophores (1/500, Jackson Immunoresearch).
Where indicated, a Zeiss LSM 510 confocal microscope was used to visualize 1 µm optical sections on immunofluorescently labeled tissue. Other samples were viewed under bright-field illumination or appropriate optical filters (immunofluorescence) using an Olympus BX41 microscope and digital camera with the Magnafire program (Optronics, Inc.). Whole-mount samples were viewed using an Olympus SZX9 dissecting microscope and photographed using a Nikon CoolPix 4300 digital camera. TIFF images from each experiment were processed equivalently for brightness and contrast in Adobe Photoshop.
β-gal activity was detected using X-gal as previously described (Wu et al., 1997). Following fixation, pancreata were washed two times for 30 minutes each in permeabilization solution. Samples were incubated overnight at room temperature in staining solution, post-fixed in 4% paraformaldehyde in PBS at 4°C for one hour, and dehydrated as above, substituting isopropanol for xylene to minimize leaching of the precipitate. Sections were counter-stained with eosin (Surgipath) for contrast.
Morphometric analysis
Endocrine Area
Sections of perinatal pancreata were immunolabeled at 250µm intervals along the length of the pancreas (at least seven sections per pancreas) for synaptophysin, a pan-endocrine marker. Total pancreatic area (identified by eosin counter-stain) was quantitated using Metamorph software (Molecular Devices). Total endocrine area was determined by manual demarcation of synaptophysin-positive cells. Average endocrine area was expressed as a percentage of total pancreatic area measured.
Recombined Area
Sections of X-gal stained pancreata were quantitated for total pancreatic area as described above. Total recombined area was quantitated by setting an RGB threshold that would cover the X-gal-positive area. Following manual demarcation of X-gal-positive islet-like clusters, endocrine area was subtracted from the total recombined area to quantitate the non-endocrine recombined area. The average non-endocrine recombined area was expressed as a percentage of total pancreatic area measured.
All statistical analyses were performed by two-tailed Student’s t-test using Excel software.
Protein extraction and western blotting
Adult pancreata were dissected and immediately placed into extraction buffer containing a protease inhibitor cocktail (0.5 mg/L TPCK, 0.5 mg/L TLCK, 0.6 µM leupeptin, and 2 µM pepstatin), DTT, and PMSF. Samples were homogenized, centrifuged, and the supernatant was quantitated by the Bio-Rad DC protein assay according to manufacturer’s instructions.
Protein was electrophoresed on 4–20% Tris-glycine gels (Invitrogen) and blotted to PVDF membrane (Amersham Bioscience). Primary antibodies were incubated at 4°C overnight using the following dilutions: rabbit anti-CTGF (1/5000); rabbit anti-MMP7 (1/1000; a gift from Dr. Barbara Fingleton, Vanderbilt Univ.); rabbit anti-p8 (1/3500; (Path et al., 2004); rabbit anti-β-actin (1:5000; Santa Cruz Biotechnology, Inc.); and rabbit anti-CFTR (1/1000; ABR-Affinity BioReagents). HRP-conjugated species-specific secondary antibodies were diluted to 1/10,000 (anti-rabbit IgG, Santa Cruz) or 1/5000 (anti-rat IgG, Amersham) in 1% non-fat milk or 1% BSA in TBS and incubated for one hour at room temperature. Protein detection was facilitated by the ECL detection system (Amersham) following the manufacturer’s instructions. Protein levels were quantitated on a Molecular Imager FX densitometer (Bio-Rad) using Quantity One 4.6 software (Bio-Rad) and normalized to β-actin, where control levels were assigned a value of 1.0.
Assessment of glucose homeostasis
Intraperitoneal glucose tolerance tests (IP-GTT) were performed on 8 week old mice as previously described (Tweedie et al., 2006). Insulin secretion in response to a glucose challenge was assessed by collecting blood retro-orbitally at fasting (0 min) and 30 minutes following glucose injection. Insulin concentration in the plasma, as well as total pancreatic insulin extracted from early postnatal and adult pancreata (acid alcohol extracts) was quantitated by solid phase radioimmunoassay as previously described (Tweedie et al., 2006).
Mice were weaned at three weeks of age and placed on high fat diet (58.7% fat, 26.7% carbohydrate, 14.8% protein, Bio-Serv) ad libitum for 8.5 weeks to induce peripheral insulin resistance. At 12 weeks of age, mice were subjected to IP-GTT and insulin secretion assays as described above.
Supplementary Material
Blood glucose levels (mg/dl) were measured at random at P2 (control n = 6; HNF6Δpanc n = 9) and 3.5 weeks of age (control n = 21; HNF6Δpanc n=10), and after a 16 hour fast at 6 weeks of age (control n = 9; HNF6Δpanc n = 6). HNF6Δpanc animals displayed elevated glucose levels at all time points compared with controls. *, p < 0.01.
Phase contrast and fluorescence images of X-gal stained P2 pancreata from control (A) and HNF6Δendo (B–E) animals. Co-localization with either insulin (A*, B*) or glucagon (A**, B**) indicates that recombined cells are able to incorporate into both hormone-expressing lineages. β cells in HNF6Δendo pancreata also express the transcription factors Pdx1 (C) and MafA (D), as well as the glucose transporter GLUT2 (E). Scale bars: 10 µm.
(A) The dorsal pancreas in HNF6Δpanc animals was poorly developed at P2 and often contained calcifications (asterisk) and twisted ducts filed with fluid or blood (arrows). The ventral pancreas appeared grossly normal. Section analysis revealed calcifications and fat necrosis (B; arrow) as well as periductular inflammation (C; arrow and arrowhead).
CTGF expression in postnatal ducts was not apparent in control pancreata (A), but was dramatically increased in ducts from HNF6Δpanc animals (B; brown).
Cytokeratin immunolabeling (A,C,E) outlines the ductal epithelial cells. Antibodies against Arl13b (B,D) or β-tubulin (F) were used to label cilia. At e18.5, control ducts were observed to contain cilia that protruded into the ductal lumen (A,B). The cilia present in morphologically normal ducts of HNF6Δpanc pancreata at e18.5 were fewer in number and did not protrude into the ductal lumen (C,D). A dramatic decrease in cilia number was observed in dilated ducts from HNF6Δpanc pancreata (E,F). The arrow in panel F indicates the single cilium observed in this particular duct. Outlines indicate border of ductal lumen. Scale bars: 20 µm.
(B) Southern blot of SpeI/EcoRV digested genomic DNA from HN6flox-neo targeted ES cells using the 5’ external probe, detected bands specific to wild type (WT; 13.3kb) and HNF6flox (fl; 11.6kb) alleles. Using 3’ the external probe, Southern blot of HpaI/SacII digested genomic DNA from HN6fl targeted ES cells detected bands specific to wild type (WT; 7.6kb) and HNF6fl (fl; 9.6kb) alleles. (C) PCR analysis of mouse tail genomic DNA using primers flanking the 5’-most loxP site resulted in amplification of a larger product (590bp) from the targeted versus wild type allele (550bp).
Acknowledgements
We would like to thank Young Ah Oh, Nikul Patel, David Lowe, Kathy D. Shelton, Xue Feng, and Venus Childress for their expert technical assistance with this project and the other members of the Gannon lab for their critical comments and suggestions throughout the course of this work. We thank Dr. Maria Pino for assistance with the BAC library screening, Dr. Doug Mortlock for assistance with BAC recombineering, and Dr. Stacey Huppert for assistance with bile duct examination. Reagents were graciously provided by Guoqiang Gu, Ray MacDonald, Maike Sander, Bernard Thorens, Chris Wright, and David Brigstock. A special thanks to Dr. Beatriz Sosa-Pineda for communicating results prior to publication. This research involved use of the Vanderbilt Transgenic Mouse/ES Cell Shared Resource, the Cell Imaging Shared Resource and the Vanderbilt Hormone Assay Core (supported in part by NIH Grants CA68485 and DK20593). Funding was provided by: NIH grant R01 DK65131-01 and JDRF Career Development Award, Grant 2-2002-583 to M. G.; U19 DK042502-17 and U01 DK072473-02 to M. A. M.; NIH Grants DK43673 and DK67166 to A. B. L.; NRSA Training Program in Developmental Biology, Grant NICHD 5-T32-HD05702 to E. T. A.; and a JDRF postdoctoral fellowship #3-2006-61 to H. Z.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
This study is dedicated to the memory of the late Robert H. Costa, Ph.D. without whom this project would not have been possible.
References
- Ahlgren U, Jonsson J, Jonsson L, Simu K, Edlund H. beta-cell-specific inactivation of the mouse Ipf1/Pdx1 gene results in loss of the beta-cell phenotype and maturity onset diabetes. Genes Dev. 1998;12:1763–1768. doi: 10.1101/gad.12.12.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Başar O, Ibiş M, Uçar E, Ertuğrul I, Yolcu O, Köklü S, Parlak E, Ulker A. Recurrent pancreatitis in a patient with autosomal-dominant polycystic kidney disease. Pancreatology. 2006;6:160–162. doi: 10.1159/000090258. [DOI] [PubMed] [Google Scholar]
- Cano DA, Murcia NS, Pazour GJ, Hebrok M. Orpk mouse model of polycystic kidney disease reveals essential role of primary cilia in pancreatic tissue organization. Development. 2004;131:3457–3467. doi: 10.1242/dev.01189. [DOI] [PubMed] [Google Scholar]
- Cano DA, Sekine S, Hebrok M. Primary Cilia Deletion in Pancreatic Epithelial Cells Results in Cyst Formation and Pancreatitis. Gastroenterology. 2006 doi: 10.1053/j.gastro.2006.10.050. [DOI] [PubMed] [Google Scholar]
- Caspary T, Larkins CE, Anderson KV. The graded response to Sonic Hedgehog depends on cilia architecture. Dev Cell. 2007;12:767–778. doi: 10.1016/j.devcel.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Crawford HC, Scoggins CR, Washington MK, Matrisian LM, Leach SD. Matrix metalloproteinase-7 is expressed by pancreatic cancer precursors and regulates acinar-to-ductal metaplasia in exocrine pancreas. J Clin Invest. 2002;109:1437–1444. doi: 10.1172/JCI15051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport J, Yoder B. An incredible decade for the primary cilium: a look at a once-forgotten organelle. Am J Physiol Renal Physiol. 2005;289:F1159–F1169. doi: 10.1152/ajprenal.00118.2005. [DOI] [PubMed] [Google Scholar]
- di Mola FF, Friess H, Martignoni ME, Di Sebastiano P, Zimmermann A, Innocenti P, Graber H, Gold LI, Korc M, Buchler MW. Connective tissue growth factor is a regulator for fibrosis in human chronic pancreatitis. Ann Surg. 1999;230:63–71. doi: 10.1097/00000658-199907000-00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- di Mola FF, Friess H, Riesle E, Koliopanos A, Buchler P, Zhu Z, Brigstock DR, Korc M, Buchler MW. Connective tissue growth factor is involved in pancreatic repair and tissue remodeling in human and rat acute necrotizing pancreatitis. Ann Surg. 2002;235:60–67. doi: 10.1097/00000658-200201000-00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dornhofer N, Spong S, Bennewith K, Salim A, Klaus S, Kambham N, Wong C, Kaper F, Sutphin P, Nacalumi R, Hockel M, Le Q, Longaker M, Yang G, Koong A, Giaccia A. Connective tissue growth factor-specific monoclonal antibody therapy inhibits pancreatic tumor growth and metastasis. Cancer Res. 2006;66:5816–5827. doi: 10.1158/0008-5472.CAN-06-0081. [DOI] [PubMed] [Google Scholar]
- Fujitani Y, Fujitani S, Boyer DF, Gannon M, Kawaguchi Y, Ray M, Shiota M, Stein RW, Magnuson MA, Wright CV. Targeted deletion of a cis-regulatory region reveals differential gene dosage requirements for Pdx1 in foregut organ differentiation and pancreas formation. Genes Dev. 2006;20:253–266. doi: 10.1101/gad.1360106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gannon M, Ray MK, Van Zee K, Rausa F, Costa RH, Wright CV. Persistent expression of HNF6 in islet endocrine cells causes disrupted islet architecture and loss of beta cell function. Development. 2000;127:2883–2895. doi: 10.1242/dev.127.13.2883. [DOI] [PubMed] [Google Scholar]
- Gannon M, Tweedie Ables E, Crawford L, Lowe D, Offield MF, Magnuson MA, Wright CV. pdx-1 function is specifically required in embryonic beta cells to generate appropriate numbers of endocrine cell types and maintain glucose homeostasis. Dev Biol. 2008;314:406–417. doi: 10.1016/j.ydbio.2007.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradwohl G, Dierich A, LeMeur M, Guillemot F. neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc Natl Acad Sci U S A. 2000;97:1607–1611. doi: 10.1073/pnas.97.4.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu G, Dubauskaite J, Melton DA. Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development. 2002;129:2447–2457. doi: 10.1242/dev.129.10.2447. [DOI] [PubMed] [Google Scholar]
- Hale MA, Kagami H, Shi L, Holland AM, Elsasser HP, Hammer RE, MacDonald RJ. The homeodomain protein PDX1 is required at mid-pancreatic development for the formation of the exocrine pancreas. Dev Biol. 2005;286:225–237. doi: 10.1016/j.ydbio.2005.07.026. [DOI] [PubMed] [Google Scholar]
- Harmon EB, Apelqvist AA, Smart NG, Gu X, Osborne DH, Kim SK. GDF11 modulates NGN3+ islet progenitor cell number and promotes {beta}-cell differentiation in pancreas development. Development. 2004;131:6163–6174. doi: 10.1242/dev.01535. Epub 2004 Nov 17. [DOI] [PubMed] [Google Scholar]
- Hogan B, Beddington R, Costantini F, Lacy E. Manipulating the Mouse Embryo. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1994. [Google Scholar]
- Holland AM, Hale MA, Kagami H, Hammer RE, MacDonald RJ. Experimental control of pancreatic development and maintenance. Proc Natl Acad Sci U S A. 2002;99:12236–12241. doi: 10.1073/pnas.192255099. Epub 2002 Sep 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemin P, Durviaux SM, Jensen J, Godfraind C, Gradwohl G, Guillemot F, Madsen OD, Carmeliet P, Dewerchin M, Collen D, Rousseau GG, Lemaigre FP. Transcription factor hepatocyte nuclear factor 6 regulates pancreatic endocrine cell differentiation and controls expression of the proendocrine gene ngn3. Mol Cell Biol. 2000;20:4445–4454. doi: 10.1128/mcb.20.12.4445-4454.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemin P, Lemaigre FP, Rousseau GG. The Onecut transcription factor HNF-6 (OC-1) is required for timely specification of the pancreas and acts upstream of Pdx-1 in the specification cascade. Dev Biol. 2003;258:105–116. doi: 10.1016/s0012-1606(03)00115-5. [DOI] [PubMed] [Google Scholar]
- Jensen J. Gene regulatory factors in pancreatic development. Dev Dyn. 2004;229:176–200. doi: 10.1002/dvdy.10460. [DOI] [PubMed] [Google Scholar]
- Jensen J, Heller RS, Funder-Nielsen T, Pedersen EE, Lindsell C, Weinmaster G, Madsen OD, Serup P. Independent development of pancreatic alpha- and beta-cells from neurogenin3-expressing precursors: a role for the notch pathway in repression of premature differentiation. Diabetes. 2000;49:163–176. doi: 10.2337/diabetes.49.2.163. [DOI] [PubMed] [Google Scholar]
- Johansson KA, Dursun U, Jordan N, Gu G, Beermann F, Gradwohl G, Grapin-Botton A. Temporal control of neurogenin3 activity in pancreas progenitors reveals competence windows for the generation of different endocrine cell types. Dev Cell. 2007;12:457–465. doi: 10.1016/j.devcel.2007.02.010. [DOI] [PubMed] [Google Scholar]
- Jonsson J, Carlsson L, Edlund T, Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371:606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- Landry C, Clotman F, Hioki T, Oda H, Picard JJ, Lemaigre FP, Rousseau GG. HNF-6 is expressed in endoderm derivatives and nervous system of the mouse embryo and participates to the cross-regulatory network of liver- enriched transcription factors. Dev Biol. 1997;192:247–257. doi: 10.1006/dbio.1997.8757. [DOI] [PubMed] [Google Scholar]
- Lannoy VJ, Decaux JF, Pierreux CE, Lemaigre FP, Rousseau GG. Liver glucokinase gene expression is controlled by the onecut transcription factor hepatocyte nuclear factor-6. Diabetologia. 2002;45:1136–1141. doi: 10.1007/s00125-002-0856-z. [DOI] [PubMed] [Google Scholar]
- Liu P, Jenkins NA, Copeland NG. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 2003;13:476–484. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowenfels A, Maisonneuve P, Cavallini G, Ammann R, Lankisch P, Andersen J, Dimagno E, Andrén-Sandberg A, Domellöf L International Pancreatitis Study Group. Pancreatitis and the risk of pancreatic cancer. N. Engl. J. Med. 1993;238:1433–1437. doi: 10.1056/NEJM199305203282001. [DOI] [PubMed] [Google Scholar]
- Lowenfels A, Maisonneuve P, Whitcomb D International Hereditary Pancreatitis Study Group. Risk factors for cancer in hereditary pancreatitis. Med Clin North Am. 2000;84:565–573. doi: 10.1016/s0025-7125(05)70240-6. [DOI] [PubMed] [Google Scholar]
- Maestro MA, Boj SF, Luco RF, Pierreux CE, Cabedo J, Servitja JM, German MS, Rousseau GG, Lemaigre FP, Ferrer J. Hnf6 and Tcf2 (MODY5) are linked in a gene network operating in a precursor cell domain of the embryonic pancreas. Hum Mol Genet. 2003;12:3307–3314. doi: 10.1093/hmg/ddg355. Epub 2003 Oct 21. [DOI] [PubMed] [Google Scholar]
- Mallo GV, Fiedler F, Calvo EL, Ortiz EM, Vasseur S, Keim V, Morisset J, Iovanna JL. Cloning and expression of the rat p8 cDNA, a new gene activated in pancreas during the acute phase of pancreatitis, pancreatic development, and regeneration, and which promotes cellular growth. J Biol Chem. 1997;272:32360–32359. doi: 10.1074/jbc.272.51.32360. [DOI] [PubMed] [Google Scholar]
- Motoo Y, Iovanna JL, Mallo GV, Su SB, Xie MJ, Sawabu N. P8 expression is induced in acinar cells during chronic pancreatitis. Dig Dis Sci. 2001;46:1640–1646. doi: 10.1023/a:1010637100933. [DOI] [PubMed] [Google Scholar]
- Offield MF, Jetton TL, Labosky PA, Ray M, Stein RW, Magnuson MA, Hogan BL, Wright CV. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development. 1996;122:983–995. doi: 10.1242/dev.122.3.983. [DOI] [PubMed] [Google Scholar]
- Path G, Opel A, Knoll A, Seufert J. Nuclear protein p8 is associated with glucose-induced pancreatic beta-cell growth. Diabetes. 2004;53:S82–S85. doi: 10.2337/diabetes.53.2007.s82. [DOI] [PubMed] [Google Scholar]
- Pictet RL, Clark WR, Williams RH, Rutter WJ. An ultrastructural analysis of the developing embryonic pancreas. Dev Biol. 1972;29:436–467. doi: 10.1016/0012-1606(72)90083-8. [DOI] [PubMed] [Google Scholar]
- Pierreux CE, Poll AV, Kemp CR, Clotman F, Maestro MA, Cordi S, Ferrer J, Leyns L, Rousseau GG, Lemaigre FP. The transcription factor hepatocyte nuclear factor-6 controls the development of pancreatic ducts in the mouse. Gastroenterology. 2006;130:532–541. doi: 10.1053/j.gastro.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Powell W, Domann FJ, Mitchen J, Matrisian L, Nagle R, Bowden G. Matrilysin expression in the involuting rat ventral prostate. Prostate. 1996;29:159–168. doi: 10.1002/1097-0045(199609)29:3<159::aid-pros2990290304>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Prado CL, Pugh-Bernard AE, Elghazi L, Sosa-Pineda B, Sussel L. Ghrelin cells replace insulin-producing {beta} cells in two mouse models of pancreas development. Proc Natl Acad Sci U S A. 2004;101:2924–2929. doi: 10.1073/pnas.0308604100. Epub 2004 Feb 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugacheva EN, Jablonski SA, Hartman TR, Henske EP, Golemis EA. HEF1-Dependent Aurora A Activation Induces Disassembly of the Primary Cilium. Cell. 2007;129:1351–1363. doi: 10.1016/j.cell.2007.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausa F, Samadani U, Ye H, Lim L, Fletcher CF, Jenkins NA, Copeland NG, Costa RH. The cut-homeodomain transcriptional activator HNF-6 is coexpressed with its target gene HNF-3 beta in the developing murine liver and pancreas. Dev Biol. 1997;192:228–246. doi: 10.1006/dbio.1997.8744. [DOI] [PubMed] [Google Scholar]
- Rodriguez CI, Buchholz F, Galloway J, Sequerra R, Kasper J, Ayala R, Stewart AF, Dymecki SM. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat Genet. 2000;25:139–140. doi: 10.1038/75973. [DOI] [PubMed] [Google Scholar]
- Schonhoff SE, Giel-Moloney M, Leiter AB. Neurogenin 3-expressing progenitor cells in the gastrointestinal tract differentiate into both endocrine and non-endocrine cell types. Dev Biol. 2004;270:443–454. doi: 10.1016/j.ydbio.2004.03.013. [DOI] [PubMed] [Google Scholar]
- Schwitzgebel VM, Scheel DW, Conners JR, Kalamaras J, Lee JE, Anderson DJ, Sussel L, Johnson JD, German MS. Expression of neurogenin3 reveals an islet cell precursor population in the pancreas. Development. 2000;127:3533–3542. doi: 10.1242/dev.127.16.3533. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Streeper R, Hornbuckle L, Svitek C, Goldman J, Oeser J, O'Brien R. Protein kinase A phosphorylates hepatocyte nuclear factor-6 and stimulates glucose-6-phosphatase catalytic subunit gene transcription. J Biol Chem. 2001;276:19111–19118. doi: 10.1074/jbc.M101442200. [DOI] [PubMed] [Google Scholar]
- Tweedie E, Artner I, Crawford L, Poffenberger G, Thorens B, Stein R, Powers AC, Gannon M. Maintenance of hepatic nuclear factor 6 in postnatal islets impairs terminal differentiation and function of beta-cells. Diabetes. 2006;55:3264–3270. doi: 10.2337/db06-0090. [DOI] [PubMed] [Google Scholar]
- Wang J, Kilic G, Aydin M, Burke Z, Oliver G, Sosa-Pineda B. Prox1 activity controls pancreas morphogenesis and participates in the production of "secondary transition" pancreatic endocrine cells. Dev Biol. 2005 doi: 10.1016/j.ydbio.2005.07.021. [DOI] [PubMed] [Google Scholar]
- Wilding Crawford L, Tweedie Ables E, Oh YA, Boone B, Levy S, Gannon M. Gene expression profiling of a mouse model of pancreatic islet dysmorphogenesis. PLoS ONE. 2008;3:e1611. doi: 10.1371/journal.pone.0001611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu KL, Gannon M, Peshavaria M, Offield MF, Henderson E, Ray M, Marks A, Gamer LW, Wright CV, Stein R. Hepatocyte nuclear factor 3beta is involved in pancreatic beta-cell- specific transcription of the pdx-1 gene. Mol Cell Biol. 1997;17:6002–6013. doi: 10.1128/mcb.17.10.6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagita M, Okuda T, Endo S, Tanaka M, Takahashi K, Sugiyama F, Kunita S, Takahashi S, Fukatsu A, Yanagisawa M, Kita T, Sakurai T. Uterine sensitization-associated gene-1 (USAG-1), a novel BMP antagonist expressed in the kidney, accelerates tubular injury. J Clin Invest. 2006;116:70–79. doi: 10.1172/JCI25445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Ackermann AM, Gusarova GA, Lowe D, Feng X, Kopsombut UG, Costa RH, Gannon M. The Foxm1 Transcription Factor is Required to Maintain Pancreatic Beta Cell Mass. Mol Endocrinol. 2006;20:1853–1866. doi: 10.1210/me.2006-0056. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Law AC, Rajagopal J, Anderson WJ, Gray PA, Melton DA. A multipotent progenitor domain guides pancreatic organogenesis. Developmental Cell. 2007;13:103–114. doi: 10.1016/j.devcel.2007.06.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Blood glucose levels (mg/dl) were measured at random at P2 (control n = 6; HNF6Δpanc n = 9) and 3.5 weeks of age (control n = 21; HNF6Δpanc n=10), and after a 16 hour fast at 6 weeks of age (control n = 9; HNF6Δpanc n = 6). HNF6Δpanc animals displayed elevated glucose levels at all time points compared with controls. *, p < 0.01.
Phase contrast and fluorescence images of X-gal stained P2 pancreata from control (A) and HNF6Δendo (B–E) animals. Co-localization with either insulin (A*, B*) or glucagon (A**, B**) indicates that recombined cells are able to incorporate into both hormone-expressing lineages. β cells in HNF6Δendo pancreata also express the transcription factors Pdx1 (C) and MafA (D), as well as the glucose transporter GLUT2 (E). Scale bars: 10 µm.
(A) The dorsal pancreas in HNF6Δpanc animals was poorly developed at P2 and often contained calcifications (asterisk) and twisted ducts filed with fluid or blood (arrows). The ventral pancreas appeared grossly normal. Section analysis revealed calcifications and fat necrosis (B; arrow) as well as periductular inflammation (C; arrow and arrowhead).
CTGF expression in postnatal ducts was not apparent in control pancreata (A), but was dramatically increased in ducts from HNF6Δpanc animals (B; brown).
Cytokeratin immunolabeling (A,C,E) outlines the ductal epithelial cells. Antibodies against Arl13b (B,D) or β-tubulin (F) were used to label cilia. At e18.5, control ducts were observed to contain cilia that protruded into the ductal lumen (A,B). The cilia present in morphologically normal ducts of HNF6Δpanc pancreata at e18.5 were fewer in number and did not protrude into the ductal lumen (C,D). A dramatic decrease in cilia number was observed in dilated ducts from HNF6Δpanc pancreata (E,F). The arrow in panel F indicates the single cilium observed in this particular duct. Outlines indicate border of ductal lumen. Scale bars: 20 µm.
(B) Southern blot of SpeI/EcoRV digested genomic DNA from HN6flox-neo targeted ES cells using the 5’ external probe, detected bands specific to wild type (WT; 13.3kb) and HNF6flox (fl; 11.6kb) alleles. Using 3’ the external probe, Southern blot of HpaI/SacII digested genomic DNA from HN6fl targeted ES cells detected bands specific to wild type (WT; 7.6kb) and HNF6fl (fl; 9.6kb) alleles. (C) PCR analysis of mouse tail genomic DNA using primers flanking the 5’-most loxP site resulted in amplification of a larger product (590bp) from the targeted versus wild type allele (550bp).