Abstract
MARTX toxins modulate the virulence of a number of Gram-negative Vibrio species. This family of toxins is defined by the presence of a cysteine protease domain (CPD), which proteolytically activates the Vibrio cholerae MARTX toxin. Although recent structural studies of the CPD have uncovered a novel allosteric activation mechanism, the mechanism of CPD substrate recognition or toxin processing is unknown. Here, we show that interdomain cleavage of MARTXVc enhances effector domain function. We also identify the first small molecule inhibitors of this protease domain and present the 2.35 Å structure of the CPD bound to one of these inhibitors. This structure, coupled with biochemical and mutational studies of the toxin, reveals the molecular basis of CPD substrate specificity and underscores the evolutionary relationship between the CPD and the clan CD caspase proteases. These studies are likely to prove valuable for devising novel anti-toxin strategies for a number of bacterial pathogens.
Bacterial toxins are critical mediators of the host-pathogen interface. Recently, a new family of toxins, the Multifunctional Autoprocessing Repeats-in-Toxins (MARTX) toxins, was identified in the genomes of Gram-negative bacterial pathogens, including bacteria of the Vibrio, Aeromonas, Photorhabdus, and Yersinia sp.1 Although only a few MARTX family members have been characterized, MARTX toxins modulate the virulence of a number of bacterial pathogens. The MARTX toxin of the marine pathogen Vibrio anguillarum induces hemolysis and is essential for virulence in Atlantic salmon2, while the MARTX toxin of the opportunistic, zoonotic pathogen Vibrio vulnificus causes cytotoxicity and is required for full virulence in mice3–5. Similarly, the MARTX toxin of Vibrio cholerae, the etiological agent of cholera, promotes colonization of mice6,7 and conserves produced by nearly all clinical and environmental isolates8,9.
MARTX toxins are large secreted proteins that are defined by specific structural features1. Glycine-rich repeat regions in the N- and C-termini of MARTX toxins likely form a pore within host cell membranes that transfers the toxin central region into the eukaryotic cytoplasm. This central region is comprised of multiple activity domains that presumably impart distinct functionalities to a given toxin. However, only the effector domains within V. cholerae MARTX toxin have been characterized. Two of these domains alter host actin dynamics: the actin crosslinking domain (ACD) covalently crosslinks actin monomers10, while the Rho-inactivating domain (RID) inhibits the function of small Rho protein GTPases11. A third domain, the cysteine protease domain (CPD)12, functions as an autoprocessing cysteine protease that is required for activation of MARTXVc toxin in eukaryotic cells12. The proteolytic function of the CPD is proposed to activate MARTX toxins by liberating MARTXVc effector domains from the plasma membrane12. Notably, the CPD is completely conserved in all MARTX family members and is always found adjacent to the C-terminal glycine-rich repeat region1.
The CPD is a novel protease that is regulated by a unique allosteric activation mechanism13. Binding of the eukaryotic-specific small molecule inositol hexakisphosphate (InsP6) to a basic cleft within the CPD induces a structural rearrangement that exposes the protease active site to its substrates. The responsiveness of the CPD to InsP6 spatially restricts MARTXVc toxin function to the eukaryotic cytosol13. Intriguingly, distantly related homologs of the CPD are found in the glucosylating toxins of Clostridium sp. (Supplementary Fig. 1)12. Similar to MARTXVc toxin, the CPD domains of Clostridial toxins are activated by InsP6, and activation of the CPD is required for Clostridium difficile Toxin B function14–16.
While the general details of CPD activation have been established, the mechanisms underlying CPD-mediated MARTXVc toxin activation, substrate recognition, and catalysis remain unknown. MARTXVc CPD exhibits weak structural similarity to clan CD proteases, including caspases and gingipain-R. This observation suggests that, in spite of their disparate mechanisms of activation, these proteases may share similar catalytic mechanisms13. However, MARTXVc CPD has proven resistant to all known inhibitors of these clan CD proteases12. In this study, we identified a series of novel inhibitors of CPD activity by screening a highly focused library of small molecule protease inhibitors. Using a combination of chemical, structural, and mutational approaches, we defined the substrate specificity of MARTXVc CPD and map multiple CPD cleavage sites within MARTXVc. These data demonstrate that the CPD cleaves exclusively after a P1 leucine within interdomain regions, an event that is required for optimal activity of a given domain. Our analyses also indicate that chemically inhibiting CPD function prevents MARTXVc toxin activation; chemical inhibition of the CPD likely occurs through a mechanism similar to that of caspases. This study furthers our understanding of protease-mediated activation of bacterial toxins, validates the CPD domain as a target for developing anti-toxin therapies, and provides a structural basis for developing improved inhibitors of this and other related virulence factors.
RESULTS
Chemical inhibitors of MARTXVc CPD
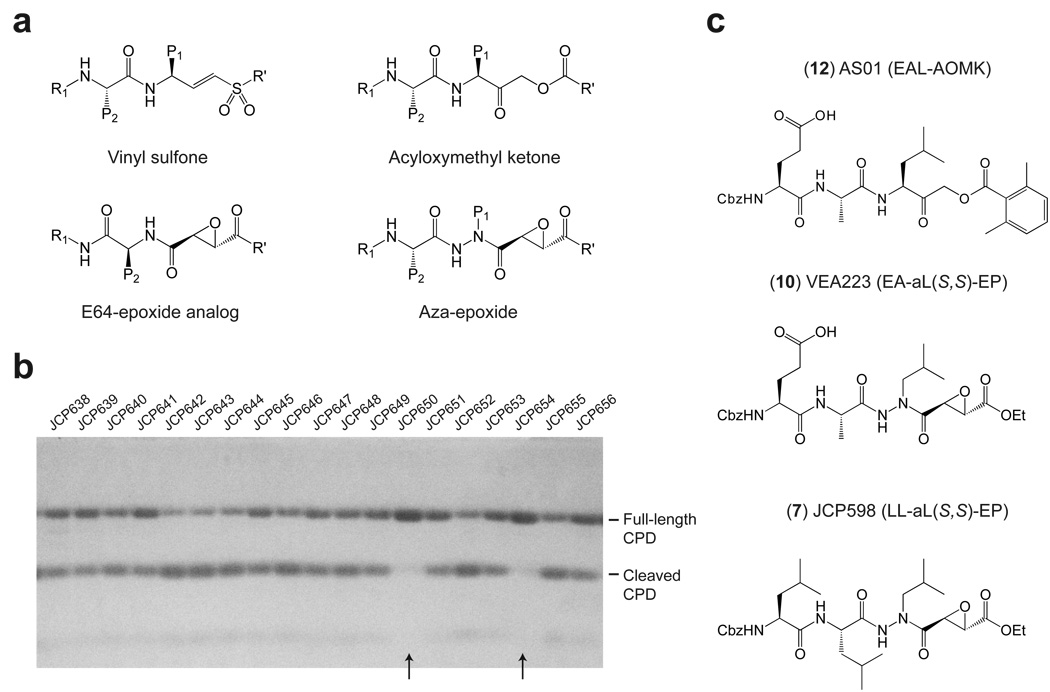
Many bacterial toxins undergo proteolytic activation upon encountering a eukaryotic cell17. Whereas most toxins are activated by host proteases, the MARTX toxin family is auto-activated by an internal cysteine protease domain12. Because genetic inactivation of the catalytic Cys of the CPD prevents V. cholerae MARTX function, we sought to chemically inhibit the protease activity of MARTXVc. To identify inhibitors of CPD function, we screened a unique library of 498 cysteine protease inhibitors18 for the ability to block recombinant CPD autoprocessing in vitro. This library is composed of cysteine protease-specific peptide vinyl sulfones, acyloxymethyl ketones (AOMKs), aza-peptide epoxides, and epoxysuccinates (Fig. 1a)19. Compounds were screened by pre-treating recombinant pro-enzyme (containing 50 residues upstream of the CPD cleavage site) with each compound then adding guanosine 5′-[γ-thio]triphosphate (GTPγS, 1) to induce activation of the protease activity. Although GTPγS is much less potent activator of the CPD than inositol hexakisphosphate13,20 (InsP6, 2), GTPγS was used as the activating compound because at the time this was the only known activator of CPD protease activity. Compounds that blocked autoprocessing were identified by SDS-PAGE analysis (Fig. 1b). This screen identified eight aza-peptide epoxides that exhibited reproducible, dose-dependent inhibitory activity in our assay (Table 1). Interestingly, all eight compounds contained leucine in the P1 position, suggesting a high degree of selectivity of this protease for the residue directly adjacent to the scissile amide bond.
Figure 1.
Identification of MARTXVc CPD autoprocessing inhibitors. (a) General structures of the main classes of covalent cysteine protease inhibitors in the library used for screening. (b) Sample gel from CPD autoprocessing inhibitor screen. Recombinant MARTXVc CPD (3391–3650 aa) was pre-treated with 100 µM inhibitor for 15 min after which GTPγS was added at 200 µM to activate autoprocessing. Cleavage reactions were resolved by SDS-PAGE and visualized by Coomassie stain. Compound numbers in the database are shown; arrowheads indicate hits in the screen (c) Structures of the most potent inhibitors of CPD-mediated autoprocessing.
Table 1.
AC50(I) values for CPD inhibitors identified in screen and for inhibitors designed to block CPD autoprocessing activity
| Number | Designation | Inhibitora | EPb | AC50 (nM) at 10 µM inhibitorc |
|---|---|---|---|---|
| – | – | No inhibitor | n/a | 0.9 ± 0.1 |
| 3 | JCP485 | Z-ALeu-EP-COO-Et | trans | 1.1 ± 0.1 |
| 4 | JCP479 | Ac-Leu-ALeu-EP-COO-Et | S,S | 21 ± 3 |
| 5 | JCP650 | Z-Leu-ALeu-EP-COO-Et | S,S | 11± 2 |
| 6 | JCP654 | Z-Leu-Leu-ALeu-EP-COO-H | S,S | 158 ± 19 |
| 7 | JCP598 | Z-Leu-Leu-ALeu-EP-COO-Et | S,S | 457 ± 80 |
| 8 | JCP657 | R,R | 5.2 ± 0.3 | |
| 9 | JCP599 | trans | 74 ± 8 | |
| 10 | VEA223 | Z-Glu-Ala-ALeu-EP-COO-Et | S,S | 429 ± 83 |
| 11 | AS04 | Z-Ala-Leu-AOMK | n/a | 187 ± 30 |
| 12 | AS01 | Z-Glu-Ala-Leu-AOMK | n/a | 529 ± 108 |
| 13 | AS02 | Z-Lys-Glu-Ala-Leu-AOMK | n/a | 290 ± 52 |
Z = Ph-CH2–O-C(O)CO–; ALeu = aza-Leu; EP = epoxide; Et = ethyl.
The trans epoxide is a mixture of S,S and R,R, while the cis epoxide is a mixture of R,S and S,R. n/a = not applicable
AC50(I) represents the concentration of InsP6 required to activate half-maximal cleavage of the CPD in the presence of 10 µM of inhibitor. Since the extent of inhibition depends on the concentration of InsP6 in the assay, the higher the AC50(I) value, the better the inhibitor. AC50(I) values were determined from triplicate experiments (± s.d.).
To compare the potencies of each inhibitor, we measured the concentration of InsP6 required to activate half-maximal cleavage of the CPD in the presence of 10 µM inhibitor (AC50(I), Table 1 and Supplementary Fig. 2). A large AC50(I) is indicative of a better CPD inhibitor, since more InsP6 is required to activate cleavage in the presence of a fixed amount of inhibitor. It should be noted that this assay only measures cis autocleavage events, as autocleavage of recombinant MARTXVc CPD in trans is strongly disfacored due to steric hindrance13. Based on these measurements, we generated a small structure-activity relationship series using the eight inhibitors identified in our screen (Table 1). Most notably, inhibitor potency correlated with peptide length: addition of a P3 Leu increased inhibitor potency by ~40-fold (11 ± 2 nM vs. 457 ± 80 nM; JCP650 vs. JCP598). Inhibitor potency was also dependent on the regio- and stereochemistry at the epoxide moiety with the order of inhibition being S,S > trans >> R,R (Table 1). Interestingly, this same preference for the trans S,S aza-peptide epoxide has been observed for the caspases21, implying that the CPD and caspases share similar mechanisms of substrate recognition.
Based on this observation, we hypothesized that functional groups previously used as caspase inhibitors might also inhibit CPD protease activity. Thus, we synthesized AOMK inhibitors19 carrying the P4-P1 (KEAL) residues of the Leu3441 cleavage site and evaluated their efficacy in the CPD autocleavage assay. We also synthesized an aza-peptide epoxide containing the P3-P1 positions of the Leu3441 cleavage site (VEA223) to directly compare the contribution of the functional group to inhibitor strength (Fig. 1c). As with the aza-Leu epoxide inhibitors, the presence of the P3 residue increased inhibitor potency (529 ± 108 nM vs. 187 ± 30 nM; AS01 vs. AS04). Addition of the P4 residue, however, did not improve inhibitor potency, perhaps because the hydrophobic Cbz (Ph-CH2-O-C(O)) group of AS01 was replaced with a basic lysine residue in AS04 (290 ± 52 nM vs. 529 ± 108 nM; AS02 vs AS01).
While the presence of P2 and P3 residues enhanced inhibitor potency, the protease exhibited a somewhat broad selectivity in these positions, since the EAaL (VEA223) and LLaL (JCP598) epoxides had similar AC50(I) values (Table 1). The clan CD-specific AOMK and aza-peptide epoxide functional groups were also equally effective at inhibiting CPD function (JCP598 vs. VEA223, Table 1). Inhibition of CPD activity was specific to these functional groups, since the proteasome inhibitors MG132 (Cbz-LLL-aldehyde, 14) and Z-L3VS (Cbz-LLL-vinyl sulfone, 15) failed to inhibit CPD function (data not shown). Taken together, our results strongly imply that optimal inhibition of CPD activity requires compounds with a P1 Leu linked to either the AOMK or aza-epoxide functional groups.
Crystal structure of inhibitor-bound, activated MARTXVc CPD
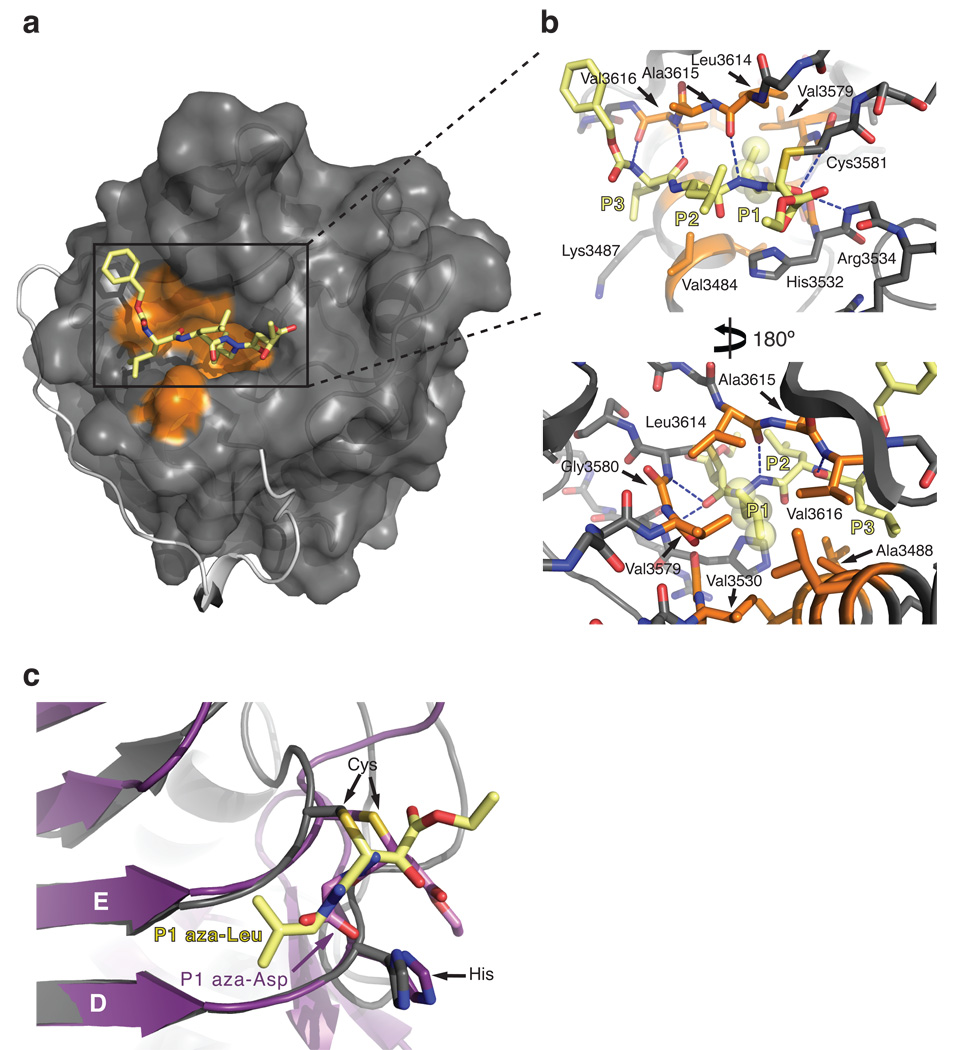
To gain insight into the mechanism of chemical inhibition of the CPD, we co-crystallized and solved the structure of activated, InsP6-bound CPD in complex with the aza-Leu epoxide inhibitor JCP598 (Fig. 2a). The overall structure of inhibitor-bound, activated CPD is nearly identical to our previous unbound structure of activated CPD (root-mean-square deviation of 0.5Å) (Supplementary Fig. 3)13. This superposition indicates that the inhibitor essentially docks into an active site cleft created upon binding of InsP6 to the CPD; no significant changes in active site topology are induced upon inhibitor binding.
Figure 2.
Structure of activated MARTXVc CPD bound to an aza-peptide epoxide inhibitor. (a) Surface topology of the CPD active site. Hydrophobic residues in the substrate binding cleft are highlighted in orange. The aza-peptide epoxide inhibitor (JCP598) is shown as a stick model bound in the substrate binding pocket. The N-terminus is shown as a grey ribbon, terminating at Ile5 and highlighting the threading of this region along the surface of the core domain. (b) Close-up “top” and “bottom” views of the S1 pocket. Hydrophobic residues in the S1 pocket are shown as orange sticks, and the side chain atoms of the P1 aza-Leu residue are shown as transparent spheres. Hydrogen bonds between the inhibitor backbone and the protein are shown as dashed lines. (c) Superposition of the D and E beta strands of caspase-3-aza-Asp epoxide (PDB ID 2C1E) and CPD-aza-Leu epoxide inhibitor structures shown as a cut-away view of the thioether inhibitor adduct bound in the S1 pocket. Caspase-3 is colored purple, while the aza-Asp inhibitor is colored pink. The MARTXVc CPD is colored grey, while the aza- Leu inhibitor is colored yellow.
As with most proteases, the substrate-binding cleft can be subdivided into multiple subsites, each consisting of residues involved in recognition of the substrate. The catalytic residues are positioned between the S1 and S1’ subsites, with subsite numbering mirroring the numbering of the corresponding substrate residues. The S1 subsite consists of a deep hydrophobic pocket that buries the side chain of the P1 Leu residue. Of the twelve residues that form this hydrophobic cleft, seven are within Van der Waals bond distance (4.4 Å) of the P1 Leu: Leu3614 and Ala3615 are contributed by the β-flap (the structural region that mediates InsP6-activation)13; Val3579 and Gly3580 are contributed by strand E; Val3530 and Gly3531 are contributed by strand D; and Ala3488 is contributed by helix 1 (Fig. 2b and 3a). Due to the covalent bond between Cys3581 and the aza-epoxide, the inhibitor is pulled in slightly towards the catalytic cysteine; in a native conformation, the P1 Leucine likely makes more diffuse contacts with both sides of the pocket.
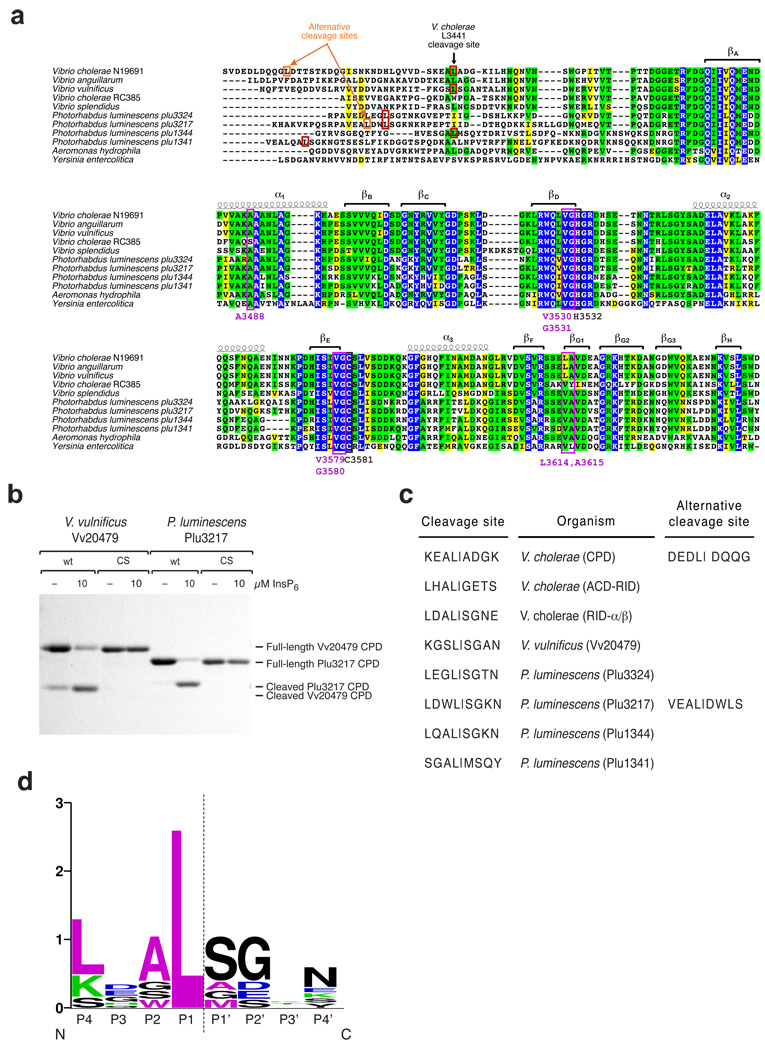
Figure 3.
MARTX CPDs cleave after a P1 Leucine. (a) Multiple sequence alignment of MARTXVc CPD with homologs from related MARTX toxins. Completely conserved identical residues are blocked in blue, conserved identical residues in green, and conserved similar residues in yellow. Secondary structure elements of the V. cholera CPD (3405–3647 aa) are shown above the amino acid sequences. The catalytic Cys and His residues are boxed and labeled in black; P1 Leu-interacting residues are boxed and labeled in purple. Cleavage site residues of MARTX CPDs from V. cholerae N16961, V. vulnificus, and P. luminescens were identified by FT-MS analysis (red boxes). Alternative cleavage site residues that occur upon mutation of the P1 Leu to Ala were mapped by FT-MS (orange boxes). (b) Activation of MARTX CPD autocleavage by InsP6. Wildtype or catalytic-dead (CS) recombinant CPDs from V. vulnificus CMCP6 VV20479 (aa 4044–4299) and P. luminescens TTO1 Plu3217 (aa 2390–2630) MARTX toxins were incubated in the presence or absence of 10 µM InsP6, and autocleavage was assessed by SDS-PAGE and Coomassie staining. (c) Comparison of P4-P4’ residues of known MARTX CPD cleavage sites. Alternative cleavage sites that result from mutation of the primary autoprocessing site are shown for V. cholerae (Leu3441) and P. luminescens Plu3217 (Leu2408). (d) Sequence logo representation of MARTX CPD consensus cleavage site based on sites shown in (c). The sequence logo was created using the website http://weblogo.berkeley.edu. The dashed line indicates the scissile bond.
C-terminal to the S1 pocket, the surface topology of the CPD is relatively flat and featureless, being composed primarily of peptide backbone atoms as well as the alkyl chain of Arg3534. On the N-terminal side, a groove is formed by helix 1 and the G1 strand of the β-flap, where the backbone atoms of the P2 and P3 inhibitor residues engage in hydrogen bond interactions with the backbone of the G1 strand (Fig. 2b). This analysis supports our prior observation that addition of P2 and P3 residues to the inhibitor scaffold increases potency irrespective of peptide sequence (Table 1). The P2 Leu residue of the inhibitor points away from the protease, tangentially interacting with Trp3631 and Glu3613, while the P3 Leu interacts with Val3484, the alkyl chain of Lys3487, Ala3488, Asn3491 and Val3616 in the S3 subsite (Fig. 2b). Based on our structure-activity analyses of the LLaL versus EAaL inhibitors (Table 1), which have similar AC50:I values, it seems likely that the surface chemistry of the S3 subsite can accommodate varied side chains at the P3 position. Taken together, the inhibitor structure provides mechanistic insight into substrate recognition and reveals how the CPD specifically recognizes a Leu in the P1 position.
Importantly, the aza-Leu epoxide inhibitor is found covalently bound to the catalytic Cys through a thioether bond in the crystal structure. Nucleophlic attack by the catalytic Cys occurs at the C3, rather than the C2, position of the epoxide; this same mechanism of catalysis is observed in the aza-peptide epoxide bound structure of caspase-322. Indeed, the catalytic Cys and His residues of the CPD and caspase-3 are similarly positioned to attack the P1-P1’ peptide bond (Fig. 2c). Furthermore, both proteases have an optimal S1 pocket for recognition of the side chain of their respective P1 substrate residues. The CPD S1 pocket, however, is considerably deeper than the caspase-3 S1 pocket, causing the JCP598 inhibitor to be buried more deeply in the CPD structure than the aza-Asp epoxide caspase inhibitor (Fig. 2c). These analyses reveal that the CPD and caspases share similar mechanisms of catalysis and substrate binding, despite differing significantly in the size and surface properties of their S1 subsites.
The substrate specificity of MARTX CPDs is conserved
Clan CD proteases are distinguished by their strict preference for specific amino acid side chains in the S1 subsite23. Thus, by inference, one would predict that all MARTX CPDs are selective for Leu in the P1 position. Indeed, multiple sequence alignment of related MARTX CPDs indicates that substrate binding pocket residues are well conserved (Fig. 3a). To directly examine the substrate specificity of related CPDs, we expressed and purified MARTX CPDs from Vibrio vulnificus and Photorhabdus luminescens, which encodes four distinct MARTX CPDs, and analyzed their autoprocessing activity in the presence of InsP6. All five MARTX CPDs tested underwent InsP6-dependent autoprocessing (Fig. 3b and Supplementary Fig. 4), indicating that MARTX CPDs exhibit a shared mechanism of activation. Analysis of the exact mass of the in vitro cleavage products by Fourier transform mass spectroscopy (FT-MS) revealed that all five MARTX CPDs were autoprocessed after leucine (Fig. 3a and 3c). When the P1 Leu residues of V. cholerae N16961 and Photorhabdus luminescens Plu3217 MARTX CPD were respectively mutated to Ala, autocleavage occurred at a previously disfavored upstream Leu residue and before a P1’ Asp (Fig. 3a and 3c). Interestingly, the CPD cleavage site mutant of V. cholerae did not cleave at the first available Leu residue (Leu3432), potentially due to the presence of a more bulky Gln residue in the P1’ position.
To generate a consensus cleavage site sequence for MARTX CPDs, we analyzed the identified cleavage sites using the WebLogo program (Fig. 3d). Although the training set is limited, these analyses suggest that, in addition to the strict requirement for Leu in the P1 position, small residues may be preferred in the P2, P1’, and P2’ positions. A small P2 residue, however, is not essential, given that substrates with Leu and Trp in the P2 position were still recognized by the CPD (Fig. 1c and 3c). No conservation was observed in the P3 position, confirming that the P3 position contributes little to substrate specificity. Taken together, these analyses demonstrate that MARTX CPDs, like other clan CD proteases, are highly selective for the P1 residue; in the case of MARTX CPDs, the P1 residue recognized is a Leucine.
The CPD processes MARTXVc toxin at multiple sites
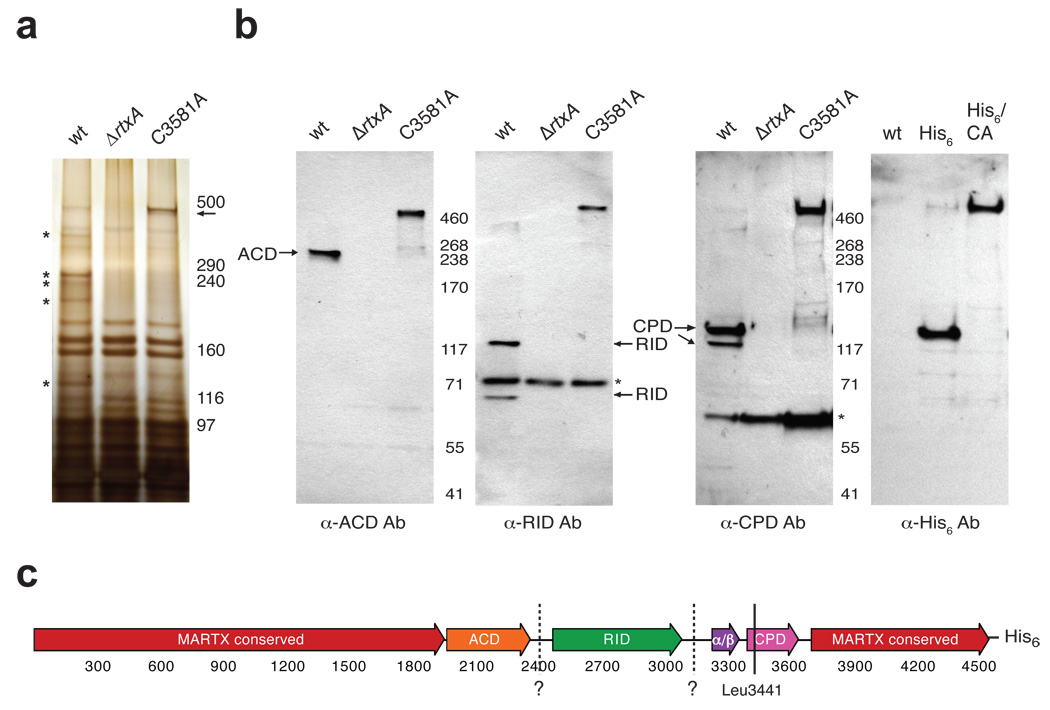
The universal conservation of CPDs in MARTX toxins suggests that these proteases play a critical role in regulating toxin function. To interrogate the role of the CPD in activating MARTX toxin, we examined the number and location of CPD-dependent processing sites within V. cholerae MARTX toxin. We first compared the secreted protein profiles of wildtype V. cholerae, an rtxA deletion strain (ΔrtxA; rtxA encodes MARTXVc), and a CPD catalytically dead strain (C3581A) to assess whether MARTXVc is cleaved at multiple sites in a CPD-dependent manner. Whereas multiple MARTXVc-specific protein bands were detected in culture supernatants of wildtype V. cholerae (Fig. 4a, asterisks), a single, predominant MARTX-specific protein the predicted size of unprocessed MARTXVc toxin (~460 kDa) was detected in culture supernatants of the C3581A mutant (Fig. 4a, arrow).
Figure 4.
MARTXVc is processed in a CPD-dependent manner (a) Silver stain of culture supernatants harvested from V. cholerae strains harboring either an intact rtxA gene (wt), a null mutation in rtxA (ΔrtxA), or point mutation in the CPD catalytic Cys (C3581A). Unprocessed MARTXVc (predicted size 460 kDa) is indicated with an arrow, while the asterisks demarcate MARTXVc-specific bands. (b) Western blot analysis of V. cholera culture supernatants used in (a) and of V. cholerae culture supernatants derived from strains expressing MARTXVc with a C-terminal His6-tag in either the wt (His6) or C3581A background (His6/CA) (far right panel). Antibodies were raised against His6-tagged ACD (aa 1964–2375), His6-tagged RID (aa 2552–3099), and His6-tagged CPD (aa 3391–3650). Background bands are indicated with asterisks. (c) Schematic of MARTXVc toxin. Conserved glycine-rich repeat regions in the N- and C-termini of MARTX toxins (MARTX conserved, red); actin crosslinking domain (ACD, orange); Rho-inactivating domain (RID, green); α/β hydrolase domain (α/β, purple), cysteine protease domain (CPD, pink). Amino acid numbering is given below. The C-terminal His6-tag encoded in strains His6 and His6/CA is shown. The known cleavage site within the CPD at Leu3441 is shown as a solid line. Putative cleavage sites inferred from Western blot analyses are shown as dashed lines and labeled with question marks.
We next analyzed the domain composition of MARTXVc cleavage products in culture supernatants by Western blot analysis using antibodies specific for the ACD, RID, and CPD domains of MARTXVc, respectively. Each antibody produced a distinct Western blot profile in wildtype culture supernatants: the anti-ACD antibody primarily detected a single protein fragment (~250 kDa); the anti-RID antibody detected two MARTXVc fragments (~110 kDa and ~65 kDa); and the anti-CPD antibody recognized two MARTXVc fragments (~120 kDa and ~110 kDa) (Fig. 4b). The ~110 kDa protein fragment was detected by both the anti-CPD and anti-RID specific antibodies, indicating that it harbors portions of both domains. The ~120 kDa fragment recognized by the anti-CPD antibody was also detected by a His6-specific antibody in culture supernatants of a V. cholerae strain harboring a MARTXVc with a C-terminal His6-tag. This result indicates that the ~120 kDa protein contains the extreme C-terminus of the CPD (Fig. 4b). In contrast, all four antibodies recognized a single ~460 kDa protein in of C3581A culture supernatant (Fig. 4b), consistent with the silver stain analysis (Fig. 4a). From these analyses, we can infer that, in addition to the previously mapped Leu3441 site within the CPD, two additional CPD-dependent processing sites are present in MARTXVc between the ACD-RID and RID-α/β junctions (Fig. 4c).
Identification of MARTXVc toxin cleavage sites
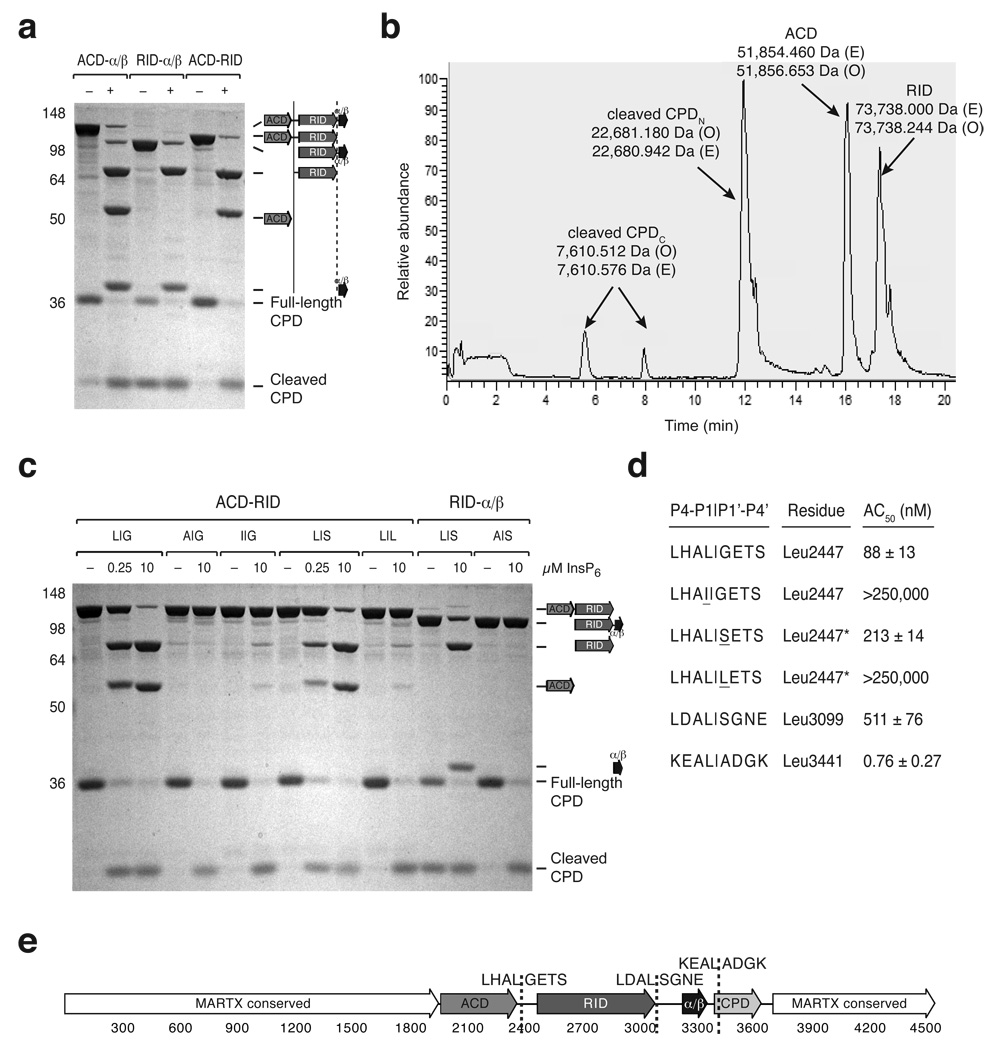
MARTXvc processing may result from direct cleavage by the CPD or from CPD-mediated activation of a second protease that sequentially cleaves MARTXVc; this latter scenario is frequently observed in viral polyprotein processing systems24,25. To distinguish between these possibilities, we tested whether InsP6-activated CPD could cleave MARTXVc-derived polypeptides in vitro. Transcleavage of recombinant ACD-α/β, ACD-RID, RID-pGap1 polypeptides by the CPD all produced an ~71 kDa fragment; CPD-mediated cleavage of the ACD-α/β and ACD-RID polypeptides produced an ~55 kDa fragment; and transcleavage of the ACD-α/β and RID-α/β fragments liberated an ~36 kDa protein (Fig. 5a). By deduction, the ~71 kDa, ~55 kDa, and ~36 kDa fragments comprise the ACD, RID, and α/β domains, respectively, indicating that the CPD directly cleaves MARTXVc between the (i) ACD and RID domains and (ii) RID and α/β hydrolase domains.
Figure 5.
Identification of MARTXVc toxin cleavage sites in vitro. (a) CPD-mediated transcleavage of MARTXVc polypeptides in vitro. Recombinant CPD (aa 3391–3650) and the indicated MARTXVc polypeptides were incubated ± InsP6, and cleavage reactions were resolved by SDS-PAGE and visualized by SDS-PAGE (schematic shown to the right). (b) Reverse-phase HPLC chromatogram of CPD-mediated transcleavage of recombinant ACD-RID. The observed masses (O) of the polypeptides detected within a given peak are indicated and were determined by FT-MS analysis; the expected masses (E) of polypeptide fragments are also shown. (c) InsP6-induced CPD-mediated transcleavage of mutant MARTXVc polypeptides. The P1 and P1’ residues of the wildtype and mutant cleavage sites are given as P1|P1’. L|G is the wildtype sequence for the ACD-RID cleavage site (Leu2447); L|S is the wildtype sequence for the RID-α/β cleavage site (Leu3099). (d) Comparison of CPD-mediated cleavage at various processing sites. The P4 to P4’ residues are shown for each cleavage site. The concentration of InsP6 at which 50% cleavage of the indicated polypeptides occurred (AC50) is shown (± s.d.). In the presence of InsP6, the ACD-RID and RID-α/β polypeptides were subjected to transcleavage with recombinant CPD, while the α/β-CPD polypeptide was subjected to autocleavage. Mutations introduced into either the P1 and P1’ sites of the ACD-RID cleavage site are underlined. (e) Schematic of MARTXVc toxin. Amino acid numbering is given below, and cleavage sites sequences (P4-P4’) are given.
In order to map specific CPD cleavage sites within recombinant MARTXVc fragments, we measured the exact mass of in vitro transcleavage products by FT-MS. While these analyses confirmed the Leu3441 CPD autoprocessing site (Fig. 5b)12, the resolution of the mass spectrometer for the larger fragments was insufficient to unequivocally identify the cleavage sites. However, given that CPD-mediated processing of MARTXVc likely occurs after Leucine, we were able to identify Leu2447 and Leu3099 as putative cleavage sites. To validate these sites, the effect on CPD-mediated transcleavage upon mutation of Leu2447 and Leu3099 to Ala was examined. Mutation of both residues to Ala abrogated processing of recombinant ACD-RID and RID-α/β polypeptides, respectively (Fig. 5c), while mutation of Leu2447 to its isomer Ile severely reduced CPD transcleavage of the ACD-RID polypeptide (Fig. 5c). These data confirm our predicted cleavage sites and support the conclusion that the CPD requires a P1 Leu residue for substrate recognition.
We next examined whether the CPD exhibited differential affinity towards MARTXVc processing sites. To this end, we measured the concentration of InsP6 required to half-maximally activate CPD cleavage (AC50) at Leu2447, Leu3099, and Leu3441 (Fig. 5d and Supplementary Fig. 5). The AC50 for the α/β-CPD junction (Leu3441) was 0.76 ± 0.27 nM, consistent with the previously measured AC50 for a recombinant CPD fragment lacking the N-terminal α/β domain (0.91 ± 0.10 nM)13. The AC50 for the ACD-RID junction (Leu2447) was 88 ± 13 nM and 511 ± 77 nM for the RID-α/β junction (Leu3099) (Fig. 5d). These results demonstrate that the CPD recognizes MARTXVc cleavage sites with differential affinity. The significantly lower AC50 for Leu3441 likely reflects the positioning of Leu3441 close to the active site13 such that the primary sequence around the Leu3441 cleavage site should not affect the CPD’s affinity for this site. In contrast, slight variations in the primary sequence around Leu2447 and Leu3099 could account for the ~6-fold difference in AC50 for these sites. To explore this possibility, we exchanged the P1’ Gly (G2448) of the Leu2447 cut site for the P1’ Ser (S3100) of the Leu3099 cleavage site. This alteration increased the AC50 of the Leu2447 cut site by ~2.5-fold (Fig. 5d), suggesting that the CPD prefers small, neutral residues to polar residues in the P1’ position. Large residues in the P1’ position are poorly tolerated by MARTXVc CPD, as mutation of Gly2448 to Leu largely abrogated CPD-mediated cleavage at Leu2447 (Fig. 5c). These substrate preferences are consistent with the observation that the S1’ subsite is flat and non-polar in the inhibitor-bound crystal structure (Fig. 2a).
CPD-mediated processing optimally activates MARTXVc function
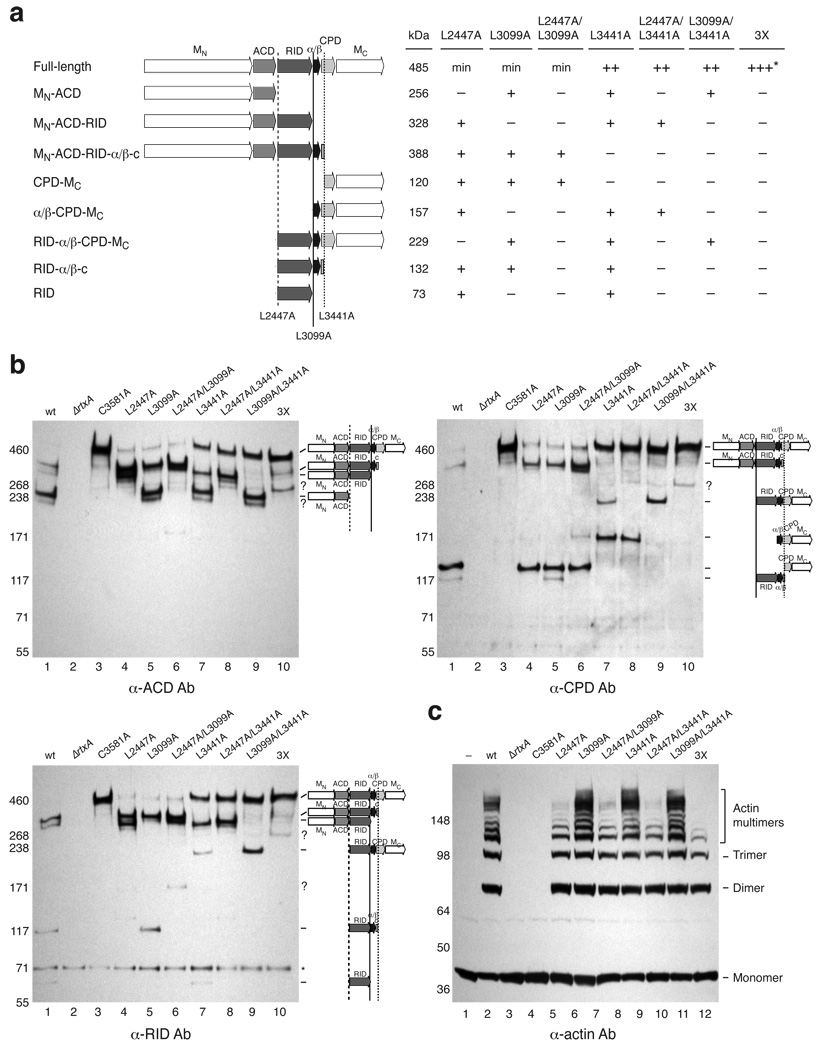
Having identified multiple MARTXVc processing sites in vitro, we sought to determine whether these cleavage sites were relevant in vivo. Thus, we introduced mutations of Leu2447, Leu3099, and Leu3441 to Ala either singly, doubly or triply into the genome of V. cholerae and assessed their effect on MARTXVc processing in culture supernatants by Western blot analysis. Cleavage of MARTXVc at these three sites theoretically should liberate eight polypeptides that can be detected by the anti-ACD, anti-RID, and anti-CPD antibodies. Indeed, all eight fragments were detected in wild-type culture supernatants (Fig. 6a and 6b). Mutation of cleavage site Leu residues to Ala prevented cleavage at these sites (Fig. 6a and 6b). For example, Leu2447A resulted in the disappearance of two polypeptides containing either a C-terminal ACD domain or N-terminal RID domain (Fig. 6a and 6b, lane 4). Likewise, culture supernatants of the L3099A mutant lacked MARTXVc fragments with either a C-terminal RID domain or an N-terminal α/β domain (Fig. 6a and 6b, lane 5). Conversely, mutation of Leu3441 caused the loss of fragments with either a C-terminal α/β domain or N-terminal CPD from culture supernatants (Fig. 6a and 6b, lane 7), as well as the accumulation of unprocessed MARTXVc relative to the L2447A and L3099A mutants. In fact, the Leu3441 mutation was epistatic to other cleavage site mutations: unprocessed MARTXVc was the most prominent species detected in culture supernatants of any strain carrying the L3441 mutation (Fig. 6b). This result suggests that processing at Leu3441 stimulates the transcleavage activity of the CPD protease. Mutation of all three cleavage sites rendered MARTXVc largely resistant to CPD-mediated processing (Fig. 6a and 6b, lane 10), since only a small amount of processing at an alternative site was observed (Fig. 6a and 6b, question marks). Given that these alternative cleavages were highly inefficient, these analyses indicate that the primary MARTXVc cleavage sites in vivo are Leu2447, Leu3099, and Leu3441.
Figure 6.
Effect of cleavage site mutations on MARTXVc processing and function. (a) Summary of Western blot analyses of MARTX toxin in V. cholerae cleavage site mutant culture supernatants. MN, N-terminal MARTX conserved region; MC, C-terminal MARTX conserved region. The predicted MWs of MARTXVc fragments are given. + indicates the presence of a given polypeptide band in culture supernatants by Western blot analysis; ++ indicates increased levels of full-length MARTXVc in mutant culture supernatants relative to the minimal amounts observed in wildtype supernatants (min). The triple mutant L2447A/L3099A/L3441A is designated as 3X. The majority of MARTXVc secreted by the 3X mutant is unprocessed, although small amounts of aberrantly processed toxin are observed (+++*). (b) Western blot analysis of V. cholera cleavage site mutant culture supernatants using antibodies specific for discrete regions of MARTXVc. Unidentifiable bands are noted with a question mark; background bands are indicated with an asterisk. Cleavage sites that affect detection of MARTXVc fragments for a given antibody are shown. (c) Actin crosslinking activity of V. cholerae cleavage site mutants. Culture supernatants harvested from strains used in (B) were incubated with HFF cells for 2 hr; HFFs were lysed, and lysates were resolved by SDS-PAGE. Actin crosslinking was visualized by Western blotting using an anti-actin antibody. The cross-linked forms of actin are labeled to the right.
To assess the role of MARTXVc processing on toxin activation, we examined whether MARTXVc cleavage site mutants exhibited reduced actin crosslinking in human foreskin fibroblast cells (HFFs). V. cholerae culture supernatants harvested from wild type, ΔrtxA, C3581A (CPD catalytic mutant), and cleavage site mutants were incubated with HFFs, and Western blot analysis was used to visualize MARTXVc-induced actin crosslinking in HFF lysates. Although all supernatants from cleavage site mutants induced actin crosslinking (Fig. 6c), supernatants from strains carrying the L2447A mutation exhibited lower amounts of actin crosslinking relative to wild type, with the triple mutant being the most attenuated. These results suggest that optimal ACD enzymatic function requires processing between the ACD-RID junction (Leu2447), although a single cleavage of MARTXVc can activate the ACD.
Chemical inhibition of MARTXVc toxin function
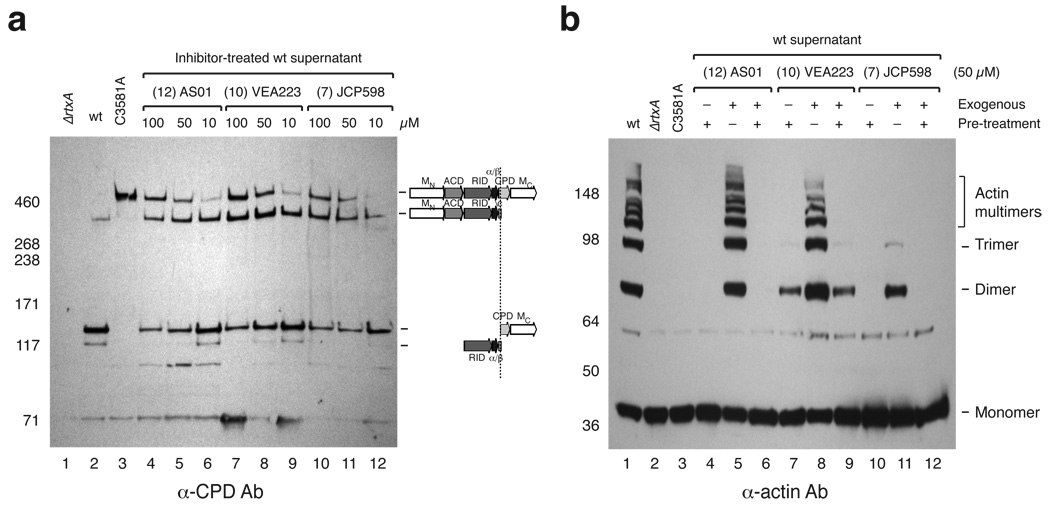
Lastly, we examined whether our small molecule CPD inhibitors could effectively disrupt MARTXVc processing in vivo. To this end, we grew wildtype V. cholerae cultures in the presence of increasing concentrations of CPD inhibitors and then measured MARTXVc processing in culture supernatants by Western blot analysis. Treatment of cultures with each of the three most potent inhibitors AS01, VEA223 and JCP598 resulted in the accumulation of unprocessed MARTXVc toxin relative to untreated wildtype culture supernatants (Fig. 7a). Although the compounds only partially blocked processing at Leu3441 even at the highest concentration of 100 µM, they completely inhibited processing at Leu3099 at concentrations greater han 50 µM. This latter result is consistent with in vitro observation that the CPD exhibits reduced affinity for the Leu3099 cleavage site relative to the Leu3441 autoprocessing site (Fig. 5d).
Figure 7.
Chemical inhibition of MARTXVc processing and toxin function. (a) Dose-dependent reduction in MARTXVc processing by CPD inhibitors. Western blot analysis of culture supernatants harvested from wildtype cultures grown in the presence or absence of inhibitor until mid-log phase using a CPD-specific antibody. (b) Effect of CPD inhibitors on MARTXVc actin crosslinking in HFFs. Pretreatment indicates that V. cholerae culture supernatants were pre-treated with 50 µM of inhibitor for 15 min. For exogenous treatment, the inhibitor was added at 50 µM to HFFs (in DMEM media) prior to addition of V. cholerae culture supernatants. HFF cells were exposed to V. cholera culture supernatants to stimulate actin crosslinking after which HFFs were lysed, and lysates were resolved by SDS-PAGE. Western blot analysis using an anti-actin antibody was used to visualize cross-linked actin species, which are indicated to the right.
The failure of CPD inhibitors to completely prevent MARTXVc processing during growth of V. cholerae in LB media could reflect their relative instability in these growth conditions. Thus, we evaluated the ability of CPD inhibitors to prevent MARTXVc toxin activation in host cells. All three compounds, AS01, VEA223 and JCP598, completely blocked the actin crosslinking activity of wildtype V. cholerae supernatants when added at concentrations of 50 µM (Fig. 7b, lanes 4,7, and 10). In contrast, addition of AS01 and VEA223 exogenously to the media of HFF cells immediately prior to adding untreated wildtype culture supernatants resulted in little inhibition of toxin function (Fig. 7b, lanes 5 and 8). Conversely, addition of JCP598 to the media of HFF cells significantly reduced MARTXVc-induced actin crosslinking (Fig 7b, lane 11). We suspect that the difference in inhibitor potencies can be attributed to differences in membrane permeability of the inhibitors. The negative charges on AS01 and VEA223 likely reduce their ability to cross host cell membranes, whereas the hydrophobic JCP598 readily passes through cell membranes to inhibit MARTXVc function even after toxin translocation. Taken together, our results validate the CPD as a target for small molecules designed to block MARTX toxin activation.
DISCUSSION
The MARTX toxin of Vibrio cholerae is autoproteolytically activated by an internal cysteine protease domain. Here, we demonstrated that MARTXVc CPD is a leucine-specific protease whose activity can be chemically inhibited to prevent MARTXVc activation. We further determined that MARTXVc processing at interdomain regions optimally activates effector domain function, since the actin crosslinking activity of the ACD was most efficient when cleavage occurred between the ACD and RID domains (Leu2447, Fig. 6c), and optimal CPD activity depended upon processing at the α/β-CPD junction (Leu3441, Fig. 6b). Based on the results of our cleavage site mapping, we propose the following model. Processing of MARTXVc by the CPD at Leu2447, Leu3099, and Leu3441 liberates the RID and α/β hydrolase domains, respectively. This cleavage profile leaves the ACD and CPD domains tethered to the membrane-bound N-and C-terminal MARTX conserved regions, respectively (Supplementary Fig. 6). Membrane localization of the CPD likely ensures that the protease can efficiently access its transcleavage substrates and may account for conservation in position of the CPD in all MARTX toxins1. Furthermore, processing at Leu3441 may additionally be required for the CPD to optimally bind its transcleavage substrates, while processing at Leu2447 may enhance ACD activity by liberating its C-terminal end.
Our study identified the first chemical inhibitors of MARTXVc CPD (Fig. 1 and Table 1) and demonstrated their utility in preventing MARTX toxin activation. (Fig. 7). These inhibitors appear to be selective, since the most potent inhibitor in our assays, JCP598, exhibits little reactivity against a wide variety of proteases in vitro21. Furthermore, these inhibitors may prevent the proteolytic activation of other MARTX toxins, since related MARTX CPDs were leucine-specific and InsP6-inducible (Fig. 3). Blocking CPD function likely represents the most effective strategy for preventing host cell intoxication by MARTX family members, which are multifunctional and heterogeneous in composition1. Additionally, the inhibitors identified in our study may block the cysteine protease activity of Clostridium sp. cytotoxins14–16, since the CPD of C. difficile Toxin B, the primary virulence factor of this nosocomial pathogen, exhibits a similar substrate specificity16.
Combined with biochemical and structural studies, the inhibitor analyses revealed that the CPD exhibits exquisite selectivity for leucine in the P1 position. Only clan CD-specific compounds with a P1 Leu had inhibitory activity (Fig. 1); all MARTX CPDs examined cleaved exclusively after a P1 Leu (Fig. 3 and Fig. 6), and mutation of the P1 Leu to Ile abrogated CPD-mediated transcleavage (Fig. 5c). These mutational studies further indicated that the P1’ residue directs the substrate specificity of the CPD. All known MARTX CPD cleavage sites contain neutral, small residues in the P1’ position (Fig. 3), and mutation of this small P1’ residue to bulkier Leu residue abrogated CPD-mediated processing (Fig. 5c), indicating that the S1’ subsite likely does not tolerate large residues. Charged residues in the P1’ position also appeared to be disfavored, since cleavage between a P1 Leu (L3415) and P1’ Asp (D3416) was only observed when Leu3441 of MARTXVc CPD was mutated to an Ala (Fig. 5c). Lastly, the P3 and P2 positions were observed to contribute little to substrate selectivity. The P3 and P2 positions are poorly conserved among known MARTXVc cleavage sites (Fig. 3c); VEA223 and JCP598 exhibited similar inhibitor potencies despite differing only in these positions (Fig. 1 and Table 1), and the P2 and P3 residues minimally interact with the CPD subsites in the crystal structure (Fig. 2b).
The crystal structure of activated CPD bound to an aza-Leu epoxide inhibitor reveals, at a molecular level, the constituents determining CPD substrate specificity. A deep, hydrophobic S1 pocket perfectly accommodates the P1 Leu of the inhibitor exclusively (Fig. 2). In contrast, the S1’ subsite consists mainly of a flat non-polar surface (Fig. 2a). The lack of recognition features in the S1’ region may explain the CPD’s preference for small residues in the P1’ position, with Gly favored over Ser (Fig. 3c), and its inability to accommodate a Leu residue in the P1’ position (Fig. 5b). Intriguingly, caspases have been shown to exhibit a similar preference for the P1’ position, favoring Gly over Ser and Ala26,27.
This observation is one of many similarities that MARTXVc CPD and caspases share in substrate recognition. Like the caspases, the CPD is sensitive to inhibition by both aza-epoxides and AOMKs (Table 1). Furthermore, MARTXVc CPD and caspase-3 exhibit the same stereoisomer preference around the epoxide group21 and react with the epoxide in a similar manner22. The most striking example of the similarity between caspases and MARTXVc CPD, however, is the observation that the active site topologies of the CPD and caspase-3 are nearly identical (Fig. 2c). Despite their weak overall structural similarity and disparate mechanisms of activation13, the catalytic residues are well aligned in a superposition of the central D and E beta strands (Fig. 2c). Furthermore, the S1 pocket of both caspase-3 and MARTXVc CPD occupies a similar position between the catalytic residues and is the primary substrate specificity determinant.
Interestingly, the distantly related clan CD protease gingipain-R shares a similar active site geometry and positioning of the S1 pocket as the caspases and MARTXVc CPD (Supplementary Fig. 7). This observation suggests that the mechanism of substrate recognition is broadly conserved among clan CD proteases. Specifically, all three proteases exhibit strict specificity for the P1 residue: MARTXVc CPD, caspases, and gingipain-R recognize hydrophobic Leu, acidic Asp23, and basic Arg28, respectively and exclusively. Accordingly, the molecular surfaces around the active sites are highly evolved to recognize their respective P1 residue: the substrate binding cleft of the CPD, caspases, and gingipain-R are neutral, basic, and acidic, respectively. The S1 binding clefts is so selective that Ile fails to functionally substitute for a P1 Leu in the CPD (Fig. 5c), and Glu fails to substitute for a P1 Asp in caspases27. Thus, MARTX CPDs, caspases, and gingipains appear to have evolved from a common structural scaffold; from this scaffold, the proteases have evolved distinct substrate recognition preferences and mechanisms of activation. These studies raise the possibility that residues in the S1 subsite of a given clan CD protease might be altered to engineer new substrate specificities for these highly specific enzymes.
Methods
Bacterial and eukaryotic cell growth conditions and strain construction
For details see Supplementary Information online.
Screen for inhibitors of MARTXVc CPD autoprocessing
Autocleavage assays were performed in 50 µL volumes containing 1 µM N-terminally His6-tagged MARTXVc CPD (aa 3391–3650) in cleavage assay buffer (60 mM NaCl, 20 mM Tris pH 7.5, 250 mM sucrose) in 96 well plates. Inhibitors were added at a final concentration of 100 µM (1:100 final dilution from a 10 mM stock) and incubated with MARTXVc CPD for 30 min at room temperature. Guanosine 5′-[γ-thio]triphosphate (GTPγS, Sigma) was then added to give a final concentration of 200 µM (1:10 dilution). Cleavage reactions were incubated at 37°C for 2 h, after which autocleavage was stopped by the addition of SDS-PAGE loading buffer. Samples were boiled for 3 min at 95°C and resolved by SDS-PAGE on 15% gels. Cleavage reactions were visualized by Coomassie staining. The screen was performed in triplicate, and hits were confirmed in a secondary screen using the autocleavage assay.
AC50:inhibitor ratios
Inhibitor potency was determined by measuring the concentration of InsP6 required to induce half-maximal cleavage of MARTXVc CPD in the presence of 10 µM inhibitor (AC50(I)). 1 µM of recombinant CPD in 50 µL cleavage assay buffer was pre-treated with 10 µM inhibitor (1:100 dilution) for 30 min at room temperature. Inositol hexakisphosphate (InsP6, Calbiochem) was added at the indicated final concentrations (1:100 dilution), and autoprocessing was allowed to proceed for 1 hr at 37°C. Cleavage reactions were resolved by SDS-PAGE and visualized by Coomassie staining. Images were quantified using the publicly available program ImageJ (http://rsb.info.nih.gov/ij/) as previously described13. The amount of autocleaved protein relative to total protein was plotted versus concentration of InsP6. The AC50(I) was determined from these plots using the Michaelis-Menten function on KaleidaGraph (Synergy Software).
Synthesis of CPD-specific inhibitors
VEA223 was synthesized in solution-phase using standard chemistries as described previously for the synthesis of JCP59821. AS01, AS02, and AS04 were generated using solid-phase synthesis as described previously29.
In vitro CPD autocleavage and transcleavage assays
Autocleavage assays were performed as described above for AC50(I) determinations except that no inhibitor was used. Transcleavage assays were identical to autocleavage assays with the exception that recombinant MARTXVc polypeptides were added to reactions at 1 µM. AC50 values were determined from triplicate assays as described above.
Protein expression and purification
Proteins for in vitro cleavage reactions and crystallization were purified as previously described13.
Crystallization and data collection
For details see Supplementary Methods online.
Structure determination and refinement
Initial phases were obtained by molecular replacement with PHASER30, using the MARTXVc CPD (PDB ID 3EEB) as a search model. The JCP598 inhibitor was constructed manually using COOT31 and the structure was refined by iterative rounds of model adjustment followed by refinement with CNS32. The final model went through TLS and restrained refinement with REFMAC533, resulting in final R and Rfree values of 22.1% and 26.5%, respectively. Ramachandran analysis with MolProbity (http://molprobity.biochem.duke.edu)34 indicated that 95.0% of residues reside in the most favorable regions, with the remaining 5.0% in additionally allowed regions. Refinement statistics can be found in Supplementary Table 1. Superposition of structures was performed with the program Superpose from the CCP4 program suite35. All structural figures were prepared with PyMol36. The final model contains four copies of the MARTXVc CPD in the asymmetric unit, each bound to one InsP6 molecule, one sodium ion, and one JCP598 molecule. Chain A is used for all figures in the paper.
Silver staining of V. cholerae culture supernatants
V. cholerae culture supernatants were prepared as described previously13 and resolved on a 3–8% Tris-acetate gel (Invitrogen). The gel was silver-stained using the SilverXpress Silver staining kit (Invitrogen).
Western blot analysis of MARTX toxin
Untreated V. cholerae supernatants were prepared and resolved as described previously13. For inhibitor treated V. cholera cultures, the indicated inhibitor was diluted 1:500 into 2 mL of LB media containing a 1:1000 dilution of overnight wildtype V. cholerae culture. Diluted, inhibitor-treated cultures were grown until mid-log phase (OD600 ~0.5, ~2.25 hr growth), and culture supernatants were TCA precipitated and resolved as described13. Polyclonal MARTX-specific antibodies were raised against recombinant ACD (1964–2375 aa, CoCalico Biologicals), RID (2552–3099 aa, CoCalico Biologicals), and CPD13, and Western blot analyses were performed as previously described13.
Fourier transform mass spectrometry
In vitro cleavage reactions were separated on a PLRPS 150 mm × 0.1 mm column (Varian, 5 µM particle size, 300 Å pore size) run at a flow rate of 700 nL/min in 0.1% trifluoroacetic acid/water (A):0.1% trifluoroacetic acid/acetonitrile (B). The column was run on a gradient of 10% B to 60% B for 25 min, 60% B to 90 % B for 2 min, held at 90% B for 2 min, then rapidly decreased to 10% B over 0.1 min, then run for 11 min at 10% B. Eluted samples were run on a Thermo LTQ-FT mass spectrometer (Thermo Fisher Scientific) using FTMS + p NSI Full MS scanning mode, mass range 400.00–2000.00, FT resolution 100,000. The MW of eluted peptides was determined by deconvolution using Isopro 3.0 (MS/MS Software). The measured mass was compared to the predicted MW of possible peptide cleavage products determined from the primary sequence of recombinant polypeptides derived from MARTXVc using PROTPARAM at http://ca.expasy.org/tools/protparam.html.
Actin crosslinking assay
For cleavage site mutant analyses, the actin crosslinking assay was performed as described previously13 using culture supernatants. In the inhibitor actin crosslinking assays, 100 µL of V. cholerae culture supernatants were pre-treated with the indicated inhibitor at a final concentration of 50 µM (1:200 dilution from a 10 mM stock). For exogenous addition, inhibitor was also added to HFFs (in 500 µL DMEM media) at a final concentration of 50 µM (1:200 dilution from a 10 mM stock).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Paul Gulig (University of Florida) for generously providing Vibrio vulnificus CMCP6 genomic DNA, Elizabeth Shank and Roberto Kolter (Harvard Medical School) for providing the Photorhabdus luminescens TTO1 strain, M. Blokech and G. Schoolnik (Stanford School of Medicine) for help with V. cholerae strain construction and providing V. cholerae genomic DNA. P.J.L. is a Damon Runyon Fellow, supported by the Damon Runyon Cancer Research Foundation. K.C.G. is supported by the Keck Foundation and the Howard Hughes Medical Institute. M.B. is supported by the Burroughs Wellcome Foundation, the Searle Scholars Program, and the NIH National Technology Center for Networks and Pathways (grant US4RR020843). Coordinates and structure factors have been deposited in the Protein Data Bank (www.rcsb.org) under accession number 3GCD.
Contributions. The inhibitor screen, synthesis of AS01, AS02, and AS04, protein expression and purification, cleavage assays, FT-MS data analysis, V. cholerae strain construction, actin crosslinking assays, MARTXVc silver staining and Western blot analyses were performed by A.S. Crystallization of the MARTX CPD/InsP6/JCP598 complex was performed by A.S. and P.J.L. P.J.L. collected the data, solved and analyzed the structure, and generated the figures of the inhibitor-bound CPD structure. J.C.P. provided the cysteine protease compound library. V.E.A. synthesized JCP598, AS01, and VEA223, guided A.S. in the synthesis of AS01, AS02, and AS04, and assessed the integrity of all compounds described in this paper. A.G. designed the conditions for running samples for FT-MS, ran the samples for FT-MS and provided advice in FT-MS analysis. Creative input and financial support for the project was provided by M.B. The manuscript was written by A.S. and M.B. with advice from P.J.L, V.E.A, K.C.G and A.G.
References
- 1.Satchell KJ. MARTX, multifunctional autoprocessing repeats-in-toxin toxins. Infect. Immun. 2007;75:5079–5084. doi: 10.1128/IAI.00525-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li L, Rock JL, Nelson DR. Identification and characterization of a repeat-in-toxin gene cluster in Vibrio anguillarum. Infect. Immun. 2008;76:2620–2632. doi: 10.1128/IAI.01308-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee BC, et al. Vibrio vulnificus rtxE is important for virulence, and its expression is induced by exposure to host cells. Infect. Immun. 2008;76:1509–1517. doi: 10.1128/IAI.01503-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee JH, et al. Identification and characterization of the Vibrio vulnificus rtxA essential for cytotoxicity in vitro and virulence in mice. J. Microbiol. 2007;45:146–152. [PubMed] [Google Scholar]
- 5.Liu M, Alice AF, Naka H, Crosa JH. The HlyU protein is a positive regulator of rtxA1, a gene responsible for cytotoxicity and virulence in the human pathogen Vibrio vulnificus. Infect. Immun. 2007;75:3282–3289. doi: 10.1128/IAI.00045-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olivier V, Haines GK, 3rd, Tan Y, Satchell KJ. Hemolysin and the multifunctional autoprocessing RTX toxin are virulence factors during intestinal infection of mice with Vibrio cholerae El Tor O1 strains. Infect. Immun. 2007;75:5035–5042. doi: 10.1128/IAI.00506-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olivier V, Salzman NH, Satchell KJ. Prolonged colonization of mice by Vibrio cholerae El Tor O1 depends on accessory toxins. Infect. Immun. 2007;75:5043–50551. doi: 10.1128/IAI.00508-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cordero CL, Sozhamannan S, Satchell KJ. RTX toxin actin cross-linking activity in clinical and environmental isolates of Vibrio cholerae. J. Clin. Microbiol. 2007;45:2289–2292. doi: 10.1128/JCM.00349-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rahman MH, et al. Distribution of genes for virulence and ecological fitness among diverse Vibrio cholerae population in a cholera endemic area: tracking the evolution of pathogenic strains. DNA Cell. Biol. 2008;27:347–355. doi: 10.1089/dna.2008.0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sheahan KL, Cordero CL, Satchell KJ. Identification of a domain within the multifunctional Vibrio cholerae RTX toxin that covalently cross-links actin. Proc. Natl. Acad. Sci. U.S.A. 2004;101:9798–9803. doi: 10.1073/pnas.0401104101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sheahan KL, Satchell KJ. Inactivation of small Rho GTPases by the multifunctional RTX toxin from Vibrio cholerae. Cell. Microbiol. 2007;9:1324–1335. doi: 10.1111/j.1462-5822.2006.00876.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sheahan KL, Cordero CL, Satchell KJ. Autoprocessing of the Vibrio cholerae RTX toxin by the cysteine protease domain. EMBO J. 2007;26:2552–2561. doi: 10.1038/sj.emboj.7601700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lupardus PJ, Shen A, Bogyo M, Garcia KC. Small Molecule-Induced Allosteric Activation of the Vibrio cholerae RTX Cysteine Protease Domain. Science. 2008;322:265–268. doi: 10.1126/science.1162403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Egerer M, Giesemann T, Jank T, Satchell KJ, Aktories K. Auto-catalytic cleavage of Clostridium difficile toxins A and B depends on cysteine protease activity. J. Biol. Chem. 2007;282:25314–25321. doi: 10.1074/jbc.M703062200. [DOI] [PubMed] [Google Scholar]
- 15.Giesemann T, Egerer M, Jank T, Aktories K. Processing of Clostridium difficile toxins. J. Med. Microbiol. 2008;57:690–696. doi: 10.1099/jmm.0.47742-0. [DOI] [PubMed] [Google Scholar]
- 16.Reineke J, et al. Autocatalytic cleavage of Clostridium difficile toxin B. Nature. 2007;446:415–419. doi: 10.1038/nature05622. [DOI] [PubMed] [Google Scholar]
- 17.Gordon VM, Leppla SH. Proteolytic activation of bacterial toxins: role of bacterial and host cell proteases. Infect. Immun. 1994;62:333–340. doi: 10.1128/iai.62.2.333-340.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arastu-Kapur S, et al. Identification of proteases that regulate erythrocyte rupture by the malaria parasite Plasmodium falciparum. Nat. Chem. Biol. 2008;4:203–213. doi: 10.1038/nchembio.70. [DOI] [PubMed] [Google Scholar]
- 19.Powers JC, Asgian JL, Ekici OD, James KE. Irreversible inhibitors of serine, cysteine, and threonine proteases. Chem. Rev. 2002;102:4639–4750. doi: 10.1021/cr010182v. [DOI] [PubMed] [Google Scholar]
- 20.Prochazkova K, Satchell KJ. Structure-function analysis of inositol hexakisphosphate-induced autoprocessing of the Vibrio cholerae multifunctional autoprocessing RTX toxin. J. Biol. Chem. 2008;283:23656–23664. doi: 10.1074/jbc.M803334200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Asgian JL, et al. Aza-peptide epoxides: a new class of inhibitors selective for clan CD cysteine proteases. J. Med. Chem. 2002;45:4958–4960. doi: 10.1021/jm025581c. [DOI] [PubMed] [Google Scholar]
- 22.Ganesan R, et al. Exploring the S4 and S1 prime subsite specificities in caspase-3 with aza-peptide epoxide inhibitors. Biochemistry. 2006;45:9059–9067. doi: 10.1021/bi060364p. [DOI] [PubMed] [Google Scholar]
- 23.Barrett AJ, Rawlings ND. Evolutionary lines of cysteine peptidases. Biol. Chem. 2001;382:727–733. doi: 10.1515/BC.2001.088. [DOI] [PubMed] [Google Scholar]
- 24.Bedard KM, Semler BL. Regulation of picornavirus gene expression. Microbes Infect. 2004;6:702–713. doi: 10.1016/j.micinf.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 25.Reed KE, Rice CM. Overview of hepatitis C virus genome structure, polyprotein processing, and protein properties. Curr. Top. Microbiol. Immunol. 2000;242:55–84. doi: 10.1007/978-3-642-59605-6_4. [DOI] [PubMed] [Google Scholar]
- 26.Schilling O, Overall CM. Proteome-derived, database-searchable peptide libraries for identifying protease cleavage sites. Nat. Biotechnol. 2008;26:685–694. doi: 10.1038/nbt1408. [DOI] [PubMed] [Google Scholar]
- 27.Stennicke HR, Renatus M, Meldal M, Salvesen GS. Internally quenched fluorescent peptide substrates disclose the subsite preferences of human caspases 1, 3, 6, 7 and 8. Biochem J. 2000;350(Pt 2):563–568. [PMC free article] [PubMed] [Google Scholar]
- 28.Eichinger A, et al. Crystal structure of gingipain R: an Arg-specific bacterial cysteine proteinase with a caspase-like fold. EMBO J. 1999;18:5453–5462. doi: 10.1093/emboj/18.20.5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kato D, et al. Activity-based probes that target diverse cysteine protease families. Nat. Chem. Biol. 2005;1:33–38. doi: 10.1038/nchembio707. [DOI] [PubMed] [Google Scholar]
- 30.McCoy AJ, et al. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 32.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta. Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 33.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta. Crystallogr. D. Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 34.Davis IW, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Potterton E, Briggs P, Turkenburg M, Dodson E. A graphical user interface to the CCP4 program suite. Acta Crystallogr. D. Biol. Crystallogr. 2003;59:1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- 36.DeLano WL. The PyMOL Molecular Graphics System. San Carlos, CA, USA: DeLano Scientific; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.