Abstract
The proto-oncogene KRAS is mutated in a wide array of human cancers, most of which are aggressive and respond poorly to standard therapies. Although the identification of specific oncogenes has led to the development of clinically effective, molecularly targeted therapies in some cases, KRAS has remained refractory to this approach. A complementary strategy for targeting KRAS is to identify gene products that, when inhibited, result in cell death only in the presence of an oncogenic allele1,2. Here we have used systematic RNA interference (RNAi) to detect synthetic lethal partners of oncogenic KRAS and found that the non-canonical IκB kinase, TBK1, was selectively essential in cells that harbor mutant KRAS. Suppression of TBK1 induced apoptosis specifically in human cancer cell lines that depend on oncogenic KRAS expression. In these cells, TBK1 activated NF-κB anti-apoptotic signals involving cREL and BCL-XL that were essential for survival, providing mechanistic insights into this synthetic lethal interaction. These observations identify TBK1 and NF-κB signaling as essential in KRAS mutant tumors and establish a general approach for the rational identification of co-dependent pathways in cancer.
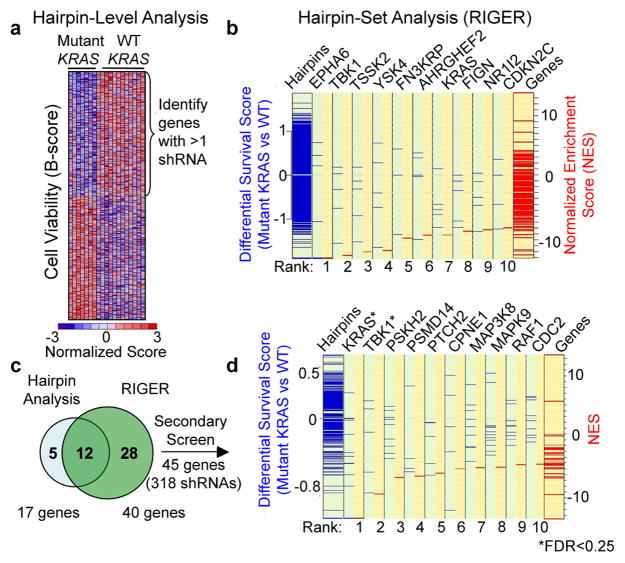
To identify essential genes in human malignant and non-transformed cell lines, we performed arrayed format RNAi screens in 19 cell lines using a short hairpin RNA (shRNA) library targeting kinases and phosphatases3 (Supplementary Fig. 1 and Table 1). We then used two methods to find genes that were selectively required in cells expressing oncogenic KRAS. First, we employed a class-discrimination feature selection method (Fig. 1a) in which normalized B-scores4 for each cell line were analyzed using a t-test statistic5 to identify the top 250 (5%) shRNAs that distinguished the cell lines that harbored mutant or wild-type (WT) KRAS. We focused on genes whose suppression by at least 2 shRNAs selectively impaired the proliferation/viability of KRAS mutant cells and identified KRAS itself (Supplementary Tables 2, 3; Supplementary Figs. 2a, b).
Fig. 1.
Meta-analysis of RNAi screens identifying KRAS synthetic lethals. (a) Supervised analysis of viability data (B-score) identified 250 shRNAs that distinguished mutant KRAS from wild-type (WT) cells, including genes targeted by multiple shRNAs. (b) Hairpin set analysis (RIGER). Genes were assigned NES (red lines) based on the KRAS mutant/WT differential survival scores (blue lines) for each shRNA. Negative values represent mutant KRAS-selectivity. (c) Union of 17 genes identified in (a) and 40 genes identified in (b). (d) Secondary screening data normalized using percent of control (POC) and analyzed using RIGER. FDR for KRAS and TBK1 was 0.04 and 0.18 respectively.
In parallel, we used RNAi Gene Enrichment Ranking (RIGER)6, a statistical approach that does not rely on arbitrary thresholds, to rank-order candidate KRAS synthetic lethal genes (Fig. 1b). RIGER considers all shRNAs for a gene as a “hairpin set”, similar to “gene sets” in gene set enrichment analysis (GSEA)7, and provides a normalized enrichment score (NES) for each gene with respect to a specific classification. Using the mutant versus WT KRAS class distinction as the classification feature, we ranked candidate KRAS synthetic lethal partners by NES and selected the top 40 genes, which included 12 of the 17 candidates identified by the individual shRNA-based analysis (Figs. 1b, c, Supplementary Tables 2, 4).
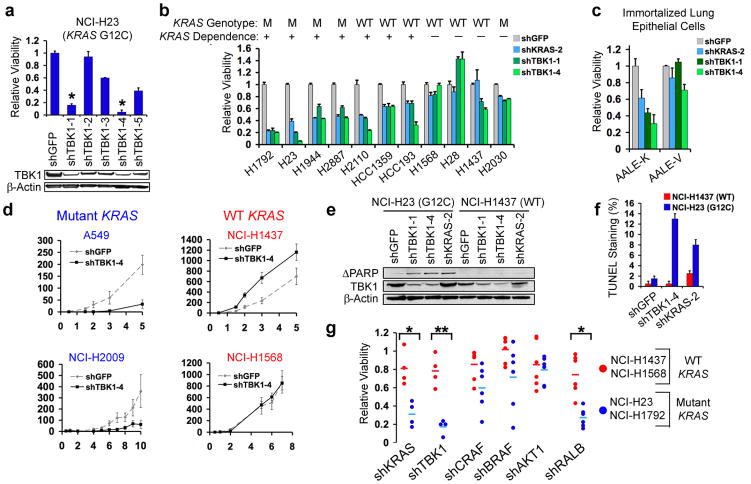
To validate the 45 candidates identified by these two approaches, we performed a secondary screen on an independent panel of mutant or WT KRAS lung adenocarcinoma cell lines (Supplementary Figs. 3a, b; 4a, b). Proliferation/viability data for each shRNA was normalized to the median value of 20 control shRNAs. Using the t-test statistic to rank shRNAs that selectively impaired proliferation/viability in mutant KRAS cells, we identified a significantly enriched subset of candidate shRNAs (p≤0.0002) (Supplementary Fig. 5a). Three KRAS-specific shRNAs were among the top four shRNAs that distinguished KRAS mutant and WT cell lines (Supplementary Figs. 3a, c; 5b). Using RIGER to rank candidate genes with respect to KRAS-selective lethality, we identified KRAS and TBK1 as the most significant genes (FDR, 0.04 and 0.18, respectively) (Fig. 1d). Although the secondary screen identified other potential KRAS synthetic lethal genes, we focused on TBK1 because it represented the top candidate after KRAS. Indeed, we found that the two top-scoring shRNAs induced TBK1 suppression and substantial cell death in NCI-H23 cells (mutant KRAS) (Fig. 2a). To confirm these findings, we introduced KRAS- or TBK1-specific shRNAs into a third set of lung cancer cell lines (Fig. 2b) and observed a strong correlation between KRAS and TBK1 dependence, even in cell lines in which KRAS mutation status and dependence were decoupled. We also used an isogenic experimental model to isolate the genetic interaction between oncogenic KRAS and TBK1. Specifically, expression of oncogenic KRAS in immortalized human lung epithelial cells (AALE-K cells)8 rendered them dependent on both KRAS and TBK1 for survival, as compared to cells expressing a control vector (AALE-V cells) (Fig. 2c). When we suppressed TBK1 in A549 or NCI-H2009 cells (mutant KRAS), tumor formation was inhibited, while suppression of TBK1 had no effect on the tumorigenicity of NCI-H1437 or NCI-H1568 cells (WT KRAS) (Fig. 2d). These observations confirm that cancer cell lines that depend on oncogenic KRAS require TBK1 expression.
Fig. 2.
TBK1 synthetic lethality with oncogenic KRAS. (a) Top-scoring TBK1-specific shRNAs (*) induced lethality and TBK1 suppression (immunoblot) in NCI-H23 cells (mutant KRAS). (b) Suppression of KRAS or TBK1 in NSCLC cell lines. HCC-1359 and HCC-193 cells expressed RAS and NF-κB signatures. (c) KRAS and TBK1 dependence of lung epithelial cells expressing oncogenic KRAS (AALE-K) or vector (AALE-V). (d) Tumor formation following TBK1 suppression. Mean and SEM of at least 11 replicates shown. (e) Immunoblot of cleaved PARP following TBK1 or KRAS suppression. (f) Percentage of TUNEL positive nuclei following TBK1 or KRAS suppression. Mean and SD shown. (g) Differential cell viability following KRAS, TBK1, CRAF, BRAF, AKT1 or RALB suppression in KRAS mutant vs. WT cell lines (t-test for comparisons). SEM of triplicate samples normalized to shGFP control vector shown.
To determine whether suppression of TBK1 in KRAS-dependent cells induced apoptosis, we found that, similar to shRNAs targeting KRAS itself (Supplementary Fig. 4b), TBK-specific shRNAs provoked an increase in PARP cleavage (Fig. 2e) and TUNEL-positive nuclei (p<0.01) (Figs. 2f, Supplementary Fig. 6a) in NCI-H23 cells (mutant KRAS) but not in NCI-H1437 cells (WT KRAS). Suppression of Tbk1 in cells derived from a KRAS-driven murine model of lung cancer (LKR-13 cells)9 also induced apoptosis (Supplementary Fig. 6b).
KRAS activates several signaling pathways including those regulated by RAF, PI3K, and RAL. We found that suppression of CRAF, BRAF or AKT1 failed to kill KRAS-dependent lung cancer cell lines selectively (Figs. 2g, Supplementary Fig. 6c). TBK1 suppression also failed to alter phospho-ERK or phospho-AKT levels (Supplementary Fig. 6d). In contrast, suppression of RALB resulted in significant selective lethality in KRAS-dependent cell lines (p<0.01, Fig. 2g). Consistent with prior work linking RALB with TBK1 activation in the setting of tumor cell survival10, this observation suggested that RALB-TBK1 signaling was required in cells that depend on oncogenic KRAS.
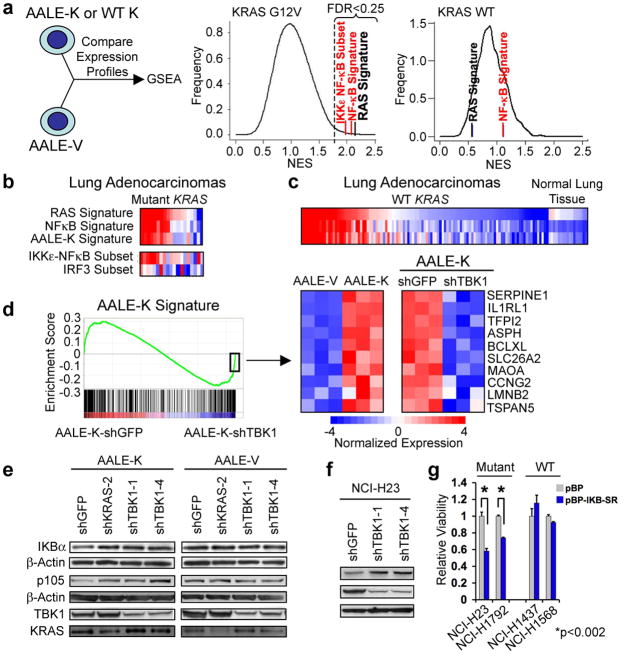
TBK1 is a non-canonical IκB kinase that regulates innate immunity through the interferon and NF-κB pathways11 and is also a component of the exocyst complex10. To examine how TBK1 contributes to survival in KRAS-dependent cell lines, we performed transcriptional profiling on AALE cells expressing a control vector (AALE-V), oncogenic KRAS (AALE-K) or WT KRAS (AALE-K WT). Using GSEA to identify gene sets from the Molecular Signatures Database (MSigDB-C2 v2)7 that were enriched in AALE-K cells, we identified a previously described oncogenic RAS signature12 as well as several NF-κB pathway activation signatures13,14 among the most significantly enriched gene sets (p≤4.5 × 10−7, hypergeometric test) (Fig. 3a, Supplementary Fig. 7a). In contrast, we failed to detect enrichment of oncogenic RAS or NF-κB signatures in AALE-K WT cells (Fig. 3a), indicating that expression of oncogenic but not WT KRAS correlated with NF-κB signaling.
Fig. 3.
Oncogenic KRAS-induced NF-κB signaling involves TBK1 (a) GSEA of AALE-V (vector), AALE-K (KRAS G12V) or AALE-WT-K (KRAS WT) cells (positive NES). A RAS oncogenic signature (black) and NF-κB signatures (red) showed significant enrichment (FDR<0.25) in AALE-K cells. N.S. = non-significant. (b) RAS signatures in mutant KRAS lung adenocarcinomas correlate with NF-κB but not IRF3 signatures (red=activation, blue=inactivation). (c) RAS and NF-κB signature expression in WT KRAS lung adenocarcinomas and normal lung tissue. (d) AALE-K signature enrichment plot following shTBK1 or shGFP expression in triplicate samples. Heatmap shows top KRAS-induced genes with negative enrichment in AALE-K-shTBK1 samples. Immunoblot of IKBα, p105, TBK1, and KRAS in AALE-K and AALE-V cells (e) or NCI-H23 cells (f) following KRAS or TBK1 suppression. (g) Cell viability after expression of control vector (pBP) or IκBα-super-repressor (pBP-IKB-SR) in mutant or WT KRAS cells. Mean and SEM of triplicate samples shown, t-test for comparisons.
To extend these observations to patient-derived tumors, we analyzed expression profiles from 128 lung adenocarcinomas15,16 for expression of the oncogenic RAS12, NF-κB13,14, and IRF317 signatures as well as a KRAS-specific signature (AALE-K) composed of the genes most significantly induced in AALE-K relative to AALE-V cells. We found that the majority of mutant KRAS tumors (14/19) showed RAS signature activation and co-expression of the NF-κB signature (p≤1.3 × 10−15, Spearman correlation test with Bonferroni adjustment) or the IKKε-regulated NF-κB gene subset (p≤0.008), but not the IRF3-regulated gene set (p≤0.18) (Fig. 3b). These observations confirm that the majority of lung cancers that harbor mutant KRAS show evidence of RAS and NF-κB pathway activation and suggest that a substantial fraction of KRAS mutant primary lung cancers may depend on TBK1 and NF-κB signaling for survival.
Consistent with recent work18, we also identified RAS and NF-κB signature co-activation in 30/109 KRAS WT tumors (Fig. 3c). These RAS and NF-κB signatures identified some but not all of the KRAS WT cell lines that exhibited KRAS dependence (Supplementary Table 1, Fig. 2b), suggesting that a subset of KRAS WT tumors depend on TBK1 and NF-κB signaling for survival. Further work will be necessary to determine whether such signatures will prove useful in predicting responsiveness to TBK1 inhibition.
Although TBK1 activates the interferon pathway through regulation of IRF3 and IRF710,11, we failed to observe increased expression of IRF3 target genes17 (Supplementary Fig. 7b) or increased IRF3 nuclear translocation (Supplementary Fig. 8a) in AALE-K cells. In addition, suppression of KRAS or TBK1 in KRAS mutant cancer cells down-regulated specific genes within the NF-κB subset, including CCND1, BCL2 and IL8, but failed to alter the expression of known interferon-responsive genes such as IFN-β1 and RANTES (Supplementary Fig. 7c). When we suppressed TBK1 in AALE-K cells, we observed that NF-κB signature components and several NF-κB targets, including the anti-apoptotic gene BCL-XL, were among the most significantly down-regulated genes (Fig. 3d). These findings confirm the importance of the NF-κB pathway in promoting survival in the setting of oncogenic RAS19 and suggest that, distinct from its role in innate immunity, TBK1 preferentially activates NF-κB signaling in tumors dependent on oncogenic KRAS.
TBK1 has been reported to regulate the stability of IκB proteins11. When we examined cytoplasmic levels of IκB family members in AALE-K cells, we found reduced levels of IκBα and p105 as compared to AALE-V cells (Fig. 3e, Supplementary Fig. 8a, b). Moreover, suppression of TBK1 in AALE-K cells or KRAS mutant NCI-H23 cells (Fig. 3e, f) returned levels to that observed in WT KRAS cells. Expression of the IκBα super-repressor (IKB-SR)20, which inhibits NF-κB activity, in AALE-K, AALE-V, or cancer cell lines expressing mutant or WT KRAS induced cell death specifically in cells harboring mutant KRAS (Fig. 3g, Supplementary Fig. 9). These findings confirm that TBK1-driven NF-κB activity promotes survival of cells that depend on mutant KRAS.
In the primary shRNA screen, we noted that one shRNA targeting the NF-κB family member c-REL scored as selectively lethal in KRAS mutant cells, albeit just below our pre-determined threshold. Suppression of c-REL but not IRF3 selectively induced apoptosis in KRAS mutant cells (p≤0.001) (Fig. 4a, Supplementary Fig. 10a). Moreover, we found that suppression of TBK1 in KRAS mutant cancer cells reduced total and nuclear c-REL levels (Fig. 4b, Supplementary Fig. 10b). Although TBK1 can phosphorylate c-REL when over-expressed21, we failed to detect an interaction between TBK1 and c-REL but confirmed that endogenous c-REL and p105 interact (Supplementary Fig. 10c)22.
Fig. 4.
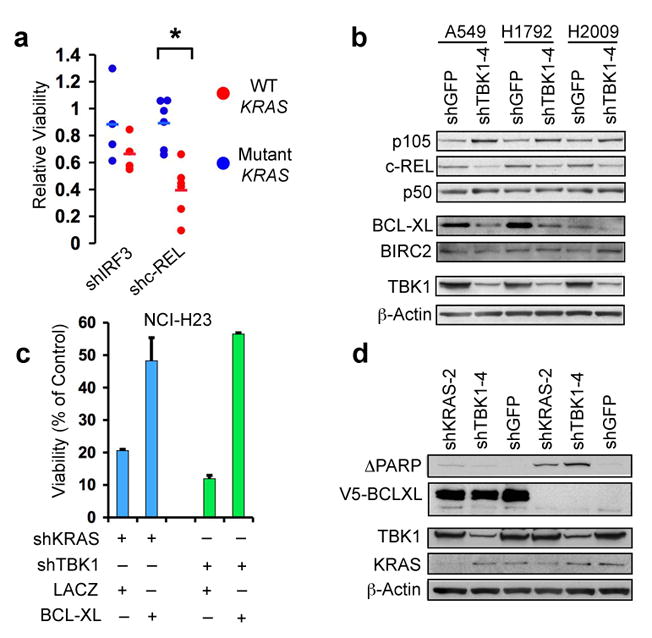
TBK1 regulates c-REL and BCL-XL in KRAS mutant cells. (a) Differential cell viability following IRF3 or cREL suppression in KRAS mutant vs. WT cell lines. (b) Immunoblot of p105, c-REL, p50, BCL-XL and BIRC2 in KRAS mutant cell lines following TBK1 suppression. (c) Cell viability following KRAS or TBK1 suppression in NCI-H23 cells expressing a control protein (LACZ) or V5-tagged BCL-XL. SEM of triplicate samples normalized to shGFP control vector shown. (d) Immunoblot showing over-expression of V5-tagged BCL-XL and inhibition of PARP cleavage.
Since BCL-XL, a known c-REL target23, was identified as a TBK1-regulated gene in AALE-K cells (Fig. 3d), we examined the expression of several anti-apoptotic genes following TBK1 suppression in KRAS mutant cancer cells and observed specific down-regulation of BCL-XL following TBK1 suppression in multiple cell lines (Fig. 4b, Supplementary Fig. 10d). Moreover, overexpression of BCL-XL rescued apoptosis induced by KRAS or TBK1 suppression in NCI-H23 cells (Fig. 4c, d) but did not significantly affect cell death induced by the suppression of survivin (Supplementary Fig. 11), confirming p105, c-REL and BCL-XL as mediators of NF-κB survival signaling downstream of TBK1 and KRAS.
In summary, we have identified TBK1 as a synthetic lethal partner of oncogenic KRAS. These findings link RALB-mediated activation of TBK110 to the generation of specific NF-κB-regulated survival signals downstream of oncogenic KRAS. Furthermore, although studies testing the effects of inhibiting TBK1 or NF-κB signaling in established tumors are necessary, TBK1 and more generally NF-κB signaling may represent an alternative method of targeting oncogenic KRAS-driven cancers. Recently, STK33 and PLK1 were identified as KRAS synthetic lethal partners through the application of RNAi screening in paired KRAS mutant and WT cell lines24,25. Both genes were also identified in our computational analyses (Supplementary Tables 3, 4), but like cREL, they fell below our initial threshold for secondary screening. We anticipate that the development of fully validated shRNA libraries coupled with the interrogation of larger numbers of cell lines will permit saturating genetic screens to identify synthetic lethal partners of KRAS as well as other oncogenes and tumor suppressor genes. More generally, this and other studies6,24–28 indicate that application of these functional and analytical approaches will facilitate the comprehensive identification of functional co-dependencies in cancer.
Methods Summary
Large-scale, arrayed format RNAi screens to identify genes essential for proliferation/viability were performed as described3,14. The effect of introducing each of the 5002 shRNAs (targeting 957 genes) was determined in 19 cell lines, and normalized using the B-score metric4. Feature selection of shRNA B-score data was performed using the Comparative Marker Application Suite in GenePattern5 and was independently analyzed using RIGER analysis6 to compute NES for each gene. Secondary screen viability data was normalized using a percent of control statistic, given the biased nature of the candidate shRNA plate. Expression profiling was used to generate a signature that correlates with KRAS activation and implicated NF-κB signaling in cell lines and tumors dependent on KRAS. Regulation of NF-κB by TBK1 was shown using biochemical and cell biological approaches. Details of the analytical methods are provided in the Full Methods.
Full Methods
RNAi Screens
Large-scale RNAi arrayed format screening was conducted using a subset of the Broad Institute RNAi Consortium (TRC) shRNA library targeting kinases, phosphatases, and other cancer-related genes3,14. ShRNA designs and protocols for high throughout lentiviral production are available at http://www.broad.mit.edu/rnai/trc/lib. Cells were seeded in quadruplicate 384 well plates on day 0, followed by infection with 8 μg/ml polybrene on day 1. Puromycin selection for duplicate plates (concentration individualized per cell line, most cell lines 2 μg/ml) was started on day 2. An ATP-based luminescence assay (Cell-Titer Glo, Promega) was used to determine cell number on day 6. Raw luminescence values from duplicate plates were averaged, and the ratio of puromycin positive to negative values was used to assess infection efficiency. Data was normalized using the B-score metric, a variant of the Z-score that uses the median absolute deviation to account for plate-to-plate variability, as well as a two-way median polish to minimize row/column effects4. After excluding shRNAs with low infection efficiency, B-score values from puromycin positive and negative replicates were averaged for each shRNA. shRNA B-score values were aligned for the 5002 shRNAs tested in all 19 cell lines. The secondary screen was conducted using a percent of control statistic instead of the B-score.
Computational Analyses
Hairpin-Level Analysis
The meta-analysis of RNAi screens was performed using complementary computational approaches. The first involved conversion of the shRNA B-score file into a .res file format for input into the Comparative Marker Selection application suite in GenePattern, along with a standard classification file to generate class distinctions5. This method uses class discrimination feature selection and ranks shRNA B-score data by the t-test statistic or the signal to noise ratio (SNR) to account for the difference in means between the two classes as well as the standard deviation across samples. Specifically, the difference in mean viability scores induced by each shRNA in the KRAS mutant class versus the WT class was normalized to its standard deviation using a t-test, and shRNAs were ranked by t-test score to determine for class selective effects. To convert shRNA data to candidate genes and to reduce the likelihood of off-target effects, the top 250 (5%) of distinguishing shRNAs in the mutant KRAS class were filtered to identify multiple shRNAs targeting the same gene. Only those genes for which at least one shRNA yielded a mean B score <-1 across the KRAS mutant class were considered. In addition, shRNAs with low infection efficiency in multiple cell lines were excluded from the analysis.
RIGER (RNAi Gene Enrichment Ranking) Analysis
Similar to “gene sets” for Gene Set Enrichment Analysis (GSEA)7, “hairpin sets” were defined as the groups of shRNAs representing a given target gene. Because the number of shRNAs in a hairpin set are relatively small compared with the number of genes, we used different input data normalization, enrichment statistics, and output formats compared with GSEA. RIGER analysis consisted of the following steps:
(1) Pre-processing
The input was the same dataset of B-scores analyzed using the individual shRNA-based method. The B-score values for each cell line were standardized with respect to the median and maximum absolute deviation of the set of control shRNAs (directed against GFP, lacZ, RFP, Luciferase) in the same cell line. This centered and rescaled the values in a sample-specific manner according to the behavior of the control hairpins. After normalization, values below −3 and above 3 were truncated to −3 and 3 respectively, to reduce the effect of outliers and were called normalized survival scores.
(2) Feature ranking
After preprocessing, each shRNA was assigned a “differential survival score,” which represented the difference in means of the normalized survival scores in the two phenotypic classes (e.g. KRAS mutant vs. WT). The difference in means was used in order to emphasize the absolute magnitude of the survival differences and not only the profile “shape.” Specifically, this favored shRNAs with strong differential killing of cells over ones that exhibited weak differential killing of cells, but had perfect discrimination profiles inside classes. The differential survival scores were computed for all the 5,002 shRNAs and sorted from high to low scores.
(3) Calculation of enrichment scores
A given gene was assigned an enrichment score according to the distribution of differential survival scores of its shRNAs within the rank list of all shRNAs using a two-sample weighted “Zhang C” statistic based on the likelihood ratio29. The Zhang C likelihood ratio statistic was used due to its greater sensitivity and better empirical results in exploratory analyses with other datasets. We adapted this method to separate positive and negative enrichment contributions and used a weighting factor based on the differential enrichment score, except that weights with absolute values under 0.5 and above −0.5 were truncated to 0.5 and −0.5 respectively, to reduce the effects of shRNAs that weakly distinguished classes. The enrichment score obtained in this manner was representative of both the extremeness of the shRNA differential survival scores for a given gene and their consistency.
(4) Normalization of enrichment scores
Since genes with different numbers of shRNAs were assigned enrichment scores on different scales, we normalized them prior to sorting the genes by using a null distribution generated by 1,000 random permutations of the locations of the shRNAs in the entire list. The normalization for negative enrichment scores was a rescaling by the absolute value of the mean of the negative values in the null distribution. This represented an effective way to place the enrichment scores on a common scale regardless of the number of shRNAs for each gene. The null distribution also provided nominal p-values for each gene enrichment score.
(5) Generation of results
The analysis resulted in a list of genes sorted by their normalized enrichment scores, and a set of complementary estimates of statistical significance such as nominal, family-wise and Bonferroni p-values plus a false discovery rate (FDR). A collection of dual-vertical plots was used to demonstrate the shRNA differential survival scores for each gene (lines in blue) and corresponding normalized enrichment scores (lines in red). Vertical plots were arranged starting with the top gene (strongest negative normalized enrichment score) on the left.
Secondary screen
Analysis of secondary screen data followed the same methodology except for the normalization of the cell proliferation/viability data. Twenty control shRNAs directed against RFP, LacZ, and Luciferase were screened in parallel with candidates. We normalized data for each shRNA in each cell line using the percent of control statistic, dividing the raw data for each shRNA by the median of control shRNA values, and taking the log10 of this number to scale values around zero. Because of the biased nature of the candidate shRNA plate, and since the number of control shRNAs was smaller in the secondary screen compared with the first, the calculation of the maximum absolute deviation was noisy and unreliable. No truncation was applied to the resulting values.
A larger set of 84 control shRNAs directed against GFP, RFP, LacZ, and Luciferase was also tested independently in all 8 cell lines chosen for the secondary screen. We normalized data for each shRNA in each cell line by dividing the raw data for each control shRNA by the plate median and taking the log10 of this number to scale values around zero. We used the t-test statistic to examine the KRAS mutant versus WT class distinction for this control plate relative to the candidate plate, restricting the analysis to shRNAs with strong effects on proliferation/viability (mutant KRAS class mean log POC<−0.2, corresponding to ~37% viability impairment) (Supplementary Fig. 4a). We used the t-test statistic threshold that was achieved by the set of control shRNAs as the boundary to identify the top 25 shRNAs KRAS synthetic lethal shRNAs that scored on the candidate plate (Supplementary Fig. 4b)
Gene Expression Profiling/GSEA
RNA was prepared from AALE cells expressing KRAS G12V or a control vector 6 d post infection and analyzed using human U133A HTA Arrays (Affymetrix). GSEA was performed using gene sets from the Molecular Signatures Database (MSigDB-C2 v2)7. In brief, the method consists of the following steps: genes are first ranked in a list, L, by the correlation between their expression and the class distinction (e.g. KRAS mutant vs. WT), using a suitable correlation metric. Given a defined set of genes S (e.g., genes members of a signaling pathway, located in the same genomic region, sharing the same GO category etc.), the goal of GSEA is to determine whether the members of S are found at the top or bottom of the list, implying they associate with the phenotypic distinction, rather than being distributed uniformly or randomly across the list. Next, to evaluate this degree of “enrichment” an Enrichment Score (ES) is calculated to quantify the degree to which a set S is over-represented at the top or bottom of the entire ranked list L. After calculation of the scores for a collection of gene sets, an empirical phenotype-based permutation test procedure is used to estimate P-values. The permutation of class labels preserves gene-gene correlations and provides an assessment of significance that is more reflective of the underlying biology. Finally an adjustment was made to the estimated significance level to account for multiple hypotheses testing (MHT). GSEA normalizes the ES for each gene set to account for the variation in set sizes, yielding a normalized enrichment score (NES) and a false discovery rate (FDR). The FDR gives an estimate of the probability that a set with a given NES represents a false positive finding; it is computed by comparing the tails of the observed and permutation-computed null distributions for the NES. The collection of gene sets used in the analysis of Figure 3a consisted of release 2.5 of the C2 (curated gene sets) sub-collection of the Molecular Signatures Database (http://www.broad.mit.edu/gsea/msigdb/). To determine the significance of identifying multiple NF-κB upregulated gene sets enriched in AALE-K cells, we used a hypergeometric test.
Gene expression profiling was also performed in triplicate in AALE-K cells (KRAS G13D) 5 passages following stable integration of the KRAS allele and compared with AALE-V cells using human U133A Arrays (Affymetrix). We created a new KRAS-specific gene signature using the mean difference in expression between triplicate samples to determine the 300 most significantly induced genes by oncogenic KRAS. In parallel, expression profiling was performed in AALE-K cells 48 hrs following expression of shGFP or shTBK1, to identify which of these genes were most significantly downregulated following TBK1 suppression.
Signature Projection Method
The RAS oncogenic signature12, AALE-K signature, NF-κB signature13, IKKε-regulated NF-κB gene set14, and IRF3 target gene set17 were projected across 38 lung adenocarcinomas derived from the Tumor Sequencing Project (TSP)15, 90 lung adenocarcinomas16, a collection of 53 lung cancer cell lines30, and 17 normal lung tissue specimens31. This was accomplished by a “single sample” extension of GSEA7 that allows one to define an enrichment score that represents the degree of absolute enrichment of a gene set in each sample within a given dataset. The gene expression values for a given sample were rank-normalized, and an enrichment score was produced using the Empirical Cumulative Distribution Functions (ECDF) of the genes in the signature and the remaining genes. This procedure is similar to GSEA but the list is ranked by absolute expression (in one sample). The enrichment score is obtained by an integration of the difference between the ECDFs. For a given signature G of size NG and single sample S, of the dataset of N genes, the genes are replaced by their ranks according the their absolute expression L={r1, r2, …, rN}. The list is then ordered from the highest rank N to the lowest 1. An enrichment score ES(G, S) is obtained by a sum (integration) of the difference between a weighted ECDF of the genes in the signature and the ECDF of the remaining genes PNG:
This calculation is repeated for each signature and each sample in the dataset. Note that the exponent of this quantity (α) is set to ¼, and adds a modest weight to the rank. In the regular GSEA a similar enrichment score is used, but the weight is typically set to 1. Also, instead of the sum over i, the enrichment score is computed according to the largest difference. This quantity is slightly more robust and more sensitive to differences in the tails of the distributions than the Kolmogorov-Smirnov statistic. It is particularly well suited to represent the activation score of gene sets based on a relatively small subset of the genes attaining high expression values. Signature values were normalized using the entire set of 128 lung adenocarcinomas and 17 normal lung specimens. p-values were calculated for the lung adenocarcinoma samples testing the hypotheses that the Spearman correlation between the RAS oncogenic signature, NF-κB gene set, and IRF3 target gene set were greater than zero.
Cell Culture
293T and A549 cells were grown in DMEM supplemented with 10% FBS, 2mM L-glutamine, penicillin (1000 U/ml), and streptomycin (1000 μg/ml). NCI-H23, NCI-H28, HCC-193, NCI-H522, HCC-1359, NCI-H1437, NCI-H1568, NCI-H1792, NCI-H1944, NCI-H1975, NCI-2009, NCI-H2030, NCI-H2110, and NCI-H2887 cells were cultured in RPMI supplemented with 10% FBS, 2mM L-glutamine, penicillin, and streptomycin. AALE cells8 were cultured in serum free SABM media with SingleQuot supplements/growth factors (Lonza). AALE cells expressing KRAS G12V or KRAS G13D alleles were generated following transduction using a pBabe retroviral vector or pLenti6.2/V5-Dest lentiviral vector (Invitrogen) respectively, as described14. Cell lines expressing the IKBα super-repressor were generated using a pBabe retroviral vector expressing the IKBα super-repressor14, and NCI-H23 cells expressing BCL-XL or LACZ were generated using pLenti6.2/V5-Dest encoding either BCL-XL or LACZ. Cells were seeded in 96-well plates for cell viability assays and in 6-well plates to prepare lysates for immunoblotting.
Low Throughput Lentiviral shRNA Production/Infection
Lentiviral vectors encoding shRNAs specific for control GFP sequences as well as KRAS, TBK1, CRAF, BRAF, AKT1, RALB, IRF3, c-REL and survivin are part of TRC shRNA library. Sequences of validated shRNAs are listed in Supplementary Table 6. Lentiviruses were produced by transfection of 293T cells with vectors encoding gene-specific shRNAs (1 μg) together with the packaging plasmids encoding Δ8.9 and VSV-G using Fugene 6 (Roche). Culture supernatants containing lentivirus were collected 48 and 72 h post-transfection. Virus was pooled and stored at −80° C. Cells were infected using a 1:11 dilution of virus in polybrene-containing media. Following centrifugation at 1000 × g for 15 min, all NSCLC lines were selected in puromycin (2 μg/ml) starting 24 h post-infection. AALE cells were treated with virus/polybrene for 4 h and selected with puromycin (1 μg/ml). Viability assays were conducted 6 d post-infection using Cell-Titer Glo (Promega) in triplicate. Lysates were harvested 72 h following shRNA expression to assess gene suppression. To determine differential viability effects in NCI-H23/NCI-H1792 (mutant KRAS) vs. NCI-H1437/NCI-H1568 (WT KRAS) cells, mean viability for each shRNA was normalized to shGFP control. Results from validated shRNAs were grouped together for each gene in KRAS mutant vs WT cell lines, and an unpaired t-test was used to determine statistical significance.
Antibodies
Immunoblotting was performed as described14. Antibodies were obtained from Cell Signaling Technology (anti-AKT1 #2967, anti-phosho-AKT Ser 473 #9271, anti-BCLXL #2762, anti-BIRC2 #4952, anti-C-RAF #9422, anti-c-REL #4727, anti-GAPDH #2118, anti-IKBα #4814, anti-Lamin A/C #2032, anti-phospho-MAPK p42/44 #9102, anti-p105/p50 #3035, anti-PARP cleaved Asp214 #9546, anti-RALB #3523, anti-survivin #2808), Santa Cruz Technology (anti-KRAS sc-30, anti-B-RAF F-7, anti-IRF3 FL-425, anti-IKBα c-15), and Upstate Biotechnologies/Millipore (anti-TBK1 clone AOW9).
Tumorigenicity Assay
Tumor xenograft experiments were performed as described14. Control GFP or TBK1-specific shRNAs were expressed in the indicated cells for 72 h, and then 2 × 106 viable cells were injected subcutaneously per site into immunodeficient mice. Mean and SEM tumor volume was plotted over time. The total number of tumors/implantations were: A549-shGFP (10/13); A549-shTBK1 (0/11); NCI-H1437-shGFP (12/12); NCI-H1437-shTBK1 (12/12), NCI-1568-shGFP (12/12); NCI-1568-shTBK1 (12/12); NCI-H2009-shGFP (9/12); NCI-H2009-shTBK1 (3/11). Tumor determination was made at 3 wks except for NCI-H2009, which were measured at 5 wks.
TUNEL Assay
NCI-H23 cells and NCI-H1437 cells were infected in 96-well plates with shRNAs specific for GFP, TBK1, and KRAS, fixed 5 d post-infection using 10% paraformaldehyde, and subjected to TUNEL staining (Roche). Nuclei were co-stained with DAPI, imaged, and counted using a Zeiss Axiovert 200 immunofluorescence microscope.
Nuclear/Cytoplasmic Fractionation
Cells were washed twice in ice cold PBS and incubated on ice for 10 min following treatment with hypotonic cytoplasmic lysis buffer (20 mM Hepes pH7.6, 10 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 1mM DTT, 0.1% NP40, 20% glycerol) + proteinase inhibitors. Nuclei were pelleted at 400 × g at 4° C for 4 min. Supernatants were collected, and the nuclear pellet was washed twice using cytoplasmic lysis buffer. Nuclear lysis buffer (same as cytoplasmic lysis buffer except 500 mM NaCl) was added to the pellet and samples were incubated for 30 min on ice. Following centrifugation at 16,000 × g at 4° C for 15 min, the supernatant was collected as the nuclear fraction.
Immunoprecipitation
Cells were washed twice with PBS, treated with lysis buffer (50 mM Tris pH 7.4, 150 mM NaCl, 0.5% NP-40, 2mM EDTA) and rotated at 4° C for 60 min. Following centrifugation at 16,000 × g, lysates were quantitated. Anti-c-REL antibody (5 μl) or buffer alone was added to lysate (750 μg) in 500 μl of lysis buffer and tubes were rotated overnight at 4° C. 50 μl of 50% protein G-sepharose beads were added, and samples were incubated for 2 h at 4° C. Following centrifugation at 800 × g for 3 min at 4° C, beads were washed 3 times in lysis buffer. Elution was performed using 2× sample buffer, and samples were divided in half and loaded onto parallel gels, along with beads only control and 1/10 input.
Real-Time Quantitative RT-PCR
Relative mRNA expression was determined using real-time quantitative PCR and normalized to GAPDH expression as an internal amplification control. Total RNA was isolated using Trizol reagent (Gibco), and 1 μg of total RNA was reverse transcribed using SuperScript First-Strand Synthesis System (Invitrogen), followed by amplification using SYBR Green PCR Master Mix (Applied Biosystems).
Supplementary Material
Acknowledgments
This work was supported in part by grants from the U.S. National Cancer Institute (R33 CA128625, R01 CA130988) (W.C.H.) and NIH T32 CA09172-33 (D.A.B., S.E.M.), the Starr Cancer Consortium (I1-A11, W.C.H., D.G.G.), the Susan Madden Fund and an ASCO YIA (D.A.B.), a Department of Defense Prostate Cancer Postdoctoral Fellowship (S.Y.K.), a Brain Science Foundation Fellowship (I.F.D), the Deutsche Krebshilfe (grant 107954) (R.K.T.), the Fritz-Thyssen-Stiftung (grant 10.08.2.175, R.K.T.) and the NGFNplus-program of the German Ministry of Science and Education (BMBF, grant 01GS08100, R.K.T.). We thank Channing Yu, Guo Wei and members of the Hahn lab for helpful discussions. High-throughput RNAi screening was conducted at the RNAi Platform of the Broad Institute of MIT and Harvard. W.C.H., M.M, and D.M.L. are consultants for Novartis Pharmaceuticals, Inc.
Footnotes
Author contributions
D.A.B., J.S.B., S.Y.K., S.E.M., and W.C.H. designed the experiments. D.A.B. and P.T. performed computational analyses. S.Y.K., I.F.D, A.C.S, P.S., C.S., S.F., P.G., J.H.R., Q.S., and R.C.W. performed primary RNAi screens; S.J.S., S.H., B.S.W., C.M. and B.A.W. assisted with data analysis. D.A.B. performed secondary screen with help from H.L. S.E.M. performed tumor xenograft experiments. E.M. performed experiments with murine cell lines. D.A.B., J.S.B., E.M.C., M.L.S., K.M., and R.K.T. performed expression-profiling experiments. S.R., D.M.L., D.M.S., E.S.L., D.G.G., T.J., D.E.R. supervised RNAi screens; M.M. and J.P.M. supervised data analysis. D.A.B. and W.C.H. wrote the manuscript. W.C.H. coordinated all aspects of the project. All authors discussed results and edited the manuscript.
Author information
All microarray data are available from the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo) under accession codes GSE17671, GSE17672, and GSE17643.
References
- 1.Hartwell LH, Szankasi P, Roberts CJ, Murray AW, Friend SH. Integrating genetic approaches into the discovery of anticancer drugs. Science. 1997;278:1064–1068. doi: 10.1126/science.278.5340.1064. [DOI] [PubMed] [Google Scholar]
- 2.Kaelin WG., Jr The concept of synthetic lethality in the context of anticancer therapy. Nat Rev Cancer. 2005;5:689–698. doi: 10.1038/nrc1691. [DOI] [PubMed] [Google Scholar]
- 3.Moffat J, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 4.Malo N, Hanley JA, Cerquozzi S, Pelletier J, Nadon R. Statistical practice in high-throughput screening data analysis. Nat Biotechnol. 2006;24:167–175. doi: 10.1038/nbt1186. [DOI] [PubMed] [Google Scholar]
- 5.Gould J, Getz G, Monti S, Reich M, Mesirov JP. Comparative gene marker selection suite. Bioinformatics. 2006;22:1924–1925. doi: 10.1093/bioinformatics/btl196. [DOI] [PubMed] [Google Scholar]
- 6.Luo B, et al. Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci U S A. 2008;105:20380–20385. doi: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lundberg AS, et al. Immortalization and transformation of primary human airway epithelial cells by gene transfer. Oncogene. 2002;21:4577–4586. doi: 10.1038/sj.onc.1205550. [DOI] [PubMed] [Google Scholar]
- 9.Wislez M, et al. Inhibition of mammalian target of rapamycin reverses alveolar epithelial neoplasia induced by oncogenic K-ras. Cancer Res. 2005;65:3226–3235. doi: 10.1158/0008-5472.CAN-04-4420. [DOI] [PubMed] [Google Scholar]
- 10.Chien Y, et al. RalB GTPase-mediated activation of the IkappaB family kinase TBK1 couples innate immune signaling to tumor cell survival. Cell. 2006;127:157–170. doi: 10.1016/j.cell.2006.08.034. [DOI] [PubMed] [Google Scholar]
- 11.Hacker H, Karin M. Regulation and function of IKK and IKK-related kinases. Sci STKE. 2006;2006:re13. doi: 10.1126/stke.3572006re13. [DOI] [PubMed] [Google Scholar]
- 12.Bild AH, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 13.Hinata K, Gervin AM, Jennifer Zhang Y, Khavari PA. Divergent gene regulation and growth effects by NF-kappa B in epithelial and mesenchymal cells of human skin. Oncogene. 2003;22:1955–1964. doi: 10.1038/sj.onc.1206198. [DOI] [PubMed] [Google Scholar]
- 14.Boehm JS, et al. Integrative genomic approaches identify IKBKE as a breast cancer oncogene. Cell. 2007;129:1065–1079. doi: 10.1016/j.cell.2007.03.052. [DOI] [PubMed] [Google Scholar]
- 15.Ding L, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takeuchi T, et al. Expression profile-defined classification of lung adenocarcinoma shows close relationship with underlying major genetic changes and clinicopathologic behaviors. J Clin Oncol. 2006;24:1679–1688. doi: 10.1200/JCO.2005.03.8224. [DOI] [PubMed] [Google Scholar]
- 17.Andersen J, VanScoy S, Cheng TF, Gomez D, Reich NC. IRF-3-dependent and augmented target genes during viral infection. Genes Immun. 2008;9:168–175. doi: 10.1038/sj.gene.6364449. [DOI] [PubMed] [Google Scholar]
- 18.Singh A, et al. A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer Cell. 2009;15:489–500. doi: 10.1016/j.ccr.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mayo MW, et al. Requirement of NF-kappaB activation to suppress p53-independent apoptosis induced by oncogenic Ras. Science. 1997;278:1812–1815. doi: 10.1126/science.278.5344.1812. [DOI] [PubMed] [Google Scholar]
- 20.Brown K, Gerstberger S, Carlson L, Franzoso G, Siebenlist U. Control of I kappa B-alpha proteolysis by site-specific, signal-induced phosphorylation. Science. 1995;267:1485–1488. doi: 10.1126/science.7878466. [DOI] [PubMed] [Google Scholar]
- 21.Harris J, et al. Nuclear accumulation of cRel following C-terminal phosphorylation by TBK1/IKK epsilon. J Immunol. 2006;177:2527–2535. doi: 10.4049/jimmunol.177.4.2527. [DOI] [PubMed] [Google Scholar]
- 22.Mercurio F, DiDonato JA, Rosette C, Karin M. p105 and p98 precursor proteins play an active role in NF-kappa B-mediated signal transduction. Genes Dev. 1993;7:705–718. doi: 10.1101/gad.7.4.705. [DOI] [PubMed] [Google Scholar]
- 23.Owyang AM, et al. c-Rel is required for the protection of B cells from antigen receptor-mediated, but not Fas-mediated, apoptosis. J Immunol. 2001;167:4948–4956. doi: 10.4049/jimmunol.167.9.4948. [DOI] [PubMed] [Google Scholar]
- 24.Scholl C, et al. Synthetic lethal interaction between oncogenic KRAS dependency and STK33 suppression in human cancer cells. Cell. 2009;137:821–834. doi: 10.1016/j.cell.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 25.Luo J, et al. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 2009;137:835–848. doi: 10.1016/j.cell.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ngo VN, et al. A loss-of-function RNA interference screen for molecular targets in cancer. Nature. 2006;441:106–110. doi: 10.1038/nature04687. [DOI] [PubMed] [Google Scholar]
- 27.Rottmann S, Wang Y, Nasoff M, Deveraux QL, Quon KC. A TRAIL receptor-dependent synthetic lethal relationship between MYC activation and GSK3beta/FBW7 loss of function. Proc Natl Acad Sci U S A. 2005;102:15195–15200. doi: 10.1073/pnas.0505114102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Silva JM, et al. Profiling essential genes in human mammary cells by multiplex RNAi screening. Science. 2008;319:617–620. doi: 10.1126/science.1149185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang J. Powerful goodness-of-fit tests based on the likelihood ratio. Journal of the Royal Statistical Society Series B (Statistical Methodology) 2002;64:281–294. [Google Scholar]
- 30.Sos ML, et al. Predicting drug susceptibility of non-small cell lung cancers based on genetic lesions. J Clin Invest. 2009;119:1727–1740. doi: 10.1172/JCI37127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhattacharjee A, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci U S A. 2001;98:13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.