SUMMARY
Balanced chromosome segregation in mitosis requires synchronous chromatid separation at anaphase and the precise coordination of anaphase with cytokinesis and mitotic exit. The mitotic spindle checkpoint monitors proper attachment and/or tension induced by microtubule binding to sister kinetochores. Within each cell, once all chromosomes achieve bipolar attachment to the spindle poles and align at the metaphase plate, the spindle checkpoint is silenced triggering anaphase onset, cytokinesis, and mitotic exit. We used a bioinformatics approach to identify a candidate protein, C13orf3/Ska3, predicted to function in mitosis. Cells in which Ska3 expression was reduced by RNAi achieved metaphase alignment but were unable to silence the spindle checkpoint and enter normal anaphase. After hours of metaphase arrest, chromatids separated but retained robust attachment to spindle microtubules. These cells remained checkpoint arrested with strong accumulation of the checkpoint protein Bub1 at kinetochores. During normal mitosis Ska3 protein accumulated on kinetochores in prometaphase after nuclear envelope breakdown. This kinetochore localization of Ska3 was dependent on Shugoshin (Sgo1), the “guardian spirit” of chromatid cohesion. In contrast, Sgo1, which accumulates at the centromeres in early prophase, was not dependent on Ska3. Although Ska3 is required for maintenance of sister chromatid cohesion and is dependent upon Sgo1, cells with reduced Sgo1 show a stronger premature chromatid separation phenotype than those with reduced Ska3. We hypothesize that Ska3 functions as a component of a network that coordinates checkpoint signaling from the microtubule binding sites within a kinetochore by laterally linking the individual binding sites. We suggest that this network plays a major role in silencing the spindle checkpoint when chromosomes are aligned at metaphase to allow timely anaphase onset and mitotic exit.
Results and Discussion
Identification of C13orf3/Ska3
A bioinformatics approach consisting of a meta-analysis of all publicly available human 2-color microarray experiments deposited in NCBI’s GEO database was conducted and guided by a literature-based analysis of the published associations of the co-regulated genes (see Supplemental Information) [1]. The approach identified C13orf3 as the highest scoring protein with positive mitotic function in concert with potential association with kinetochores. Very recently, three other reports have identified C13orf3 as a binding partner of the Ska1/Ska2 complex [2–4] and another identified it as a novel kinetochore protein [5]. The Ska1/Ska2 (Spindle and Kinetochore Associated) complex was itself first identified by Hanisch et al. who found that siRNA mediated knockdown resulted in a temporary metaphase delay [6]. Three of the recent reports on C13orf3 (now called Ska3) suggest a primary role for Ska3 and the Ska complex in kinetochore-microtubule attachment and chromosome movement [2, 4, 5] while the third implicates the Ska complex in interacting with protein phosphatase PP2A and maintenance of chromatid cohesion, the spindle checkpoint, and spindle bipolarity [3].
Reduced Ska3 expression induces mitotic arrest
We targeted Ska3 expression using pSiren shRNA vectors that co-express dsRed in Hela cells expressing GFP-histone H2B and analyzed the results by long term, time-lapse microscopy. In the initial screening over 70% of cells targeted by one of two different shRNA sequences to Ska3 showed mitotic defects. The most common phenotype was a transient metaphase arrest lasting up to a few hours, followed by scattering of chromosomes on the spindle and long term mitotic arrest (Fig. 1A and Supplemental Video 1). We confirmed the effects using a “smart pool” consisting of 4 siRNA duplexes targeting Ska3 expression. In cultures treated with Ska3 siRNA, the majority of cells exhibited the phenotype of prolonged metaphase followed by scattering of chromosomes and mitotic arrest (Supplemental Fig. 1A and Supplemental Video 2). In some cases cells exhibited a prolonged metaphase and then entered anaphase. The majority of chromosomes in Ska3-depleted cells moved to the metaphase plate within a time frame similar to controls. However, during prometaphase, one or a few chromosomes in Ska3-depleted cells often temporarily moved away from the spindle equator delaying complete metaphase alignment, and in a few cases some chromosomes failed to align (Supplemental Fig 1B).
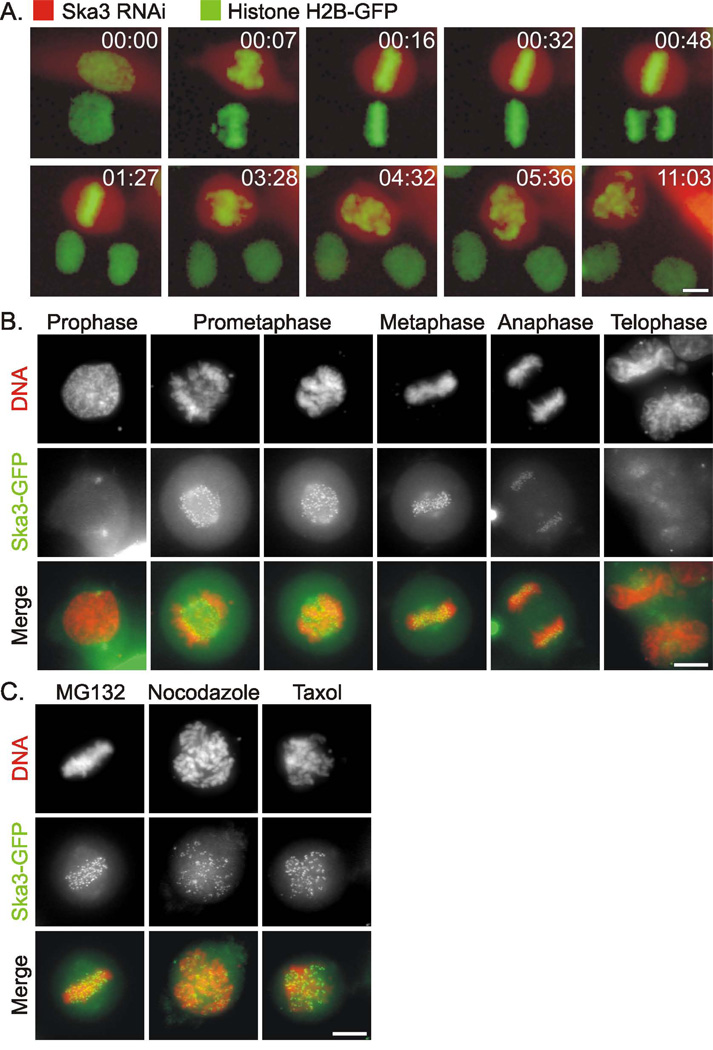
Figure 1. Mitotic Defects in Cells Depleted of Ska3 and Kinetochore Localization of Ska3-GFP.
(A) Fluorescence images of a Hela H2B-GFP cell 30 hours after transfection with shRNAi plasmid targeting Ska3 (red cytoplasm) adjacent to a non-transfected control cell in the same field. The transfected cell arrests at metaphase for approximately three hours after which chromosomes begin to scatter. The control cell proceeds normally through mitosis. Time is hours:minutes. (B) Ska3-GFP is diffusely distributed in prophase Hela cells with some concentration on centrosomes. After nuclear envelope breakdown, Ska3 concentrates strongly on kinetochores and to a lesser extent along spindles. The kinetochore concentration diminishes during anaphase and is lost in telophase. (C) Ska3 remains at the kinetochores of cells treated with the proteasome inhibitor, MG132, with the microtubule disruptor, Nocodazole, and with the microtubule stabilizer, Taxol. Bars, 5 µm.
Ska3 associates with kinetochores after nuclear envelope breakdown
We generated plasmids encoding Ska3-GFP fusion proteins and introduced these into Hela cells (Fig. 1B). Ska3-GFP was diffusely cytoplasmic during interphase and prophase. In prophase some association with the centrosomes was apparent. Ska3-GFP became strongly concentrated on kinetochores after nuclear envelope breakdown and remained highly concentrated there through metaphase. During anaphase Ska3 concentration at the kinetochores diminished and was lost at telophase. In addition Ska3-GFP also showed some association with the mitotic spindle throughout mitosis. Ska3-GFP remained concentrated at kinetochores in cells arrested at metaphase with the proteasome inhibitor, MG132, in cells treated with Nocodazole to disrupt microtubules, or in cells treated with Taxol to induce microtubule hyperstabilization (Fig. 1C). Immunolabeling with an antibody we prepared to bacterially expressed Ska3 showed a distribution identical to that seen with Ska3-GFP (Supplemental Fig. 2A). Ska3 was localized to the region of the outer kinetochore coincident with the Ndc80 complex protein Nuf2 and slightly outside the region labeled by human auto-immune anti-centromere antibody (Supplemental Fig. 2B).
Ska-3 depleted cells arrest in mitosis with separated chromatids and robust attachment of kinetochores to spindle microtubules
Immunolabeling of kinetochores in Ska3 depleted cells indicated that many cells arrested in mitosis with separated chromatids (Fig. 2A). This was confirmed in chromosome spreads where we detected a mixture of cells with paired chromatids, presumably derived from metaphase-arrested cells, and those with separated chromosomes, derived from cells with the scattered phenotype (Fig. 2B). To determine whether Ska3 depletion led to loss of stable kinetochore attachment to microtubules, we treated cells with cold prior to fixation. This treatment preserves stable kinetochore-attached microtubules but leads to destabilization of other free microtubules [7]. Ska3-depleted cells with scattered chromosomes showed robust kinetochore fibers, and quantification of spindle microtubules showed an enhancement of cold-stable spindle microtubules compared to metaphase controls (Fig. 2C). This finding conflicts with recent two reports suggesting that depletion of Ska complex proteins results in a decrease in spindle microtubules [2, 5]. The reasons for this discrepancy are unknown.
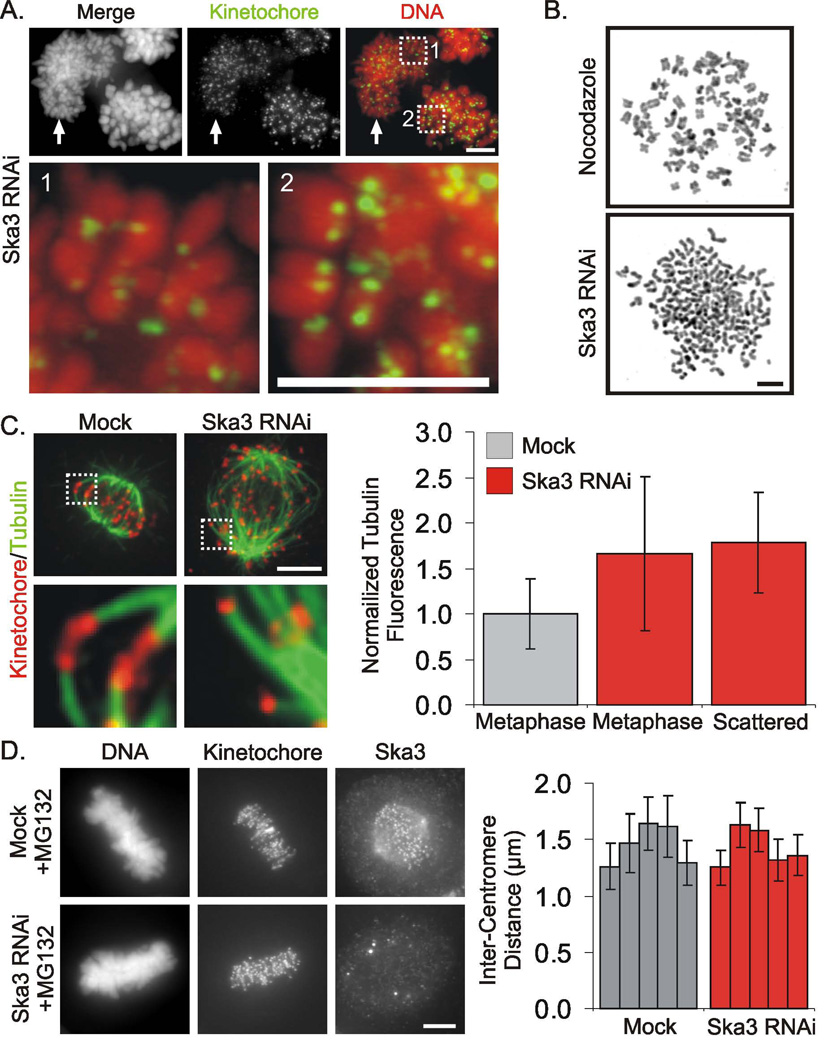
Figure 2. Ska3 Depleted Cells Separate Chromatids but Retain Microtubule Attachment.
(A) Cells arrested in mitosis by siRNA to Ska3 were hypotonically swollen, lysed and centrifuged onto coverslips before fixation and labeling. A mixture of cells showing single chromatids (arrow and inset 1) and cells with paired chromatids (inset 2) were found. Presumably the former derive from cells with scattered chromosomes arrested for longer periods with the latter derive from cells arrested at metaphase. (B) Chromosome spreads reveal loss of chromatid cohesion in Ska3-depleted cells. (C) Microtubule bundles extending to kinetochores are retained in Ska3-depleted cells after cold treatment to destabilize non-kinetochore microtubules. Total levels of cold stable spindle microtubules (averages ± SD) are increased somewhat the spindles of Ska3-depleted cells both at metaphase and after chromosome scattering in comparison with mock transfected cells at metaphase. (D) Mock treated and Ska3 siRNA-treated Hela cells were incubated with the proteasome inhibitor MG132 for 35 min, fixed, and labeled with human anti-kinetochore and anti-Ska3 antibodies. Five cells at metaphase in both the mock and RNAi-treated cultures were selected and inter-kinetochore distances were measured (>160 sister chromatid pairs for each). The inter-kinetochore distances were plotted indicating average separation for each cell ± SD. No significant differences in the inter-kinetochore distances between control and Ska3 siRNA-treated metaphase cells were detected. Bars, 5 µm.
To test the pulling forces on kinetochores in Ska3-depleted cells, we measured the average separation distances of sister kinetochores of metaphase cells in mock and Ska3-depleted cultures treated with MG132 for 35 min to induce metaphase arrest in control cells (Fig 2D). We found no significant differences in interkinetochore distances in the two cell populations indicating that kinetochore-microtubule attachment and the poleward pulling forces of kinetochores are not significantly compromised by depletion of Ska3.
Ska-3 depleted cells arrest in mitosis through activation of the spindle checkpoint
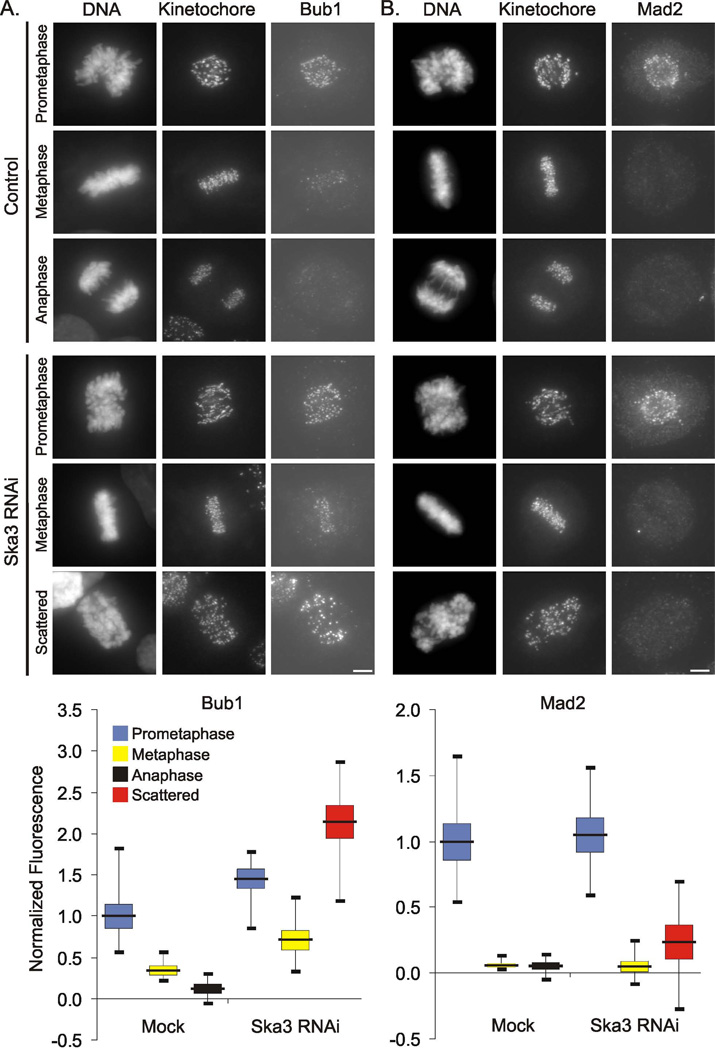
We found that the checkpoint kinase Bub1 showed enhanced concentration at kinetochores of Ska3-depleted cells at metaphase and most notably in cells with scattered chromosomes (Fig. 3A). On average, kinetochore accumulation of the checkpoint proteins Mad2 and BubR1 in Ska3-depleted cells were near those of control cells at metaphase (Fig 3B and Supplemental Fig. 3). Mad2 labeling in Ska3-depleted cells at metaphase was slightly increased on some kinetochores as indicated by the greater range in the Mad2 labeling measurements. Mad2 and BubR1 levels became modestly more increased in cells with scattered chromosomes. To test if spindle checkpoint activation was responsible for mitotic arrest of Ska3-depleted cells, we inhibited checkpoint signaling in two ways. First, we treated cells with ZM447439, an Aurora kinase inhibitor that overrides the spindle checkpoint [8]. In Ska3 depleted cells, ZM447439 induced mitotic exit, an event that was blocked by addition of the proteasome inhibitor MG132 (Supplemental Fig. 4A). Second, depletion of the checkpoint protein BubR1 also induced mitotic exit in cells depleted of Ska3 (Supplemental Fig. 4B and Supplemental Video 3).
Figure 3. Ska3 Depletion Activates the Spindle Checkpoint Inducing Strong Kinetochore Accumulation of Bub1.
(A) Control cells and cells treated with Ska3 siRNA were fixed and labeled with anti-kinetochore serum and anti-Bub1 antibody. Bub1 labeling is increased at all stages in Ska3-depleted cells but most enhanced in the arrested cells showing scattered chromosomes. (B) Metaphase cells in mock treated and Ska3-depleted cultures show on average comparable levels of kinetochore-associated Mad2 although the range of Mad2 at kinetochores in Ska3-depleted cells is greater. Ska3-dpeleted cells with scattered chromosomes show a modest increase in the average kinetochore concentration of Mad2. In the graphs, the black horizontal bars indicate averages; the colored boxes indicate standard errors and the thin vertical “whiskers” indicate ranges. Bars, 5 µm.
Comparison of Ska3 and Sgo1 in maintaining chromatid cohesion
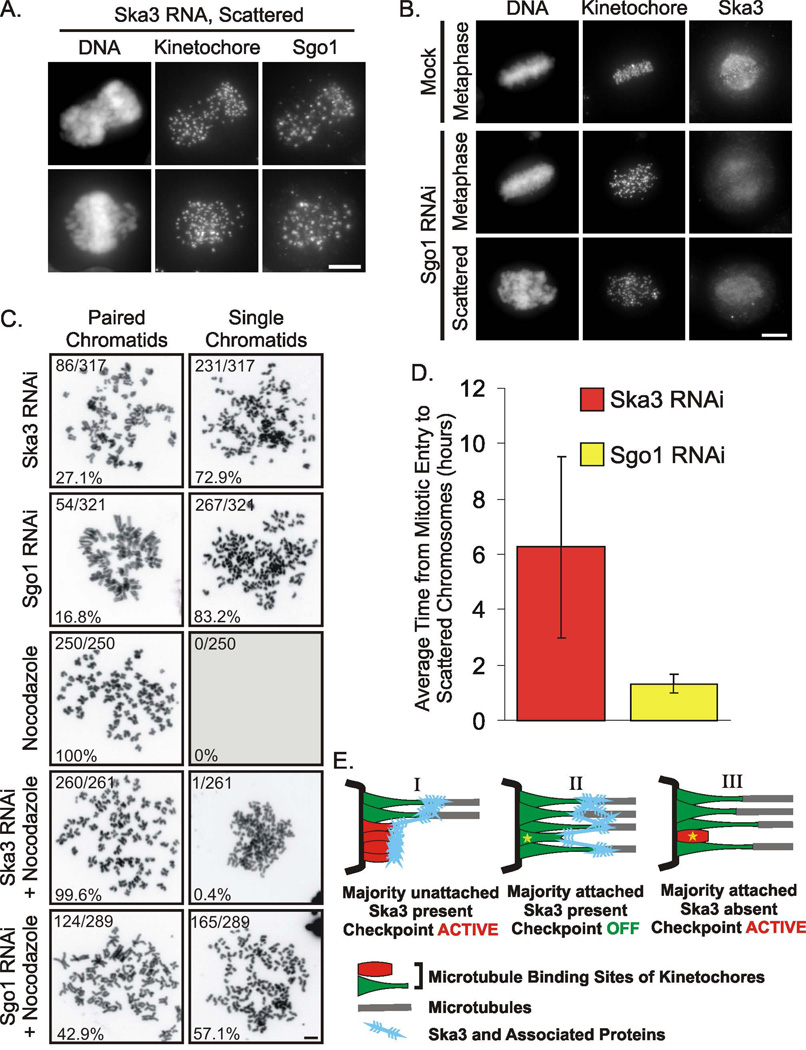
The mitotic arrest accompanied by chromatid separation in Ska3-depeleted cells was reminiscent of the phenotype obtained from siRNA repression of Sgo1 [9–12]. Therefore we examined the possible connection of Sgo1 to the Ska3 depletion phenotype. Sgo1 was present on kinetochores of chromosomes and scattered chromatids depleted of Ska3 (Fig. 4A). In contrast, Sgo1 depletion by siRNA resulted in loss of Ska3 accumulation at kinetochores both before and after chromatids separated, although the weak association of Ska3 with the mitotic spindle persisted (Fig 5C). Ska3’s dependency on Sgo1 localization and the lack of the reciprocal dependency is consistent with the fact that Sgo1 associates with the centromere region in early prophase while Ska3 does not concentrate at kinetochores until after nuclear envelope breakdown. Unlike the results reported in a recent study [3], we found that Ska3 protein levels were not reduced by Sgo1 depletion (Supplemental Fig. 5).
Figure 4. Ska3 Accumulation at Kinetochores is Dependent upon Shugoshin and May Coordinate Checkpoint Signaling and Chromatid Separation.
(A) Sgo1 remains associated with kinetochores in cells depleted of Ska3. (B) Depletion of Sgo1 blocks kinetochore but not spindle association of Ska3. (C) In the presence of intact microtubules both Ska3-depleted and Sgo1-depleted cells show high levels of chromatid separation in chromosome spreads. In Nocodazole alone, no chromatid separation occurs. The disruption of microtubules with Nocodazole nearly eliminates chromatid separation in Ska3-depleted cells but only partially decreases chromatid separation in Sgo1-depleted cells. Bars, 5 urn. (D) Ska3-depleted cells arrest at metaphase for substantially longer times than do Sgo1-depleted cells before undergoing chromosome scattering. For Ska3, n = 84; for Sgo1, n = 53. (E) Model for Ska3 in integration of spindle checkpoint signaling within the kinetochore. (Diagram I) At kinetochores where most end-on microtubule binding sites are unattached, checkpoint signaling is active. (Diagram II). At kinetochores where most end-on microtubule binding sites are attached checkpoint signaling is inactive. Ska3 and associated proteins provide lateral attachments to inhibit checkpoint signaling in the face of momentary loss of some end-on microtubule attachments (star) that occurs as a consequence of microtubule dynamics. (Diagram III). In the absence of the Ska3 meshwork, release of an end-on attachment immediately results in binding site collapse (star) and reactivation of checkpoint signaling.
We examined the question of whether the loss of chromatid cohesion caused by Sgo1 depletion might be fully explained by loss of kinetochore-associated Ska3. Quantifying results from chromosome spreads we found that Ska3 or Sgo1 depletion led to high levels of chromatid separation in cells with intact spindles. However, when cells were incubated in nocodazole to disrupt microtubules, chromatids very rarely separated in Ska3-depleted cells but still did so in a large proportion of Sgo1-depleted cells (Fig. 4C). The fact that spindle pulling forces are necessary to generate chromatid separation in Ska3-depleted cells is consistent with an interpretation that Ska3’s role in chromatid cohesion is less central than that of Sgo1.
Using video microscopy we compared the timing of chromatid scattering in Sgo1- and Ska3-depleted cells. Whereas cells depleted of Ska3 generally arrested for several hours at metaphase before chromatid scattering Sgo1-depleted cells arrested at metaphase average of one hour before chromatid scattering (Fig. 4D). While differences in protein stability, synthesis, and efficiency of siRNA make strict comparisons difficult, these results again indicate a more central role for Sgo1 in maintaining chromosome cohesion beyond simply targeting the Ska complex to kinetochores. Conversely these finding suggest that Ska3 and its partners play a more indirect role in maintaining cohesion.
A model for Ska3 in spindle checkpoint signaling
The interplay between mechanical tension, stability of microtubule-kinetochore attachment, chromatid separation, and signaling of the spindle checkpoint is complex and controversial [13–19]. In Ska3-depleted cells, the spindle checkpoint is activated but the kinetochores of the arrested cells maintain robust bundles of kinetochore-associated microtubules and show particularly strong accumulation of the checkpoint protein Bub1. In three recent studies Ska3 and the other members of the Ska complex were proposed to play their major roles as mediators of microtubule attachment to kinetochores and in chromosome movement [2, 4, 5]. A fourth recent study detected the chromatid separation phenotype of Ska3 depletion and proposed roles for the Ska complex in the maintenance of spindle bipolarity, in spindle checkpoint activation, and in metaphase to anaphase progression [3]. We observed that Ska3-depleted cells retain robust kinetochore-microtubule attachment and that chromosomes readily align at the spindle equator, albeit occasionally with slight delays. While our data are consistent with the Ska complex as an auxiliary element aiding chromosome movement, our findings argue against this as a primary function.
Sister kinetochores in Ska3-depleted cells show vigorous stretching of the interkinetochore distance comparable to that of controls. However, in these cells the spindle checkpoint is not extinguished, and thus the cells do not undergo normal anaphase onset and mitotic exit. Instead they remain arrested for long periods at metaphase before chromatids begin to separate. Once separated, chromatids exhibit strong and persistent checkpoint signaling thus trapping the cells in an essentially permanent mitotic arrest. It is noteworthy that Bub1 accumulation at kinetochores is markedly increased by Ska3 depletion. During mitosis in human cells and in budding yeast Bub1 is required for the centromere localization of Sgo1 [20–22]. In human cells Sgo1 functions to protect cohesin at centromeres from premature dissociation [21, 22]. In budding yeast, Sgo1 and the kinase domain of Bub1 are not essential for spindle checkpoint activation upon loss of kinetochore-microtubule attachment. However, they are required for checkpoint arrest in response to loss of mechanical tension at kinetochores [20, 23]. We speculate that the Ska complex may be important in coupling the sensing of mechanical tension with checkpoint silencing and the normally synchronous release of centromere cohesion at anaphase.
While the full details of the pathways linking these functions remain uncertain, we here propose a model suggesting that a major function for Ska3 and its associated proteins is integrating spindle checkpoint signaling for the multiple microtubule-binding sites within a single kinetochore. Accumulated evidence indicates that end-on binding of microtubules to microtubule-binding sites within kinetochores requires the Ndc80 complex [7, 24–26]. We suggest a model in which the Ska complex physically couples unoccupied microtubule-binding sites to the microtubules embedded in adjacent binding sites (Fig. 4E). We propose that the Ska complex forms part of a filament network that coordinates checkpoint activation of each microtubule binding site at a kinetochore with the overall status of microtubule attachment and tension. Thus, for a chromosome with both kinetochores attached to bundles of microtubules from each pole at metaphase, the loss of single microtubule attachments due to microtubule dynamic instability would not immediately reactivate checkpoint signaling. With this “majority vote” mechanism, the Ska complex would allow sustained silencing of checkpoint signaling from kinetochores at metaphase without requiring that every microtubule binding site be constantly occupied. Given our findings, we speculate that Bub1 checkpoint signaling may play a particularly key role in this process. In the absence of the Ska complex, complete silencing of the checkpoint becomes more difficult and many cells arrest at metaphase with aligned chromosomes but unable to enter anaphase. Eventually under the pulling forces of the spindle, chromatid cohesion becomes compromised and chromatids begin to separate. The resulting unpaired kinetochores reignite strong checkpoint signals leading to inescapable arrest in M phase. It has been reported that mitotic arrest for several hours at metaphase induced by treatment with proteasome inhibitor results in loss of centromere but not arm cohesion [27]. How Ska3 depletion and the accompanying metaphase arrest result in eventual and complete loss of both centromere and chromatid arm cohesion remains to be investigated.
Supplementary Material
Acknowledgments
We thank Dr. Todd Stukenberg for advice, discussion, and anti-BubR1 antibody. We thank Dr. Steve Taylor for anti-Bub1, Dr. Ted Salmon for anti-Mad2, and Dr. Hongtao Yu for anti-Sgo1. We thank Dr. Susannah Rankin for advice, discussion, and for use of microscopy equipment. We also thank the members of the Gorbsky lab and members of the Cell Cycle & Cancer Biology program at the OMRF for discussion. This work was supported by a grant from the National Institute of General Medical Sciences (GM50412) and by the McCasland Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wren JD. A global meta-analysis of microarray expression data to predict unknown gene functions and estimate the literature-data divide. Bioinformatics. 2009 doi: 10.1093/bioinformatics/btp290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gaitanos TN, Santamaria A, Jeyaprakash AA, Wang B, Conti E, Nigg EA. Stable kinetochore-microtubule interactions depend on the Ska complex and its new component Ska3/C13Orf3. EMBO J. 2009;28:1442–1452. doi: 10.1038/emboj.2009.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Theis M, Slabicki M, Junqueira M, Paszkowski-Rogacz M, Sontheimer J, Kittler R, Heninger AK, Glatter T, Kruusmaa K, Poser I, et al. Comparative profiling identifies C13orf3 as a component of the Ska complex required for mammalian cell division. EMBO J. 2009;28:1453–1465. doi: 10.1038/emboj.2009.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Welburn JP, Grishchuk EL, Backer CB, Wilson-Kubalek EM, Yates JR, 3rd, Cheeseman IM. The human kinetochore Ska1 complex facilitates microtubule depolymerization-coupled motility. Dev Cell. 2009;16:374–385. doi: 10.1016/j.devcel.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raaijmakers JA, Tanenbaum ME, Maia AF, Medema RH. RAMA1 is a novel kinetochore protein involved in kinetochore-microtubule attachment. J Cell Sci. 2009 doi: 10.1242/jcs.051912. [DOI] [PubMed] [Google Scholar]
- 6.Hanisch A, Sillje HH, Nigg EA. Timely anaphase onset requires a novel spindle and kinetochore complex comprising Ska1 and Ska2. EMBO J. 2006;25:5504–5515. doi: 10.1038/sj.emboj.7601426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeLuca JG, Moree B, Hickey JM, Kilmartin JV, Salmon ED. hNuf2 inhibition blocks stable kinetochore-microtubule attachment and induces mitotic cell death in HeLa cells. J Cell Biol. 2002;159:549–555. doi: 10.1083/jcb.200208159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ditchfield C, Johnson VL, Tighe A, Ellston R, Haworth C, Johnson T, Mortlock A, Keen N, Taylor SS. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J Cell Biol. 2003;161:267–280. doi: 10.1083/jcb.200208091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salic A, Waters JC, Mitchison TJ. Vertebrate shugoshin links sister centromere cohesion and kinetochore microtubule stability in mitosis. Cell. 2004;118:567–578. doi: 10.1016/j.cell.2004.08.016. [DOI] [PubMed] [Google Scholar]
- 10.Kitajima TS, Kawashima SA, Watanabe Y. The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature. 2004;427:510–517. doi: 10.1038/nature02312. [DOI] [PubMed] [Google Scholar]
- 11.Tang Z, Sun Y, Harley SE, Zou H, Yu H. Human Bub1 protects centromeric sister-chromatid cohesion through Shugoshin during mitosis. Proc Natl Acad Sci U S A. 2004;101:18012–18017. doi: 10.1073/pnas.0408600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McGuinness BE, Hirota T, Kudo NR, Peters JM, Nasmyth K. Shugoshin prevents dissociation of cohesin from centromeres during mitosis in vertebrate cells. PLoS Biol. 2005;3:e86. doi: 10.1371/journal.pbio.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 14.Pinsky BA, Biggins S. The spindle checkpoint: tension versus attachment. Trends Cell Biol. 2005;15:486–493. doi: 10.1016/j.tcb.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 15.Tanaka T, Fuchs J, Loidl J, Nasmyth K. Cohesin ensures bipolar attachment of microtubules to sister centromeres and resists their precocious separation. Nat Cell Biol. 2000;2:492–499. doi: 10.1038/35019529. [DOI] [PubMed] [Google Scholar]
- 16.Pinsky BA, Kung C, Shokat KM, Biggins S. The Ipl1-Aurora protein kinase activates the spindle checkpoint by creating unattached kinetochores. Nat Cell Biol. 2006;8:78–83. doi: 10.1038/ncb1341. [DOI] [PubMed] [Google Scholar]
- 17.Huang H, Hittle J, Zappacosta F, Annan RS, Hershko A, Yen TJ. Phosphorylation sites in BubR1 that regulate kinetochore attachment, tension, and mitotic exit. J Cell Biol. 2008;183:667–680. doi: 10.1083/jcb.200805163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elowe S, Hummer S, Uldschmid A, Li X, Nigg EA. Tension-sensitive Plk1 phosphorylation on BubR1 regulates the stability of kinetochore microtubule interactions. Genes Dev. 2007;21:2205–2219. doi: 10.1101/gad.436007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ahonen LJ, Kallio MJ, Daum JR, Bolton M, Manke IA, Yaffe MB, Stukenberg PT, Gorbsky GJ. Polo-like kinase 1 creates the tension-sensing 3F3/2 phosphoepitope and modulates the association of spindle-checkpoint proteins at kinetochores. Curr Biol. 2005;15:1078–1089. doi: 10.1016/j.cub.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 20.Fernius J, Hardwick KG. Bub1 kinase targets Sgo1 to ensure efficient chromosome biorientation in budding yeast mitosis. PLoS Genet. 2007;3:e213. doi: 10.1371/journal.pgen.0030213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kitajima TS, Hauf S, Ohsugi M, Yamamoto T, Watanabe Y. Human Bub1 defines the persistent cohesion site along the mitotic chromosome by affecting Shugoshin localization. Curr Biol. 2005;15:353–359. doi: 10.1016/j.cub.2004.12.044. [DOI] [PubMed] [Google Scholar]
- 22.Tang Z, Shu H, Qi W, Mahmood NA, Mumby MC, Yu H. PP2A is required for centromeric localization of Sgo1 and proper chromosome segregation. Dev Cell. 2006;10:575–585. doi: 10.1016/j.devcel.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 23.Indjeian VB, Stern BM, Murray AW. The centromeric protein Sgo1 is required to sense lack of tension on mitotic chromosomes. Science. 2005;307:130–133. doi: 10.1126/science.1101366. [DOI] [PubMed] [Google Scholar]
- 24.Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- 25.McCleland ML, Gardner RD, Kallio MJ, Daum JR, Gorbsky GJ, Burke DJ, Stukenberg PT. The highly conserved Ndc80 complex is required for kinetochore assembly, chromosome congression, and spindle checkpoint activity. Genes Dev. 2003;17:101–114. doi: 10.1101/gad.1040903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vorozhko VV, Emanuele MJ, Kallio MJ, Stukenberg PT, Gorbsky GJ. Multiple mechanisms of chromosome movement in vertebrate cells mediated through the Ndc80 complex and dynein/dynactin. Chromosoma. 2008;117:169–179. doi: 10.1007/s00412-007-0135-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee J, Kitajima TS, Tanno Y, Yoshida K, Morita T, Miyano T, Miyake M, Watanabe Y. Unified mode of centromeric protection by shugoshin in mammalian oocytes and somatic cells. Nat Cell Biol. 2008;10:42–52. doi: 10.1038/ncb1667. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.