Abstract
5-Lipoxygenase (5-LOX), an enzyme involved in the metabolism of arachidonic acid, participates in the modulation of the proliferation and differentiation of neural stem cells and cerebellar granule cell (CGC) precursors. Since epigenetic mechanisms including DNA methylation regulate 5-LOX expression and have been suggested as possible modulators of stem cell differentiation and aging, using primary cultures of mouse CGC (1, 5, 10, 14, 30 days in vitro; DIV), we studied DNA methylation patterns of the 5-LOX promoter and 5-LOX mRNA levels. We also measured the mRNA and protein content of the DNA methyltransferases DNMT1 and DNMT3a. 5-LOX, DNMT1, and DNMT3a mRNA levels were measured by real-time PCR. We observed that 5-LOX expression and the expression of maintenance DNMT1 is maximal at 1 DIV (proliferating neuronal precursors), whereas the expression of the de novo DNA methyltransferase DNMT3a mRNA increased in aging cultures. We analyzed the methylation status of the 5-LOX promoter using the methylation-sensitive restriction endonucleases AciI, BstUI, HpaII, and HinP1I, which digest unmethylated CpGs while leaving methylated CpGs intact. The 5-LOX DNA methylation increased with the age of the cells. Taken together, our data show that as cultured CGC mature and age in-vitro, a decrease in 5-LOX mRNA content is accompanied by an increase in the methylation of the gene DNA. In addition, an increase in DNMT3a but not DNMT1 expression accompanies an increase of 5-LOX methylation during in-vitro maturation.
Keywords: 5-Lipoxygenase, epigenetic, aging, cerebellar granule, DNMT1, DNMT3a
The ALOX5 gene, which encodes 5-lipoxygenase (5-LOX; EC 1.13.11.34), the key enzyme in the biosynthesis of the inflammatory leukotrienes and the anti-inflammatory lipoxins (Radmark et al., 2007, Serhan et al., 2008), is regulated by epigenetic modifications of DNA methylation of the 5-LOX promoter (Radmark et al., 2007; Uhl et al., 2002, 2003; Zhang et al., 2004). Epigenetic mechanisms play a significant role in neuronal development (MacDonald and Roskams, 2009; Wen et al., 2009) and aging (Siegmund et al., 2007). The expression of 5-LOX in mouse (Wada et al., 2006) and rat (Uz et al., 2001) neural stem cells and the production of 5-LOX metabolites have been shown to play a role in regulating neural stem cell proliferation and differentiation. On the other hand, the brain expression of 5-LOX increases during aging (Chinnici et al., 2007; Uz et al., 1998) and has been associated with the pathobiology of Alzheimer's disease (Firuzi et al., 2008; Ikonomovic et al., 2008) and possibly with mechanisms of co-morbid cardiovascular and neuropsychiatric disorders (Manev and Manev, 2007; Chu and Pratico, 2009).
Both increases and decreases in DNA methylation occur during neurodevelopment (Wen et al., 2009) and with aging (Ono et al., 1993; Rath and Kanungo, 1989). Furthermore, aging-altered epigenetically regulated gene expression in the central nervous system (CNS) may contribute to aging-associated brain pathologies; for example, Tohgi et al. (1999) reported that cytosines, particularly those at −207 to approximately −182 in the promoter region of the amyloid precursor protein (APP) gene, are frequently methylated and suggested that their demethylation during aging may favor the development of Aβ deposition in the aged brain. It has been suggested that the modulation of gene methylation could be a strategy for preventing Alzheimer’s disease (AD) (Morrison et al., 1996; Scarpa et al., 2003). However, it has to be stressed that changes in the CNS DNA methylation pattern in aging-associated neurodegenerative disorders do not appear to be general. Further, it was proposed that their putative pathobiological role might be limited to certain genes and/or cell types (Barrachina and Ferrer, 2009). Recent data indicate that 5-LOX DNA methylation assayed in mouse brain samples increases with aging (Manev et al., 2008).
Interpretation of DNA methylation data obtained from analyses of tissue samples, such as different brain regions, are complicated by the fact that these samples contain a mixture of various cell types that could be differentially susceptible to epigenetic modifications. Alternatively, in-vitro experiments provide the advantage of studying uniform cell populations. Xiong et al. (2004) established a model of cerebellar granule cell (CGC) cultures as a tool to study the effects of neuronal maturation and in-vitro aging (i.e., cultures maintained for up to 1 month). These cultures are prepared from the postnatal neuronal precursor grown in a uniform neuronal population of CGC. In this model, 5-LOX participates in the regulation of neuronal precursor proliferation (Uz et al., 2001). We used a model of in-vitro differentiation and aging of CGC to investigate 5-LOX expression along with the characterization of 5-LOX DNA methylation. Since DNA methylation depends on DNA-methyltransferases (DNMTs), we investigated the effect of in-vitro differentiation/aging on the CGC content of mRNAs for DNMT1 and DNMT3a, which are the maintenance and de novo DNMTs, respectively (Sharma et al., 2008).
EXPERIMENTAL PROCEDURES
Preparation of primary cerebellar granule cell (CGC) cultures
Primary CGC cultures were prepared from 5-day-old C57BL/6J mouse pups as described previously (Uz et al., 2001). The experimental protocol was approved by the Institutional Animal Care and Ethics Committee of the University of Illinois at Chicago. CGC were plated in 20 µg/ml poly-D-lysine- (Sigma, St. Louis, MO) coated 3.5-cm-dishes at the density of 0.6 million/ml. Cells were grown either in a serum-free medium (Neurobasal medium, GibcoBRL, Rockville, MD) containing the B27 supplement (GibcoBRL) or in basal modified Eagle’s medium (BME, GibcoBRL) supplemented by 10% heat inactivated fetal bovine serum (GibcoBRL). To prevent the growth of non-neuronal cells in the BME cultures, 10 µM Cytosine-β-D-arabinofuranoside was added to the BME medium 18–24 h after plating. Both media contained 2 mM L-glutamine, 50 mg/ml gentamicin, and 25 mM KCl (Sigma). Cultures were kept at 37 °C and 5% CO2 and maintained up to 30 days by adding 5 mM D-glucose (Sigma) to the growth medium on the seventh day in vitro (DIV) followed by additional glucose supplementation every fourth day in addition to adding some water to compensate for loss due to evaporation.
Real-time PCR
Total RNA was extracted using the TRIzol® reagent (Invitrogen, Carlsbad, CA) following the manufacturer's instructions (3 dishes were pooled for each individual sample). To eliminate possible DNA contamination, RNA samples were treated with a DNase reagent, DNA-free™ (Ambion, Inc., Austin, TX). Total RNA (3 µg) was reverse-transcribed with 200U of cloned Moloney Murine Leukemia Virus reverse transcriptase (Gibco BRL, Carlsbad, CA). Real-time RT-PCR was performed in a Mx 3005P Real-time PCR System (Stratagene, La Jolla, CA) using Maxima™ SYBR Green pPCR Master Mix (Fermentas, Glen Burnie, MD) in a two-step cycling protocol as described by the manufacturer. The initial 5-min denaturing step was followed by 40 cycles of denaturing at 95°C for 15 s and annealing/elongation at 60°C for 1 min. Table 1 shows the sequences of primers used to detect 5-LOX, DNMT1, DNMT3a, and cyclophilin mRNA (primers were purchased from Integrated DNA Technologies, Inc., Coralville, IA). Reactions were performed using three different concentrations (12.5, 25, 50 ng) of reverse transcribed material (duplicated for each sample). Real-time PCR results were normalized against the corresponding cyclophilin mRNA signals. Data are presented as units (i.e., the coefficient of variation; Schmittgen and Livak, 2008). In addition, the amplification products of all genes studied (5-LOX, DNMTs, and cyclophilin) were verified using an agarose gel assay that revealed a single band at the expected size for each product.
Table 1.
The sequences of primers used for real-time PCR.
| Gene Bank Accession Number |
Primers | Fragment size (bp) |
|
|---|---|---|---|
| 5-LOX | NM_009662 | F: 5’-ATTGCCATCCAGCTCAACCAAACC-3’ R: 5’-TGGCGATACCAAACACCTCAGACA-3’ |
178 |
| DNMT1 | NM_010066 | F: 5’-AGTGCAAGGCGTGCAAAGATATGG-3’ R: 5’-TGGGTGATGGCATCTCTGACACAT-3’ |
150 |
| DNMT3a |
NM_007872 NM_153743 |
F: 5’-ACAAGAATGCTACCAAAGCAGCCG -3’ R: 5’-TGAGAACTTGCCATCTCCGAACCA-3’ |
200 |
| Cyclophilin | X52803 | F: 5’- AGCATACAGGTCCTGGCATCTTGT –3’ R: 5’- AAACGCTCCATGGCTTCCACAATG -3’ |
153 |
5-LOX DNA methylation assay
For the DNA methylation assay, we targeted the promoter-5'UTR of the mouse 5-LOX gene (Table 2), which contains 14 CpG dinucleotides located upstream from the ATG translation start codon. In preliminary studies, we also investigated the 27 CpG dinucleotides located downstream in the first exon-intron region of the 5-LOX gene and found that in CGC cultures their methylation status was not affected by different days in vitro. Here, we investigated the methylation rate of the 5-LOX promoter with the aid of four methylation-sensitive endonucleases – AciI (C↓CGC), BstUI (CG↓CG), HinP1I (G↓CGC), and HpaII (C↓CGG) (their respective recognition sequence cutting sites are shown in parentheses). For the extraction of genomic DNA, cells were collected in saline and resuspended in homogenizing buffer (100 mM NaCl, 10 mM Tris-HCl pH 8.0, 25 mM EDTA, 0.5% SDS, 0.1 mg/ml proteinase K) and incubated 4–5 h at 37°C. The DNA was purified with phenol, phenol-chloroform (pH 8.0), precipitated with isopropanol, and dissolved in H2O. To measure 5-LOX DNA methylation levels, we used restriction digest-quantitative PCR (SYBR Green RD-qPCR) - the methylation-sensitive endonucleases digest only unmethylated recognition sites and do not act on sites with methylated cytosine. As a result, in methylated DNA samples greater amounts of templates are available for the action of Taq DNA polymerase. The assay was performed as follows: 1 µg of genomic DNA was used in a restriction digest reaction for each of the four endonucleases. Digested DNA samples were diluted with water and an aliquot (100 ng DNA) was used for qPCR (Stratagene) with the Maxima™ SYBR Green qPCR Master Mix (Fermentas) according to the manufacturer’s protocol. The following primers were used: forward 5’-agagaaggatgcgttggaaggt-3’, reverse 5’-gactccgggcaagtgagtgct -3’. These primers amplify the 238 nt region upstream of the first ATG translation start codon. This region contains two recognition sites for each of 4 endonucleases selected for the assay. For input control, we used the 394 nt region in the first intron because this region does not contain recognition sites for the selected methylation-sensitive endonucleases. This region was amplified with the following primers: forward 5’- tgatgtggctggcctcttatgtga-3’, reverse 5’-actgggactgagtgcaggaaatgt-3’. qPCR reactions were run with 2 different primer sets (target and input) in separate tubes and the coefficient of variation (CV) for the relative amount of a target sequence was calculated.
Table 2.
The sequence of the mouse 5-LOX promoter-5’UTR analyzed for methylation states with methylation-sensitive endonucleases AciI (C↓CGC), BstUI (CG↓CG), HinP1I (G↓CGC), and HpaII (C↓CGG).
 |
The endonuclease recognition sites are highlighted in black (two for each endonuclease). ud are primers used in real-time PCR - forward 5’-agagaaggatgcgttggaaggt-3’, reverse 5’-gactccgggcaaagtgct-3’. Position 1 indicates the translation start codon (ATG).
Quantitative Western blotting
Pooled CGC samples from three dishes were homogenized in RIPA lysis buffer [50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 4% (w/v) sodium deoxycholate, 1% (w/v) SDS (sodium dodecylsulfate), 1% (v/v) NP-40, and 1mM EDTA (ethylenediamine tetraacetic acid)] supplemented with protease inhibitors (2 mM dithiothreitol, 0.2 units/ml aprotinin; 1.45 µM pepstatin A, 2.1 μM leupeptin, 5.7 mM phenylmethylsulfonylfluoride) immediately before homogenizing the samples. Protein concentrations were measured using a BCA protein assay kit (Pierce, Rockford, IL). Ten and 20 µg proteins per sample were run on polyacrylamide Thermo Scientific Precise Proteins Gels [4–20% (w/v), Thermo Scientific, Rockford, IL] in a running solution (100 mM Tris-base, 100 mM HEPES, 3 mM SDS, pH 8, Thermo Scientific). The proteins were transferred electrophoretically overnight to a nitrocellulose membrane (Amersham Piscataway, NJ) in transfer buffer [25 mM Tris–base, 192 mM glycine, and 20% (v/v) methanol]. The non-specific binding sites were blocked by a solution containing 5% (w/v) non-fat dry milk, 10% (v/v) 10 × TBST buffer (100 mM Tris–base, 1.5 M NaCl, 5 ml/l Tween 20), 0.002% (v/v) Igepal, and 0.02% (w/v) SDS, pH 8 for 30 min. The membranes were incubated overnight at 4°C with an anti-DNMT1 primary antibody (1:1000; Sigma) and subsequently incubated for 3 h at room temperature with anti-rabbit IgG horseradish-peroxidase-linked secondary antibody (1:1000; Amersham). An ECL Plus Kit (Amersham) was used for band visualization. Blots were scanned and the bands were visualized using a Storm 860 Phosphor Imager (GE Helthcare, Piscataway, NJ). The membranes were stripped with a Western Blot Recycling Kit (Alpha Diagnostic International, San Antonio, TX) and re-probed with anti-DNMT3a primary antibody (1:300; overnight at 4 °C, Imgenex, San Diego, CA) and with an anti-mouse IgG horseradish-peroxidase-linked secondary antibody (1:1000, Amersham). To normalize the signal for DNMT1 and DNMT3a, the amount of β-actin was measured on the same blot using an anti-β-actin antibody (1:5000, Sigma). Band intensities were analyzed by ImageQuant software (GE Helthcare).
Statistics
For statistical analysis, we used SPSS software (version 12.0). Data were analyzed by either one-way ANOVA followed by Dunnett’s multiple comparison test or by an independent sample t-test. Results are expressed as the mean ± standard error mean (SEM). The p<0.05 values were accepted as statistically significant.
RESULTS
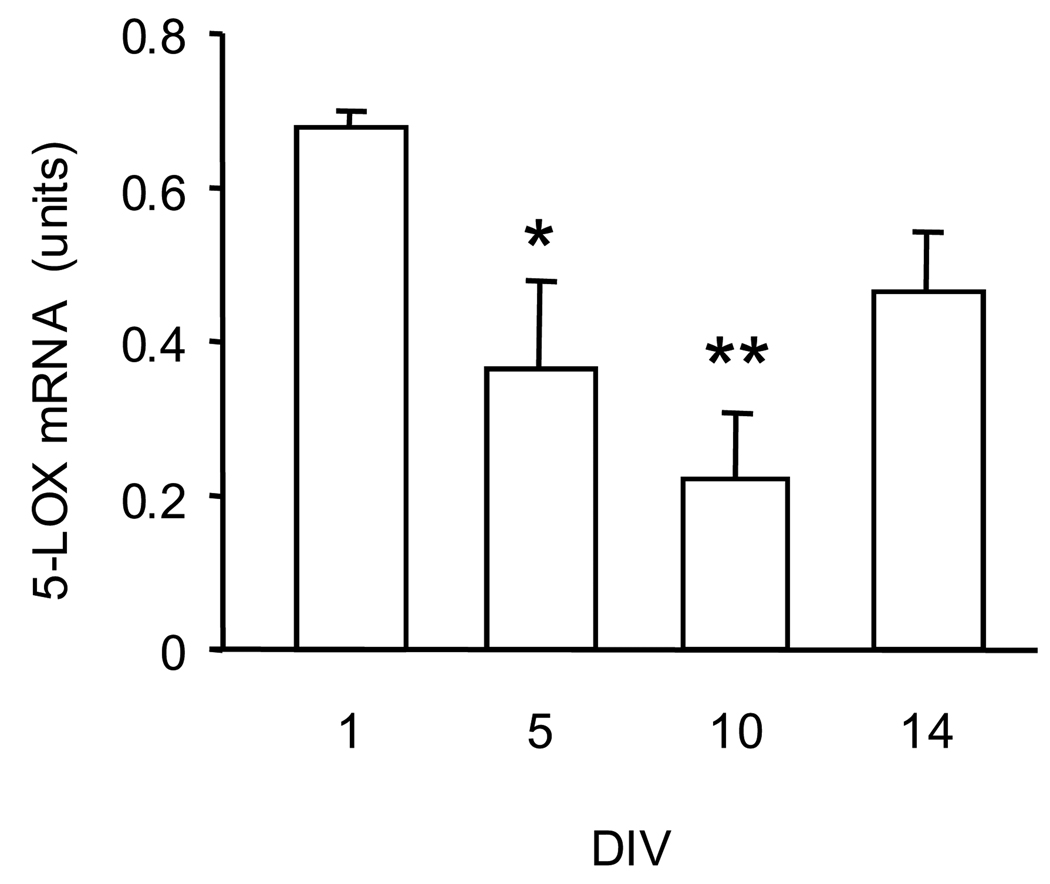
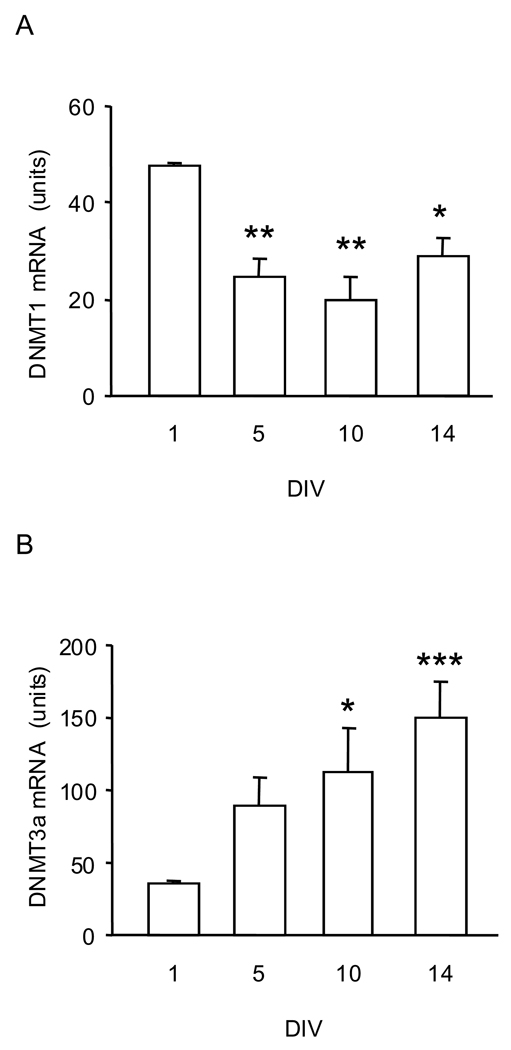
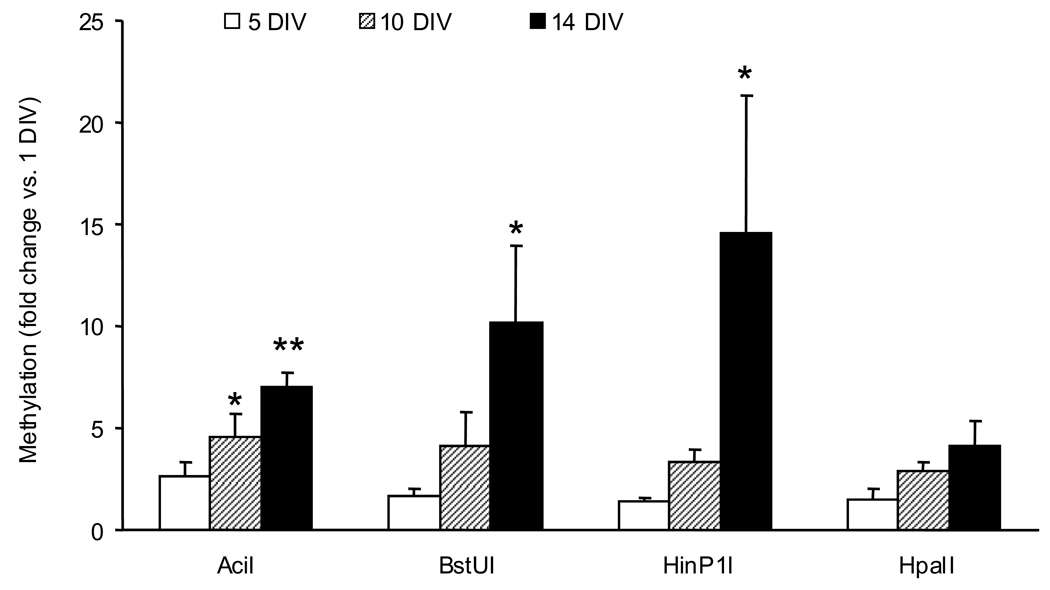
The expression of 5-LOX mRNA in CGC cultures is significantly lower than it is in leukocytes, the cell type considered one of the main sources of 5-LOX (Fig. 1). Furthermore, the levels of CGC 5-LOX mRNA content decreased significantly as cultures transitioned from their immature state (1 DIV) to mature neurons (e.g., 10 DIV, which is typically reported in the literature as mature CGC) (Fig. 2). The levels of DNMT1 mRNA changed in a similar pattern - they decreased as the cultures matured (Fig. 3A). On the other hand, the levels of DNMT3 mRNA increased significantly from 1 DIV to 14 DIV (Fig. 3B). Using four methylation-sensitive endonucleases – AciI, BstUI, HinP1I, and HpaII – to assay DNA methylation status of the 5-LOX promoter, we found that maturation of CGC grown in a serum-free medium was accompanied by increased DNA methylation. This increase was most significant at the AciI site. Furthermore, across all four sites, the methylation increase was most consistent after 14 DIV (Fig. 4).
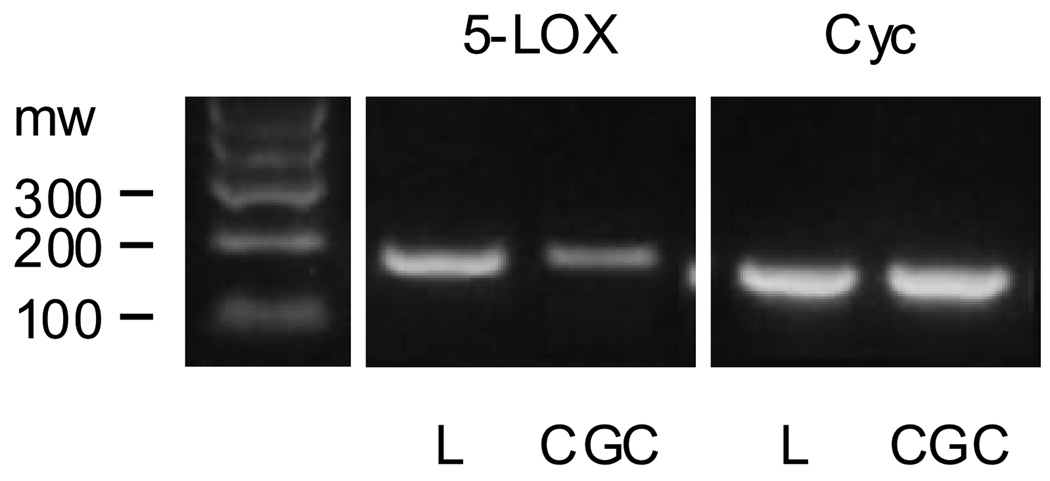
Fig. 1.
5-LOX mRNA expression in mouse leukocytes (L) and cerebellar granule cells (CGC). Leukocytes were extracted from whole blood as described elsewhere (Mohammadi and Saberivand, 2009). The signals for 5-LOX and cyclophin (Cyc; a loading control) mRNAs were obtained by RT-PCR using specific primers (see Table 1). The amplification step lasted for 30 cycles with denaturation (94°C, 30 sec), annealing (57°C, 30 sec), and elongation (72°C, 30 sec). Shown is an example of gel electrophoresis of the PCR products (bands at the expected size for 5-LOX and Cyc, i.e., 178 and 153 bp respectively). Note the stronger 5-LOX mRNA signal in leukocytes compared to CGC whereas the cyc signals were comparable in both samples. A real-time PCR assay of these samples revealed that leukocytes express about 400 times more 5-LOX mRNA than CGC [5-LOX mRNA (units): leukocyte = 87, CGC = 0.22; n = 2].
Fig. 2.
5-LOX mRNA levels in CGC matured in-vitro in a serum-free medium (containing the B27 supplement). Total RNA was extracted at 1, 5, 10, and 14 days in vitro (DIV). 5-LOX mRNA was measured by real-time PCR. The results were normalized against the corresponding cyclophilin contents and are presented as units (coefficient of variation arbitrarily multiplied by a factor of 1000, see text). Data are expressed as mean ± SEM of 4 different cell preparations and were evaluated by one-way ANOVA followed by Dunnett’s multiple comparison test. * P<0.05; ** P<0.01 compared to 1 DIV.
Fig. 3.
DNMT mRNA levels in CGC maturing in-vitro in a serum-free medium. (A) DNMT1 and (B) DNMT3a mRNA levels measured by real-time PCR in CGC at 1, 5, 10, and 14 DIV. The results were normalized against the corresponding cyclophilin contents and are presented as units (see text). Data are expressed as mean ± SEM of 4 different cell preparations and were evaluated by one-way ANOVA followed by Dunnett’s multiple comparison test. * P<0.05; ** P<0.01; *** P<0.001 compared to 1 DIV.
Fig. 4.
Methylation of the 5-LOX promoter – 5’UTR region in maturing CGC (serum-free medium) assayed at the methylation-sensitive endonuclease (AciI, BstUI, HinP1, and HpaII) sites. Data obtained in samples from 5, 10, and 14 DIV cultures are expressed as a fold change compared to the methylation level in 1 DIV cultures. Data are expressed as mean ± SEM of 4 different cell preparations and were evaluated by one-way ANOVA followed by Dunnett’s multiple comparison test. *P<0.05, **P<0.001 vs. corresponding 1 DIV.
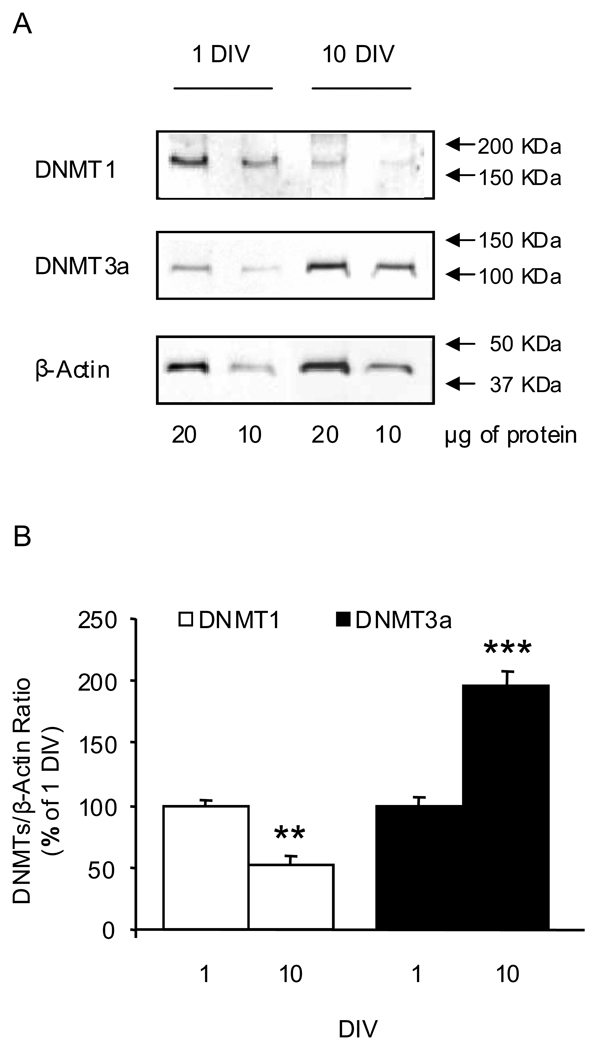
We confirmed that the maturation-associated changes in CGC DNMT1 and DNMT3a mRNA levels were accompanied by corresponding alterations in DNMT protein content. Thus, a quantitative Western blotting assay revealed lower DNMT1 protein content in 10 DIV CGC compared with 1 DIV CGC, whereas the content of the de novo DNA-methylating enzyme DNMT3a increased in 10 DIV CGC (Fig. 5).
Fig. 5.
DNMT1 and DNMT3a protein levels in immature (1 DIV) and mature (10 DIV) CGC grown in serum-free conditions. Panel A shows representative Western immunoblots assayed at two different concentrations of proteins (20 and 10 µg). DNMT1 and DNMT3a signals were normalized by the corresponding β-actin signals measured on the same blot. Panel B shows the quantitative data (open bars DNMT1; closed bars DNMT3a). The ratio for each sample at two different protein concentrations was similar and the average of these two ratios was used as single value. Data (mean ± SEM from 3–4 different cell preparations) are presented as a percentage of the corresponding 1 DIV and were evaluated by independent sample t-test. ** P<0.01; *** P<0.001.
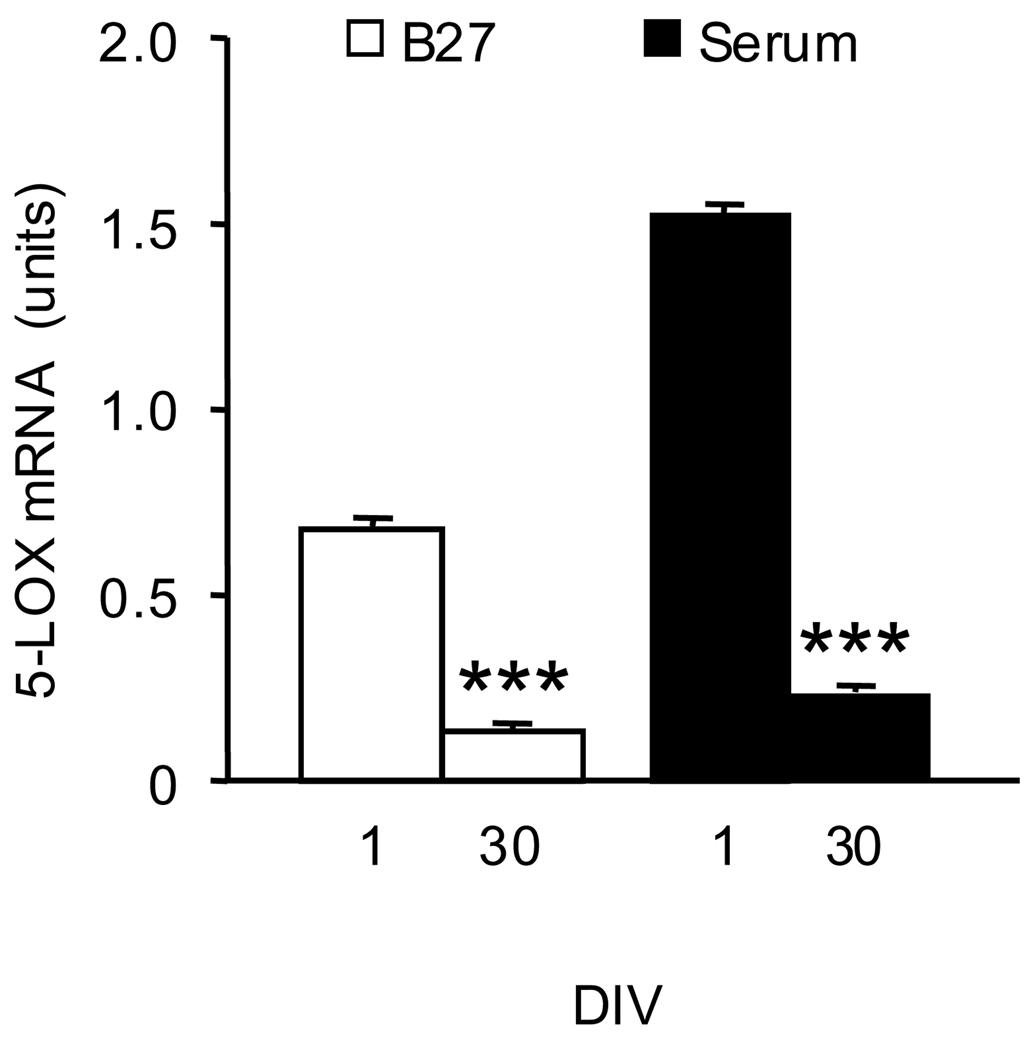
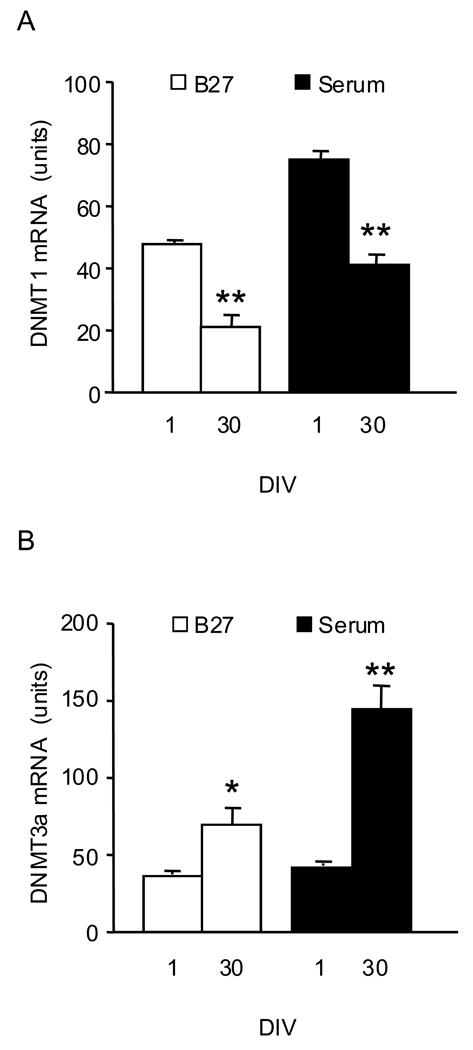
In studies of the effects of in-vitro aging, we used 1 DIV cultures and their corresponding sister cultures (i.e., from the same cell preparation) maintained in-vitro for 30 days. Since previous work has demonstrated that the addition of serum to the culture medium increases the expression of CGC 5-LOX (Manev and Uz, 1999), we performed these experiments both in cultures grown in a serum-free medium and in cultures grown in the presence of serum. Although the 5-LOX mRNA content was greater in 1 DIV cultures grown in the presence of serum compared to cultures grown in the B27-containing medium, aging significantly reduced 5-LOX mRNA content in both culture conditions (Fig. 6). Also, in both culture conditions the content of DNMT1 mRNA was lower and the content of DNMT3a mRNA was higher in 30 DIV CGC compared to 1 DIV cultures (Fig. 7).
Fig. 6.
5-LOX mRNA content decreases in old (30 DIV) CGC compared to 1 DIV cultures grown both in the serum-free (B27 supplement) and serum-containing medium. Total RNA was extracted from CGC grown in a serum-free medium (B27, open bars) and a serum-supplemented medium (Serum, closed bars) at 1 and 30 DIV. 5-LOX mRNA levels were measured by real-time PCR. The results were normalized against the corresponding cyclophilin contents and are presented as units (see text). Data are expressed as mean±SEM from 3 different cell preparations and were evaluated by independent sample t-test. *** P<0.001 compared to 1 DIV.
Fig. 7.
DNMT1 mRNA content decreases while DNMT3a mRNA increases in aged (30 DIV) compared to 1 DIV CGC grown both in serum-free (B27 supplement) and serum-containing conditions. The mRNA levels of DNMT1 (panel A) and DNMT3a (panel B) were measured by real-time PCR. The results were normalized against the corresponding cyclophilin contents and are presented as units (see text). Data are expressed as mean ± SEM from 3 different cell preparations and were evaluated by independent sample t-test. * P<0.05; ** P<0.01 compared to 1 DIV.
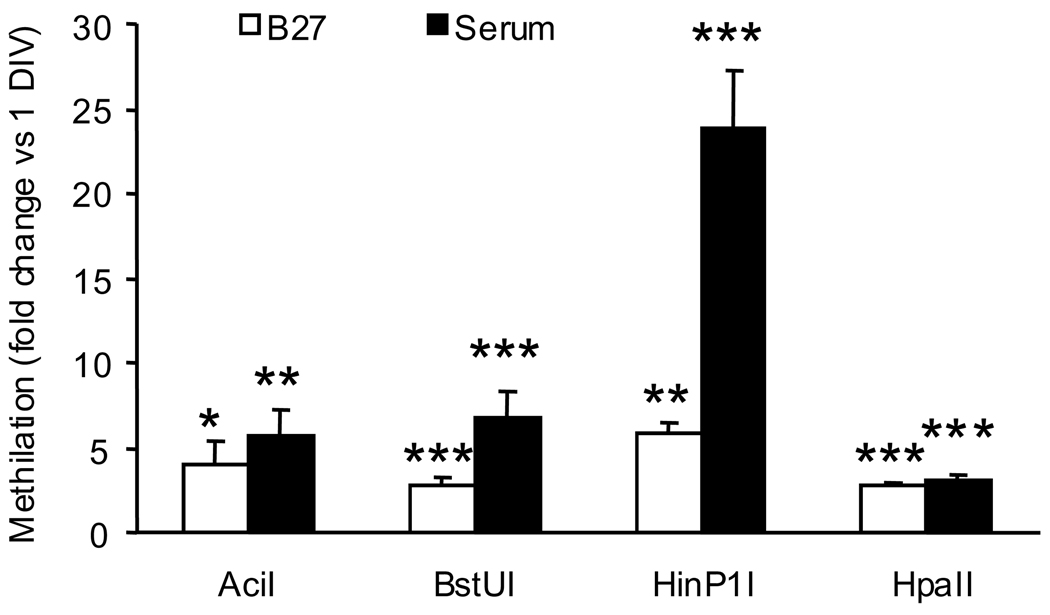
The methylation-sensitive endonuclease assay revealed that in both culture conditions, in-vitro aging was accompanied by a significant increase in 5-LOX DNA methylation at all four sites (AciI, BstUI, HinP1I, and HpaII) (Fig. 8)
Fig. 8.
Methylation of the 5-LOX promoter – 5’UTR region in young (1 DIV) and old (30 DIV) CGC assayed at methylation-sensitive endonuclease (AciI, BstUI, HinP1, and HpaII) sites. Data obtained in samples from 30 DIV cultures are expressed as a fold change compared to the methylation level in 1 DIV cultures (open bars, serum-free; closed bars, serum-containing medium). Data are expressed as mean ± SEM of 5 different cell preparations and were evaluated by t-test. *P<0.05, **P<0.01 ***P<0.001 vs. corresponding 1 DIV.
DISCUSSION
Using primary cultures of mouse CGC, we found that this in-vitro model of neuronal maturation and aging revealed consistent maturation and aging-associated changes in 5-LOX mRNA expression and 5-LOX promoter DNA methylation. As expected, we found that the content of 5-LOX mRNA in CGC cultures is several orders of magnitude lower than in leukocytes, cells known for their abundant 5-LOX expression. Consistent with previous findings in rat CGC cultures (Manev and Uz, 1999; Uz et al., 2001), we found that the content of 5-LOX mRNA in mouse CGC was the highest at 1 DIV and decreased significantly as cultures matured and aged. Furthermore, similar to rat CGC (Manev and Uz, 1999), the mouse CGC cultures grown in the presence of serum also expressed higher levels of 5-LOX compared to cultures grown in the absence of serum (in a B27 supplemented medium). Furthermore, the presence of serum in the culture medium did not interfere with the aging-associated (i.e., 30 DIV) decrease in 5-LOX mRNA levels observed in serum-free conditions. Previously, we found that CGC cultures contain less than 5% glia cells (Favaron et al., 1988). Since we did not observe any differences in glia content in cultures grown in different culture conditions (visually inspected), it appears that the observed mRNA changes are of a neuronal origin.
In these in-vitro conditions, the maturation/aging-decreased 5-LOX mRNA expression in mouse CGC cultures was consistently accompanied by increased methylation of CpG dinucleotides in the promoter region of the 5-LOX gene. DNA methylation at the sites of CpG dinucleotides is a known regulator of gene expression, and an aging-associated rise in the DNA methylation levels of 5' CpG islands of certain CNS genes frequently accompanies their decreased mRNA levels (Siegmund et al., 2007). Uhl et al. (2002) demonstrated the importance of 5-LOX promoter methylation for 5-LOX expression in human cell lines. Thus, a hypermethylation of the 5-LOX promoter in certain human tumor cell lines was associated with the complete silencing of this gene. Compared to the human 5-LOX gene (on chromosome 10), the mouse 5-LOX gene (on chromosome 6) lacks the typical CpG islands - the mouse promoter-5’ UTR sequence contains two recognition sites for each of the four endonucleases used in our assay to assess the promoter methylation. All these sites were hypermethylated in older cultures compared to1 DIV CGC cultures.
If this DNA methylation change contributed to decreased 5-LOX expression in older cultures, one possible mechanism involved may be the methylation-sensitive binding of regulatory molecules (e.g., transcription factors) to the DNA region of the 5-LOX promoter. By analyzing the mouse 5-LOX promoter sequence (Table 2) with software that searches transcription factor binding sites in transcription factor database TRANSFAC R.3.4. (Tsunoda and Takagi, 1999), we identified the following putative transcription factors: AHR/ARNT, NMYC, E2F, AP4, AP2, USF, and SP1. In ongoing studies, we plan to directly examine the role of DNA methylation and also to look at these factors in regulating the activity of the mouse 5-LOX promoter.
Multiple enzymatic mechanisms are involved in determining the pattern of DNA methylation, including DNA-demethylases (Hamm et al., 2008) and DNA-methyltransferases - DNMTs (MacDonald and Roskams, 2009). DNMT1, which is involved in the maintenance of DNA methylation in proliferating cells, is also expressed in post-mitiotic neurons but its role in these cells is unclear. It is possible that a pathological expression of neuronal DNMT1 may contribute to the pathobiology of psychiatric disorders such as schizophrenia (Costa et al., 2007). It appears that in our in-vitro CGC model, significantly greater expression of DNMT1 was associated with the proliferation of neuronal precursors (Uz et al., 2001) at 1 DIV and that this was accompanied by lower 5-LOX promoter methylation and greater 5-LOX mRNA expression. Hence, the changes in 5-LOX promoter methylation we observed during neuronal maturation cannot be explained by altered DNMT1. On the other hand, the increase in the expression of DNMT3a was associated with neuronal maturation and also with an increased methylation of 5-LOX promoter accompanied by decreased 5-LOX mRNA levels. Also in-vivo, the expression of neuronal DNMT3a increases during the first 3 weeks of postnatal development (Feng et al., 2005). Collectively, these data are consistent with the proposed role for DNMT3a in neuronal maturation (Watanabe et al., 2006).
Furthermore, our results indicate that epigenetic regulation of 5-LOX expression may be important during neural development. Previously, it was shown that leukotrienes and lipoxins produced via the 5-LOX pathway play a significant role in the regulation of neural stem cell (NSC) proliferation and differentiation (Wada et al., 2006). These studies demonstrated that in addition to 5-LOX, NSCs express leukotriene and lipoxin receptors and also that these receptors directly regulate proliferation and differentiation of NSCs. Also in proliferating CGC precursors, i.e., at 1 DIV, inhibitors of 5-LOX were capable of inhibiting their proliferation (Uz et al., 2001). The results we obtained in this study suggest that epigenetic suppression of 5-LOX expression in neural precursors, possibly via the activity of DNMT3a, may be involved in the transition of these proliferating cells from precursors into differentiated neurons.
CONCLUSION
The main finding in this work is that in-vitro CGC maturation involves epigenetic alterations of the 5-LOX promoter, i.e., its increased methylation that was accompanied by decreased 5-LOX mRNA levels. The concomitant differential changes in expression of DNMT1 and DNMT3a suggest that DNMT3a but not DNMT1 is responsible for maturation-associated epigenetic 5-LOX silencing. Future studies, for example in different culture models such as glia cells are needed to evaluate how findings from this in-vitro model, consisting of a population of a single neuronal type, i.e., CGC, apply to studies in brain samples, which typically contain multiple cell types.
Acknowledgments
This work was supported by a grant from the National Institute on Aging R01AG015347-05A2 (H.M.).
List of abbreviations
- AD
Alzheimer's disease
- APP
amyloid precursor protein
- CGC
cerebellar granule cells
- CNS
central nervous system
- DNMT
DNA-methyltransferase
- 5-LOX
5-lipoxygenase
- NSC
neural stem cell
- SEM
standard error mean
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Barrachina M, Ferrer I. DNA methylation of Alzheimer disease and tauopathy-related genes in postmortem brain. J Neuropathol Exp Neurol. 2009;68:880–891. doi: 10.1097/NEN.0b013e3181af2e46. [DOI] [PubMed] [Google Scholar]
- Chinnici CM, Yao Y, Praticò D. The 5-lipoxygenase enzymatic pathway in the mouse brain: young versus old. Neurobiol Aging. 2007;28:1457–1462. doi: 10.1016/j.neurobiolaging.2006.06.007. [DOI] [PubMed] [Google Scholar]
- Chu J, Praticò D. The 5-lipoxygenase as a common pathway for pathological brain and vascular aging. Cardiovascular Psychiatry and Neurology. 2009;vol. 2009 doi: 10.1155/2009/174657. Article ID 174657, 5 pages, 2009. doi:10.1155/2009/174657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa E, Dong E, Grayson DR, Guidotti A, Ruzicka W, Veldic M. Reviewing the role of DNA (cytosine-5) methyltransferase overexpression in the cortical GABAergic dysfunction associated with psychosis vulnerability. Epigenetics. 2007;2:29–36. doi: 10.4161/epi.2.1.4063. [DOI] [PubMed] [Google Scholar]
- Favaron M, Manev H, Alho H, Bertolino M, Ferret B, Guidotti A, Costa E. Gangliosides prevent glutamate and kainate neurotoxicity in primary neuronal cultures of neonatal rat cerebellum and cortex. Proc Natl Acad Sci U S A. 1988;85:7351–7355. doi: 10.1073/pnas.85.19.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Chang H, Li E, Fan G. Dynamic expression of de novo DNA methyltransferases Dnmt3a and Dnmt3b in the central nervous system. J Neurosci Res. 2005;79:734–746. doi: 10.1002/jnr.20404. [DOI] [PubMed] [Google Scholar]
- Firuzi O, Zhuo J, Chinnici CM, Wisniewski T, Praticò D. 5-Lipoxygenase gene disruption reduces amyloid-beta pathology in a mouse model of Alzheimer's disease. FASEB J. 2008;22:1169–1178. doi: 10.1096/fj.07-9131.com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamm S, Just G, Lacoste N, Moitessier N, Szyf M, Mamer O. On the mechanism of demethylation of 5-methylcytosine in DNA. Bioorg Med Chem Lett. 2008;18:1046–1049. doi: 10.1016/j.bmcl.2007.12.027. [DOI] [PubMed] [Google Scholar]
- Ikonomovic MD, Abrahamson EE, Uz T, Manev H, Dekosky ST. Increased 5-lipoxygenase immunoreactivity in the hippocampus of patients with Alzheimer's disease. J Histochem Cytochem. 2008;56:1065–1073. doi: 10.1369/jhc.2008.951855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald JL, Roskams AJ. Epigenetic regulation of nervous system development by DNA methylation and histone deacetylation. Prog Neurobiol. 2009;88:170–183. doi: 10.1016/j.pneurobio.2009.04.002. [DOI] [PubMed] [Google Scholar]
- Manev H, Manev R. 5-lipoxygenase as a possible biological link between depressive symptoms and atherosclerosis. Arch Gen Psychiatry. 2007;64:1333. doi: 10.1001/archpsyc.64.11.1333. [DOI] [PubMed] [Google Scholar]
- Manev H, Uz T. Primary cultures of rat cerebellar granule cells as a model to study neuronal 5-lipoxygenase and FLAP gene expression. Ann N Y Acad Sci. 1999;890:183–190. doi: 10.1111/j.1749-6632.1999.tb07994.x. [DOI] [PubMed] [Google Scholar]
- Manev H, Dzitoyeva S, Imbesi M, Uz T. Neuroscience Meeting Planner. Washington, DC: Society for Neuroscience; 2008. DNA methylation-related sites in the mouse 5-lipoxygenase gene. Program no. 144.25/V12 2008. Online. [Google Scholar]
- Mohammadi G, Saberivand A. Simple method to extract DNA from mammalian whole blood. J Mol Genet. 2009;1:7–10. [Google Scholar]
- Morrison LD, Smith DD, Kish SJ. Brain S-adenosylmethionine levels are severely decreased in Alzheimer’s disease. J Neurochem. 1996;67:1328–1331. doi: 10.1046/j.1471-4159.1996.67031328.x. [DOI] [PubMed] [Google Scholar]
- Ono T, Uehara Y, Kurishita A, Tawa R, Sakurai H. Biological significance of DNA methylation in the ageing process. Age Ageing. 1993;22:S34–S43. doi: 10.1093/ageing/22.suppl_1.s34. [DOI] [PubMed] [Google Scholar]
- Rådmark O, Werz O, Steinhilber D, Samuelsson B. 5-Lipoxygenase: regulation of expression and enzyme activity. Trends Biochem Sci. 2007;32:332–341. doi: 10.1016/j.tibs.2007.06.002. [DOI] [PubMed] [Google Scholar]
- Rath PC, Kanungo MS. Methylation of repetitive DNA sequences in the brain during aging of the rat. FEBS Lett. 1989;244:193–198. doi: 10.1016/0014-5793(89)81191-3. [DOI] [PubMed] [Google Scholar]
- Scarpa S, Fuso A, D'Anselmi F, Cavallaro RA. Presenilin 1 gene silencing by S-adenosylmethionine: a treatment for Alzheimer disease? FEBS Lett. 2003;541:145–148. doi: 10.1016/s0014-5793(03)00277-1. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protocols. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Serhan CN, Yacoubian Y, Yang R. Anti-inflammatory and proresolving lipid mediators. Annu Rev Pathology. 2008;3:279–312. doi: 10.1146/annurev.pathmechdis.3.121806.151409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma RP, Tun N, Grayson DR. Depolarization induces downregulation of DNMT1 and DNMT3a in primary cortical cultures. Epigenetics. 2008;3:74–80. doi: 10.4161/epi.3.2.6103. [DOI] [PubMed] [Google Scholar]
- Siegmund KD, Connor CM, Campan M, Long TI, Weisenberger DJ, Biniszkiewicz D, Jaenisch R, Laird PW, Akbarian S. DNA methylation in the human cerebral cortex is dynamically regulated throughout the life span and involves differentiated neurons. PLoS One. 2007;2(9):e895. doi: 10.1371/journal.pone.0000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohgi H, Utsugisawa K, Nagane Y, Yoshimura M, Genda Y, Ukitsu M. Reduction with age in methylcytosine in the promoter region −224 approximately −101 of the amyloid precursor protein gene in autopsy human cortex. Brain Res Mol Brain Res. 1999;70:288–292. doi: 10.1016/s0169-328x(99)00163-1. [DOI] [PubMed] [Google Scholar]
- Tsunoda T, Takagi T. Estimating transcription factor bindability on DNA. Bioinformatics. 1999;15:622–630. doi: 10.1093/bioinformatics/15.7.622. [DOI] [PubMed] [Google Scholar]
- Uhl J, Klan N, Rose M, Entian KD, Werz O, Steinhilber D. The 5-lipoxygenase promoter is regulated by DNA methylation. J Biol Chem. 2002;277:4374–4379. doi: 10.1074/jbc.M107665200. [DOI] [PubMed] [Google Scholar]
- Uhl J, Klan N, Rose M, Entian KD, Werz O, Steinhilber D. DNA methylation regulates 5-lipoxygenase promoter activity. Adv Exp Med Biol. 2003;525:169–172. doi: 10.1007/978-1-4419-9194-2_35. [DOI] [PubMed] [Google Scholar]
- Uz T, Pesold C, Longone P, Manev H. Aging-associated up-regulation of neuronal 5-lipoxygenase expression: putative role in neuronal vulnerability. FASEB J. 1998;12:439–449. doi: 10.1096/fasebj.12.6.439. [DOI] [PubMed] [Google Scholar]
- Uz T, Manev R, Manev H. 5-Lipoxygenase is required for proliferation of immature cerebellar granule neurons in vitro. Eur J Pharmacol. 2001;418:15–22. doi: 10.1016/s0014-2999(01)00924-4. [DOI] [PubMed] [Google Scholar]
- Wada K, Arita M, Nakajima A, Katayama K, Kudo C, Kamisaki Y, Serhan CN. Leukotriene B4 and lipoxin A4 are regulatory signals for neural stem cell proliferation and differentiation. FASEB J. 2006;20:1785–1792. doi: 10.1096/fj.06-5809com. [DOI] [PubMed] [Google Scholar]
- Watanabe D, Uchiyama K, Hanaoka K. Transition of mouse de novo methyltransferases expression from Dnmt3b to Dnmt3a during neural progenitor cell development. Neuroscience. 2006;142:727–737. doi: 10.1016/j.neuroscience.2006.07.053. [DOI] [PubMed] [Google Scholar]
- Wen S, Li H, Liu J. Epigenetic background of neuronal fate determination. Prog Neurobiol. 2009;87:98–117. doi: 10.1016/j.pneurobio.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Xiong J, Camello PJ, Verkhratsky A, Toescu EC. Mitochondrial polarisation status and [Ca2+]i signalling in rat cerebellar granule neurones aged in vitro. Neurobiol Aging. 2004;25:349–359. doi: 10.1016/S0197-4580(03)00123-4. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Chen CQ, Manev H. DNA methylation as an epigenetic regulator of neural 5-lipoxygenase expression: evidence in human NT2 and NT2-N cells. J Neurochem. 2004;88:1424–1430. doi: 10.1046/j.1471-4159.2003.02275.x. [DOI] [PubMed] [Google Scholar]