Abstract
A diverse series of mammalian two-hybrid technologies for the detection of protein–protein interactions have emerged in the past few years which complement the established yeast two-hybrid approach. Given the mammalian background in which they operate, these assays open new avenues to study the dynamics of mammalian protein interaction networks, i.e. the temporal, spatial and functional modulation of protein–protein associations. In addition, novel assay formats are available that enable high-throughput mammalian two-hybrid applications, facilitating their use in large-scale interactome mapping projects. Finally, as they can be applied in drug discovery and development programs, these techniques also offer exciting new opportunities for biomedical research.
Interactome mapping: a work in progress
Protein interactomics is a fast growing field of research that aims to add sense to the wealth of genomic data generated over the last decade by mapping “who sees who” in the cellular proteome. Such an approach should help us to understand how proteins are organized and how they function. Interaction mapping also is particularly relevant for human health; many diseases can be linked to aberrations in protein interaction patterns which rewire signalling networks and cause disease phenotypes [1–3].
The two main approaches which have been applied in high-throughput efforts to chart protein (sub-) network maps in a variety of model organisms have generated complementary datasets. Biochemical technologies involve affinity purification of protein complexes from cell lysates followed by mass spectrometry-based identification of their constituents [4]. Genetic methods, by contrast, are based on reconstitution or activation of a reporter upon interaction of genetically fused ‘bait’ and ‘prey’ proteins in living cells, and mostly generate binary interactions [5]. Although many genetic interaction mapping technologies have been developed, large-scale binary mapping efforts largely rely on the ‘classic’ yeast two-hybrid (Y2H) system as it is one of the only genetic screening platforms that provides the throughput necessary to create large-scale interaction maps (Box 1). To challenge the widespread scepticism surrounding the quality of these datasets, a recently published series of papers describing large-scale Y2H-based interactome analysis in yeast [6], worm [7,8] and human [9] applied a number of mammalian two-hybrid methods as validation tools, demonstrating that state-of-the-art Y2H technologies can generate high-quality datasets. The idea of experimental quality assessment through retesting was further elaborated by the development of a confidence score for binary protein interactions using a panel of orthogonal methods [10]. Interestingly, cross-comparison revealed that sensitivity and specificity of the mammalian technologies are comparable to that of the Y2H system. In addition, Y2H and mammalian approaches are highly complementary with regard to the subset of interactions they are able to detect. Although the application of mammalian methods has often been limited to validating (high-throughput) interactomics data, the data generated by most mammalian techniques is ‘richer’, and the additional information they provide could help to reveal the biological significance of particular protein interactions. Therefore, a more widespread application of these technologies seems justified. In this review, we zoom-in on the mammalian genetic complementation methods, or mammalian two-hybrids in the broad sense, focussing on recent applications that demonstrate their added value in comparison to established yeast-based technologies.
Box 1. The Y2H system.
The Y2H principle was originally developed by Stanley Fields [53] and was based on the observation that eukaryotic transcription factors have a modular structure, consisting of DNA-binding domain (DB) and a transactivation domain (AD) separated by a linker (Figure Ia). The DB recognizes and binds a specific sequence in the promoter region of a gene, and the AD interacts with the RNA polymerase II enzyme (Pol) to stimulate gene transcription. The flexibility of the linker enables the transcription factor to regulate gene transcription even when binding a DNA sequence remotely positioned relative to the gene’s transcription start site. Importantly, neither of these subdomains alone can induce transcription.
In the Y2H system (Figure Ib), the DB and AD of a yeast transcription factor (e.g. GAL4) each are tethered to a protein of interest (X and Y), resulting in two hybrid proteins (X-DB and Y-AD). Physical association between the proteins X and Y in yeast cells genetically engineered to express both hybrid proteins brings the DB and AD into proximity, thereby reconstituting the transcription factor. The DB of this functionally complemented transcription factor can bind a recognition site in the reporter gene promoter region, and the AD interacts with RNA polymerase II, driving transcription of one or more reporter genes. The produced reporter proteins enable selection of those cells that harbor a pair of interacting proteins.
For nearly two decades, Y2H has been the workhorse of interactome mapping. The system has been applied to chart the first drafts of genome-wide protein networks in a broad range of organisms [6,8,54–56]; at the same time it has been used for focused studies of (disease-related) subnetworks [1,2]. Currently, large-scale Y2H analyses to systematically map the estimated 130,000 binary interactions in the human interactome are ongoing and this screening effort is expected to be completed within the next few years [9,10].
Figure I. Outline of the Y2H concept.
(a) Modularity of eukaryotic transcription factors. DB and AD are coupled by a flexible linker, thereby allowing simultaneous binding of the DB to its cognate promoter region and the AD with RNA polymerase II (Pol). Both events are required to activate gene transcription,
(b) The Y2H assay. Interaction of proteins X and Y reconstitutes transcription factor activity by complementing its DB and AD, respectively, and activating reporter gene transcription.
From static to dynamic interactomes
Interactomes are dynamic protein networks in which many protein–protein interactions are context-dependent and require specific modifications and/or structural alterations of the interacting partners which only occur under the appropriate cellular conditions [11,12]. In contrast to yeast systems where cofactors or regulatory proteins are often absent, mammalian two-hybrids allow the assayed proteins to undergo the proper modifications in their native cellular context. Additionally, this native background provides the necessary adaptor proteins to bridge the association of indirectly interacting proteins. Thus, not only should mammalian methods enable detection of a subset of interactions that might remain hidden using yeast-based approaches, they should also allow protein interactions to be tracked as a function of time, space (subcellular distribution) and physiological context (activation or inactivation of a cellular process induced by natural or synthetic stimuli). Adding information that describes changes in connectivity to the static interactome scaffolds, as currently obtained using Y2H, should provide insight into the functional dynamics of the interactome, extending the interactomics paradigm to “who sees who, when, where and how”. Thus, whereas Y2H methods likely will remain unsurpassed in throughput and coverage, mammalian technologies could become essential tools for focused studies on the dynamics of (subsets of) the interactome. In the following paragraphs, we review the most prominent technologies that have been successfully applied to capture some of the plasticity of protein interaction networks in living mammalian cells.
Mammalian two-hybrid sensu stricto
The most straightforward adaptation of the Y2H approach to mammalian cells is a conceptual replica of the original yeast technique, in which the physical association of two proteins coupled to the DNA-binding domain (DB) and trans-activating domain (AD) of a transcription factor, respectively, elicits transcriptional activation of a reporter gene. The reconstituted transcription factor in this ‘mammalian two-hybrid sensu stricto’ (M2H s.s.), typically consists of the yeast GAL4 DB and the AD of the herpes simplex virus VP16 protein, a transcriptional activator in mammalian cells. An early account where this approach was applied to analyze the context-dependency of protein–protein interactions described the transforming growth factor-β (TGF-β)-dependent association of Smad2 and Smad4 with CREB binding protein (CBP). Smad2–Smad4 heterodimers mediate TGF-β signalling, and this study showed that their interaction with the transcriptional co-activator CBP is essential for TGF-β-induced transcriptional responses [13]. More recently, the M2H s.s. method was applied to monitor the effect of protein kinase A (PKA) activation and treatment with the anti-cancer drug tamoxifen on the interaction between estrogen receptor α (ERα ) and the transcriptional co-activator steroid receptor coactivator-1 (SRC-1) thereby unraveling the mechanism behind PKA-induced tamoxifen resistance [14].
Split β-galactosidase
Based on the success of the two-hybrid assay principle, the use of transcription factor fragment complementation has been extended to utilize other proteins whose reconstitution generates a quantifiable signal. In some cases the reconstituted protein itself is a reporter, e.g. the split β-galactosidase assay. Here, two proteins of interest are fused to subunits of the Escherichia coli β-galactosidase enzyme, modified to inhibit spontaneous association. Interaction between the proteins of interest brings the β-galactosidase subunits into close proximity, reconstituting a functional enzyme which can be detected with a suitable substrate (Figure 1a). This technique was applied to dissect ligand-dependent interactions between epidermal growth factor receptor (EGFR) family members, a number of which have been associated with breast cancer malignancy (EGFR and ErbB2), thereby providing insight into the mechanism of action of the anti-ErbB2 antibody Herceptin which is widely used in cancer therapy [15]. The addition of EGF-type ligands to mouse fibroblasts which express fusion proteins in which the intracellular domain of the receptor chain is replaced by a fragment of the β-galactosidase reporter, revealed that ErbB2 can associate with the EGFR and ErbB3 upon stimulation, but does not spontaneously form homodimers. In a field that is troubled by conflicting reports, these results support the notion that ErbB2 cannot signal on its own, but instead requires interactions with other EGFR family members. Using the same fusion proteins, Herceptin action was revealed mainly to disrupt EGFR–ErbB2 heterodimers, indirectly resulting in reduced EGFR cell surface expression; these results could explain the antibody’s strong anti-tumour effect [15]. In two other reports, the same β-galactosidase complementation assay was inventively applied to monitor ligand-induced activation of G-protein-coupled receptors (GPCRs), a therapeutically important receptor family whose activation has been difficult to assess with existing methodologies [16,17]. Although GPCRs comprise an extensive family of receptors involved in diverse physiological processes, two universal protein–protein interactions associated with GPCR activation were exploited to develop two independent split β-galactosidase-based receptor activation assays. Dose-dependent responses to GPCR agonists and antagonists agreed between the different assays. Further, using a luminescence readout, (ant)agonist-dependent GPCR activity was followed in live mice implanted with cells engineered to express components of the complementation assay [17]. In addition, the assays were amenable to compound screening in a pilot experiment to identify molecules which interfere with GPCR activation [16].
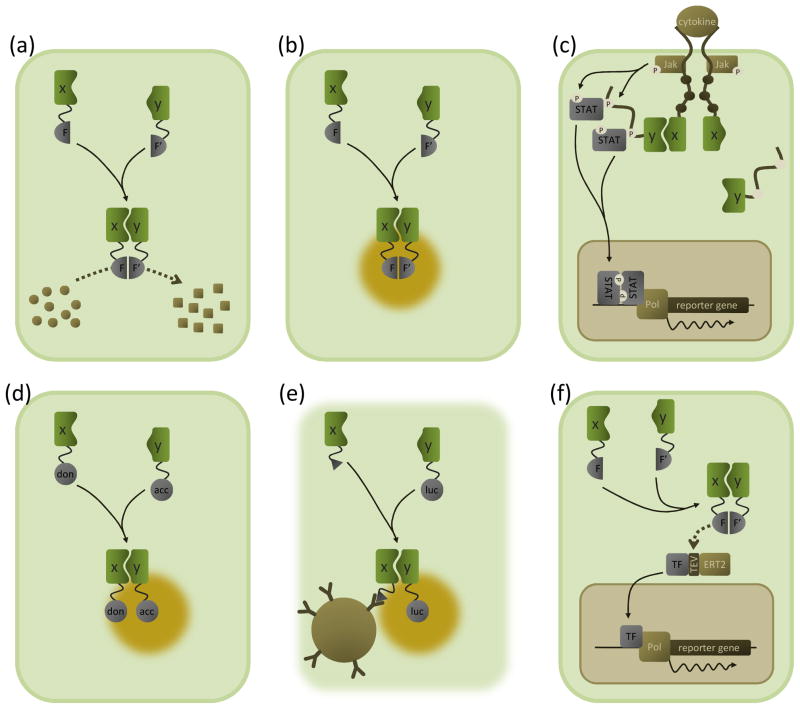
Figure 1. Overview of M2H assay principles.
Schematic outline of the M2H assays discussed in the text. The interacting proteins X and Y are depicted in green; complemented protein fragments or proteins are grey. Please note that the M2H (sensu stricto) assay is not depicted here as it is conceptually identical to the Y2H system (Box 1).
(a) Enzyme complementation systems. The interaction between two proteins of interest (X and Y) brings together two fragments of a reporter enzyme (F and F′); this reconstitutes the enzyme’s catalytic activity, converting a substrate (circles) into a detectable product (squares). The enzyme fragments can be pre-folded subunits (e.g. the split β-galactosidase assay) or fragments that fold only upon complementation (e.g. the β-lactamase and luciferase protein fragment complementation assays).
(b) BiFC. Association of proteins X and Y reconstitutes a fluorescent protein from two fragments (F and F′), resulting in fluorescent emission upon excitation at a suitable wavelength (yellow halo).
(c) MAPPIT. Bait X is coupled to a signalling-deficient cytokine receptor (via substituting tyrosine residues in the receptor tail which are critical for recruiting signal transducer and activator of transcription (STAT) proteins (brown dots)). The prey Y is tethered to a portion of another receptor containing intact recruitment sites. Upon bait–prey interaction, a functional receptor is reconstituted which can be activated by the appropriate cytokine ligand. Upon ligand binding, pre-associated Janus kinases (Jak) are activated by cross-phosphorylation (P). Activated Jaks phosphorylate (P) tyrosine residues in the receptor fragment coupled to the prey (white dots), which then act as docking sites for STATs. Recruited STATs are in turn phosphorylated by the Jaks, leading to their activation and subsequent dissociation and translocation to the nucleus. In the nucleus, STAT dimers induce STAT-dependent reporter gene transcription. In the MASPIT setup, dihydrofolate reductase (DHFR) is coupled to the signalling deficient receptor chain and a chemical compound is fused to methotrexate, a molecule that binds to DHFR with very high affinity. Addition of such a methotrexate fusion compound to cells expressing the DHFR-coupled receptor results in the chemical compound being displayed as a bait.
(d) FRET and BRET. Association of proteins X and Y brings into proximity an energy donor (don) and acceptor protein (acc). In FRET, (intact) fluorescent proteins are used as donor and acceptor. Excitation of the donor fluorophore causes nonradiative energy transfer to occur between the two, resulting in fluorescent emission of the acceptor fluorophore (yellow halo). Instead of a fluorescent protein, a luciferase enzyme is used as a donor in BRET. Enzymatic oxidation of a luciferase substrate generates bioluminescence, and energy transfer to the acceptor protein produces a fluorescent signal.
(e) LUMIER. Interaction between a Flag-tagged (triangle) bait protein X and a prey protein Y fused to Renilla luciferase is detected by bioluminescence measurements (yellow halo) of immunoprecipitates obtained from cell lysates using anti-Flag antibodies coupled to sepharose beads (brown sphere).
(f) Split TEV assay. Association between proteins X and Y complements a functional TEV protease from fragments F and F′. At the site of a TEV recognition sequence, the protease cleaves a chimeric protein consisting of a transcription factor (TF) coupled to an ERT2 domain, thus retaining the transcription factor in the cytosol. After cleavage, the released transcription factor migrates into the nucleus where it induces reporter gene transcription. Note that the split TEV assay is a flexible system and that the setup shown here is only one of the possible choices.
β-lactamase protein-fragment complementation
The split β-galactosidase assay exploits the natural occurrence of defined and spontaneously associating subdomains in the native enzyme that can be separated and which reconstitute enzymatic activity upon complementation. Although the assay uses mutant enzyme domains to prevent spontaneous assembly, a weak affinity remains, leading to undesired background signal. This source of background is avoided in a number of other complementation assays by using complementary protein fragments whose folding is proximity-dependent. This strategy is generally referred to as protein-fragment complementation assay (PCA) and applies to a variety of reporter proteins (including β-lactamase, dihydrofolate reductase (DHFR), ubiquitin, different luciferase enzymes and various fluorescent proteins) and is used in a range of cellular backgrounds (reviewed in [18,19]; Boxes 2 and 3).
Box 2. Working with hybrid proteins: some cautionary remarks.
In contrast to protein interaction mapping methods based on affinity purification/mass spectrometry, where a tagged bait pulls down complexes of endogenously expressed proteins, two-hybrid technologies rely on exogenous expression of hybrid bait and prey proteins. These hybrid proteins consist of a genetic fusion between the protein (domain) of interest and a component of the complementation system (e.g. a subdomain of a transcription factor or a fragment of a fluorescent protein). The use of exogenously expressed fusion proteins entails a number of risks that could generate false positive (interactions which do not occur between the corresponding endogenous proteins) or false negative (endogenous interactions which are missed) signals. The points discussed below apply to two-hybrid technologies in general, regardless of the cellular background they are used in. However, certain methods are more sensitive to some of the potential pitfalls than others.
Steric hindrance
Depending on the nature of the complementation fragments being coupled to the tested protein pair and on the length and flexibility of the linker between these fragments and the tested protein, the complementation fragments can hinder association of the tested proteins, resulting in a false negative interaction. Moreover, the size and orientation of the tested proteins can preclude reporter fragment complementation, similarly leading to false negatives.
Conformational changes
In some cases, fusion proteins adopt altered conformations, either inhibiting normally occurring interactions or facilitating non-native interactions. Conformational perturbation can also disrupt context-dependency, e.g. by exposing an interaction domain that is normally hidden in the protein structure unless a context-dependent protein modification occurs.
Altered localization
Fusion proteins can localize to cellular regions where the endogenous protein normally never exists, thus enabling non-native interactions. Such interactions are sometimes termed ‘pseudo-interactions’ [10].
Artificial co-expression
In two-hybrid assays, the two chimeric proteins are co-expressed, whereas the corresponding endogeneous proteins might never be present concurrently. Such non-native interactions are also referred to as ‘pseudo-interactions’.
Over-expression
Exogenous expression of the bait and prey fusions creates the risk that the tested proteins interact merely due to their high concentration in the cell. To avoid this pitfall, in theory one can titrate expression levels of the fusion proteins to approximate those of the endogenous proteins, but owing to the requirement for case-by-case optimization, this is not routinely done.
Box 3. Mammalian two-hybrids: features and opportunities.
Aside from the requirement of operating in a mammalian background, the ideal technology for studying mammalian protein interaction dynamics would combine high sensitivity and specificity with high spatial and temporal resolution, causing minimal cell perturbation and offering a choice of readouts. In reality, however, the available methods exhibit specific pros and cons. Here we review some of the important features related to tracking protein interaction dynamics.
Signal amplification
Different levels of signal amplification contribute to an assay’s sensitivity. For example, reconstituted enzymatic activity is an amplification step as it can constitutively process substrate into a signal (e.g. β-galactose, β-lactamase and luciferase PCAs). In reporter gene assays, the transcriptional activation amplifies the signal, and when the reporter gene encodes an enzyme, additional signal amplification is obtained (e.g. M2Hs.s.). Enzymatic reporter genes can be further combined with another amplification level, e.g. in MAPPIT, where transcription factors are constitutively activated by a complemented cytokine receptor. In a number of assays there is no signal amplification at all, e.g. BiFC, FRET and BRET.
Signal reversibility
To allow dynamic tracking of protein interactions, signals must be reversible. This property depends on a number of factors. First, the complemented reporter protein fragments must rapidly separate such that dissociation triggers reporter activity loss. This requirement presents a problem in BiFC, as fluorescent protein folding is irreversible [21,22]. Second, in reporter gene assays producing a stable enzyme, the accumulated enzyme remains active even when reporter gene activation ends following disruption of the protein interaction. Clearly, a trade-off exists between signal intensity and reversibility, and consequently between sensitivity and the ability to monitor interaction dynamics.
Kinetics of signal generation
The speed at which a signal is generated determines the temporal resolution of the assay. In PCAs, which depend on folding of a reporter protein, folding speed is a critical factor. Fluorescent proteins generally fold slowly [21,22] whereas some luciferases fold particularly fast [25,26]. Reporter gene assays also exhibit a lower temporal resolution because they depend on transcription kinetics. The fastest on- and off-rate is obtained with FRET and BRET which do not rely on folding, transcription or substrate conversion, thereby resulting in real-time kinetics [30,31].
Ability to visualize the signal
Some techniques generate a visible signal which allows subcellular localization of the interacting proteins. The highest spatial resolution is obtained with BiFC and FRET; DHFR andRenillaluciferase PCAs and BRET can obtain less detailed data [21,22,26,30,31,57].
One of the first studies to apply PCA to analyze context-dependent interactions in mammalian cells used complementation of β-lactamase enzymatic activity to track the phosphorylation-dependent association between cAMP response element binding protein (CREB) and CBP [20]. Two rationally dissected β-lactamase fragments were fused to the CBP KIX domain and the CREB kinase inducible domain (KID), and interaction-mediated reporter activity was measured by fluorescence microscopy. The CREB–CBP association relies on phosphorylation of a specific serine residue within the CREB KID. This PKA-dependent phosphorylation is activated by increased intracellular cAMP levels. Interestingly, the β-lactamase complementation assay was able to reproduce the context-dependency of the CREB–CBP interaction as signals significantly increased upon exposure to forskolin, an activator of cellular adenylyl cyclase, and to the cell-permeable cAMP analog CPT-cAMP. In addition, time-lapse fluorescence microscopy was used to track the signal, as a function of time, upon forskolin and CPT-cAMP stimulation, and the results were in accordance with the reported kinetics of endogenous CREB phosphorylation [20].
Bimolecular fluorescence complementation
A remarkably versatile class of PCAs with regard to the spatial component of context-dependent protein interaction mapping are those that make use of a split fluorescent protein reporter, also referred to as bimolecular fluorescence complementation (BiFC; Figure 1b). A wide variety of spectral variants of the green fluorescent protein (GFP) have been engineered into PCA-type protein interaction assays to allow direct subcellular visualisation of protein association and translocation of protein complexes, and multicolour applications allow simultaneous monitoring of different protein interactions (reviewed in [21,22]). Apart from delineating the subcellular site of protein complex localization, which already adds a layer of functional information onto an interaction network, BiFC approaches can also track the effects of changes in the cellular physiology. One example applied PCA to investigate functional cross-talk between TGF-β and insulin signalling pathways [23]. A split β-lactamase assay identified a novel physical link between protein kinase B (PKB) and Smad3. Next, a YFP fragment complementation assay revealed that PKB–Smad3 interaction is induced by insulin and inhibited by TGF-β; moreover, these complexes were localized at the plasma membrane and cytosol, but not in the nucleus of insulin-treated cells. In addition, wortmannin, a phosphoinositide-3-kinase (PI3K) inhibitor that impedes PKB phosphorylation, prevented insulin-induced PKB–Smad3 association. These results support the model that insulin can counteract TGF-β-mediated apoptosis by inducing phospho-PKB-mediated Smad3 sequestration [23].
Although fluorescence-based PCAs potentially enable analysis of every aspect (space, time and functional modulation) of protein interaction dynamics, two features limit their application to mainly the spatial component: the slow and irreversible nature of reconstituted fluorescent reporter protein folding. Maturation of most complementation-competent fluorescent proteins takes minutes if not hours, precluding accurate time-resolved analysis of complex association. Once folded, the interacting protein pair anchored to the fluorescent protein is stabilized and cannot dissociate, thereby limiting the analysis of dynamic interactions. Next generation fluorescent proteins with reduced maturation times and reversible folding would greatly extend application of the BiFC technology. Nevertheless, it should be mentioned that this irreversibility also has a bright side: it traps interactions that are too weak and/or transient to detect with other methods. For instance, GFP PCA can identify weak interactions with a Kd of 1mM [24].
Luciferase complementation
Two recently reported PCAs are based on reconstitution of Gaussia and Renilla luciferase fragments [25,26]. Owing to the absence of cellular background luminescence, these reporter proteins exhibit high sensitivity when compared to fluorescent PCA reporters which suffer from cellular autofluorescence. Importantly, folding of the complemented luciferase enzymes is very fast (in the order of seconds) and reversible, enabling their use for near real-time analysis of protein interaction dynamics. Indeed, the Gaussia PCA was used to quantitatively analyse functional modulation of the PKB–Smad3 association upon insulin and/or TGFβ stimulation [25]. The Renilla PCA was used to monitor association and dissociation of the regulatory and catalytic subunits of PKA, which reflect the enzyme’s inactivation and activation, respectively. The assay faithfully reported rapid changes in PKA (dis)assembly induced by treatment with (ant)agonists of endogenous GPCRs [26]. As the wavelengths associated with the ‘classical’ luciferase from firefly are longer than those produced through the Gaussia and Renilla enzymes, a property which facilitates deeper tissue penetration, firefly luciferase complementation is particularly useful for protein interaction analysis in living animals. By example, this technique enabled the visualization of dose-dependent rapamycin-induced interaction between FK506-binding protein 12 (FKBP12) and rapamycin target protein (FRB) in live mice [27].
Split TEV assay
In the complementation systems described thus far, a designated reporter protein is reconstituted upon protein–protein interaction. Each of these reporters combines a set of specific features that determine the sensitivity and flexibility of the assay: incorporation of an enzymatic signal amplification step, a requirement for exogenous substrates and availability of substrates compatible with different read-outs (colorimetric, luminometric, fluorometric, microscopic). In a highly flexible approach, a proteolytic activity that can drive a reporter system of choice, rather than the reporter itself, is reconstituted. In the split TEV assay, protein association complements the tobacco etch virus (TEV) protease, which then cleaves a specific recognition sequence, either proteolytically liberating a reporter enzyme or releasing a transcription factor that shuttles to the nucleus where it can activate a reporter gene [28] (Figure 1f). The latter option allows the introduction of an additional transcriptional amplification step, in addition to the enzymatic amplification, in the event that an enzymatic reporter is chosen. Although the irreversibility of reporter activation by TEV cleavage and the different levels of amplification contribute to the sensitivity of the assay and its ability to identify weak and transient interactions, these features at the same time compromise the assay’s ability to capture interaction kinetics and dynamics. Nevertheless, a transcription factor-coupled split TEV firefly luciferase reporter assay was able to monitor neuregulin-induced ErbB2–ErbB4 heterodimerization [28] and phosphorylation-dependent ErbB4–adaptor protein binding [29].
Resonance energy transfer systems
In the broad definition, mammalian two-hybrids also include complementation methods in which signalling relies on fluorescence resonance energy transfer (FRET) or bioluminescence resonance energy transfer (BRET) between two hybrid proteins (Figure 1d). As in BiFC, the FRET signal can be generated by a fluorescent protein, but not by complementation of fragments of the fluorescent protein. Instead, in FRET the proteins of interest are fused to intact fluorescent proteins with overlapping emission and excitation spectra. When brought into close proximity, nonradiative energy transfer between the excited ‘donor’ (e.g. CFP) and ‘acceptor’ fluorophore (e.g. YFP) elicits fluorescence emission from the latter [30]. As this complementation strategy does not rely on protein folding or maturation, it is fast and reversible, allowing true real-time analysis of protein association and dissociation with high spatial resolution. In BRET, a luciferase enzyme (typically from Renilla), instead of a fluorescent protein, is used as energy donor, thereby eliminating the need for exogenous excitation and avoiding some of the related problems, e.g. photobleaching. Likewise, BRET allows real-time analysis of protein complex formation. Compared to FRET, the subcellular resolution is limited, but BRET allows protein–protein interactions to be monitored for several hours [31]. Both FRET and BRET approaches have been used to monitor the dose-dependent ligand-induced interaction between GPCRs and their cognate G-proteins during GPCR activation as well as subsequent desensitization with high temporal, and in the case of FRET also spatial, resolution [32,33]. Although both FRET and BRET are conceptually very well suited for studying protein interaction dynamics, their widespread application is hampered because both methods are technically demanding. In addition, as ~100Å proximity between the donor and acceptor groups is required for significant energy transfer to occur, design of the fusion proteins cannot be generalized and requires dedicated case-by-case optimization.
Capturing protein interaction dynamics using Y2H
Although the previous paragraphs described the significance and potential utility of mammalian approaches in charting modulation of protein interaction networks, it should be noted that Y2H can capture some aspects of mammalian protein interaction dynamics. In cases where the bait itself is subject to context-dependent modification, but where no additional context-specific factors are required, Y2H can identify and analyse context-dependent modulation of protein association. As an example, Y2H was used to identify nitric oxide (NO)-dependent procaspase-3 interaction partners [34]. Via nitrosylation, NO functionally regulates proteins. To determine whether NO also controls the protein interaction spectrum of nitrosylated targets, a Y2H screen was carried out to find interaction partners for procaspase-3, an apoptosis-related protein whose activity is regulated by (de-)nitrosylation [35]. After adapting the yeast background to permit NO signalling (by deleting the endogenous NO-scavenger flavohemoglobin), a prey cDNA library from murine macrophages was screened in the presence of NO. A number of interacting proteins were recovered and their NO-dependent interactions with caspase-3 were confirmed via Y2H and immunoprecipitation assays in mammalian cells.
Increasing the throughput
In most of the examples described above, functional regulation of a single or a limited set of protein interactions was monitored. Simultaneous evaluation of the dynamic properties of larger subsets of the interactome, or ultimately the proteome-wide systematic mapping of interactome changes, will require application of these approaches on a larger scale. Mammalian assays are much less amenable to high-throughput analyses than yeast technologies such as Y2H. However, current lab automation standards have proven adequate to scale-up interaction assays in mammalian cells thereby facilitating interrogation of larger parts of the interactome space. LUMIER (luminescence-based mammalian interactome mapping), a method which combines aspects of both genetic and biochemical approaches, has generated one of the most elaborate mammalian interactome subnetworks obtained thus far [36] (Figure 1e). In LUMIER, two hybrid proteins are co-expressed: one contains a Flag-tag to enable immunoprecipitation and the other is fused to a Renilla luciferase reporter to enable luminescent detection of the interaction. The assay was automated in 96-well format and used to map the TGFβ protein interaction network. A set of baits comprising key members of the TGFβ signalling pathway was combined with a library of 518 prey proteins, and interaction was assayed in the absence or presence of TGFβ, revealing a dynamic interaction pattern for many of the tested baits and identifying novel, physiologically validated protein associations.
cDNA library screening provides a generic approach to interrogate a large pool of proteins for interaction with a particular bait. A number of two-hybrid methods have been adapted for the screening of complex prey cDNA libraries in mammalian cells, successfully identifying novel protein associations (e.g. [37]). Although cDNA libraries offer a wide coverage of the proteome, including different splice variants as well as full length proteins and protein fragments, these approaches are generally time-consuming and require the tedious process of identifying the prey associated with cell clones recovered in the screening process. One way to avoid this identification step involves the use of reverse transfected cell arrays. In this approach, arrayed prey plasmids are reverse transfected with a bait-expressing cell pool, thereby allowing rapid screening of a fixed collection of prey (Box 4). As the array format allows miniaturization and robotic handling and because prey identity is easily determined by its position in the array, both efficiency and throughput can be increased tremendously. This cell array concept has been successfully applied to MAPPIT (mammalian protein–protein interaction trap), a mammalian two-hybrid developed in our own lab where bait and prey are tethered to fragments of cytokine receptors whose functions are restored upon their complementation [38] (Figure 1c). One of the strengths of this methodology is that activation of the readout relies not only on bait–prey interaction but also requires addition of the proper cytokine ligand, thus adding an extra level of control over false positives. Together with LUMIER and a YFP-based complementation assay, MAPPIT is one of the mammalian two-hybrid approaches that was incorporated into the validation tool-kit used to retest Y2H data resulting from the ongoing proteome-wide human interactome mapping effort [10]. In a pilot study where MAPPIT was coupled to the cell array format, a 384-well microplate array of ~1900 full length human preys was used to identify novel proteins that associate with SKP1 (S-phase-kinase-associated protein-1) or Elongin C, subunits of multi-protein-type E3 ubiquitin ligase complexes [39] (Box 4). Downscaling the cell array approach to glass slides, as recently illustrated in a small-scale experiment using M2H s.s. [40], should further increase efficiency.
Box 4. Reverse transfected cell arrays.
Several steps are required to produce reverse transfected cell arrays, i.e. arrays of small cell populations that are reverse transfected with a different nucleic acid (Figure Ia). First, a collection of nucleic acids (plasmid DNAs, PCR products, siRNAs, shRNAs) are mixed with a transfection agent and gelatin and are spotted on a carrier, which can be a glass slide or a microtiter plate. Next, cells are seeded on the carrier, forming a lawn on top of the nucleic acid spots. Owing to the presence of a transfection agent, the nucleic acids are taken up by the cells growing on the spots. The gelatinous matrix immobilizes the nucleic acids, spatially restricting the nucleic acid transfer. This localized reverse transfection results in an array of cell clusters each containing a distinct nucleic acid. When siRNAs are used, a different gene is targeted for silencing in each cluster of the cell array. Similarly, if expression plasmids are utilized, each spot of the cell array expresses a different gene. The stability of the nucleic acid-transfection agent mixes allows repeated and/or parallel testing of large collections of nucleic acids. The use of glass slides as a carrier renders the assay economical with regard to reagent consumption both for array production (transfection reagents) and for signal readout. High-density reverse transfection arrays can contain several thousand spots on a single slide, each accommodating the transfection of 30–500 cells depending on spot size and cell type. This level of miniaturization, however, requires strong signals and/or sensitive detection methods. Microtiter plate arrays commonly generate more robust signals, at the expense of reduced cost-effectiveness and throughput.
Reverse transfected cell arrays were first developed by the Sabatini group; their original approach arrayed a collection of expression plasmids on glass slides and screened the resulting cell microarray for proteins involved in tyrosine kinase signalling, apoptosis and cell adhesion [58]. More recently, the assay primarily has been used to screen arrays of siRNAs, enabling rapid identification of genes involved in a particular phenotype [59].
The MAPPIT approach incorporates the array format into a mammalian two-hybrid assay (Figure Ib [39]). Here, a cell array is generated starting from a 384-well microtiterplate array which contains a collection of prey plasmids mixed with a luciferase reporter gene. These arrays are overlaid with cells expressing a particular bait, yielding a different bait-prey combination in each well of the array. This setup allows rapid screening of a large collection of preys for novel bait interaction partners.
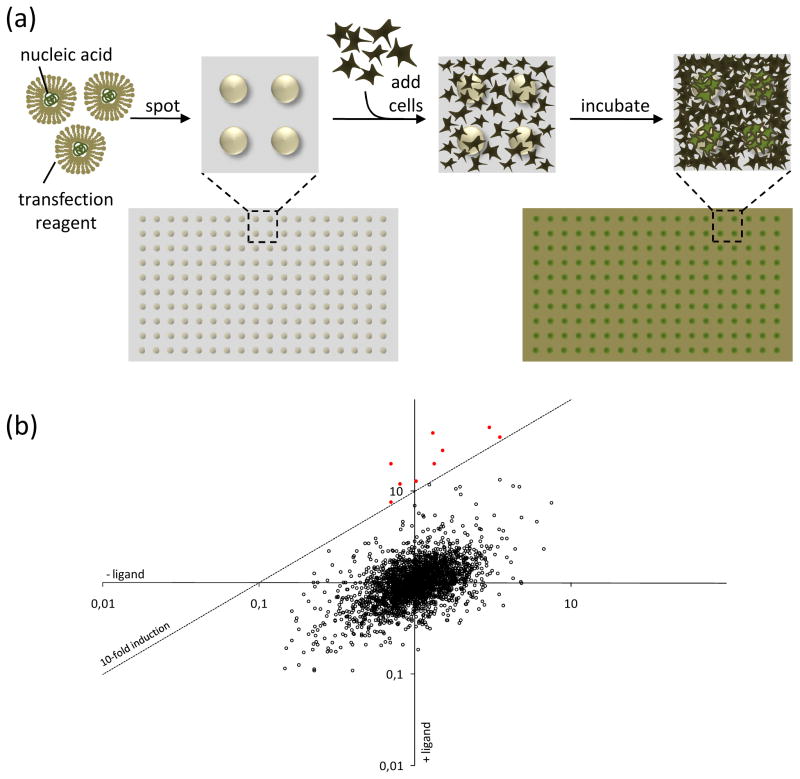
Figure I. Reverse transfected cell arrays and their application in MAPPIT.
(a) Generation of a cell array. Reverse transfection mixtures containing nucleic acids complexed with a transfection reagent are spotted onto a carrier surface. After drying, these arrays are overlaid with cells and incubated. Cells growing on top of the spots are (reverse) transfected with the nucleic acids present in the dried reverse transfection complexes, resulting in an array of transfected cells.
(b) Result of an array MAPPIT screen. MAPPIT is an inducible assay that requires stimulation with an appropriate cytokine ligand. To exploit this additional level of control, luciferase signals obtained from samples treated with ligand are corrected for the background signal from samples that were not ligand-stimulated. The resulting induction factor is used to score interactions. In the conceptual example depicted here, each dot represents a different prey being combined with the bait that is being screened. For each bait–prey combination the normalized luciferase activity obtained with and without addition of the ligand are plotted against each other. Bait–prey combinations yielding a signal above a user-defined cut-off (dashed line; here corresponding to 10-fold induction of ligand treated versus untreated) score positive (red dots).
The increase in throughput that can be obtained by using cell arrays should make proteome-wide interaction mapping feasible in mammalian cells. Such a goal seems sensible in light of the complementarity in interactome coverage that has been observed in benchmarking experiments comparing Y2H with mammalian two-hybrids, i.e. no single approach is able to uncover a complete protein interaction network [10]. In addition, the increased scale should allow rapid evaluation of interaction networks under varying physiological conditions induced by differential treatment of parallel sets of an array, helping to map interaction network dynamics.
Applications in drug discovery
The majority of marketed drugs target single proteins, mainly enzymes and membrane receptors. These protein classes are very attractive for drug development because they exhibit well-defined catalytic pockets or ligand binding domains that can readily be targeted with small molecular weight compounds. A notable issue with these classical drug targets is their limited number. Compared to single protein targets, protein–protein interactions represent a nearly unlimited resource for therapeutic intervention. On the other hand, protein interactions also represent a greater challenge for manipulation: interaction interfaces often consist of large and flat surfaces that are difficult to disrupt with small organic molecules. However, the identification within such seemingly featureless surfaces of compact ‘hot spots’ that significantly contribute to the affinity of the association between proteins boosted the interest in protein–protein interactions as drug targets [41–43]. As a result, a number of protein–protein interaction disruptors are currently proceeding through clinical trials, e.g. nutlins (which disrupt the p53–Mdm2 interaction [44]) and ABT-737 (which targets the interface between Bcl-2 family members [45]).
As they operate in intact mammalian (human) cells, mammalian two-hybrid technologies provide excellent opportunities to assist in the quest for compounds which target protein–protein interactions. Apart from their application in the identification of interacting protein targets, most available approaches can detect compounds that directly interfere with protein–protein associations by disruption of the interaction interface or through allosteric effects. To date, however, such examples are limited to exemplary cases (e.g. [25,28,46–49]). Although details about large-scale compound screening campaigns are unlikely to find their way into the public literature, a number of reports describe the successful application of mammalian two-hybrids in screening large compound collections. A recent example used a β-galactosidase complementation assay to identify molecules that inhibit inducible nitric oxide synthase (iNOS) homodimerization, which is essential for activation of this NO synthesizing enzyme. A screen of ~800,000 compounds identified several candidate iNOS inhibitors [50]. Another widely recognized challenge in drug development is restricting a drug’s action spectrum to the intended target in order to minimize side-effects. When a drug binds its (un)intended target, a biochemical pathway is perturbed, leading to the (un)desired phenotype. As biochemical pathways are characterized by sets of protein–-protein interactions, tracking these interactions can be an efficient way to monitor cellular drug effects. Mammalian two-hybrid technologies are particularly useful for this purpose as they can assess (changes in) protein interaction networks in living cells. A recent report confirmed the validity of this concept using YFP-based protein-fragment complementation in human cells [51]. PCAs were developed for 49 protein pairs that represent well-characterized protein interactions in 10 different cellular processes or pathways. These intracellular ‘sensors’ were individually expressed in a standard cell line and the effect of 107 drugs was assessed both quantitatively and qualitatively using fluorescence microscopy. Clustering of the resulting pharmacological profiles, together with knowledge of drug-dependent phenotypes, pointed to unexpected effects for a number of the drugs tested. In addition, the interaction pattern of a subset of 25 protein pairs proved to be highly predictive of an antiproliferative phenotype, thus demonstrating the prognostic power of this approach [51].
A more direct way to explain or predict the (off-target) effects of drugs is offered by a particular adaptation of the MAPPIT technique. In MASPIT (mammalian small molecule-protein interaction trap), a three-hybrid setup displays a chemical compound as bait, enabling direct identification of its protein targets [52]. A cDNA library screen for proteins which bind the ABL tyrosine kinase inhibitor PD173955 identified not only ABL, but also a number of other, known and novel, kinase targets. This approach also should prove particularly useful for cases in which no cellular target is known.
Concluding remarks
Numerous mammalian two-hybrid technologies, which offer ample opportunities to study mammalian interactome dynamics, are reaching maturity. Most of these methods are highly complementary not only in the subset of interactions they can detect, but also in the nature (spatial, temporal, and contextual) and level of detail of the information they generate. As the throughput at which these approaches are being applied increases, we can expect a growing amount of qualitative interactome data to supplement the protein network scaffold that is currently being generated by proteome-wide Y2H- and affinity purification-based screening campaigns. Further integration with other ‘omics’ datasets, including those derived from large-scale gene and protein expression and metabolomics studies, should support systems biology efforts to capture cellular processes in network models. Such models will be of tremendous value to our basic understanding of the complexity of biological processes and as a predictive tool in disease monitoring and drug development.
Acknowledgments
Due to space limitations, we could not cover all developments in the field of mammalian two-hybrids. Therefore we apologize to colleagues whose research was not cited.
Our work is supported by grants from the Fund for Scientific Research – Flanders (G.0031.06N), Ghent University (GOA 12051401), the IUAP-6 (N° P6:28) and the NIH (R01 HG001715-11). I.L. is a postdoctoral fellow with the Fund for Scientific Research – Flanders.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Goehler H, et al. A protein interaction network links GIT1, an enhancer of huntingtin aggregation, to Huntington’s disease. Mol Cell. 2004;15:853–865. doi: 10.1016/j.molcel.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 2.Lim J, et al. A protein-protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell. 2006;125:801–814. doi: 10.1016/j.cell.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 3.Taylor IW, et al. Dynamic modularity in protein interaction networks predicts breast cancer outcome. Nat Biotechnol. 2009;27:199–204. doi: 10.1038/nbt.1522. [DOI] [PubMed] [Google Scholar]
- 4.Kocher T, Superti-Furga G. Mass spectrometry-based functional proteomics: from molecular machines to protein networks. Nat Methods. 2007;4:807–815. doi: 10.1038/nmeth1093. [DOI] [PubMed] [Google Scholar]
- 5.Suter B, et al. Two-hybrid technologies in proteomics research. Curr Opin Biotechnol. 2008;19:316–323. doi: 10.1016/j.copbio.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 6.Yu H, et al. High-quality binary protein interaction map of the yeast interactome network. Science. 2008;322:104–110. doi: 10.1126/science.1158684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boxem M, et al. A protein domain-based interactome network for C. elegans early embryogenesis. Cell. 2008;134:534–545. doi: 10.1016/j.cell.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simonis N, et al. Empirically controlled mapping of the Caenorhabditis elegans protein-protein interactome network. Nat Methods. 2009;6:47–54. doi: 10.1038/nmeth.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Venkatesan K, et al. An empirical framework for binary interactome mapping. Nat Methods. 2009;6:83–90. doi: 10.1038/nmeth.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Braun P, et al. An experimentally derived confidence score for binary protein-protein interactions. Nat Methods. 2009;6:91–97. doi: 10.1038/nmeth.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seet BT, et al. Reading protein modifications with interaction domains. Nat Rev Mol Cell Biol. 2006;7:473–483. doi: 10.1038/nrm1960. [DOI] [PubMed] [Google Scholar]
- 12.Wilkins MR, Kummerfeld SK. Sticking together? Falling apart? Exploring the dynamics of the interactome. Trends Biochem Sci. 2008;33:195–200. doi: 10.1016/j.tibs.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 13.Topper JN, et al. CREB binding protein is a required coactivator for Smad-dependent, transforming growth factor beta transcriptional responses in endothelial cells. Proc Natl Acad Sci U S A. 1998;95:9506–9511. doi: 10.1073/pnas.95.16.9506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zwart W, et al. PKA-induced resistance to tamoxifen is associated with an altered orientation of ERalpha towards co-activator SRC-1. EMBO J. 2007;26:3534–3544. doi: 10.1038/sj.emboj.7601791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wehrman TS, et al. A system for quantifying dynamic protein interactions defines a role for Herceptin in modulating ErbB 2 interactions. Proc Natl Acad Sci U S A. 2006;103:19063–19068. doi: 10.1073/pnas.0605218103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hammer MM, et al. A novel enzyme complementation-based assay for monitoring G-protein-coupled receptor internalization. FASEB J. 2007;21:3827–3834. doi: 10.1096/fj.07-8777com. [DOI] [PubMed] [Google Scholar]
- 17.von Degenfeld G, et al. A universal technology for monitoring G-protein-coupled receptor activation in vitro and noninvasively in live animals. FASEB J. 2007;21:3819–3826. doi: 10.1096/fj.07-9597com. [DOI] [PubMed] [Google Scholar]
- 18.Michnick SW, et al. Universal strategies in research and drug discovery based on protein-fragment complementation assays. Nat Rev Drug Discov. 2007;6:569–582. doi: 10.1038/nrd2311. [DOI] [PubMed] [Google Scholar]
- 19.Remy I, Michnick SW. Application of protein-fragment complementation assays in cell biology. Biotechniques. 2007;42:137–145. doi: 10.2144/000112396. [DOI] [PubMed] [Google Scholar]
- 20.Spotts JM, et al. Time-lapse imaging of a dynamic phosphorylation-dependent protein-protein interaction in mammalian cells. Proc Natl Acad Sci U S A. 2002;99:15142–15147. doi: 10.1073/pnas.232565699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kerppola TK. Visualization of molecular interactions by fluorescence complementation. Nat Rev Mol Cell Biol. 2006;7:449–456. doi: 10.1038/nrm1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kerppola TK. Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells. Annu Rev Biophys. 2008;37:465–487. doi: 10.1146/annurev.biophys.37.032807.125842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Remy I, et al. PKB/Akt modulates TGF-beta signalling through a direct interaction with Smad3. Nat Cell Biol. 2004;6:358–365. doi: 10.1038/ncb1113. [DOI] [PubMed] [Google Scholar]
- 24.Magliery TJ, et al. Detecting protein-protein interactions with a green fluorescent protein fragment reassembly trap: scope and mechanism. J Am Chem Soc. 2005;127:146–157. doi: 10.1021/ja046699g. [DOI] [PubMed] [Google Scholar]
- 25.Remy I, Michnick SW. A highly sensitive protein-protein interaction assay based on Gaussia luciferase. Nat Methods. 2006;3:977–979. doi: 10.1038/nmeth979. [DOI] [PubMed] [Google Scholar]
- 26.Stefan E, et al. Quantification of dynamic protein complexes using Renilla luciferase fragment complementation applied to protein kinase A activities in vivo. Proc Natl Acad Sci U S A. 2007;104:16916–16921. doi: 10.1073/pnas.0704257104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luker KE, et al. Kinetics of regulated protein-protein interactions revealed with firefly luciferase complementation imaging in cells and living animals. Proc Natl Acad Sci U S A. 2004;101:12288–12293. doi: 10.1073/pnas.0404041101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wehr MC, et al. Monitoring regulated protein-protein interactions using split TEV. Nat Methods. 2006;3:985–993. doi: 10.1038/nmeth967. [DOI] [PubMed] [Google Scholar]
- 29.Wehr MC, et al. Analysis of transient phosphorylation-dependent protein-protein interactions in living mammalian cells using split-TEV. BMC Biotechnol. 2008;8:55. doi: 10.1186/1472-6750-8-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Piston DW, Kremers GJ. Fluorescent protein FRET: the good, the bad and the ugly. Trends Biochem Sci. 2007;32:407–414. doi: 10.1016/j.tibs.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 31.Pfleger KD, Eidne KA. Illuminating insights into protein-protein interactions using bioluminescence resonance energy transfer (BRET) Nat Methods. 2006;3:165–174. doi: 10.1038/nmeth841. [DOI] [PubMed] [Google Scholar]
- 32.Gales C, et al. Real-time monitoring of receptor and G-protein interactions in living cells. Nat Methods. 2005;2:177–184. doi: 10.1038/nmeth743. [DOI] [PubMed] [Google Scholar]
- 33.Hein P, et al. Dynamics of receptor/G protein coupling in living cells. EMBO J. 2005;24:4106–4114. doi: 10.1038/sj.emboj.7600870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsumoto A, et al. Screening for nitric oxide-dependent protein-protein interactions. Science. 2003;301:657–661. doi: 10.1126/science.1079319. [DOI] [PubMed] [Google Scholar]
- 35.Mannick JB, et al. Fas-induced caspase denitrosylation. Science. 1999;284:651–654. doi: 10.1126/science.284.5414.651. [DOI] [PubMed] [Google Scholar]
- 36.Barrios-Rodiles M, et al. High-throughput mapping of a dynamic signaling network in mammalian cells. Science. 2005;307:1621–1625. doi: 10.1126/science.1105776. [DOI] [PubMed] [Google Scholar]
- 37.Remy I, Michnick SW. Regulation of apoptosis by the Ft 1 protein, a new modulator of protein kinase B/Akt. Mol Cell Biol. 2004;24:1493–1504. doi: 10.1128/MCB.24.4.1493-1504.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eyckerman S, et al. Design and application of a cytokine-receptor-based interaction trap. Nat Cell Biol. 2001;3:1114–1119. doi: 10.1038/ncb1201-1114. [DOI] [PubMed] [Google Scholar]
- 39.Lievens S, et al. Array MAPPIT: high-throughput interactome analysis in mammalian cells. J Proteome Res. 2009;8:877–886. doi: 10.1021/pr8005167. [DOI] [PubMed] [Google Scholar]
- 40.Fiebitz A, et al. High-throughput mammalian two-hybrid screening for protein-protein interactions using transfected cell arrays. BMC Genomics. 2008;9:68. doi: 10.1186/1471-2164-9-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arkin MR, Wells JA. Small-molecule inhibitors of protein-protein interactions: progressing towards the dream. Nat Rev Drug Discov. 2004;3:301–317. doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]
- 42.Domling A. Small molecular weight protein-protein interaction antagonists: an insurmountable challenge? Curr Opin Chem Biol. 2008;12:281–291. doi: 10.1016/j.cbpa.2008.04.603. [DOI] [PubMed] [Google Scholar]
- 43.Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature. 2007;450:1001–1009. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- 44.Vassilev LT, et al. In vivo activation of the p 53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 45.Oltersdorf T, et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 46.Eyckerman S, et al. Reverse MAPPIT: screening for protein-protein interaction modifiers in mammalian cells. Nat Methods. 2005;2:427–433. doi: 10.1038/nmeth760. [DOI] [PubMed] [Google Scholar]
- 47.Galarneau A, et al. Beta-lactamase protein fragment complementation assays as in vivo and in vitro sensors of protein protein interactions. Nat Biotechnol. 2002;20:619–622. doi: 10.1038/nbt0602-619. [DOI] [PubMed] [Google Scholar]
- 48.Pattyn E, et al. MAPPIT (MAmmalian Protein-Protein Interaction Trap) as a tool to study HIV reverse transcriptase dimerization in intact human cells. J Virol Methods. 2008;153:7–15. doi: 10.1016/j.jviromet.2008.06.021. [DOI] [PubMed] [Google Scholar]
- 49.Wehrman T, et al. Protein-protein interactions monitored in mammalian cells via complementation of beta -lactamase enzyme fragments. Proc Natl Acad Sci U S A. 2002;99:3469–3474. doi: 10.1073/pnas.062043699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mallinder PR, et al. Identification of iNOS inhibitors using InteraX. J Biomol Screen. 2009;14:263–272. doi: 10.1177/1087057109331476. [DOI] [PubMed] [Google Scholar]
- 51.MacDonald ML, et al. Identifying off-target effects and hidden phenotypes of drugs in human cells. Nat Chem Biol. 2006;2:329–337. doi: 10.1038/nchembio790. [DOI] [PubMed] [Google Scholar]
- 52.Caligiuri M, et al. MASPIT: three-hybrid trap for quantitative proteome fingerprinting of small molecule-protein interactions in mammalian cells. Chem Biol. 2006;13:711–722. doi: 10.1016/j.chembiol.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 53.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 54.Rual JF, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 55.Stelzl U, Wanker EE. The value of high quality protein-protein interaction networks for systems biology. Curr Opin Chem Biol. 2006;10:551–558. doi: 10.1016/j.cbpa.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 56.Stelzl U, et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 57.Remy I, Michnick SW. Visualization of biochemical networks in living cells. Proc Natl Acad Sci U S A. 2001;98:7678–7683. doi: 10.1073/pnas.131216098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ziauddin J, Sabatini DM. Microarrays of cells expressing defined cDNAs. Nature. 2001;411:107–110. doi: 10.1038/35075114. [DOI] [PubMed] [Google Scholar]
- 59.Wheeler DB, et al. Cell microarrays and RNA interference chip away at gene function. Nat Genet. 2005;37(Suppl):S25–30. doi: 10.1038/ng1560. [DOI] [PubMed] [Google Scholar]