Abstract
Proteases are widely studied as they are integral players in cell cycle control and apoptosis. We report a new approach for the design of a family of genetically encoded turn-on protease biosensors. In our design, an auto-inhibited coiled-coil switch is turned on upon proteolytic cleavage, which results in the complementation of split-protein reporters. Utilizing this new auto-inhibition design paradigm, we present the rational construction and optimization of three generations of protease biosensors, with the final design providing a 1000 fold increase in bioluminescent signal upon addition of the TEV protease. We demonstrate the generality of the approach utilizing two different split-protein reporters, firefly luciferase and beta-lactamase, while also testing our design in the context of a therapeutically relevant protease, caspase-3. Finally, we present a dual-protease sensor geometry that allows for the use of these turn-on sensors as potential AND logic gates. Thus these studies potentially provide a new method for the design and implementation of genetically encoded turn-on protease sensors while also providing a general auto-inhibited coiled-coil strategy for controlling the activity of fragmented proteins.
Introduction
The cleavage of specific amide bonds by proteases are implicated in numerous biological pathways including cell-cycle regulation and programmed cell death. 1–4 Thus there is much interest in developing probes for measuring protease activities both in vitro and in vivo. Peptide substrates with attached self-quenched fluorescent probes are widely utilized for in vitro measurement of protease activity and substrate specificities. 5 However, many chemical probes containing large peptidic inserts often lack cell permeability and are not ideal for studying proteases such as caspases or viral proteases in their natural intracellular context. 6 To this end genetically encoded sensors 7 based upon tethered green fluorescent protein (GFP) variants have been designed, which detect protease activity through changes in Forster energy transfer or changes in fluorescence cross-correlation, both requiring sensitive instrumentation. More recently, a sensitive yet practical luminescent turn-on biosensor was reported by Wood and co-workers, where they discovered a genetically encoded turn-on luciferase based protease sensor through screening a library of cyclically permutated firefly luciferase (FLuc) mutants 8 with embedded protease cleavage sites. However, to our knowledge there are no general and rational approaches (non-selection based) for designing turn-on biosensors for proteases that rival the elegant design principles central to generating self-quenched small molecular probes. Towards the long-term goal of made to order turn-on protease biosensors, we detail our progress towards a new design paradigm utilizing an auto-inhibitory coiled-coil design architecture embedded in user-defined split-proteins (Figure 1).
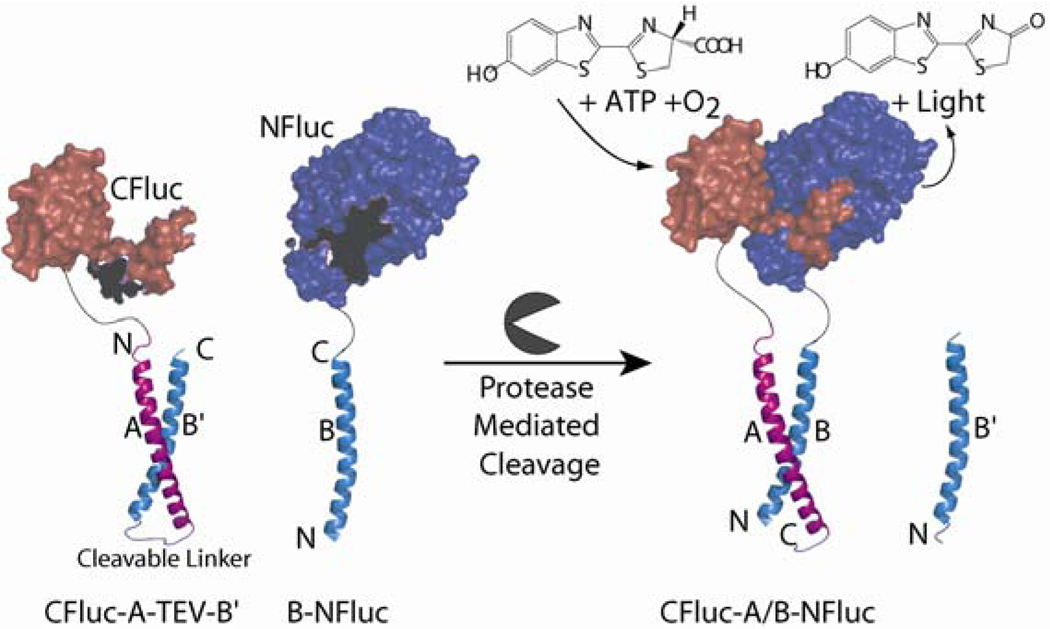
Figure 1.
Turn-on protease biosensor design: auto-inhibited intramolecular coiled-coil cleavage dependent split-protein complementation. CFluc; C-terminal fragment of firefly luciferase (Fluc), residues 398–550, NFluc; N-terminal fragment of Fluc, residues 2–416. Helices A and B’ comprise an intramolecular dimeric coiled-coil connected by a tobacco etch virus (TEV)-protease cleavage site. In the first generation design the coiled-coils, B and B’ have identical amino acid sequences. Upon incubation of the bipartite split-protein sensor (CFluc-A-TEV-B’ and B-NFluc) with TEV-protease, the linker is cleaved and the two firefly luciferase halves can potentially reassemble firefly luciferase that catalyzes the mono-oxygenation of luciferin to produce light.
Our new biosensor design relies both on sensitive split-protein signal generating domains as well as designed coiled-coil. Split-protein or protein fragment complementation methods, starting with split-ubiquitin, 9 have emerged as a promising approach for studying a wide range of biomolecular interactions. Numerous signal generating split-protein sensors have been developed based upon careful fragmentation of the green fluorescent protein 10–12 dihydrofolatereductase, 13 β-lactamase, 14–16 firefly luciferase, 17, 18 and Gaussia Luciferase. 19 At the same time, in another protein-design arena, natural and designed parallel and anti-parallel coiled-coils have emerged as a widely utilized modular domain 20–22 for applications in synthetic biology. Coiled-coils have been designed to interrogate the relation between sequence space and structure/stability of proteins,23 for testing novel amino acids,24, 25 in the design of self replicating systems,26, 27 in the construction of biomaterials,28–30 and perhaps most widely as made to order dimerization/trimerization motifs. 31–33 What is also of note for our sensor design strategy is that nature has utilized auto-inhibited coiled-coils for allosteric control. For example, in calmodulin dependent protein kinase-II, a dimeric coiled-coil domain blocks substrate and ATP binding to the otherwise active kinase domains.34 To the best of our knowledge, coiled-coil auto-inhibition, whether natural or designed, and the subsequent relief of auto-inhibition by protease action has not been utilized as a means for driving the reassembly of split-proteins as a direct read-out for protease activity (Figure 1). 8, 34
We envisioned that our general biosensor architecture for measuring protease activity would comprise three modular domains; a split reporter protein, an anti-parallel heterodimeric coiled coil and a protease cleavable linker (Figure 1). One split-protein reporter half would be attached to one of the coiled coil partners, B, whereas the other split-protein reporter half would be attached to the cognate coiled coil, A, and its binding partner, B’ (identical amino acid sequence as B) through a protease cleavable linker (Figure 1). Thus initially, the complementation of the split reporter halves is auto-inhibited since the coiled coils A and B’ prefer to interact intra-molecularly due to favorable entropy. However, once auto-inhibition is relieved by cleavage of the linker by a specific protease, the split reporter halves can potentially complement each other through intermolecular coiled-coil interactions, which will restore the activity of the split-protein reporter. Herein, we successfully demonstrate the feasibility and generality of our turn-on biosensor design strategy and also provide several iteratively redesigned systems with split-firefly luciferase and split-β-lactamase to achieve high sensitivity (~1000 fold S/N) as well as specificity (>15 fold ). We utilize a cell free in vitro translation system to test our methodology, 18 which in addition to providing an extremely rapid assay platform also enables the study of proteases in a lysate environment that approximates a complex in vivo setting.
Results and Discussion
Firefly Luciferase Complementation Utilizing Anti-parallel Coiled-Coils
Since our proposed design requires the use of dimeric anti-parallel coiled coils, we first tested two different designed coiled coil pairs namely, the EE/RR pair designed by Vinson and co-workers and the Acid /Base (A/B) coiled-coil pair designed by Oakley and co-workers. 35, 36–38 These peptides were fused to either the N- or C- termini of split-luciferase reporter protein fragments (B-NFluc/CFluc-A and RR-NFluc/CFluc-EE) through flexible (GGGS)3 linkers (supporting information, Figure S1 and Figure S2). If the coiled-coils dimerize and complement firefly luciferase assembly, we expect that the functional luciferase will catalyze the mono-oxygenation of luciferin with the concomitant production of light.39–40 As a first test, proteins were produced by co-translation of mRNA corresponding to B-NFluc/CFluc-A and RR-NFluc/CFluc-EE in Rabbit Reticulocyte Lysate system (Promega) followed by the addition of the luceferin reagent (SteadyGlo, Promega), which resulted in >60 and 1500 fold increase in luminescence respectively, when compared to the translated individual halves alone (Figure 2b, 2e) verifying that the coiled-coils under study are amenable for reassembling a functional split-luciferase enzyme.
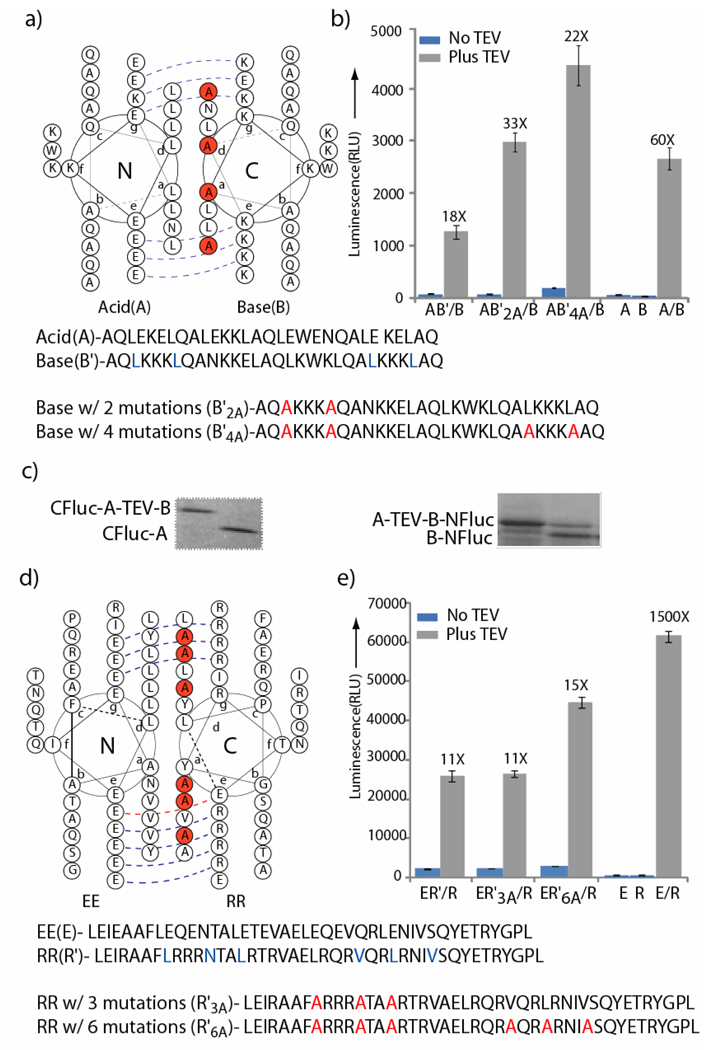
Figure 2.
First and second generation turn-on protease biosensors. a) Helical wheel diagram depicting the A/B coiled-coil, with sites for mutations shown in orange. b) TEV protease cleavage dependent change in luciferase activity in the singly inhibited A/B coiled-coil split-luciferase sensor. The translated proteins were incubated in the presence or absence of TEV (40 pmols) for 3 hrs and luminescence was measured. AB’/B refers to the B-NFluc and CFluc-ATEV-B’ sensor, AB’2A/B refers to the B-NFluc and CFluc-A-TEV-B’2A sensor and AB’4A/B refers to the B-NFluc and CFluc-A-TEV-B’4A sensor. c) SDS-PAGE analysis of the TEV protease cleaved translation products using 35S methionine after 3 hrs. d) Helical wheel diagram depicting the EE/RR coiled-coil with sites for mutations shown in orange. e) TEV protease cleavage dependent change in luciferase activity in the singly inhibited EE/RR coiled-coil split reporter. The translated proteins were incubated in the presence or absence of TEV (40 pmols) for 3 hours and luminescence was measured. ER’/R refers to the RR-NFluc and CFlucEE-TEV-RR’ sensor, EERR’3A/RR refers to the RR-NFluc and CFluc-EE-TEV-RR’3A sensor, and EERR’6A/B refers to the RR-NFluc and CFluc-EE-TEV-RR’6A sensor. Please see text for details of the design.
A First Generation Singly Inhibited Protease Biosensor
To test our proposed sensor design strategy, the coiled-coil, B’ and RR’ was fused to the CFluc-A and CFluc-EE constructs through a tobacco etch virus (TEV) protease cleavable linker (GGGGENLYFQ-GGKLGGGG) to yield CFluc-A-TEV-B’ and CFluc-EE-TEV-RR’ respectively. The mRNA corresponding to B-NFluc and CFluc-A-TEV-B’ (AB’/B system) and CFluc-EE and CFluc-EE-TEV-RR’ (ER/R’ system) were then co-translated with subsequent incubation in the presence or absence of TEV 42, 43 protease. The AB’/B system provided an 18 fold enhancement while the ER/R’ system provided an 11 fold enhancement in luminescence respectively (Figure 2b, 2e). This to our knowledge is the first demonstration of a rationally designed split-protein system that produces a turn-on signal upon enzymatic cleavage. To test if switching the coiled-coil attachment with respect to the split-protein halves influences signal to noise (S/N), experiments with A’-TEV-B-NFluc and CFluc-A were also carried out and resulted in almost identical enhancements in signal upon addition of TEV protease as the original design (Supporting Information, Figure S3). To directly confirm that the increase in enhancement correlated with TEV cleavage, translations were carried out in the presence of 35S labelled methionine. The products of translation, in the presence and absence of TEV protease, were separated by SDS PAGE and visualized by autoradiography (Figure 2c), which confirmed TEV protease dependent cleavage (please see Supporting Information for a more complete time-course, Figure S4 and Figure S5). Thus these results help validate that the production of CFluc-A and B-NFluc mediated by TEV cleavage is most likely responsible for the observed enhancement in split-luciferase activity.
A Second Generation Protease Biosensor: Optimization of Biosensor Response by Coiled-coil Redesign
In order to further optimize the response of our first generation turn-on biosensor systems ( AB’/B and ER’/R), we first chose to modulate the affinities of the coiled-coil, B’ and RR’, which are intramolecularly attached to CFluc-A and CFluc-EE. We reasoned that post-proteolytic cleavage, if the newly redesigned B’ and RR’, possess weaker affinity to CFluc-A and CFluc-EE than the parent coiled-coil, B and RR, attached to NFluc, this would increase the relative population of CFluc-A/B-NFluc and CFluc-EE/RR-NFluc, resulting in the functional complementation of the split-luciferase fragments. Thus systematic destabilizing mutations were introduced (Figure 2a, 2d) in both coiled coil systems. For the AB’/B system the first redesign incorporated two alanines in place of leucines in the first heptad (AB’2A) and the second redesign incorporated four leucine to alanine mutations (AB’4A) (Figure 2a). For the ER’/R system the first redesign incorporated two leucines and an asparagine mutations to alanines in the second heptad (ER’3A/R) and the second redesign incorporated three more leucines to alanines mutations in the fourth and fifth heptads (ER’6A/R). Previous studies44, 45 with coiled-coils suggests that each leucine to alanine mutation is expected to decrease affinity by (~0.5 – 2 kcal/mol) while the asparagine mutation in EE/RR was introduced so as to neutralize the preference for parallel orientation.35 Utilizing these new coiled-coils, we repeated our TEV protease assay with both AB’/B and ER’/R systems (Figure 2b, 2e). We observed a 2.5 fold increase in overall signal and no significant change in background signal (no added protease) for the AB’2A/B system leading to an overall S/N ratio of 33. A 1.5 fold increase in signal was observed for the AB’4A/B system as compared to the AB’2A/B system but a concomitant 2 fold increase in the background signal led to a overall S/N ratio of 22. Overall, the AB’2A/B system showed a ~1.8 fold increase in signal over background ratio than our original AB’/B system. In contrast, the ER’3A/R system showed the same S/N noise ratio as the original ER’/R system, which somewhat surprisingly suggests that two leucine to alanine mutations are not sufficiently destabilizing to afford an appreciable change in the relative populations of the EE/RR’3A and EE/RR’ coiled-coil pairs at the nanomolar concentration regime of the experiments. However, with the incorporation of further destabilizing mutations, the ER’6A/R system exhibited a 1.5 fold increase in signal to noise ratio over the parent ER’/R system. It is also noteworthy that the protease biosensors derived from the EE/RR coiled coil exhibited a much higher background as compared to the protease biosensor derived from A/B coiled coil, which may arise from formation of low affinity three-helix bundles or higher assemblies, which are possible with designed and natural coiled-coil systems 46–48 (Supporting Information, Figure S6). Since the Acid-Base coiled-coil systems afforded a much higher signal to noise ratio upon proteolytic cleavage, we chose to use this coiled-coil pair for application in other split-protein sensors as well as for further systematic redesigns.
A Turn-on Split-β-lactamase Protease Biosensor
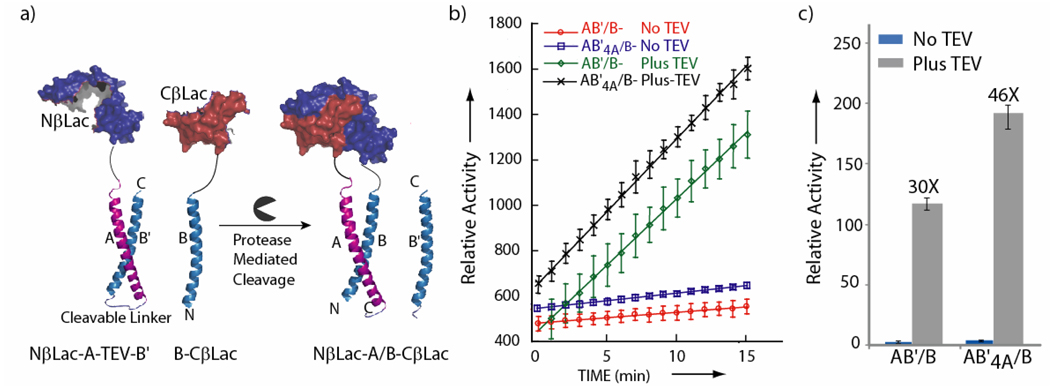
To investigate the generality of our auto-inhibitory design strategy we chose a second split-protein reporter, β-lactamase (Figure 3a), which when reassembled can catalyze the cleavage of a lactam ring in a designed substrate, Fluorocilin Green (Invitrogen), resulting in fluorescence. To construct the singly inhibited split-β-lactamase protease biosensor (AB’/B system) Acid-TEV-Base was attached to the N-terminus of NβLac (NβLac-A-TEV-B’) and Base was attached to the C-terminus of CβLac (B-CβLac). The mRNAs corresponding to NβLac-A-TEV-B’and B-CβLac were co-translated with subsequent incubation in the presence or absence of TEV protease. The observed enhancement in signal from TEV protease mediated proteolysis utilizing the new split-β-lactamase coiled-coil systems were very similar to that observed with our split-luciferase biosensor. We observed a 30 fold enhancement in S/N upon addition of TEV protease and a subsequent 1.6 fold enhancement in S/N upon introduction of the second generation AB’4A/B coiled-coil (Figure 3b, 3c). Thus these studies demonstrate that the auto-inhibited coiled-coil strategy is general and can be applied to other available split-protein systems.
Figure 3.
Turn-on Protease Biosensor utilizing Split-β-Lactamase. a) Schematic presentation of the protease biosensor design based upon the singly inhibited β-lactamase split-protein reporter with either no (B’) or four mutations (B’4A), b) Rate of hydrolysis of the Fluorocillin Green, β-Lactamase substrate, (Invitrogen) subsequent to the addition of TEV as a function of time. AB’/B refers to assays performed with co-translation of NβLac-A-TEV-B and B-CβLac, AB’4A/B refers to co-translation of NβLac-A-TEV-B4A and B-CβLac. c) The slope of the hydrolysis data from (b) represented as a bar graph for the indicated split-β-lactamase sensors in the presence and absence of TEV protease for comparison to the split-luciferase derived sensors.
Third Generation Protease Sensors: Doubly Inhibited Coiled-coil Systems
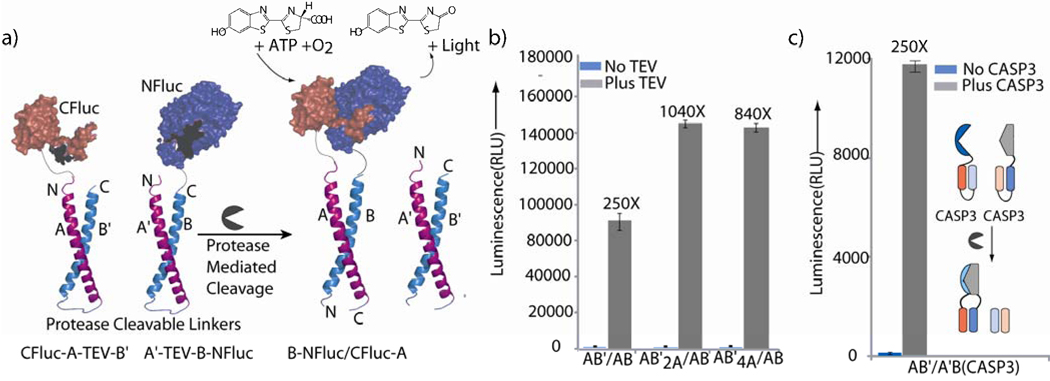
We next chose to test a third generation designed system (Figure 4a), where two auto-inhibited coiled-coils, A–B, are attached to the split-protein halves. It is worth noting that the split-luciferase system has been previously shown to be reversible and under thermodynamic control18 as opposed to kinetic control observed for some split-GFP sensors.49 The off-state requires that the intramolecular coiled-coils attached to the two halves of luciferase be thermodynamically favored, primarily through favourable entropy, when compared to the alternate extended coiled-coil system which can potentially result in undesirable complementation (Supporting Information, Figure S7). To test such a doubly inhibited system, mRNAs corresponding to A’-TEV-B-NFluc and CFluc-A-TEV- B’ were co-translated and an impressive 250 fold increase in firefly luciferase activity was observed upon addition of TEV protease (Figure 4b). Moreover, the incorporation of the designed mutations as in the second generation singly inhibited system, to afford CFluc-A-TEV-B’ 2A and CFluc-A-TEV-B’4A, led to a signal to noise ratio of 1040 and 840 for the AB’2A/A’B and AB’4A/A’B systems respectively. The overall signal obtained from the doubly inhibited system was surprisingly 5 fold higher than that obtained in the singly inhibited system, which we speculate may possibly arise from an increase in folding competent split-protein halves in the translation system.
Figure 4.
Third generation doubly inhibited protease biosensors and application to caspase-3. a) Schematic representation of the doubly inhibited luciferase sensor, Helices A and A’ have identical amino acid sequences and have been labelled differently for clarity. b) TEV protease dependent change in luminescence signal in the doubly inhibited coiled coil-split reporter system. AB’/A’B, AB’2A/A’B and AB’4A/A’B refer to co-translations of the A’-TEV-B-NFluc half with either CFluc-A-TEV-B’, Cfluc-A-TEV-B’2A, or CFluc-A-TEV-B’4A, to yield the three different sensors. c) Caspase-3 dependent activation of doubly inhibited firefly luciferase split reporter system in the presence and absence of added caspases-3. AB’/A’B(CASP3) refers to the CFluc-A-CASP3-B’ and A’-CASP3-B-NFluc containing sensor.
A Turn-on Biosensor for Caspase-3 and Assessment of Specificity
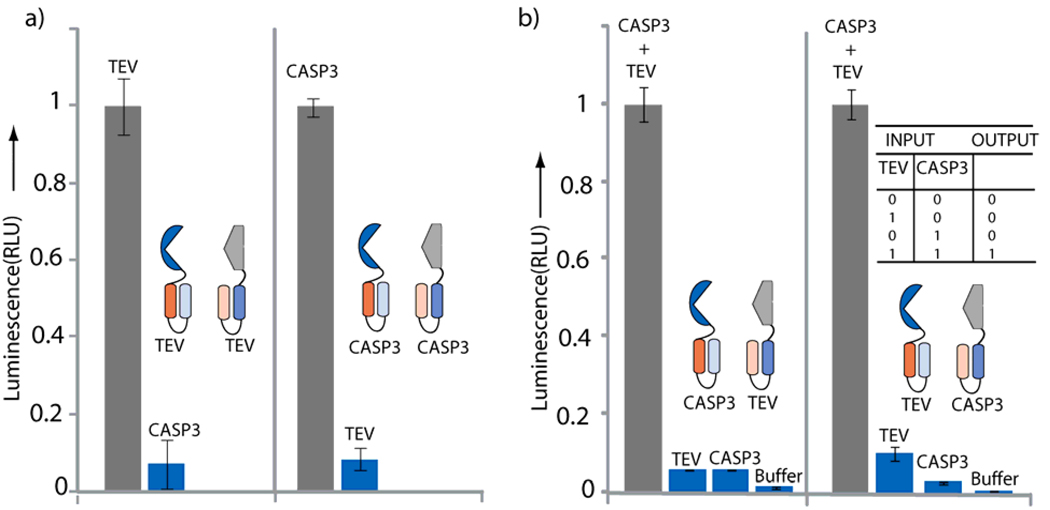
Having established three iterative redesigns of our auto-inhibited coiled-coil approach, we next turned to establishing the utility of our new turn-on biosensor for a biologically relevant protease, caspase-3.50 To this end, we incorporated a caspase-3 cleavable linker (GGGGDEVD-GKLGGGG) within the split-luciferase AB’/A’B system. This new caspases-3 sensor also afforded a ~250 fold increase (Figure 4c) in signal upon addition of caspase-3. Notably, our rationally designed TEV and caspase-3 biosensors utilizing split-luciferase provide comparable S/N as that afforded by selection strategies 8 with the possibility of further rational redesign or incorporation of selection steps51, 52. Further, to establish the specificity of these biosensors, we investigated TEV versus caspase-3 dependent signal for both AB’/A’B coiled-coil based protease sensors. We observed >15 fold specificity (Figure 5a), where the TEV sensor selectively responds to addition of TEV and not caspases-3, while the caspase-3 sensor responds to caspase-3 and not TEV. Thus these results further demonstrate the generality of the design strategy and it potential application to biologically relevant intracellular proteases.
Figure 5.
Design of AND logic gates utilizing dual protease bionsensors. a) Protease specific activation of the TEV and caspase-3 turn-on biosensors. For the split-luciferase TEV protease biosensor, mRNA corresponding to A’-TEV-B-NFluc was co-translated with CFluc-A-TEV-B’ and either TEV or Caspase-3 was added. For the split-luciferase caspase-3 protease biosensor, mRNA corresponding to A’-CASP3-B-NFluc was co-translated with CFluc-A-CASP3-B’ and either caspase-3 or TEV was added and luminescence recorded. b) Dual protease biosensors; mRNA corresponding to either A’-TEV-B-NFluc and CFluc-A-CASP3-B’ or A’-CASP3-B-NFluc and CFluc-A-TEV-B’ were co-translated and appropriate proteases and buffers were added separately or together and luminescence recorded. The biosensors are fully activated only upon the addition of both TEV and caspase-3. A truth table for the AND logic gate (inset), wherein, an output of 1 is obtained only when both inputs equal 1.
A Dual Protease Biosensor and its Application as an AND Logic Gate
Finally, we envisioned a mixed protease biosensor utilizing the doubly inhibited system, wherein the biosensor is fully activated only when two different proteases are present. To test such a system mRNAs corresponding to A’-TEV-B-NFluc and CFluc-A-CASP3-B’ were co-translated and the response of the biosensor was monitored after addition of either TEV, Caspase-3, TEV + Caspase-3, or no protease. The results (Figure 5b) clearly demonstrate that the dual protease biosensor was fully active only when both proteases were added to the system. Similarly, if we utilized A’-CASP3-B-NFluc and CFluc-A-TEV-B’ we also observed a 30 fold increase in signal compared to the presence of each individual proteases. These new dual-protease sensors systems function as molecular logic gates, producing an output of 1 when both of its inputs are 1, and an output of 0 if either or both inputs are 0. Thus this mixed protease activated sensors may perhaps be useful for designing a new class of molecular logic gates and also for targeted detection/therapy in a situation where both proteases are present in a diseased state but not in a normal state allowing for selectively turning on a previously split-enzyme.
Conclusion
We have developed a suite of genetically encoded protease biosensors utilizing an auto-inhibited coiled-coil strategy utilizing the complementation of split-proteins as our read out. The generality of the protease biosensor had been demonstrated using both a fluorescent (β-lactamase) and bioluminescent (luciferase) split-protein reporter as well as two different proteases. Our results confirm that it is possible to rationally design turn-on split-protein sensors for proteases utilizing an auto-inhibited coiled-coil design strategy with up to 1000 fold increase in S/N. The modular nature of our auto-inhibitory design will likely allow for further improvements and its application to numerous split-protein systems 8 and proteases of interest. The currently described turn-on sensors provide a method for directly detecting protease activity in complex lysate mixtures. Future research will focus upon testing the potential of our new class of split-protein protease biosensors for the more challenging measurement of protease activity in living cells. The described system may also be of potential utility in investigating the design of functional coiled-coil assemblies, which are of much interest as components of key circuits in synthetic biology. Further, the protease activated doubly inhibited systems presented herein have the potential to be utilized for the selective activation of split-toxins domains at the cell-surface similar to protease dependent specific delivery of toxic payloads.6, 53 Finally, the design of molecular logic gates as demonstrated by the mixed protease biosensor may be further refined for designing more elaborate cellular logic systems that can function in a cellular context. Thus these studies not only provide turn-on protease sensors but also provide a general auto-inhibited coiled-coil methodology for controlling the activity of fragmented proteins, which may be of general utility.
Materials and Methods
General Materials
Chemicals were purchased from Sigma unless otherwise noted. Restriction enzymes were obtained from NEB, in vitro transcription and translation products were purchased from Promega. Oligonucleotide primers were obtained from IDT. Caspase-3 was purchased from CalBioChem.
Plasmid construction and mRNA production
The plasmid constructs used in this study are shown in the supporting information (Figure S1, Figure S2) and were generated by overlapping PCR with appropriate primers and subsequently cloned into a pETDUET-1 prsfDuet (Novagen) or pMAL-c2x (NEB) vector containing the desired reporter protein fragments as described18 using standard techniques. All sequences were verified by dideoxynucleotide sequencing. For mRNA synthesis, PCR fragments corresponding to the desired fusion constructs were generated as previously described.18 The PCR products were subsequently used as templates for in vitro transcription using the RiboMAX Large Scale RNA Production System-T-7(Promega) following the manufacturer’s protocols.
Protease Sensing utilizing split-firefly luciferase
For the singly inhibited firefly luciferase TEV biosensor assay, 25 µL translation was carried out in Flexi Rabbit Reticulocyte Lysate(Promega) according to the manufacturer’s protocol using 1 pmol of mRNA encoding for B-NFluc (4–416) and 0.5 pmol of mRNA corresponding to one of the following; CFluc-A, CFluc-A-TEV-B’, CFluc-A-TEV-B’2A, CFluc-A-TEV-B’4A. For the doubly inhibited TEV biosensor, 1 pmol of mRNA encoding for A’-TEV-B-NFluc was co-translated with 1 pmol of mRNA corresponding to either CFluc-A-TEV-B’, CFluc-A-TEV-B’2A or CFluc-A-TEV-B’4A. For the doubly inhibited firefly luciferase capsase-3 biosensor, 1 pmol of mRNA encoding for A’-CASP3-B-NFluc was co-translated with 1 pmol of mRNA corresponding to CFluc-A-CASP3-B’. After translation for 90 minutes at 30 °C, the translation sample was divided into two lots of 12.5 µL each. To one sample, treated as control, 7.5 µl of appropriate protease buffer was added and to the other sample either 7.5 µl of TEV (40 pmols) or 200 units (2 µl) of caspase-3 (Calbiochem) was added. The samples were subsequently incubated for three hours at 30°C. To perform the assay 10 µL of translation solution was mixed with 40 µL of Steady-Glo Luciferase Assay System (Promega). Light emission was monitored, immediately after mixing Steady Glow containing luceferin, using a Turner Biosystems 20/20 luminometer with a 3s delay and a 10 s integration time. The single tube luminometer is equipped with a photomultiplier tube (PMT) with a spectral (photon collection) window of 350 – 650 nm. The TEV used in all the assays was expressed and purified as described by Doudna and co-workers.39 There is a ~1–2 fold difference in the absolute signal depending upon the lysate lot obtained from Promega Lysate, however the overall fold differences are conserved. Experiments with CFluc-EE-TEV-RR’, CFluc-EE-TEV-RR’ 3A or CFluc-EE-TEV-RR’6A with the corresponding EE-TEV-RR-NFluc luciferase halves were also carried out as essentially described above.
Reassembly of β-lactamase
For each β-lactamase assay, 25 µL translations, in duplicate, were carried out in Wheat Germ Extract Plus (Promega) according to manufacturer’s protocol using 1 pmol mRNA encoding for B-CβLac(198–290) and 0.0625 pmols of mRNA encoding for either NβLac-A-TEV-‘B or NβLac-A-TEV-B’4A. After translation for 90 minutes at 30°C, the translation sample was divided into two halves of 12.5 µL each. To one sample, treated as control, 7.5 µl TEV buffer was added, while to the other 7.5 µl (40 pmols) of TEV was added. The samples were subsequently incubated for three hours at 30°C. To measure lactamase activity, 20 µL of translation sample was combined with 60 µL of PBS buffer containing a final concentration of 10 µM Fluorocilin Green soluble β-lactamase substrate (Invitrogen). The rate of Fluorocillin Green hydrolysis was determined by exciting at 495 nm and monitoring emission at 525 nm with a 515 nm emission cutoff using a Spectra Max Gemini plate reader (Molecular Devices). Emission was read every min for 15 min.
Protease specific activation of protease biosensors
For the TEV biosensor, 1 pmol of mRNA encoding for A’-TEV-B-NFluc was co-translated with 1 pmol of mRNA corresponding to CFluc-A-TEV-B’ and for the caspase-3 biosensor 1 pmol of mRNA encoding for A’-CASP3-B-NFluc was co-translated with 1 pmol of mRNA corresponding to CFluc-A-CASP3-B’. After translation for 90 minutes, to both protease biosensors either TEV (40 pmols) or Caspase-3 (5 units from Calbiochem), was added and the samples were incubated for three hours at 30° C. To perform the assay 10 µL of translation solution was mixed with 40 µL of Steady-Glo Luciferase Assay System (Promega). Light emission was monitored, immediately after mixing Steady Glow, using a Turner Biosystems 20/20 luminometer with a 3s delay and a 10 s integration time.
Dual protease biosensor
50 µL translations, in duplicate were carried out in Flexi Rabbit Reticulocyte Lysate(Promega) according to the manufacturer’s protocol using 2 pmol of mRNA encoding for A’-TEV-B-NFluc and 2 pmol of mRNA encoding for CFluc-A-CASP3-B’. After translation for 90 minutes, the sample was divided into four 12.5 µl aliquots and either i) TEV (7.5 µl, 40pmols), ii) caspase-3 (2 µl, 200 units), iii) TEV (7.5 µl, 40pmols), and caspase-3 (2 µl, 200 units), or iv) TEV buffer (5 µl ) and Caspase-3 buffer (2 µl) was added and the samples were incubated for three hours at 30° C. Mixed protease biosensor with mRNA corresponding to A’-Casp3-Base-NFluc and CFluc-Acid-TEV-Base was performed in an identical manner as described. To perform the assay 10 µL of translation solution was mixed with 40 µL of Steady-Glo Luciferase Assay System (Promega). Light emission was monitored, immediately after mixing Steady Glow, using a Turner Biosystems 20/20 luminometer with a 3s delay and a 10s integration time.
35S labeled protein translations and auto-radiography
Two separate translations were carried out using 1 pmol of mRNA corresponding to CFluc-A-TEV-B’ and A-TEV-B-NFluc constructs as described in the manufacturer’s protocol (Promega). The specific activity of 35S used was 10 µCi/µl. After translation for 1.5 h at 30° C the translation sample was divided into two halves of 12.5 µL each. To one sample, treated as control, 7.5 µl of TEV protease buffer was added and to the other 7.5 µl of TEV (40 pmols) added. The samples were subsequently incubated at 30° C and 5 µl aliquots were taken out every hour and stored at −20° C overnight. The samples (5 µl) were mixed with 15 µl of loading dye (50 Mm Tris-HCl, 2% SDS, 0.1 % bromophenol blue, 10% glycerol, BME) and heated at 70° C for 15 min and subsequently run on a 15% SDS-PAGE gel. The PAGE gels were subsequently fixed (50% methanol, 10% glacial acetic acid, 40% water) for three hours and dried at 80° C under vacuum and exposed to a phosphor screen overnight with imaging on a Typhoon Imager (GE Healthcare).
Supplementary Material
Acknowledgment
The authors thank members of the Ghosh lab for helpful discussions as well as the NSF (CHE-0548264) and NIH (R01AI068414) for partial support of this research. We thank Professor Roy Parker for the use of the Typhoon Imager. J.R.P was supported by an NIH training grant and A.S by a NASA training grant.
Footnotes
Supporting Information Available: Details of cloning, assay conditions and protocols are available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Earnshaw WC, Martins LM, Kaufmann SH. Annu. Rev. Biochem. 1999;68:383–424. doi: 10.1146/annurev.biochem.68.1.383. [DOI] [PubMed] [Google Scholar]
- 2.Borgono CA, Diamandis EP. Nat. Rev. Cancer. 2004;4:876–890. doi: 10.1038/nrc1474. [DOI] [PubMed] [Google Scholar]
- 3.Jesenberger V, Jentsch S. Nat. Rev. Mol. Cell Biol. 2002;3:112–121. doi: 10.1038/nrm731. [DOI] [PubMed] [Google Scholar]
- 4.Thornberry NA, Lazebnik Y. Science. 1998;281:1312–1316. doi: 10.1126/science.281.5381.1312. [DOI] [PubMed] [Google Scholar]
- 5.Weissleder R, Tung CH, Mahmood U, Bogdanov A. Nat. Biotechnol. 1999;17:375–378. doi: 10.1038/7933. [DOI] [PubMed] [Google Scholar]
- 6.Jiang T, Olson ES, Nguyen QT, Roy M, Jennings PA, Tsien RY. Proc. Natl. Acad. Sci. U. S. A. 2004;101:17867–17872. doi: 10.1073/pnas.0408191101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luo KQ, Yu VC, Pu YM, Chang DC. Biochem. Biophys. Res. Commun. 2001;283:1054–1060. doi: 10.1006/bbrc.2001.4896. [DOI] [PubMed] [Google Scholar]
- 8.Fan F, Binkowski BF, Butler BL, Stecha PF, Lewils MK, Wood KV. ACS Chem. Biol. 2008;3:346–351. doi: 10.1021/cb8000414. [DOI] [PubMed] [Google Scholar]
- 9.Johnsson N, Varshavsky A. Proc. Natl. Acad. Sci. U. S. A. 1994;91:10340–10344. doi: 10.1073/pnas.91.22.10340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghosh I, Hamilton AD, Regan L. J. Am. Chem. Soc. 2000;122:5658–5659. [Google Scholar]
- 11.Hu CD, Kerppola TK. Nat. Biotechnol. 2003;21:539–545. doi: 10.1038/nbt816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.(a) Stains CI, Porter JR, Ooi AT, Segal DJ, Ghosh I. J. Am. Chem. Soc. 2005;127:10782–10783. doi: 10.1021/ja051969w. [DOI] [PubMed] [Google Scholar]; (b) Furman JL, Badran AH, Shen S, Stains CI, Hannallah J, Segal DJ, Ghosh I. Bioorg. Med. Chem. Lett. 2009;19:3748–3751. doi: 10.1016/j.bmcl.2009.04.141. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Ghosh I, Stains CI, Ooi AT, Segal DJ. Mol. BioSyst. 2006;2:551–560. doi: 10.1039/b611169f. [DOI] [PubMed] [Google Scholar]
- 13.Pelletier JN, Campbell-Valois FX, Michnick SW. Proc. Natl. Acad. Sci. U. S. A. 1998;95:12141–12146. doi: 10.1073/pnas.95.21.12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galarneau A, Primeau M, Trudeau LE, Michnick SW. Nat. Biotechnol. 2002;20 doi: 10.1038/nbt0602-619. 619-12141-12146. [DOI] [PubMed] [Google Scholar]
- 15.Ooi AT, Stains CI, Ghosh I, Segal DJ. Biochemistry. 2006;45:3620–3625. doi: 10.1021/bi0517032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Porter JR, Stains CI, Segal DJ, Ghosh I. Anal. Chem. (Washington, DC, U. S.) 2007;79:6702–6708. doi: 10.1021/ac071163+. [DOI] [PubMed] [Google Scholar]
- 17.Luker KE, Smith MCP, Luker GD, Gammon ST, Piwnica-Worms H, Piwnica-Worms DP. Proc. Natl. Acad. Sci. U. S. A. 2004;101:12288–12293. doi: 10.1073/pnas.0404041101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Porter JR, Stains CI, Jester BW, Ghosh I. J. Am. Chem. Soc. 2008;130:6488–6497. doi: 10.1021/ja7114579. [DOI] [PubMed] [Google Scholar]
- 19.Remy I, Michnick SW. Nat. Methods. 2006;3:977–979. doi: 10.1038/nmeth979. [DOI] [PubMed] [Google Scholar]
- 20.Lupas A. TIBS. 1996;21:375–382. [PubMed] [Google Scholar]
- 21.Bromley EHC, Chanoon K, Moutevelis E, Woolfson DN. ACS Chem. Bio. 2008;3:38–50. doi: 10.1021/cb700249v. [DOI] [PubMed] [Google Scholar]
- 22.Mason J, Arndt KM. ChemBioChem. 2004;5:170–176. doi: 10.1002/cbic.200300781. [DOI] [PubMed] [Google Scholar]
- 23.Kohn WD, Kay CM, Hodges RS. Prot. Sci. 1995;4:237–250. doi: 10.1002/pro.5560040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bilgicer B, Fichera A, Kumar K. J. Am. Chem. Soc. 2001;123:4393–4399. doi: 10.1021/ja002961j. [DOI] [PubMed] [Google Scholar]
- 25.Schnarr NA, Kennan AJ. J. Am. Chem. Soc. 2004;126:14447–14451. doi: 10.1021/ja047496v. [DOI] [PubMed] [Google Scholar]
- 26.Lee DH, Granja JR, Martinez JA, Severin K, Ghadiri MR. Nature. 1996;382:525–528. doi: 10.1038/382525a0. [DOI] [PubMed] [Google Scholar]
- 27.Yao S, Ghosh I, Zutshi R, Chmielewski J. Nature. 1998;396:447–450. doi: 10.1038/24814. [DOI] [PubMed] [Google Scholar]
- 28.(a) Ryadnov MG, Woolfson DN. Nat. Mat. 2003;2:329–332. doi: 10.1038/nmat885. [DOI] [PubMed] [Google Scholar]; (b) Zhou M, Bently D, Ghosh I. J. Am. Chem. Soc. 2004;126:734–735. doi: 10.1021/ja036634y. [DOI] [PubMed] [Google Scholar]; (c) Zhou M, Ghosh I. Organic Lett. 2004;6:3561–3564. doi: 10.1021/ol0485262. [DOI] [PubMed] [Google Scholar]
- 29.Wang C, Stewart RJ, Kopecek J. Nature. 1999;397:417–420. doi: 10.1038/17092. [DOI] [PubMed] [Google Scholar]
- 30.Petka WA, Harden JL, McGrath KP, Wirtz D, Tirrell DA. Science. 1998;281:389–392. doi: 10.1126/science.281.5375.389. [DOI] [PubMed] [Google Scholar]
- 31.Yang X, Florin L, Farzan M, Kolchinsky P, Kwong PD, Sodroski J, Wyatt R. J. Viro. 2000;74:4746–4754. doi: 10.1128/jvi.74.10.4746-4754.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.(a) Arndt KM, Muller KM, Pluckthun A. J. Mol. Bio. 2001;312:221–228. doi: 10.1006/jmbi.2001.4915. [DOI] [PubMed] [Google Scholar]; (b) Meyer SC, Shomin CD, Gaj T, Ghosh I. J. Am. Chem. Soc. 2007;129:13812–13813. doi: 10.1021/ja076197d. [DOI] [PubMed] [Google Scholar]
- 33.Rosenberg OS, Deindl S, Sung RJ, Nairn AC, Kuriyan J. Cell (Cambridge, MA, U. S.) 2005;123:849–860. doi: 10.1016/j.cell.2005.10.029. [DOI] [PubMed] [Google Scholar]
- 34.Kanno A, Yamanaka Y, Hirano H, Umezawa Y, Ozawa T. Angew.Chem. Int. Ed. 2007;46:7595–7599. doi: 10.1002/anie.200700538. [DOI] [PubMed] [Google Scholar]
- 35.Moll JR, Ruvinov SB, Pastan I, Vinson C. Prot. Sci. 2001;10:649–655. doi: 10.1110/ps.39401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oakley MG, Kim PS. Biochemistry. 1998;37:12603–12610. doi: 10.1021/bi981269m. [DOI] [PubMed] [Google Scholar]
- 37.McClain DL, Woods HL, Oakley MG. J. Am. Chem. Soc. 2001;123:3151–3152. doi: 10.1021/ja004099l. [DOI] [PubMed] [Google Scholar]
- 38.Kim BM, Oakley MG. J. Am. Chem. Soc. 2002;124:8237–8244. doi: 10.1021/ja020275+. [DOI] [PubMed] [Google Scholar]
- 39.Seliger HHH, McElroy WD. Arch. Biochem. Biophys. 1960;88:136–141. doi: 10.1016/0003-9861(60)90208-3. [DOI] [PubMed] [Google Scholar]
- 40.White EH, Rapaport E, Hopkins TA, Seliger HH. J. Am. Chem. Soc. 1969;91:2178–2180. doi: 10.1021/ja01036a093. [DOI] [PubMed] [Google Scholar]
- 41.White EH, Branchini BR. J. Am. Chem. Soc. 1975;97:1243–1245. doi: 10.1021/ja00838a049. [DOI] [PubMed] [Google Scholar]
- 42.Lucast LJ, Batey RT, Doudna JA. BioTechniques. 2001;30:544–554. doi: 10.2144/01303st06. [DOI] [PubMed] [Google Scholar]
- 43.Kapust RB, Tozser J, Fox JD, Anderson DE, Cherry S, Copeland TD, Waugh DS. Prot. Eng. 2001;14:993–1000. doi: 10.1093/protein/14.12.993. [DOI] [PubMed] [Google Scholar]
- 44.Hadley EB, Gellman SH. J. Am. Chem. Soc. 2006;128:16444–16445. doi: 10.1021/ja067178r. [DOI] [PubMed] [Google Scholar]
- 45.Acharya A, Ruvinov SB, Gal J, Moll JR, Vinson C. Biochemistry. 2002;41:14122–14131. doi: 10.1021/bi020486r. [DOI] [PubMed] [Google Scholar]
- 46.Grigoryan G, Keating AE. Curr. Opin. Struct. Biol. 2008;18:477–483. doi: 10.1016/j.sbi.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harbury PB, Kim PS, Alber T. Nature. 1994;371:80–83. doi: 10.1038/371080a0. [DOI] [PubMed] [Google Scholar]
- 48.Nautiyal S, Alber T. Prot. Sci. 1999;8:84–90. doi: 10.1110/ps.8.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Magliery TJ, Wilson CJM, Pan W, Mishler D, Ghosh I, Hamilton A, Regan L. J. Am. Chem. Soc. 2005;127:146–157. doi: 10.1021/ja046699g. [DOI] [PubMed] [Google Scholar]
- 50.Ganesan R, Mittl PRE, Jelakovic S, Grutter MG. J. Mol. Biol. 2006;359:1378–1388. doi: 10.1016/j.jmb.2006.04.051. [DOI] [PubMed] [Google Scholar]
- 51.Tisi LC, White PJ, Squirrell DJ, Murphy MJ, Lowe CRR, Murray JAH. Anal. Chim. Acta. 2002;457:115–123. [Google Scholar]
- 52.Hida N, Awais M, Takeuchi M, Ueno N, Tashiro M, Takagi C, Singh T, Hayashi M, Ohmiya Y, Ozawa T. PLOS ONE. 2009;4:1–12. doi: 10.1371/journal.pone.0005868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu S, Redeye V, Kuremsky JG, Kuhnen M, Molinolo A, Bugge TH, Leppla SH. Nat. Biotechnol. 2005;23:725–730. doi: 10.1038/nbt1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.