SUMMARY
The polyomavirus JC (JCV) causes the demyelinating disease progressive multifocal leukoencephalopathy (PML). Infection by JCV is very common in childhood after which the virus enters a latent state, which is poorly understood. Under conditions of severe immunosuppression, especially AIDS, JCV may reactivate to cause PML. Expression of JC viral proteins is regulated by the JCV non-coding control region (NCCR), which contains an NF-κB binding site previously shown to activate transcription. We now report that C/EBPβ inhibits basal and NF-κB-stimulated JCV transcription via the same site. Gel shift analysis showed C/EBPβ bound to this region in vitro and ChIP assays confirmed this binding in vivo. Further, a ternary complex of NF-κB/p65, C/EBPβ-LIP and JCV DNA could be detected in co-immunoprecipitation experiments. Mutagenesis analysis of the JCV NCCR indicated p65 and C/EBPβ-LIP bound to adjacent but distinct sites and that both sites regulate basal and p65-stimulated transcription. Thus C/EBPβ negatively regulates JCV, which together with NF-κB activation, may control the balance between JCV latency and activation leading to PML. This balance may be regulated by proinflammatory cytokines in the brain.
Keywords: C/EBPβ, NF-κB, JC Virus, Progressive multifocal leukoencephalopathy, cytokines, viral latency
INTRODUCTION
1.1. The polyomavirus JC and progressive multifocal leukoencephalopathy
The high prevalence of antibodies in human sera against the human polyomavirus JC, also called JC virus (JCV), indicates that JCV infection is widespread in the human population worldwide (Padgett and Walker, 1973; Walker and Padgett, 1983). It is generally accepted that JCV infects most people in childhood and then remains in a persistent but dormant state known as latency (Hou and Major, 2000; Khalili et al., 2006). The molecular mechanisms that underlie latency are poorly understood but latency can be defined as a state of asymptomatic, chronic, persistent infection where viral DNA may be detectable by PCR but expression of viral proteins cannot be detected. Many tissues have been reported to harbor latent JC virus including kidneys (Yogo et al., 1990; Zhong et al, 2006), tonsils (Monaco et al., 1996), GI tract (Del Valle et al., 2005; Ricciardiello et al., 2000) and brain (Elsner and Dorries, 1992; Greenlee et al., 2005; Mori et al., 1992; Perez-Liz et al., 2008; White et al., 1992). In most individuals, the level of JCV replication remains low and infection is asymptomatic. However in the context of severe immunosuppression, especially AIDS, JCV becomes reactivated in the CNS and replicates in oligodendrocytes leading to the fatal demyelinating disease, progressive multifocal leukoencephalopathy, PML (Berger, 2003; Khalili et al, 2008). The high prevalence of PML in patients infected with HIV-1 makes it an AIDS-defining disease (Holman et al., 1998). PML has also recently been observed in patients receiving the immunomodulatory drugs natalizumab (Berger and Houff, 2006; Khalili et al., 2007), rituximab (Berger, 2007; Carson et al., 2009) and efalizumab (FDA Public Health Advisory, 2009).
The clinical signs of patients with PML depend on the location of the demyelinated lesions but common symptoms include headaches, limb weakness, and cognitive impairments (Khalili et al., 2008). While the demyelinating lesions are generally thought to be caused by the lytic destruction of oligodendrocytes, which produce myelin, it is clear that JCV can also replicate in astrocytes as judged by immunohistochemical labeling for viral capsid protein and the presence of virions observed by electron microscopy (Del Valle et al., 2008; Mázló et al., 2001).
The early events involved in JCV reactivation may involve indirect effects of immunosuppression, e.g., changes in cytokine profiles, or direct effects of HIV-1 on JCV transcription, e.g., the action of the HIV-1 transactivator protein Tat (reviewed by Khalili et al., 2006, 2008). Expression of JCV early and late genes is controlled by the non-coding control region (NCCR) of the circular viral genome. The NCCR contains the origin of DNA replication and is bidirectional and so regulates both early and late gene expression. The NCCR contains binding sites for cellular transcription factors and some of these are regulated by signaling pathways that lie downstream of cell surface receptors. Thus, JCV gene expression and reactivation may be regulated by extracellular cytokines and immunomodulators. In particular, earlier work indicated a role for the NF-κB pathway and proinflammatory cytokines, such as TNF-α, in the activation of JCV transcription (Mayreddy et al., 1996; Ranganathan and Khalili, 1993; Safak et al., 1999). The unique site for NF-κB is located in the NCCR on the early side of the origin of replication (Ranganathan and Khalili, 1993). Previous studies demonstrated that this site is a functional NF-κB binding site and activates JCV gene expression in response to PMA (Ranganathan and Khalili, 1993) and TNF-α (Mayreddy et al., 1996).
1.2. Transcription factor NF-κB
NF-κB is an inducible transcription factor that regulates the expression of many cellular and viral genes (Nabel and Baltimore, 1987; West et al., 2001). NF-κB exists in cells as a hetero- or homodimer consisting of the Rel family of proteins which is comprised of RelA (p65), RelB, c-Rel, p50/p105 and p52/p100. These are normally complexed in the cytoplasm with the inhibitor protein IκB (Ghosh and Karin, 2002; Phelps et al, 2000). Upon stimulation by cytokines, upstream protein kinases are activated and IκB becomes phosphorylated and targeted for ubiquitination and degradation. This releases NF-κB to translocate to the nucleus where it activates gene expression (Karin and Ben-Neriah, 2000).
1.3. Transcription factor C/EBPβ
Another family of transcription factors that are modulated by cytokines is comprised of the CAAT/enhancer binding proteins (C/EBPs). The C/EBP family contains six members (α, β, γ, δ, ε, and ζ), which contain a C-terminal DNA binding domain, a leucine zipper domain that mediates homo- and heterodimerization and an N-terminal transactivation domain (Ramji and Foka, 2002). Further diversity is provided by the generation by the use of alternate translation initiation sites resulting in production of shortened isoforms with different N-termini from the same mRNA. For example, in addition to full-length (38 kD) C/EBPβ, two smaller forms of C/EBPβ exist, liver-enriched transcriptional-activator protein (C/EBPβ-LAP, 35 kD) and liver-enriched transcriptional-inhibitory protein (C/EBPβ-LIP, 20 kD), which have common C-termini containing the leucine-zipper and DNA-binding domains but different N-termini resulting in changes to the transactivation domain (Ossipow et al, 1993). C/EBP proteins are regulated by cytokines and play important roles in many cellular processes (Ramji and Foka, 2002).
Direct physical and functional association can occur between members of NF-κB and C/EBP proteins involving interaction of the Rel domain of NF-κB with the bZIP domain of C/EBP (Stein et al., 1993). Through such interactions, C/EBP can co-operate with NF-κB to regulate cellular promoters, e.g., amyloid A (Shimizu and Yamamoto, 1994), IL-6 (Ray and Ray, 1995), and IL-8 (Stein and Baldwin, 1993) and viral promoters, e.g., avian leukosis virus (Bowers et al., 1996), HIV-1 (Mukerjee et al., 2007) and BK virus (BKV)(Gorrill and Khalili, 2005). The juxtaposition of p65 and C/EBPβ binding sites is important for their co-operation at the HIV-1 LTR (Mukerjee et al., 2007) and BKV promoter (Gorrill and Khalili, 2005). A possible role for C/EBPβ in the regulation of JCV has not been previously addressed.
1.4. NF-κB, C/EBPβ and JCV reactivation
As described in Section 1.1, JCV transcription is activated by NF-κB via a p65 binding site that lies on the early side of the NCCR (Ranganathan and Khalili, 1993) and is conserved in closely related BKV where NF-κB co-operates with C/EBPβ to stimulate transcription (Gorrill and Khalili, 2005). We have now investigated the corresponding DNA region in JCV and find that NF-κB and C/EBPβ regulate JCV transcription but that, unlike BKV, C/EBPβ has a negative effect.
MATERIALS AND METHODS
2.1. Cell culture, transfection and plasmids
U-87 MG human glioblastoma and SVGA (SV40 T-antigen transformed human glial cells) were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and were transfected using the calcium phosphate precipitation method (Graham and van der Eb, 1973) except for the ChIP experiments where Fugene6 was used according to the manufacturer’s instruction (Roche). Reporter constructs, JCVE-CAT and JCVL-CAT contained the JCV promoter from the Mad-1 strain linked to the chloramphenicol acetyltransferase (CAT) gene in the early and late orientations respectively (Chen and Khalili, 1995). The JCVE-CAT promoter mutants m1 and m2 were made by site-directed mutagenesis using the QuickChange II XL Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) with double stranded oligonucleotides with the sequences 5’-aaaaacaactcaatttccctggcctcc-3’ and 5’-aaaaacaagggactctccctggcctcc-3’ respectively. The expression plasmids pCMV-p65, pCMV-CHOP and pCMV-C/EBPβ were described previously (Gorrill and Khalili, 2005; Mameli et al, 2007). pCMV-LIP and pCMV-LAP were kindly provided by Dr. Ueli Schibler, University of Geneva.
2.2. Antibodies
Mouse monoclonal anti-C/EBPβ (H7, sc-7962, Santa Cruz Biotechnology, Inc., Santa Cruz, CA) which recognizes all three C/EBPβ isoforms and rabbit polyclonal anti-p65 (C-20, sc-372, Santa Cruz) were used for immunoprecipitation and Western blots.
2.3. Transient transfection assays
U-87 MG or SVGA cells were transfected with the JCVE-CAT, m1JCVE-CAT, m2JCVE-CAT or JCVL-CAT reporter constructs alone (5 µg) or in combination with pCMV-C/EBPβ, pCMV-LIP, pCMV-LAP and/or pCMV-p65 plasmid(s). The total amount of transfected DNA was normalized with empty vector DNA. When p65 siRNA was used, 50 pmol of Smartpool p65 siRNA (Dharmacon, Lafayette, CO) was included in the transfection. When PMA was used, 100 ng/ml tetradecanoyl phorbol acetate (PMA) was added to the cultures 90 min before harvesting. Chloramphenicol acetyl transferase (CAT) activity of samples was determined as previously described (Safak et al., 2001; Romagnoli et al., 2008).
2.4. Adenoviral vector expressing siRNA directed against C/EBPβ
To construct an adenoviral vector to deliver siRNA to silence CEBPβ, a target sequence described by Goransson et al. (2005) was used. This target sequence was 5’-GAAGACCGTGGACAAGCAC-3’. This sequence was used to design top and bottom strands of small hairpin RNA (shRNA) using the Clontech online designer tool (http://bioinfo.clontech.com). A unique NheI restriction site was introduced to facilitate identification of recombinant clones. The sequence of top and the bottom strands were: 5'-gatccgaagaccgtggacaagcacttcaagagagtgcttgtccacggtcttcttttttgctagcg-3' and 5'-aattcgctagcaaaaaagaagaccgtggacaagcactctcttgaagtgcttgtccacggtcttcg-3' respectively. These two strands were annealed and cloned into the BamHI and EcoRI site downstream of the human U6 promoter sites of the RNAi ready plasmid pSIREN-DNR (BD Biosciences, San Jose, CA). Recombinant clones were screened using the restriction enzyme NheI and the resulting plasmid was used to clone the C/EBPβ shRNA-expressing cassette into the BD Adeno-X™ Expression System 1 by following procedures recommended in the User Manual, PT3739-1 (BD Biosciences). Adenoviral vector was grown and purified by cesium chloride density gradient centrifugation. Ad-null control virus was generated in a similar fashion using empty shuttle plasmid with no transgene as previously described (White et al, 2006). In U-87 MG cell transfection experiments, adenovirus was added to cultures at an moi of five, three days prior to transfection, which was found to be optimal for downregulation of C/EBPβ protein level.
2.5. Gel Shift Assays
Cells were transfected with and without the expression plasmids for 48 hours and harvested. Nuclear proteins were then extracted and 10 µg were incubated with 50,000 cpm of a [32P]-labeled double-stranded oligodeoxyribonucleotide probe as previously described (Raj and Khalili, 1994; Romagnoli et al., 2008). The following JCV-specific probes were used in these gel shift experiments. Nucleotide numbers refer to the Mad-1 reference strain of JCV (GenBank # NC_001699, formerly J02226), which is a 5130 base pair circular DNA genome. Note that the ORI oligonucleotide spans the numerical origin.
κB: 5’ - aaaacaagggaatttccctggcctc - 3’ (5052-5078).
ORI: 5' - ggaggcggaggcggcctcggcctcc - 3' (5118 to 2).
CR1: 5’ - cctgtatatataaaaaaaagggaag - 3’ (11 to 35)
CR2: 5’ - ggatagctgccagccaagcatgagc - 3’ (36 to 60)
CR3: 5’ - tcatacctagggagccaaccagcta - 3’ (61 to 85)
CR4: 5’ - acagccagtaaacaaagcacaaggc - 3’ (86 to 110)
The following mutated forms of the κB region were also used:
m1: 5’ - aaaaacaaCTCaatttccctggcctcc - 3’ m2: 5’ - aaaaacaagggaCTCtccctggcctcc - 3’
m3: 5’ - aaaaacaagggaattGAActggcctcc - 3’
For gel shifts in the presence of antibody, 2 µl of 1 mg/ml of antibody described above or non-immune mouse or rabbit serum was added.
2.6. Co-immunoprecipitation and Western blots assay
Co-immunoprecipitation and Western blot assays were performed as previously described (White et al., 2006). U-87 MG cells were transfected with C/EBPβ-LIP and p65 expression plasmids and after 48 hours cells were lysed and 300 µg cell extract incubated with anti-p65 or anti-C/EBPβ antibody or control normal rabbit serum for 2 h at 4°C in the presence or absence of 500 ng of the κB double-stranded oligonucleotide described above. Immunocomplexes were precipitated with protein ASepharose beads (Pharmacia, Peapack NJ) for an additional 45 min, washed, resolved by SDS-10% PAGE and analyzed by Western blotting as we have previously described (White et al., 2006).
2.7. ChIP assay
106 U-87 MG cells were transfected with 3 µg of plasmid containing the JCV genome (pBluescript-Mad-1) in combination with 3 µg of expression plasmid using Fugene6 according to the manufacturer’s instructions (Roche). ChIP was performed 48 hours after transfection as we have previously described (Amini et al., 2005) using the ChIP assay kit (Upstate Cell Signaling Solutions). Briefly, cross-linking was performed with formaldehyde and the DNA sheared by sonication. The cells were lysed and immunoprecipitation was performed with antibodies as indicated. DNA was extracted the following primers spanning the JCV NCCR used for PCR:
5’- cctccctattcagcactttgtcc - 3’ (Mad-1, 4989 to 5011), 5’ - ggccagctggtgacaagcc - 3’ (276-258).
PCR was 30 cycles (94°C for 30 sec, 55°C for 30 sec, 72°C for 30 sec) then 72°C for 7 min.
RESULTS
3.1. Effects of C/EBPβ isoforms on transcription by the JCV early and late promoters: analysis of ectopic C/EBPβ expression
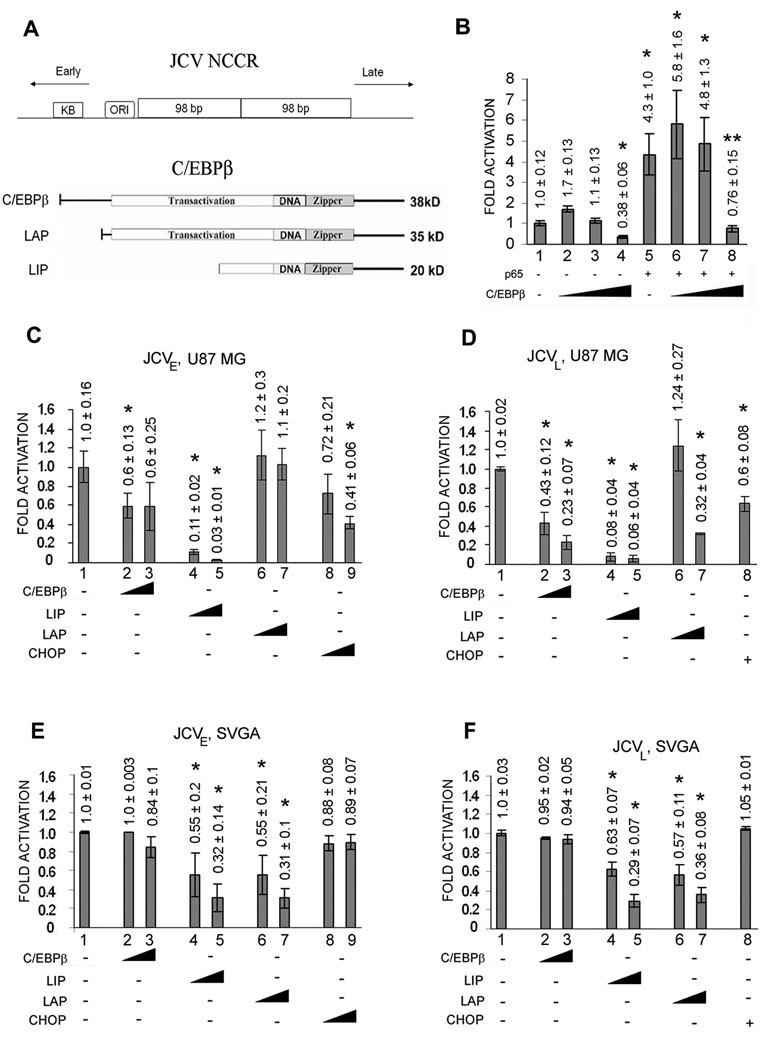
As described in the introduction, our earlier studies showed that Mad-1 JCV contains a single functional NF-κB site located in the NCCR between the origin of replication and the start codon for the early coding region. The upper part of Figure 1A schematically illustrates the location of the NF-κB binding site (labeled κB) relative to the early and late JCV transcription units and the origin of viral DNA replication (ORI). In the present study, we now investigate the interaction of NF-κB and C/EBPβ at this site and the effect of this interaction on JCV transcription. In the first experiment, we transfected U-87MG glioblastoma cells with the JCVE-CAT JCV early reporter plasmid in the presence or absence of p65 expression plasmid and tested the effect of inclusion of increasing amounts of C/EBPβ expression plasmid. As shown in Fig. 1B, p65 alone stimulated CAT expression about 4–5-fold (compare lanes 1 and 5). Expression of C/EBPβ did not affect CAT expression at low concentrations (lanes 2 and 3) but at a higher concentration, C/EBPβ inhibited the JCV early promoter (compare lanes 1 and 4). The asterisks in each panel denote lanes that are statistically significantly (P < 0.05) different from control (lane 1). The simulation by p65 was unaffected by low concentrations of C/EBPβ (lanes 5–7) but at a higher concentration of C/EBPβ (lane 8), p65 stimulation was significantly reversed and CAT activity was reduced to a similar level as the control (lane 1).
Figure 1. Effects of C/EBPβ, LIP, LAP and CHOP on the JCV early and late promoters in the absence and presence of T-antigen.
A. Upper Panel. Diagrammatic representation of the JCV NCCR DNA region showing the relative locations and orientations of the early and late promoters, the κB region containing the binding sites for NF-κB and C/EBPβ, the origin of viral DNA replication (ORI) and the 98 base-pair repeats. Lower Panel. Diagrammatic representation of the three protein isoforms of C/EBPβ produced by alternative translation initiation showing the transactivation domain, DNA-binding domain and leucine zipper domain. B. U-87 MG cells were transfected with the JCVE-CAT reporter plasmid in the presence or absence of pCMV-p65 and increasing amounts of pCMV-C/EBPβ (0, 0.05, 0.25, 1.25 µg). CAT activity was normalized to JCVE-CAT alone (lane 1) and presented as a histogram. The error bars encompass one standard deviation. The asterisks indicate lanes that are significantly different (P < .05) from the control (lane 1). C. U-87 MG cells were transfected with the JCVE-CAT reporter plasmid alone or with increasing amounts (0.25, 1.25 µg) of pCMV-C/EBPβ, LIP, LAP or CHOP. CAT activity was normalized to JCVE-CAT alone (lane 1) and presented as a histogram. The error bars encompass one standard deviation. D. As for C, except using JCVL-CAT instead of JCVE-CAT. 0.25 µg of pCMV-CHOP was used in lane 8. E. As for C, except using SVGA cells, which express SV40 T-antigen, in place of U-87 MG cells. F. As for C except using JCVL-CAT and SVGA. 0.25 µg of pCMV-CHOP was used in lane 8.
To further investigate the effect of C/EBPβ on JCV transcription, we undertook a comprehensive analysis of the effect of all three C/EBPβ isoforms on the JCV early and late promoters. The upper part of Figure 1A schematically illustrates the relationship of full-length C/EBPβ protein to its LAP and LIP isoforms which arise by the usage of alternative translation initiation sites within the same mRNA (Ossipow et al, 1993). We also included C/EBP-homologous protein (CHOP) in this analysis. CHOP is a protein that can interact with C/EBPβ and LAP and functions to modulate their effects on gene transcription (Fornace et al., 1988; Ron and Haebner, 1992). The effect of C/EBPβ, its isoforms (LIP and LAP) and CHOP on JCV expression in U-87 MG cells was examined for the early promoter (Fig. 1C) and late promoter (Fig. 1D). Results indicate inhibition by C/EBPβ (lanes 2 and 3) and a strong inhibition (92–97%, lanes 4 and 5) of both JCV early and late transcription by the C/EBPβ LIP isoform. To investigate these effects in the presence of T-antigen, the experiments were repeated in SVGA cells, which express SV40 large T-antigen and are permissive for JCV replication (Figs. 1E and 1F). Inhibition of transcription was observed with the both the LIP (lanes 4 and 5) and LAP (lanes 6 and 7) isoforms of C/EBPβ but to a lesser extent than seen in the absence of T-antigen. The asterisks denote lanes that are statistically significantly (P < 0.05) different from control (lane 1 in each panel). These data indicate that C/EBPβ LIP inhibited transcription from both the JCV early and late promoters in both the presence and absence of T-antigen and that this inhibition was stronger in the absence of T-antigen. For this reason, we chose the LIP isoform of C/EBPβ to be used in subsequent experiments.
3.2. Effects of C/EBPβ isoforms on transcription by the JCV early and late promoters: analysis of p65 expression, PMA treatment and RNA interference
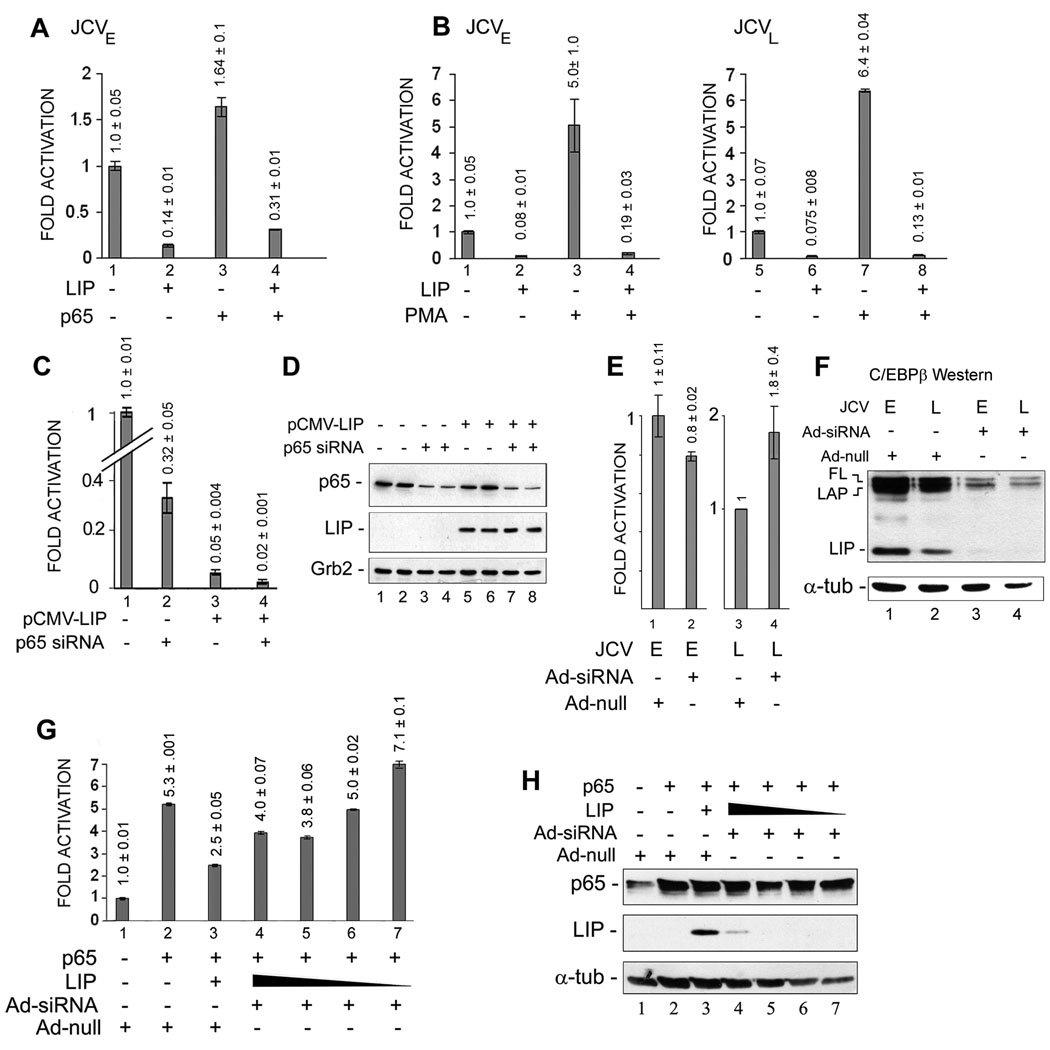
In the next series of experiments, we assessed the interplay of C/EBPβ LIP and p65 in regulating JCV transcription. Early transcription, both basal and p65-stimulated, was strongly inhibited by expression of C/EBPβ-LIP (Fig. 2A, compare lane 1 to 2 and lane 3 to 4). In the next experiments, we utilized the phorbol ester TPA, which activates a number of signal transduction pathways including the induction of NF-κB. As shown in Fig. 2B, PMA treatment stimulated transcription of both the early (5-fold) and late (6.4-fold) promoters. However inhibition of transcription was observed in cells expressing C/EBPβ-LIP for both the early promoter (left panel) and the late promoter (right panel), whether untreated or PMA-treated, to give a level of transcription that was between 0.075-0.19-fold of basal. When p65 was down-regulated by RNA interference, transcription of the early promoter was down-regulated to about one third of basal (Fig. 2C, compare lanes 1 and 2) while expression was down-regulated to about 5% of basal in cells expressing C/EBPβ LIP (Fig. 2C, compare lanes 1 and 3). When p65 was down-regulated by RNA interference in cells that were also expressing C/EBPβ-LIP, early promoter activity was down-regulated to about 2% of basal (Fig. 2C, compare lanes 1 and 4). All of these changes are statistically significant. Downregulation of p65 by siRNA and expression of C/EBPβ LIP in the experiment shown in Fig. 2C were verified by Western blot (Fig. 2D).
Figure 2. C/EBPβ-LIP reverses the transcriptional activation of p65.
A. U-87 MG cells were transfected with the JCVE-CAT reporter plasmid in the presence or absence of pCMV-p65 and/or pCMV-LIP. CAT activity was normalized to JCVE-CAT alone (lane 1) and presented as a histogram. The error bars encompass one standard deviation. The differences between all four bars are statistically significant. B. U-87 MG cells were transfected with the JCVE-CAT or JCVL-CAT reporter plasmid in the presence or absence of pCMV-LIP and either untreated or treated with PMA to induce NF-κB. The error bars encompass one standard deviation. The differences between all four bars in each of the histograms are statistically significant. C. U-87 MG cells were transfected with the JCVE-CAT in the presence or absence of pCMV-LIP and/or siRNA to p65. The specificity of this siRNA to interfere with p65 expression was demonstrated in previous studies (Gorrill et al., 2006; Mukerjee et al., 2007). The error bars encompass one standard deviation. The differences between all four bars are statistically significant. D. Downregulation of p65 expression and expression of C/EBPβ-LIP was verified by Western blot for the extracts used in Panel C with Grb2 serving as a loading control. E. U-87 MG cells were infected with either adenovirus expressing siRNA to C/EBPβ (Ad-siRNA) or control adenovirus (Ad-null) and transfected with either the JCVE-CAT or JCVL-CAT reporter plasmid as described in Materials and Methods, Section 2.4. The error bars encompass one standard deviation. The differences between the two bars each of the histograms are NOT statistically significant. F. Expression levels of the C/EBPβ isoforms, LIP LAP and full-length (FL) were measured by Western blot for the extracts used in Panel E with α-tubulin (α-tub) serving as a loading control. G. U-87 MG cells were infected with either adenovirus expressing siRNA to C/EBPβ (Ad-siRNA) or control adenovirus (Ad-null) and transfected with the JCVL-CAT reporter plasmid in the presence or absence of pCMV-p65 (1 µg) and/or pCMV-LIP (2, 1, 0.5 and 0.25 µg) as described in Materials and Methods, Section 2.4. The error bars encompass one standard deviation. The differences between each bar of the histogram are statistically significant. H. Expression of p65 and LIP was measured by Western blot for the extracts used in Panel G with α-tubulin (α-tub) serving as a loading control.
To further explore the role of C/EBPβ, we next employed an RNA interference approach. Since the downregulation of C/EBPβ by transfection of siRNA was not efficient (data not shown), we constructed an adenoviral vector that expressed siRNA directed against C/EBPβ (Ad-siRNA) and an empty adenoviral vector as a control (Ad-null), which were used in transduction experiments to downregulate C/EBPβ. As shown in Fig. 2E, downregulation had only a small effect on early and late transcription (compare lane 1 to 2 and lane 3 to 4, respectively) and these small differences were not statistically significant. This may indicate that a complementary or redundant transcription factor, perhaps another C/EBP family member, is able to compensate for the lack of endogenous C/EBPβ. The downregulation of endogenous C/EBPβ was verified by Western blot (Fig. 2F) showing that the LIP C/EBPβ isoform became was undetectable and the LAP and full-length (FL) isoforms were greatly reduced following the Ad-siRNA transduction (Fig. 2F, lanes 3 and 4). In addition, transduction experiments were also performed in the presence of transfected expression plasmids (Fig. 2G). In the presence of Ad-null, expression of the JCV late promoter was induced by p65 and this was reversed by C/EBPβ LIP as expected (Fig. 2G, lanes 1–3). In the presence of Ad-siRNA, the expression of the JCV late promoter was increased (lane 4) and was also further increased when the amount of C/EBPβ LIP plasmid was decreased (lanes 4–7) until it reached a level that was higher than that with p65 plasmid alone (compare lanes 2 and 7). Downregulation of C/EBPβ LIP by siRNA and expression of p65 were verified by Western blot for this experiment (Fig. 2H).
3.3. Binding of C/EBPβ to the JCV non-coding control region (NCCR)
3.3.1 Electrophoretic mobility shift assays (gel shift assays)
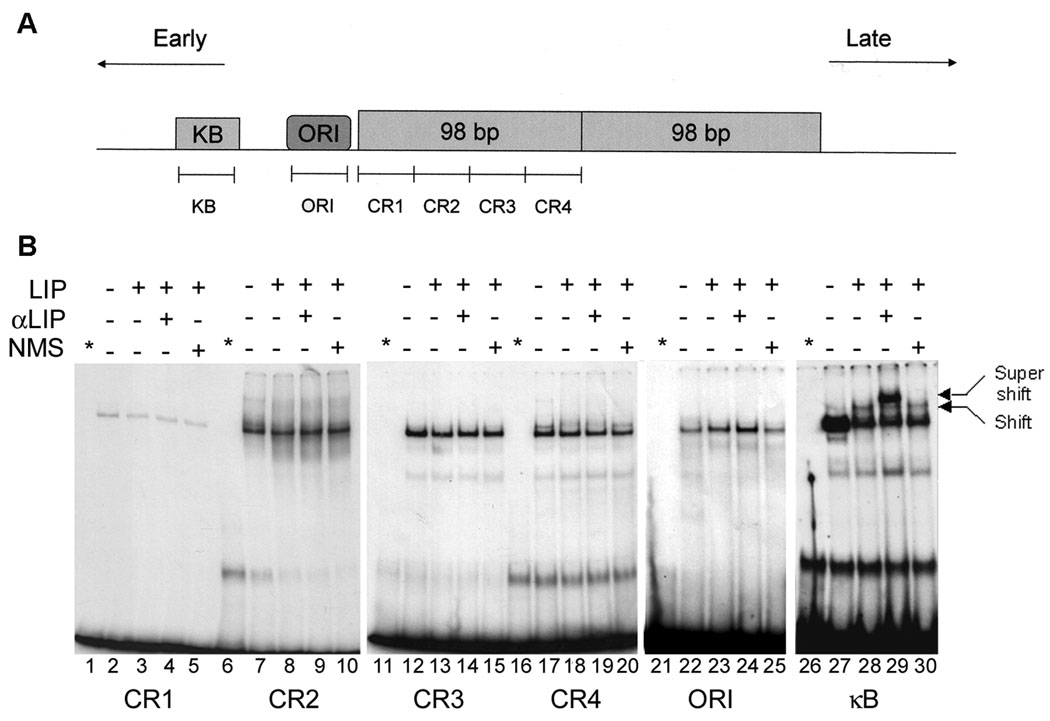
To define binding of C/EBPβ LIP to the JCV NCCR, a series of double-stranded oligonucleotides (Fig. 3A) were employed to perform of gel shift experiments using nuclear extracts from U-87 MG cells that had been transfected with or without the C/EBPβ-LIP expression plasmid. No binding of C/EBPβ LIP was detected to the oligonucleotides CR1-CR4 which span the 98 base-pair repeat region (Fig. 3B, lanes 1–20) nor to the origin of replication (Fig. 3B, lanes 21–25). A gel shift was detected with the oligonucleotide designated κB (compare lanes 27 and 28) which was supershifted with antibody to C/EBPβ (lane 29) but not by normal mouse serum (lane 30). The position of the supershift band in lane 29 indicates that it migrates more slowly, i.e., the αC/EBPβ antibody has bound to the complex demonstrating the specificity of the gel shift for C/EBPβ LIP. The band is also more intense suggesting that binding of the αC/EBPβ antibody may stabilize the interaction of C/EBPβ LIP with DNA. These data confirm the presence of an authentic binding site for the LIP isoform of C/EBPβ in the vicinity of the NF-κB site and shows that C/EBPβ-LIP does not bind to the other regions within the JCV NCCR.
Figure 3. C/EBPβ-LIP binds to the κB region but not other regions within the JCV NCCR.
A. Diagrammatic representation of the JCV NCCR showing the relative locations of the oligonucleotides used in the gel shift experiments. B. U-87 MG cells were transfected with or without pCMV-LIP (LIP) and nuclear extracts prepared. Gel shifts were performed with these extracts in the presence and absence of antibody to C/EBPβ (α-LIP) or nonimmune mouse serum (NMS). The positions of the Shift and Supershift bands are indicated. The asterisk indicates labeled probe alone.
3.3.2. Chromatin Immunoprecipitation assays (ChIP assays)
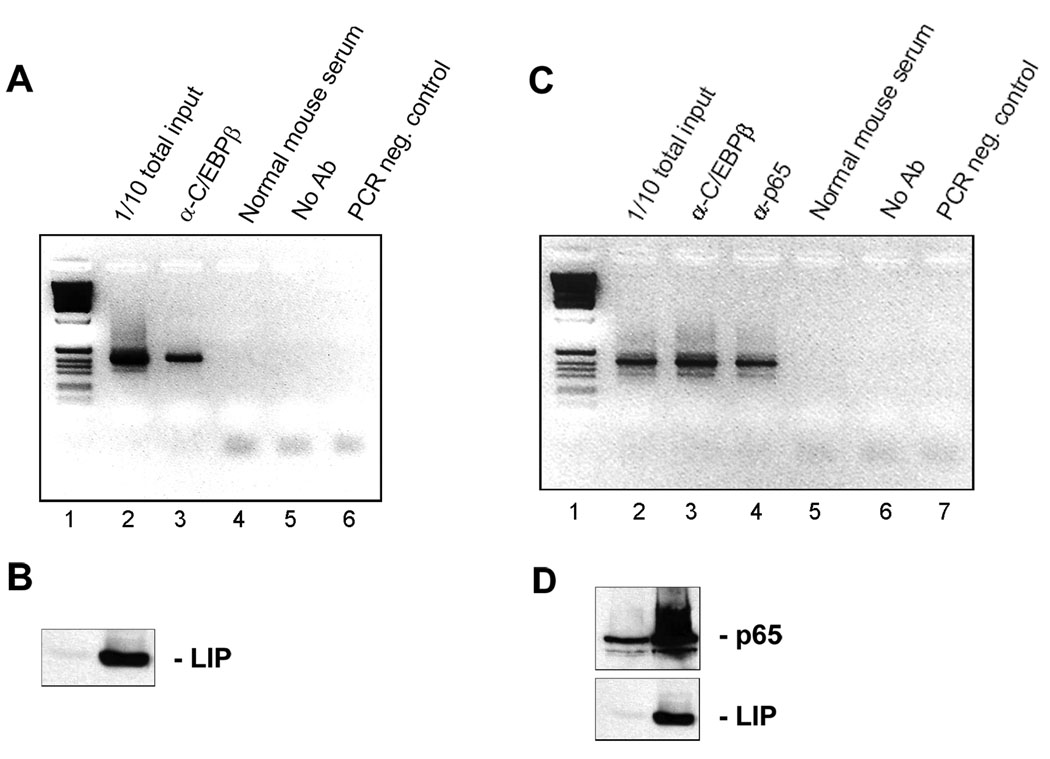
To establish that p65 and C/EBPβ LIP bound to the JCV NCCR in vivo, we performed ChIP assays using PCR primers that flank the NCCR. When U-87 MG cells were transfected with C/EBPβ LIP expression plasmid, cross-linked and immunoprecipitated, a band corresponding to the NCCR could be amplified in samples immunoprecipitated with the α-C/EBPβ antibody (Fig. 4A, lane 3) but not with normal mouse serum control (lane 4). Expression of C/EBPβ LIP in these cells was verified by Western blot (Fig. 4B). Similarly, when U-87 MG cells were transfected with both C/EBPβ-LIP and p65, a band corresponding to the NCCR could be amplified in samples immunoprecipitated with the α-C/EBPβ antibody (Fig. 4C, lane 3) and with the antibody to p65 (lane 4) but the antibody controls were negative (lanes 5 and 6).
Figure 4. C/EBPβ LIP and p65 bind to the JCV NCCR in vivo.
A. U-87 MG cells were transfected with pCMV-LIP, cross-linked and ChIP assay performed using antibody to C/EBPβ, normal mouse serum or no antibody. Lane 1 - molecular weight markers. B. Expression of C/EBPβ-LIP in A was verified by Western blot. C. U-87 MG cells were transfected with pCMV-LIP and pCMV-p65, cross-linked and ChIP assay performed using antibody to C/EBPβ, antibody to p65, normal mouse serum or no antibody. Lane 1 - molecular weight markers. D. Expression of p65 and C/EBPβ-LIP in C was verified by Western blot.
Next, we sought to define the molecular basis of the interaction of p65, C/EBPβ LIP and the κB DNA sequence. As described below, we first performed co-immunoprecipitation experiments to test for an association between p65 and C/EBPβ-LIP (Fig. 5). Then we created a series of mutations in the κB DNA sequence and tested them in gel shift experiments for binding of C/EBPβ LIP (Fig. 6A) and p65 (Fig. 6B).
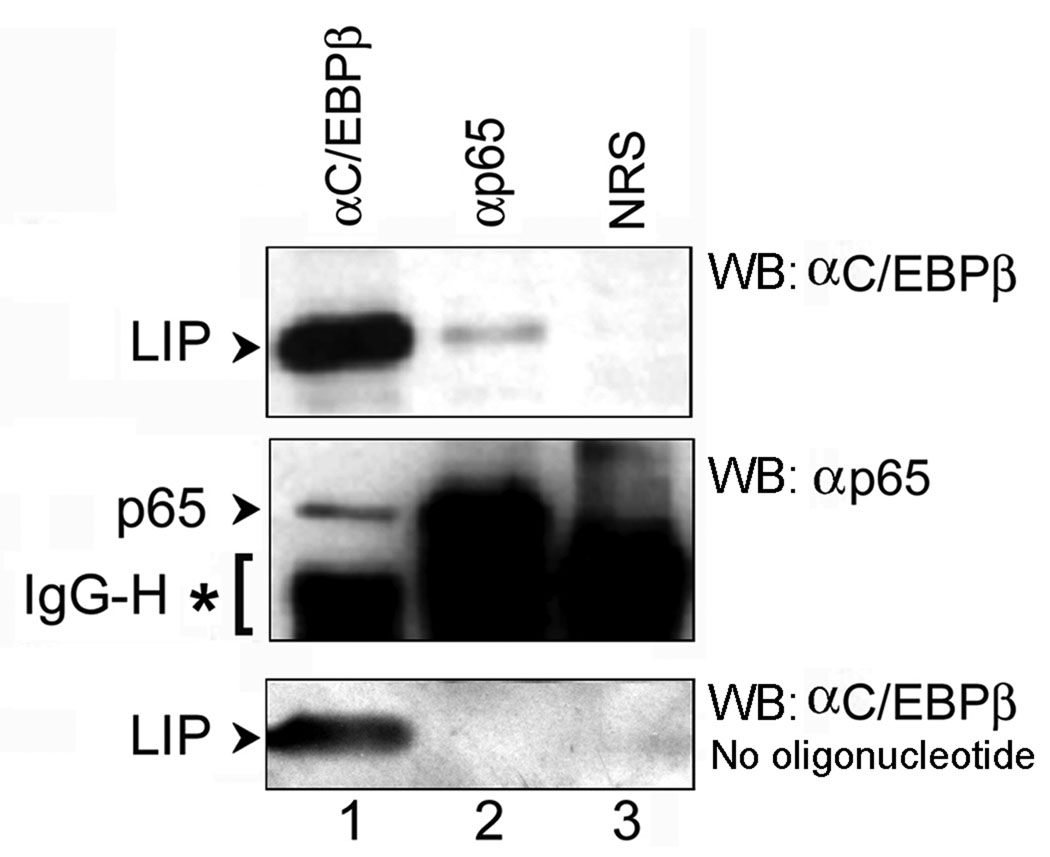
Figure 5. p65 and C/EBPβ LIP co-immunoprecipitate in the presence of κB oligonucleotide.
U-87 MG cells were transfected with expression plasmids for p65 and C/EBPβ LIP, cell extracts were prepared, incubated in the presence of κB oligonucleotide and immunoprecipitated (IP) with antibody to C/EBPβ (αC/EBPβ), p65 (αp65) or normal rabbit serum (NRS). After electrophoresis and transfer to membrane, Western blot (WB) was performed for C/EBPβ (top panel) and p65 (middle panel). Arrows indicate the bands corresponding to C/EBPβ-LIP and p65 in each panel. The band corresponding to the immunoglobulin heavy chain (IgG-H) is labeled with an asterisk. The immunoprecipitations were also performed in the absence of κB oligonucleotide lower panel).
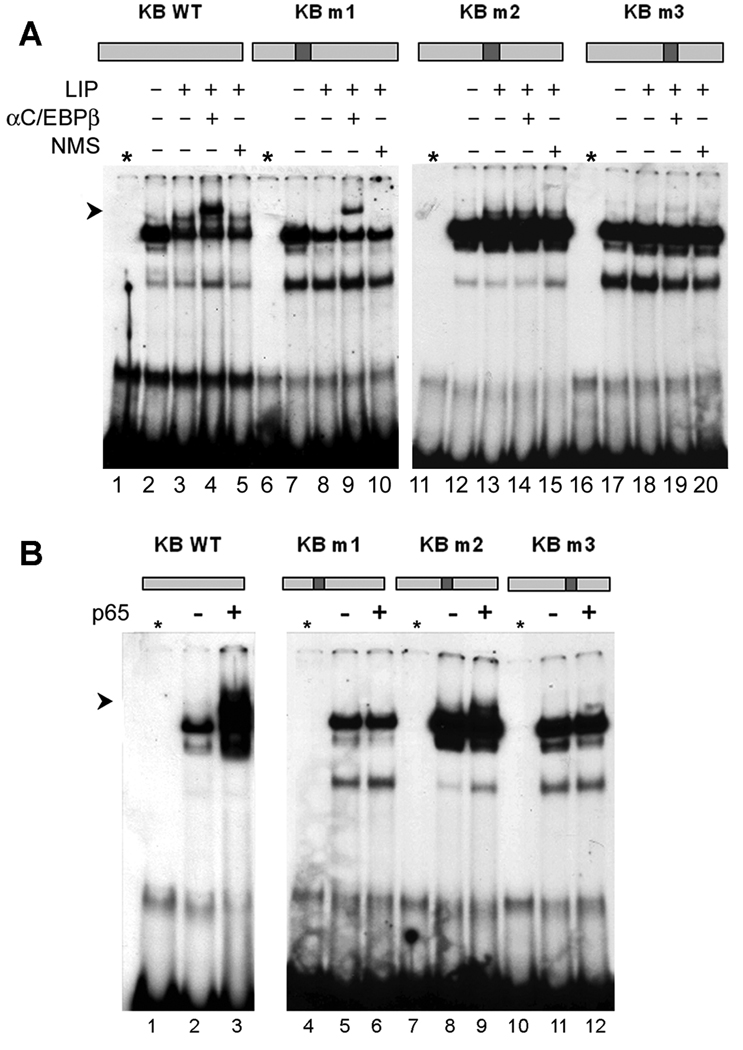
Figure 6. Mapping of C/EBPβ LIP and p65 binding sites within the κB region by mutagenesis.
Three mutations (m1, m2, m3) were introduced into the κB oligonucleotide and their positions are shown at the top of each panel. The sequences of all oligonucleotides are given in the Methods section. A. U-87 MG cells were transfected with and without pCMV-LIP and gel shifts performed with the wild-type κB and each of the mutant oligonucleotides as described in the legend to Figure 4. B. U-87 MG cells were transfected with and without pCMV-p65 and gel shifts performed with the wild-type κB and each of the mutant oligonucleotides.
3.3.3. Co-immunoprecipitation assays
To investigate the interaction of p65 and C/EBPβ-LIP with each other, we performed co-immunoprecipitation experiments. Extracts for co-immunoprecipitations were prepared from cells that had been co-transfected with p65 and C/EBPβ-LIP expression plasmids. Initial co-immunoprecipitation experiments with protein extracts alone did not indicate the occurrence of a p65-C/EBPβ-LIP complex (shown in the bottom panel of Fig. 5). To investigate the interaction of p65 and C/EBPβ-LIP in the presence of the κB DNA sequence, we next included a double-stranded oligonucleotide corresponding to the κB DNA sequence. Thus, the immunoprecipitation step in this experiment included the presence of the κB double-stranded oligonucleotide DNA and employed α-C/EBPβ, anti-p65 antibodies or normal rabbit serum as a negative control. Immune complexes were isolated and separated by electrophoresis followed by Western blotting for p65 and C/EBPβ. As shown in Fig. 5, C/EBPβ-LIP was precipitated not only by anti-C/EBPβ antibody which recognizes the LIP C/EBPβ isoform (top panel, arrow, lane 1), but also was co-precipitated by the anti-p65 antibody (top panel, arrow, lane 2). Similarly, p65 was precipitated not only by anti-p65 antibody (middle panel, arrow, lane 2), but also was co-precipitated by the anti-C/EBPβ antibody (middle panel, arrow, lane 1). The strong band that is found below p65 in all lanes is the immunoglobulin heavy chain and is indicated by an asterisk. It is important to note that the interaction between p65 and C/EBPβ LIP was observed only in the presence of double-stranded κB oligonucleotide DNA and not in its absence, where antibody to p65 failed to co-immunoprecipitate C/EBPβ-LIP (bottom panel, lane 2). Since co-immunoprecipitation required the presence of κB oligonucleotide, these data suggest that p65, C/EBPβ-LIP and the κB oligonucleotide form a ternary complex. However, it should be noted that the relative stoichiometry of the ternary complex relative to C/EBPβ LIP/DNA and p65/DNA binary complexes remains to be established.
3.3.4. Mutagenesis of κB binding site and gel shift assays
To further define the binding sites for the transcription factor C/EBPβ LIP on the κB oligonucleotide, we synthesized a series of mutant forms of the double-stranded κB oligonucleotide and tested them for binding of C/EBPβ-LIP by gel shift assay as shown in Fig. 6A. The exact sequences of these mutants, designated m1, m2 and m3 are given in the Materials and Methods, Section 2.5 and the positions of the mutations in the κB oligonucleotide are shown diagrammatically above the respective gel shift experiment in Fig. 6A. A band corresponding to a C/EBPβ LIP supershift (labeled with an arrow) is clearly evident for wild-type and mutant m1 (lanes 4 and 9). This band was abrogated in the case of m2 and m3 (lanes 14 and 19). Thus the C/EBPβ-LIP binding site lies within the region containing the sites of mutations m2 and m3.
The mutant κB oligonucleotides m1-m3 were also used to define the binding site for p65 (Fig. 6B). A band corresponding to a p65 gel shift (labeled with an arrow) was seen for wild-type κB oligonucleotide (lane 3) but not for mutant m1 (lane 6). This band was present for the other mutants, m2 and m3 (lanes 9, 12, 15 and 18). Thus the p65 binding site lies within the region containing the site of the mutation m1, i.e., to the left of the binding site for C/EBPβ-LIP.
3.4. Mutagenesis of κB binding site and promoter activity assays
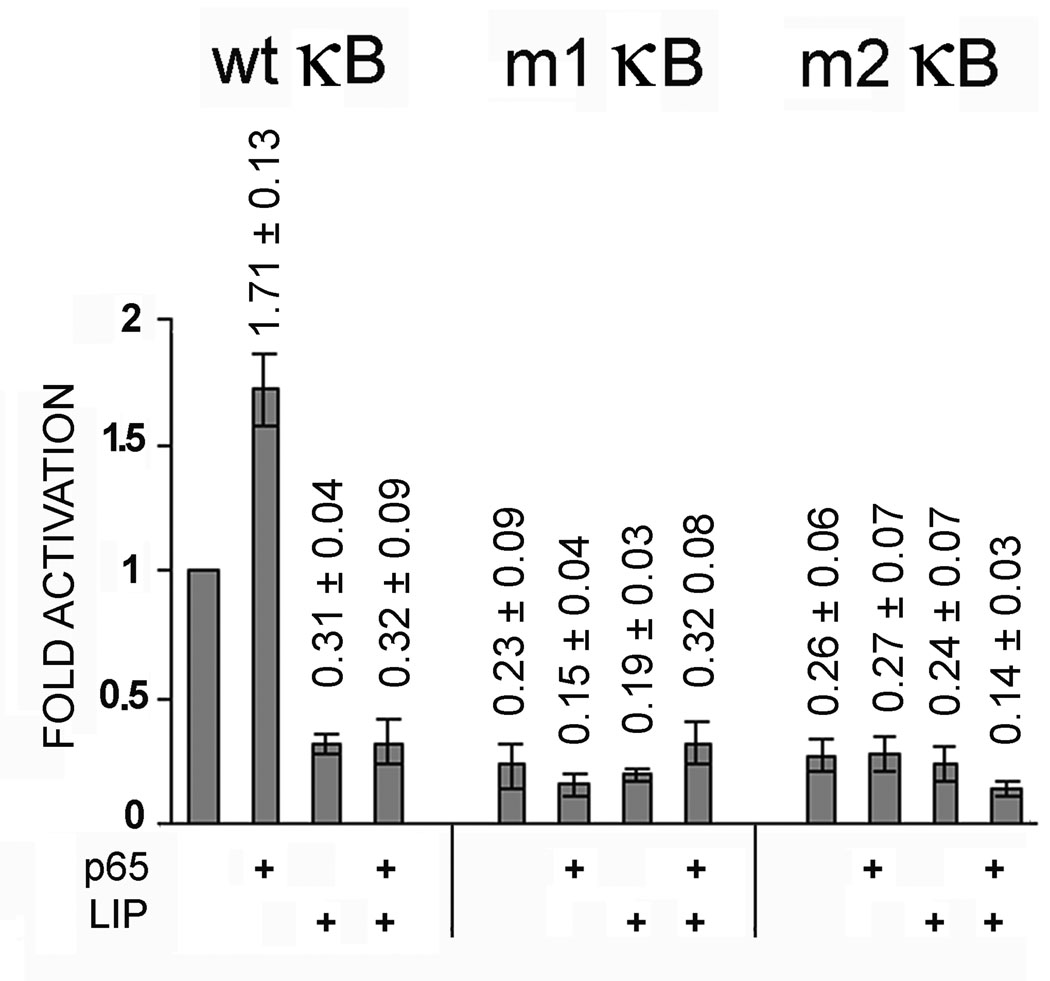
As shown above, κB oligonucleotide mutant m1 binds C/EBPβ-LIP but does not bind p65 and m2 binds p65 but does not bind C/EBPβ-LIP. To examine the effects of these mutations on the function on the JCV early promoter, mutations m1 and m2 were introduced into JCVE-CAT by site-directed mutagenesis as described in Materials and Methods, Section 2.1. Wild-type JCVE-CAT, m1 JCVE-CAT or m2 JCVE-CAT were transfected into U-87 MG cells in the presence or absence of expression plasmid for p65 or C/EBPβ LIP. As shown in Fig. 7, wild-type JCVE-CAT was activated by p65 and inhibited by C/EBPβ-LIP as expected. However, in the case of both m1 JCVE-CAT and m2 JCVE-CAT, the CAT activity was only 10–20% of the activity of wild-type JCVE-CAT under all conditions. In other words, abolition of either the NF-κB binding site or the C/EBPβ-LIP binding site caused a large reduction in basal transcription and abrogated the ability of the promoter to respond to p65.
Figure 7. Mutations m1 and m2 impair basal and p65-induced transcription of the JCV early promoter.
The mutation m1, which prevents p65 binding, was introduced into JCVE-CAT to create m1-JCVE-CAT. Similarly, the mutation m2, which prevents C/EBPβ LIP binding, was introduced into JCVE-CAT to create m2-JCVE-CAT. U-87 MG cells were transfected with wild-type JCVE-CAT, m1-JCVE-CAT or m2-JCVE-CAT in the presence or absence of PCMV-p65 and/or pCMV-LIP. CAT activity was normalized to wild-type JCVE-CAT alone (lane 1) and presented as a histogram. The error bars encompass one standard deviation.
DISCUSSION
We have analyzed the effects of p65 and C/EBPβ on the transcription of JCV and find that while expression of p65 enhances transcription, expression of C/EBPβ isoforms, especially C/EBPβ-LIP, inhibits transcription even when p65 is co-expressed. The effects of p65 and C/EBPβ were confirmed by RNA interference experiments and are mediated through a site that is located between the viral origin of replication and the start site of the early region. Earlier work had indicated a role for this unique NF-κB site in the activation of JCV transcription by proinflammatory cytokines, such as TNF-α via the NF-κB signaling pathway (Mayreddy et al., 1996; Rangathan and Khalili, 1993; Safak et al., 1999). Results from gel shift experiments showed that C/EBPβ-LIP also binds to this site in vitro as does NF-κB p65 while ChIP assays indicate that the binding of C/EBPβ-LIP and p65 to this site occurs in living cells.
The DNA region that binds p65 and C/EBPβ-LIP is conserved in the closely related polyomavirus, BKV (White et al. 2009). However, in the case of BKV, the interaction between p65 and C/EBPβ stimulates rather than inhibits transcription (Gorrill and Khalili, 2005). Perhaps differences in cis-acting promoter DNA elements between JCV and BKV modify the effects of NF-κB and C/EBPβ on transcription or alternatively trans-acting cell type-specific transcription factors may be involved, since the experiments with BKV were performed in the CV-1 and Vero kidney cell lines (Gorrill and Khalili, 2005; Gorrill et al., 2006) while our experiments with JCV used the U-87 MG and SVGA glial cell lines.
Gel shift studies utilizing mutant oligonucleotides show that the binding sites for C/EBPβ-LIP and p65 in the JCV NCCR are adjacent but not identical. The C/EBPβ-LIP binding site lies slightly closer to the JCV origin than the p65 binding site. Surprisingly, mutations that removed either the p65 or C/EBPβ-LIP binding sites diminished basal promoter activity when introduced into the early reporter plasmid (m1JCVE-CAT and m2JCVE-CAT respectively) and also removed the ability of the promoter to respond to either transcription factor. This may be because mutations in this region of the JCV genome are not well tolerated and indeed this DNA region is conserved between almost all isolates of JCV unlike other NCCR DNA regions which are frequently rearranged (Pietropaolo et al., 2003). It is also possible that the mutations in this region might affect the binding of other factors, e.g., Manley et al. (2006) have suggested that NFAT4 binds to this site.
Our data suggest that interplay between the NF-κB and C/EBPβ transcription factors may regulate the life cycle of JCV. Since both p65 and C/EBPβ isoforms are regulated by signal transduction pathways that lie downstream of cytokines and immunomodulators, cross-communication between these two transcription factors may be important in controlling the balance of JCV latency and reactivation that occurs in response to immunosuppression. For example, we have previously reported that TNF-α stimulates JCV transcription through the action of NF-κB at this site (Mayreddy et al., 1996; Rangathan and Khalili, 1993; Safak et al., 1999). The present data suggest that C/EBPβ may also be involved. Regulation of C/EBPβ occurs at a number of levels, including gene transcription, translation initiation site selection, protein-protein interactions and phosphorylation-dependent changes in DNA-binding activity, activation potential and subcellular localization (Buck et al., 2001a,b; Hu et al, 2004; Piwien-Pilipuk et al., 2002; Ramji and Foka, 2002). Note, all three isoforms of C/EBPβ (full-length, LAP and LIP) are expressed in human astrocytic and oligodendroglial cells, which are permissive for JCV replication.
From these considerations, we postulate that cytokines modulating NF-κB and C/EBPβ activities control JCV reactivation. Indeed, there have been several reports of the presence of latent JCV in the brain (Elsner and Dorries, 1992; Greenlee et al., 2005; Mori et al., 1992; Perez-Liz et al., 2008; White et al., 1992). For example, we have detected the presence of JCV DNA, but not viral protein expression, in the brains of normal individuals who died of non-neurological conditions. Importantly, Laser Capture Microdissection showed that this JCV DNA is present in oligodendrocytes and astrocytes, but not in neurons, in these brains (Perez-Liz et al., 2008). We hypothesize that latent virus in oligodendrocytes and astrocytes can be activated by proinflammatory cytokines allowing expression of viral proteins and viral replication. In highly immunosuppressed individuals, the virus may then be able to infect neighboring cells leading to the spread of virus, because of the lack of an adequate antiviral immune response, and leading to the development of a PML lesion.
Conclusions
In conclusion, our data indicate a novel inhibitory function for C/EBPβ isoforms in the regulation of JCV transcription. Interplay between the positive effects of NF-κB and the negative effects of C/EBPβ upon JCV transcription may be a key factor in the balance of JCV latency and reactivation. The observation that the LIP isoform of C/EBPβ strongly inhibits JCV transcription may have therapeutic implications for the treatment of PML.
ACKNOWLEDGMENTS
We thank past and present members of the Center for Neurovirology for their insightful discussion and sharing of ideas and reagents. We also wish to thank C. Schriver for editorial assistance and Jennifer Gordon, Ilker Sariyer, Bassel Sawaya and Armine Darbinyan for their helpful advice. This work was supported by grants awarded by the NIH to MKW, MS and KK.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Amini S, Mameli G, Del Valle L, Skowronska A, Reiss K, Gelman BB, White MK, Khalili K, Sawaya BE. p73 Interacts with human immunodeficiency virus type 1 Tat in astrocytic cells and prevents its acetylation on lysine 28. Mol. Cell. Biol. 2005;25:8126–8138. doi: 10.1128/MCB.25.18.8126-8138.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger JR. Progressive multifocal leukoencephalopathy in acquired immunodeficiency syndrome: explaining the high incidence and disproportionate frequency of the illness relative to other immunosuppressive conditions. J. Neurovirol. 2003;9 Suppl 1:38–41. doi: 10.1080/13550280390195261. [DOI] [PubMed] [Google Scholar]
- Berger JR. Progressive multifocal leukoencephalopathy. Curr. Neurol. Neurosci. Rep. 2007;7:461–469. doi: 10.1007/s11910-007-0072-9. [DOI] [PubMed] [Google Scholar]
- Berger JR, Houff S. Progressive multifocal leukoencephalopathy: lessons from AIDS and natalizumab. Neurol. Res. 2006;28:299–305. doi: 10.1179/016164106X98198. [DOI] [PubMed] [Google Scholar]
- Bowers WJ, Baglia LA, Ruddel A. Regulation of avian leukosis virus long terminal repeat-enhanced transcription by C/EBP-Rel interactions. J. Virol. 1996;70:3051–3059. doi: 10.1128/jvi.70.5.3051-3059.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck M, Poli V, Hunter T, Chojkier M. C/EBPbeta phosphorylation by RSK creates a functional XEXD caspase inhibitory box critical for cell survival. Mol. Cell. 2001a;8:807–816. doi: 10.1016/s1097-2765(01)00374-4. [DOI] [PubMed] [Google Scholar]
- Buck M, Zhang L, Halasz NA, Hunter T, Chojkier M. Nuclear export of phosphorylated C/EBPbeta mediates the inhibition of albumin expression by TNF-alpha. EMBO J. 2001b;20:6712–6723. doi: 10.1093/emboj/20.23.6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson KR, Evens AM, Richey EA, Habermann TM, Focosi D, Seymour JF, Laubach J, Bawn SD, Gordon LI, Winter JN, Furman RR, Vose JM, Zelenetz AD, Mamtani R, Raisch DW, Dorshimer GW, Rosen ST, Muro K, Gottardi-Littell NR, Talley RL, Sartor O, Green D, Major EO, Bennett CL. Progressive multifocal leukoencephalopathy following rituximab therapy in HIV negative patients: a report of 57 cases from the Research on Adverse Drug Event and Reports (RADAR) project. Blood. 2009;113:4834–4840. doi: 10.1182/blood-2008-10-186999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen NN, Khalili K. Transcriptional regulation of human JC polyomavirus promoters by cellular proteins YB-1 and Pur alpha in glial cells. J. Virol. 1995;69:5843–5848. doi: 10.1128/jvi.69.9.5843-5848.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Valle L, Gordon J, Enam S, Delbue, Croul S, Abraham S, Radhakrishnan S, Assimakopoulou M, Katsetos CD, Khalili K. Expression of human neurotropic polyomavirus JCV late gene product agnoprotein in human medulloblastoma. J. Natl. Cancer Inst. 2002;94:267–273. doi: 10.1093/jnci/94.4.267. [DOI] [PubMed] [Google Scholar]
- Del Valle L, White MK, Enam S, Pina-Oviedo S, Bromer MQ, Thomas RM, Parkman HP, Khalili K. Detection of JC virus DNA sequences and expression of viral T antigen and agnoprotein in esophageal carcinoma. Cancer. 2005;103:516–527. doi: 10.1002/cncr.20806. [DOI] [PubMed] [Google Scholar]
- Del Valle L, White MK, Khalili K. Potential mechanisms of the human polyomavirus JC in neural oncogenesis. J. Neuropathol. Exp. Neurol. 2008;67:729–740. doi: 10.1097/NEN.0b013e318180e631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsner C, Dorries K. Evidence of human polyomavirus BK and JC infection in normal brain tissue. Virology. 1992;191:72–80. doi: 10.1016/0042-6822(92)90167-n. [DOI] [PubMed] [Google Scholar]
- FDA Public Health Advisory. Updated Safety Information about Raptiva (efalizumab) 2009 http://www.fda.gov/cder/drug/advisory/efalizumab.htm.
- Fornace A, Alamo I, Hollander M. DNA damage-inducible transcripts in mammalian cells. Proc. Natl. Acad. Sci. USA. 1988;85:8800–8804. doi: 10.1073/pnas.85.23.8800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Karin M. Missing pieces in the NF-kappaB puzzle. Cell. 2002;109:S81–S96. doi: 10.1016/s0092-8674(02)00703-1. [DOI] [PubMed] [Google Scholar]
- Göransson M, Elias E, Ståhlberg A, Olofsson A, Andersson C, Aman P. Myxoid liposarcoma FUS-DDIT3 fusion oncogene induces C/EBP beta-mediated interleukin 6 expression. Int. J. Cancer. 2005;115:556–560. doi: 10.1002/ijc.20893. [DOI] [PubMed] [Google Scholar]
- Gorrill TS, Khalili K. Cooperative interaction of p65 and C/EBPbeta modulates transcription of BKV early promoter. Virology. 2005;335:1–9. doi: 10.1016/j.virol.2005.02.006. [DOI] [PubMed] [Google Scholar]
- Gorrill T, Feliciano M, Mukerjee R, Sawaya BE, Khalili K, White MK. Activation of early gene transcription in polyomavirus BK by human immunodeficiency virus type 1 Tat. J. Gen. Virol. 2006;87:1557–1566. doi: 10.1099/vir.0.81569-0. [DOI] [PubMed] [Google Scholar]
- Graham FL, van der Eb AJ. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Greenlee JE, Clawson SA, Carney HC, O-Neill FJ. Detection of JC virus early region sequences of brains of individuals without progressive multifocal leukoencephalopathy. Ann. Neurol. 2005;28(S9):S61. [Google Scholar]
- Holman RC, Torok TJ, Belay ED, Janssen RS, Schonberger LB. Progressive multifocal leukoencephalopathy in the United States, 1979–1994: increased mortality associated with HIV infection. Neuroepidemiology. 1998;17:303–309. doi: 10.1159/000026184. [DOI] [PubMed] [Google Scholar]
- Hou J, Major EO. Progressive multifocal leukoencephalopathy: JC virus induced demyelination in the immune compromised host. J. Neurovirol. 2000;6 Suppl 2:S98–S100. [PubMed] [Google Scholar]
- Hu B, Wu Z, Jin H, Hashimoto N, Liu T, Phan SH. CCAAT/enhancer-binding protein beta isoforms and the regulation of alpha-smooth muscle actin gene expression by IL-1 beta. J. Immunol. 2004;173:4661–4668. doi: 10.4049/jimmunol.173.7.4661. [DOI] [PubMed] [Google Scholar]
- Karin M, Ben-Neriah Y. Phosphorylation meets ubiquitination: the control of NF-[kappa]B activity. Annu. Rev. Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- Khalili K, Gordon J, White MK. The polyomavirus, JCV, and its involvement in human disease. Adv. Exp. Med. Biol. 2006;577:274–287. doi: 10.1007/0-387-32957-9_20. [DOI] [PubMed] [Google Scholar]
- Khalili K, White MK, Lublin F, Ferrante P, Berger JR. Reactivation of JC virus and development of PML in patients with multiple sclerosis. Neurology. 2007;68:985–990. doi: 10.1212/01.wnl.0000257832.38943.2b. [DOI] [PubMed] [Google Scholar]
- Khalili K, Safak M, Del Valle L, White MK. JC virus molecular biology and the human demyelinating disease, progressive multifocal leukoencephalopathy. In: Shoshkes Reiss C, editor. Neurotropic virus infections. Cambridge, UK: Cambridge University Press; 2008. pp. 190–211. [Google Scholar]
- Mameli G, Deshmane SL, Ghafouri M, Cui J, Simbiri K, Khalili K, Mukerjee R, Dolei A, Amini S, Sawaya BE. C/EBPbeta regulates human immunodeficiency virus 1 gene expression through its association with cdk9. J. Gen. Virol. 2007;88:631–640. doi: 10.1099/vir.0.82487-0. [DOI] [PubMed] [Google Scholar]
- Manley K, O'Hara BA, Gee GV, Simkevich CP, Sedivy JM, Atwood WJ. NFAT4 is required for JC virus infection of glial cells. J. Virol. 2006;80:12079–12085. doi: 10.1128/JVI.01456-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayreddy RP, Safak M, Razmara M, Zoltick P, Khalili K. Transcription of the JC virus archetype late genome: importance of the kappa B and the 23-base-pair motifs in late promoter activity in glial cells. J. Virol. 1996;70:2387–2393. doi: 10.1128/jvi.70.4.2387-2393.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mázló M, Ressetar HG, Stoner GL. The neuropathology and pathogenesis of progressive multifocal leukoencephalopathy. In: Khalili K, Stoner GL, editors. Human polyomaviruses: molecular and clinical perspectives. New York: Wiley-Liss, Inc.; 2001. pp. 257–335. [Google Scholar]
- Monaco MC, Atwood WJ, Gravell M, Tornatore CS, Major EO. JC virus infection of hematopoietic progenitor cells, primary B lymphocytes, and tonsillar stromal cells: implications for viral latency. J. Virol. 1996;70:7004–7012. doi: 10.1128/jvi.70.10.7004-7012.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori M, Aoki N, Shimada H, Tajima M, Kato K. Detection of JC virus in the brains of aged patients without progressive multifocal leukoencephalopathy by the polymerase chain reaction and Southern hybridization analysis. Neurosci. Lett. 1992;141:151–155. doi: 10.1016/0304-3940(92)90883-9. [DOI] [PubMed] [Google Scholar]
- Mukerjee R, Sawaya BE, Khalili K, Amini S. Association of p65 and C/EBPbeta with HIV-1 LTR modulates transcription of the viral promoter. J. Cell. Biochem. 2007;100:1210–1216. doi: 10.1002/jcb.21109. [DOI] [PubMed] [Google Scholar]
- Nabel G, Baltimore D. An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature. 1987;326:711–713. doi: 10.1038/326711a0. [DOI] [PubMed] [Google Scholar]
- Ossipow V, Descombes P, Schibler U. CCAAT/enhancer-binding protein mRNA is translated into multiple proteins with different transcription activation potentials. Proc. Natl. Acad. Sci. USA. 1993;90:8219–8223. doi: 10.1073/pnas.90.17.8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett BL, Walker DL. Prevalence of antibodies in human sera against JC virus, an isolate from a case of progressive multifocal leukoencephalopathy. J. Infect. Dis. 1973;127:467–470. doi: 10.1093/infdis/127.4.467. [DOI] [PubMed] [Google Scholar]
- Perez-Liz G, Del Valle L, Gentilella A, Croul S, Khalili K. Detection of JC virus DNA fragments but not proteins in normal brain tissue. Ann Neurol. 2008;64:379–387. doi: 10.1002/ana.21443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelps CB, Sengchanthalangsy LL, Malek S, Ghosh G. Mechanism of kappa B DNA binding by Rel/NF-kappa B dimers. J. Biol. Chem. 2000;275:24392–24399. doi: 10.1074/jbc.M003784200. [DOI] [PubMed] [Google Scholar]
- Pietropaolo V, Videtta M, Fioriti D, Mischitelli M, Arancio A, Orsi N, Degener AM. Rearrangement patterns of JC virus noncoding control region from different biological samples. J. Neurovirol. 2003;9:603–611. doi: 10.1080/13550280390246507. [DOI] [PubMed] [Google Scholar]
- Piwien-Pilipuk G, MacDougald O, Schwartz J. Dual regulation of phosphorylation and dephosphorylation of C/EBPbeta modulate its transcriptional activation and DNA binding in response to growth hormone. J. Biol. Chem. 2002;277:44557–44565. doi: 10.1074/jbc.M206886200. [DOI] [PubMed] [Google Scholar]
- Raj GV, Khalili K. Identification and characterization of a novel GGA/C-binding protein, GBP-i, that is rapidly inducible by cytokines. Mol. Cell. Biol. 1994;14:7770–7781. doi: 10.1128/mcb.14.12.7770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramji DP, Foka P. CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem. J. 2002;365:561–575. doi: 10.1042/BJ20020508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranganathan PN, Khalili K. The transcriptional enhancer element, kappa B, regulates promoter activity of the human neurotropic virus, JCV, in cells derived from the CNS. Nucleic Acids Res. 1993;21:1959–1964. doi: 10.1093/nar/21.8.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A, Ray BK. Lipopolysaccharide-mediated induction of the bovine interleukin-6 gene in monocytes requires both NF-kappa B and C/EBP binding sites. DNA Cell. Biol. 1995;14:795–802. doi: 10.1089/dna.1995.14.795. [DOI] [PubMed] [Google Scholar]
- Ricciardiello L, Laghi L, Ramamirtham P, Chang CL, Chang DK, Randolph AE, Boland CR. JC virus DNA sequences are frequently present in the human upper and lower gastrointestinal tract. Gastroenterology. 2000;119:1228–1235. doi: 10.1053/gast.2000.19269. [DOI] [PubMed] [Google Scholar]
- Romagnoli L, Sariyer IK, Tung J, Feliciano M, Sawaya BE, Del Valle L, Ferrante P, Khalili K, Safak M, White MK. Early growth response-1 protein is induced by JC virus infection and binds and regulates the JC virus promoter. Virology. 2008;375:331–341. doi: 10.1016/j.virol.2008.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D, Habener JF. CHOP, a novel developmentally regulated nuclear protein that dimerizes with transcription factors C/EBP and LAP and functions as a dominant negative inhibitor of gene transcription. Genes Dev. 1992;6:439–453. doi: 10.1101/gad.6.3.439. [DOI] [PubMed] [Google Scholar]
- Safak M, Gallia GL, Khalili K. A 23-bp sequence element from human neurotrophic JC virus is responsive to NF-kappa B subunits. Virology. 1999;262:178–189. doi: 10.1006/viro.1999.9886. [DOI] [PubMed] [Google Scholar]
- Safak M, Barrucco R, Darbinyan A, Okada Y, Nagashima K, Khalili K. Interaction of JC virus agno protein with T antigen modulates transcription and replication of the viral genome in glial cells. J. Virol. 2001;75:1476–1486. doi: 10.1128/JVI.75.3.1476-1486.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu H, Yamamoto K. NF-kappa B and C/EBP transcription factor families synergistically function in mouse serum amyloid A gene expression induced by inflammatory cytokines. Gene. 1994;149:305–310. doi: 10.1016/0378-1119(94)90166-x. [DOI] [PubMed] [Google Scholar]
- Stein B, Baldwin AS. Distinct mechanisms for regulation of the interleukin-8 gene involve synergism and cooperativity between C/EBP and NF-kappa B. Mol. Cell. Biol. 1993;13:7191–7198. doi: 10.1128/mcb.13.11.7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein B, Cogswell PC, Baldwin AS. Functional and physical associations between NF-kappa B and C/EBP family members: a Rel domain-bZIP interaction. Mol. Cell. Biol. 1993;13:3964–3974. doi: 10.1128/mcb.13.7.3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker DL, Padgett BL. The epidemiology of human polyomaviruses. Prog. Clin. Biol. Res. 1983;105:99–106. [PubMed] [Google Scholar]
- West MJ, Lowe AD, Karn J. Activation of human immunodeficiency virus transcription in T cells revisited: NFkB p65 stimulates transcriptional elongation. J. Virol. 2001;75:8524–8537. doi: 10.1128/JVI.75.18.8524-8537.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White FA, Ishaq M, Stoner GL, Frisque RJ. JC virus DNA is present in many human brain samples from patients without progressive multifocal leukoencephalopathy. J. Virol. 1992;66:5726–5734. doi: 10.1128/jvi.66.10.5726-5734.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MK, Skowronska A, Gordon J, Del Valle L, Deshmane SL, Giordano A, Khalili K. Analysis of a mutant p53 protein arising in a medulloblastoma from a mouse transgenic for the JC virus early region. Anticancer Res. 2006;26:4079–4092. [PubMed] [Google Scholar]
- White MK, Safak M, Khalili K. Regulation of gene expression in primate polyomaviruses. J Virol. 2009 doi: 10.1128/JVI.00542-09. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yogo Y, Kitamura T, Sugimoto C, Ueki T, Aso Y, Hara K, Taguchi F. Isolation of a possible archetypal JC virus DNA sequence from nonimmunocompromised individuals. J. Virol. 1990;64:3139–3143. doi: 10.1128/jvi.64.6.3139-3143.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Zheng HY, Suzuki M, Chen Q, Ikegaya H, Aoki N, Usuku S, Kobayashi N, Nukuzuma S, Yasuda Y, Kuniyoshi N, Yogo Y, Kitamura T. Age-Related Urinary Excretion of BK Polyomavirus by Non-immunocompromised Individuals. J. Clin. Microbiol. 2006;45:193–198. doi: 10.1128/JCM.01645-06. [DOI] [PMC free article] [PubMed] [Google Scholar]