Abstract
With the rapid growth of complex heterogeneous biological molecules, effective techniques that are capable of rapid, characterization of biologics are essential to ensure the desired product characteristics. To address this need, we have developed a method for analysis of intact glycoproteins based on high resolution capillary electrophoretic separation coupled to an LTQ-FT mass spectrometer. We evaluated the performance of this method on the alpha subunit of mouse cell line-derived recombinant human chorionic gonadotrophin (r-αhCG), a protein that is glycosylated at two sites and is part of the clinically-relevant gonadotrophin family. Analysis of r-αhCG, using capillary electrophoresis (CE) with a separation time under 20 minutes, resulted in the identification of over 60 different glycoforms with up to nine sialic acids. High resolution CE/FT-MS allowed separation and analysis of not only intact glycoforms with different numbers of sialic acids but also intact glycoforms that differed by the number and extent of neutral monosaccharides. The high mass resolution of the FT-MS enabled a limited mass range to be targeted for the examination of the protein glycoforms, simplifying the analysis without sacrificing accuracy. In addition, the limited mass range resulted in a fast scan speed that enhanced the reproducibility of the relative quantitation of individual glycoforms. The intact glycoprotein analysis was complemented with the analysis of the tryptic glycopeptides and glycans of r-αhCG to enable the assignment of glycan structures to individual sites, resulting in a detailed characterization of the protein. Samples of r-αhCG obtained from a CHO cell line were also analyzed and briefly shown to be significantly different from the murine cell line product. Taken together, the results suggest that the CE coupled to high resolution FT-MS can be one of the effective tools for in-process monitoring, as well as for final product characterization.
Introduction
Biotherapeutics, a class of pharmaceuticals which is primarily comprised of glycoproteins, are used for a wide variety of clinical indications, including inflammatory and immunomodulatory disorders, such as rheumatoid arthritis, multiple sclerosis, and cancer. Unlike typical small molecules, the structural elucidation of biotherapeutics, especially glycoproteins, and their manufacture and control is a difficult task. First, glycoprotein pharmaceutical agents are significantly more complex in their structure than small molecules. Second, and more importantly, while the active pharmaceutical ingredient in a typical small molecule therapeutic is one or, at most, several forms, a glycoprotein therapeutic can be a mixture of tens or hundreds or more of individual forms.
Analysis of intact glycoproteins to survey individual forms in a sample is typically conducted using isoelectric focusing1. Since the introduction of capillary electrophoresis coupled to mass spectrometry (CE-MS) in the late 1980s, advances in electrophoretic separations and the development of accurate high mass resolution MS has greatly improved the characterization of complex biological molecules, including intact glycoproteins2. High resolution separations coupled to high resolution MS has the potential to be a powerful tool for the analysis and quantitation of individual intact glycoforms of glycoproteins. For example, CE-MS can be used for rapid characterization of glycoprotein properties without the need for laborious sample preparation3–6. Indeed, Neusuβ and coworkers recently characterized different isoforms of erythropoietin, including oxidated and acetylated variants of the glycoforms, as well as other glycoproteins, using a CE/qTOF-MS with a mass resolution of 10,000 to 12,0007–10.
To move beyond composition to specific glycan structures, determination of released glycans is necessary. Various methods have been applied for the analysis of labeled glycans including hydrophilic interaction LC11 and capillary electrophoresis12. These separation techniques also provide quantitative analysis and can be performed in combination with exoglycosidase digestion, leading to the determination of monosaccharide linkages13, 14. In addition, mass spectrometry can be applied, with infusion or with separation,15 on a mixture of unlabeled glycans16, or labeled by, for example, permethylation17, 18. In the case of glycoproteins with multiple glycosylation sites, the assignment of individual sites is typically performed after proteolytic digestion followed by LC-MS19.
Due to the potential glycan complexity, we sought to build upon the above studies to (1) develop a rapid, robust and reproducible CE-FT-MS methodology that is amenable to very high mass resolution (55,000) of intact glycoproteins while maintaining high throughout; (2) combine the accuracy and resolution of this technology with glycan and glycopeptides analysis; and (3) assess the capability of the technology to determine differences between materials derived from different cell lines. For our studies, we selected the alpha subunit of recombinant human chorionic gonadotrophin (r-αhCG) 20, obtained from a murine cell line, as a representative example of a challenging glycoprotein therapeutic. The results of r-αhCG from this cell line were then compared to a CHO cell line, revealing significant differences It is shown in this work that the high resolving power of CE/FT-MS allows detection of over 60 glycoforms. The method is demonstrated to be a powerful tool for profiling of intact glycoproteins.
Experimental
Recombinant-αhCG
Recombinant-αhCG, expressed in a mouse cell line, was obtained from Sigma-Aldrich (St. Louis, MO). The protein was dissolved in 50 mM ammonium bicarbonate (final protein concentration 4 µg/µL), aliquoted and stored at −80 °C. Recombinant-αhCG, expressed in a CHO cell line, was obtained from Feldan Bio (Hamilton, NJ). Due to the large amount of sucrose and phosphoric acid in the latter sample, the protein required purification. A 50 µg sample of r-αhCG from the CHO cell line was dissolved in 200 µL of water and desalted using a Microcon Ultracel YM-3 centrifugal filter device (Millipore, Billerica, MA). The filter was washed twice with water, followed by 50 mM ammonium bicarbonate solution. The solution was concentrated to a final volume of 20 µL, leading to an estimated concentration of 2.5 µg/µL and stored at −80 °C.
Chemicals
CE-MS System
The home-made CE-MS system, schematically shown in Figure S1, consisted of a 20 cm long, 50 µm i.d., 360 µm o.d., PVA coated separation capillary (Agilent Technologies, Santa Clara, CA) attached to a pressurized liquid junction interface. The reservoir, containing background electrolyte at the injection end of the capillary, was fitted with an HPLC PEEK union (Upchurch, Oak Harbor, WA) to allow sample injection while providing airtight connection during separation. The second reservoir was connected to one arm of a liquid junction cross made from a polypropylene block. The additional three arms, were connected to (i) the separation capillary, (ii) a syringe to allow rapid flushing of the intercross volume and (iii) a 3.5 cm long, 50 µm i.d., 280 µm o.d. stainless steel ESI needle (New Objective, Woburn, MA). All machined plastic parts of the CE system that came in contact with liquids were made of highly chemical resistant polypropylene to prevent leaching into the MS. Reservoirs, made airtight using silicon O-rings, were connected to a chamber pressurized with nitrogen from a gas cylinder. A needle valve was used to control the pressure, which was monitored using a digital manometer (model DM8215, MSC Direct, Melville, NY). In a typical experiment, the pressure was maintained at 10 cm of H2O (~1 kPa), resulting in a flow rate at the ESI tip of roughly 200 nL/min. Since both reservoirs were maintained at the same pressure, there was no hydrodynamic flow in the separation capillary. The background electrolyte reservoir was equipped with a platinum electrode while the ESI voltages was applied directly to the ESI metal tip. To provide independent control of the separation and ESI voltage, two high voltage power supplies (CZE1000A, Spellman, Hauppauge, NY) were employed. A 50 MOhm resistor was added to the ESI electrical circuit to provide sufficient current drain to maintain the ESI voltage constant.
The CE-MS system was coupled to an LTQ-FT MS (Thermo Scientific) using a PicoView interface (New Objective) that allowed precise positioning of the ESI tip in front of the MS orifice. The LTQ-FT MS was operated in the MS only mode using a combination of a low resolution LTQ MS scan, followed by a high resolution FT scan in a limited mass range window. The target number of ions in the FT cell was set at 106, and the mass resolution of the FT MS was estimated to be roughly 55,000 at m/z = 1800.
Intact r-αhCG was separated using 2% acetic acid (pH = 2.5) as the background electrolyte with 8 kV as separation voltage. Samples were injected hydrodynamically with a height difference of 10 cm for 30 seconds. The injected amount was estimated to be roughly 100 ng, i.e. 5 pmol of the intact protein (4 µg/µL). The ESI solution consisted of 2% (v/v) acetic acid in 20 % (v/v) aqueous/acetonitrile solution.
See Supplementary Material for detailed description of the following methods: Deglycosylation and Analysis of Released Glycans, Trypsin Digestion of r-αhCG Expressed in a Murine Cell Line and LC-MS Analysis of r-αhCG Tryptic Digest.
Data Analysis
Accurate masses of intact glycoforms from CE-MS were calculated as follows. First, the r-αhCG sequence was converted to its elemental composition and corrected for the presence of 5 disulfide bridges, i.e. subtraction of 10 hydrogens. Then, masses of glycoproteins were generated by adding the elemental composition of individual glycans, followed by calculation of isotopic distribution using Protein Prospector 21. The mass of the most intense isotope in the isotopic cluster was used to confirm the correct assignment of a particular structure to the experimental mass with a mass tolerance of ±50 ppm. Average rather than monoisotopic masses are reported throughout in the manuscript. A set of in-house developed perl scripts was used to assign glycan compositions, match glycans to intact protein glycoforms and calculate theoretical combinations of glycans corresponding to a particular intact protein glycoform. Since glycan structures were not determined, glycans are referred to as: HexNAc, Hexose, Sialic acid (NeuAc) and sulfated glycan (+SO4).
Abundances of individual intact protein glycoforms were estimated from the data acquired using the linear ion trap with Qualbrowser (Thermo Scientific) based on peak areas derived from extracted ion electropherograms on the 9+ charge state of m/z values calculated with the average mass with the mass tolerance of ±0.5 Da. Similarly, abundances of glycopeptides were determined using the same method but with average masses for the 3+ and 4+ charge states.
Results and Discussion
The aim of this work was to develop a rapid method for profiling intact glycoproteins based on high resolution capillary electrophoretic separation coupled to high mass resolution FT mass spectrometer and apply this method to analysis of recombinant human chorionic gonadotrophin (r-αhCG) produced in a murine cell line. The results are then briefly compared to the glycoforms produced from a CHO cell line. A second goal was to integrate glycan and glycopeptide analysis to identify possible glycosylation structures for individual glycoforms. The power and effectiveness of the CE/MS high resolution method are demonstrated.
Intact Protein Analysis
A CE/MS system was constructed in-house in which the CE was coupled to an LTQ-FT mass spectrometer using a low-volume pressurized liquid junction interface (Figure S1). The interface provided rapid analysis, preserved the high resolution of the CE and allowed independent tuning of the separation and ESI conditions. The high mass resolution of the FT - MS allowed direct determination of the charge states of ions and accurate mass measurement of the intact protein glycoforms. To provide high resolution and reproducible separation, interaction of analytes with the capillary wall was prevented by employing polyvinylalcohol (PVA) as a permanent capillary coating. The neutral coating also suppressed the electroosmotic flow. In addition, unlike a dynamic coating, the permanent coating did not require regeneration after analysis.
In preliminary experiments using CE/LTQ-MS without the FT, in the m/z range of 1000–3000, it was found that most r-αhCG protein glycoforms were observed in the 8+, 9+ and 10+ charge states, with the 9+ charge state generally displaying the highest intensity. Based on the results from these initial experiments, the m/z range was restricted to 1400–2000 upon transfer of the method to the LTQ-FT-MS. The high resolving power of the FT-MS in the limited mass range allowed the accurate intact protein mass to be determined without the requirement to analyze multiple charge states. Restricting the m/z window in the FT-MS had a number of additional benefits including an increase in the acquisition speed and sensitivity of the analysis.
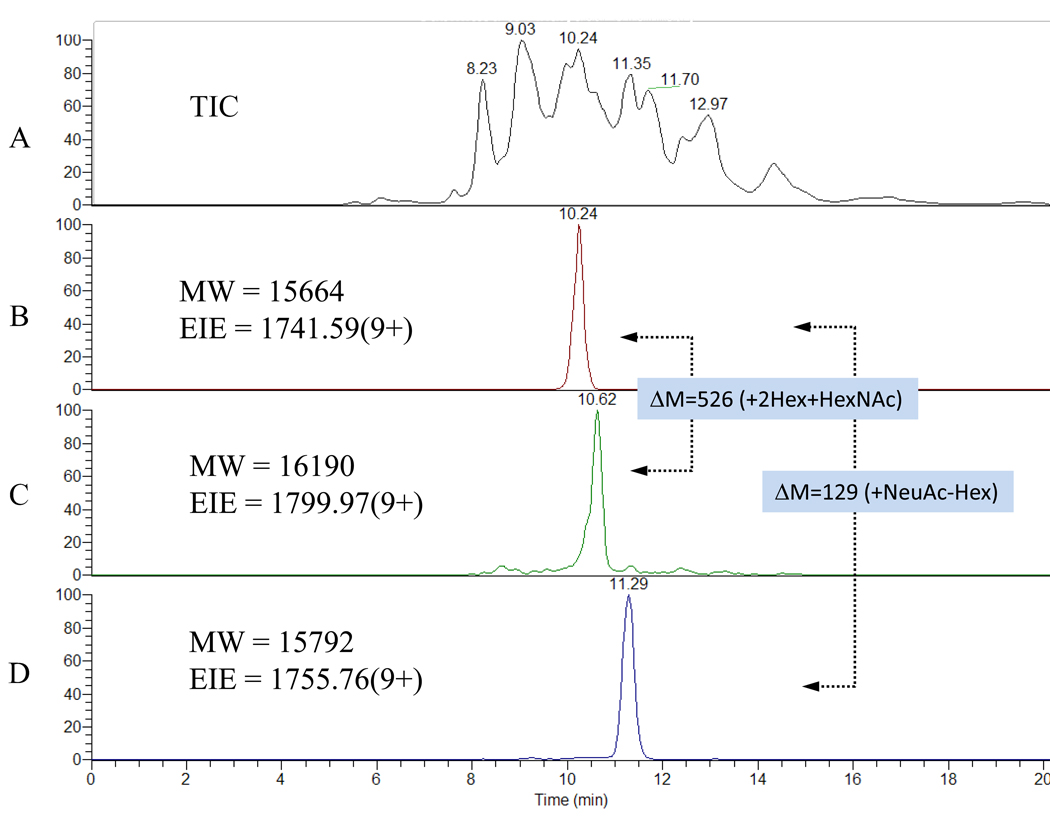
Figure 1 shows the total ion electropherogram for r-αhCG derived from the murine cell line, as well as examples of extracted ion electropherograms (EIE) for three different glycoforms. A background electrolyte consisting of 2% acetic acid (pH=2.5) was found to lead to high resolution separation by CE in the rapid analysis time of under 20 minutes. The electrophoretic widths for EIE peaks at half-height were only 12 sec, further demonstrating the resolving power of CE. Since each peak consisted of multiple isomeric forms, the actual peak widths of individual species were likely even narrower.
Figure 1. Demonstration of the high resolution separation of intact r-αhCG derived from a murine cell line using CE/MS.
A) Total ion current, (B)–(D) selected extracted ion electropherograms. See text for more details.
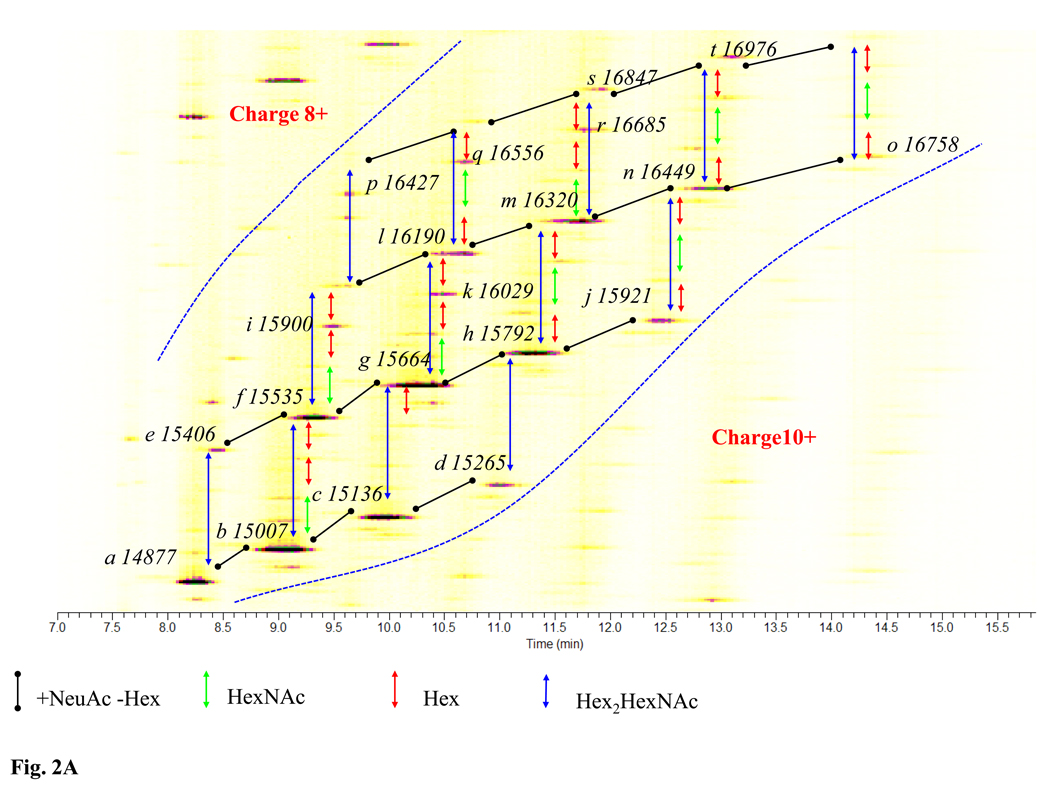
Figure 2A presents an overall separation pattern of CE-MS for r-αhCG in the form of a heat map to demonstrate the high number of glycoforms that can be observed. The twenty most intense forms are labeled with their corresponding masses, and the differences in glycan composition are highlighted using color-coded arrows. Due to the broad dynamic range of the amounts for individual glycoforms, only the abundant forms can be clearly observed. In order to reveal the true complexity of the r-αhCG glycoforms, Figure 2B shows the separation pattern plotted on a log intensity scale to emphasize lower abundance forms. The individual bands corresponding to the glycoforms with the same number of sialic acids (listed in the figure) are connected with a dashed line. The figure also shows the distribution of glycoforms in different charge states separated by the dotted lines.
Figure 2. CE/MS analysis of r-αhCG produced from a murine cell line.
A) Annotation of mass differences between high intensity glycoforms are indicated. High intensity glycoforms are labeled with a letter (Table 1) and an average molecular weight; B) log-intensity plot of separation. Numbers indicate the number of sialic acids for the glycoforms.
Dotted lines show approximate boundaries for 8+,9+ and 10+ charge states.
Mass measurement by FT-MS allowed direct determination of the exact molecular mass of the observed glycoforms. Table 1 lists the masses of the 20 most abundant peaks, labeled in Figure 2A, along with a summary of their glycan compositions, relative migration times and relative peak areas, which will be discussed later. To assign glycan compositions accurately to individual glycoforms, deglycosylated intact r-αhCG (PNGase F) was analyzed by CE/MS, leading to an average experimental molecular mass of 10,196 Da, which was within the expected mass error of the predicted value (data not shown). It should be noted that the compositions in Table 1 correspond to the sum of glycan structures on the two glycosylation sites.
Table 1.
Twenty most abundant glycoforms for r-αhCG from murine cell line determined by CE/MS and their peak area measurements obtained in three CE/MS runs.
| Symboli | Average Mass, Da |
Composition (HexNAc, Hex, NeuAc) |
Rel. migration timeii |
Rel. areaiii Run 1 % |
Rel. areaiii Run 2 % |
Rel. areaiii Run 3 % |
CV % |
|---|---|---|---|---|---|---|---|
| a | 14,877 | 9,14,2 | 0.92 | 54.1 | 54.2 | 61.5 | 7.4 |
| b | 15,006 | 9,13,3 | 1.00 | 100.0 | 100.0 | 100.0 | -- |
| c | 15,135 | 9,12,4 | 1.09 | 59.8 | 56.6 | 57.8 | 2.8 |
| d | 15,265 | 9,11,5 | 1.20 | 17.4 | 17.2 | 17.8 | 1.6 |
| e | 15,405 | 10,16,2 | 0.94 | 12.8 | 12.8 | 13.0 | 1.0 |
| f | 15,534 | 10,15,3 | 1.03 | 56.1 | 57.8 | 52.6 | 4.8 |
| g | 15,663 | 10,14,4 | 1.12 | 93.8 | 86.9 | 86.9 | 4.5 |
| h | 15,792 | 10,13,5 | 1.23 | 56.3 | 57.8 | 58.2 | 1.7 |
| i | 15,899 | 11,16,3 | 1.05 | 12.5 | 11.6 | 13.0 | 5.5 |
| j | 15,921 | 10,12,6 | 1.35 | 21.5 | 18.7 | 21.0 | 7.4 |
| k | 16,028 | 11,15,4 | 1.15 | 17.3 | 16.4 | 15.7 | 4.7 |
| l | 16,190 | 11,16,4 | 1.17 | 29.2 | 26.8 | 22.5 | 11.0 |
| m | 16,319 | 11,15,5 | 1.28 | 46.1 | 44.9 | 42.4 | 4.3 |
| n | 16,449 | 11,14,6 | 1.40 | 30.3 | 30.9 | 30.5 | 1.0 |
| o | 16,578 | 11,13,7 | 1.56 | 11.4 | 12.5 | 11.6 | 4.8 |
| p | 16,427 | 12,18,3 | 1.06 | 8.3 | 7.0 | 8.1 | 8.7 |
| q | 16,556 | 12,17,4 | 1.17 | 13.6 | 14.0 | 13.6 | 1.7 |
| r | 16,685 | 12,16,5 | 1.28 | 16.8 | 17.0 | 15.9 | 3.5 |
| s | 16,847 | 12,17,5 | 1.29 | 12.4 | 11.8 | 11.4 | 4.0 |
| t | 16,976 | 12,17,5 | 1.42 | 15.6 | 15.5 | 14.3 | 4.7 |
Symbol as used in Figure 2A
Relative migration time with respect to peak b
Peak areas normalized to the abundance of the most intense glycoform (peak b)
Upon comparison of the calculated glycan compositions with the results in Figure 2, it can be seen that, in agreement with others8, there is a clear separation pattern in terms of the number of sialic acids per glycoform, as a result of the negative charges provided by the sialic acids. To illustrate the separation power of capillary electrophoreisis, peaks labeled g and h in Fig. 2A, differing by an addition of one sialic acid and subtraction of a hexose, i.e. ΔM=129 Da, were found to be baseline separated by CE, see Figures 1B and D. Moreover, glycoforms with the same number of sialic acids, such as peaks g and l in Figure 2A with ΔM =526 Da, corresponding to the addition of two hexoses and one HexNAc, could also be resolved, see Figure 1B and C. Even more interesting is the partial separation of peaks k and l with ΔM=162 Da, which differ by only one hexose8. These examples clearly illustrate the resolving power of capillary electrophoresis, with separations even for forms of the same charge but small differences in neutral glycans on the intact protein. Finally, it should be noted that since all analyzed r-αhCG glycoforms that differed in the number of sialic acids were at least partially resolved by capillary electrophoresis, the ESI ionization process did not lead to loss of sialic acid.
Relative Abundance of r-αhCG Glycoforms
Analysis of the relative abundance of individual glycoforms is one of the potential applications of CE/MS for intact protein profiling. We focused first on evaluation of the reproducibility of the relative abundance of individual glycoforms since such forms could be easily matched based on their accurate masses. We evaluated the consistency of glycoform relative quantitations measurements by comparing peak areas for selected forms determined from the extracted ion electropherograms. It should be noted that, due to potential differences in ionization efficiencies of individual glycoforms, the relative peak areas may not correspond directly to in-solution abundances of glycoforms. In addition, the distribution of the electrospray charge states for different glycoforms may change with the mass and/or the number of sialic acids, further complicating quantitative analysis. Thus, samples of intact forms should be compared on a relative basis, e.g. from batch to batch or within a process.
The CE/MS analysis of the mouse-derived intact r-αhCG glycoforms was repeated 3 times, and the peak areas of the 20 most intense peaks, using linear ion trap data, were measured and compared, see Experimental Section. In order to prevent a potential bias during the sample introduction, all analyses were performed using hydrodynamic injection, and relative run-to-run comparisons were determined. Finally, repeatability rather than reproducibility, i.e. a replicate analysis of the same sample, by the same operator, on the same instrument and the same day, was performed. The results, including the glycoform mass, glycan composition, relative migration time normalized to the glycoform with the highest intensity, and peak areas normalized again to the most intense glycoform, are summarized in Table 1. It can be seen that the average relative coefficient of variance of the peak areas is less than 10% for all but one glycoform, a relativity low CV for such an analysis. Moreover, it is expected that further improvements in sample injection and automation of operation would reduce the CV values to even lower levels. In addition, inclusion of internal standards would allow direct estimation of the abundance of a specific glycoforms. Nevertheless, even with the simple experiments conducted in this work, the variation in quantitation is quite good.
Assuming equal ionization efficiencies, it was estimated, from the sum of all peak areas, that the 20 highest intensity glycoforms accounted for over 90% of the total amount of the glycoprotein. The abundance of the additional roughly 40 different glycoforms, representing the remaining 10%, were not estimated. However, if the abundances of these forms were distributed uniformly, the average of each species would approximate to less than 1% of the mass of the total glycoprotein.
Analysis of the Released Glycans
We next examined of individual glycan structures, after enzymatic release, to better understand the potential structures of the intact glycoforms. The LC-MS/MS analysis of the 2-AB labeled N-glycan pool (see Supplementary Material) again revealed extensive glycosylation complexity of r-αhCG expressed in the murine cell line, Figure S2A. The data from the glycan analysis, including MS/MS spectra and exoglycosidase digestion, was used to construct a list of structures for the higher as well as the lower abundance glycans (Table S1). The compositions of many of the species were tentatively assigned as those commonly found in complex-type glycans of the form HexNAcNHexN+1 with variable numbers of NeuAc. Fragments were linked to the most likely structure based on the minimum number of bond cleavages. Interestingly, a number of glycans containing the terminal Gal-α1–3–Gal structure, an important immunogenic epitope were found. Sulfated glycans as well as pentasialylated species were also identified. See Supplementary Material for more details.
Glycopeptide Analysis
Since recombinant-αhCG is a glycoprotein with than two glycosylation sites, the analysis of the intact glycoprotein will determine the overall glycan composition, but not the specific glycan structures attached to individual sites. Analysis at the glycopeptide level is necessary to associate a particular glycan with a specific glycosylation site. A theoretical tryptic digest of r-αhCG revealed that the glycosylation sites were found on two separate tryptic fragments, allowing a straightforward determination of the glycans associated with a particular site.
A tryptic digest of r-αhCG was analyzed by nano LC-MS/MS in the data dependent mode on the LTQ MS. Retention times of glycopeptides were first determined by analysis of the deglycosylated tryptic peptides which generally have similar retention times to the corresponding glycopeptides in reversed phase LC19. Compositions of glycans associated with a specific glycosylation site are presented in Table S2, based on the glycopeptide molecular weight and the list of identified glycans in Table S1. MS/MS spectra of glycopeptides were analyzed to confirm the assignment of a given glycan structure.
Peak areas for glycopeptides were calculated from extracted ion chromatograms for masses corresponding to all determined glycans with >1% abundance in Table S2 (a total of 18). The peak areas were normalized to represent the percent abundance of a specific glycopeptide. As discussed for the intact glycoprotein, peak areas represent an approximation of the real abundances of glycopeptides since ionization efficiencies and distribution of charge states will likely be dependent on the specific glycopeptide. As seen in Table S2, the majority of glycans can be found on both glycosylation sites, though the differences in abundance can be substantial. For example, the glycan with composition HexNAc5Hex8NeuAc represents roughly 30% of all forms for glycopeptide NVTSESTCCVAK (site N76) but only 2% for VENHTACHCSTCYYHK (site N102). In addition to the glycopeptides, resulting from the 18 most abundant glycans, potential glycopeptide forms with sulfated or pentasialylated glycans (Table S2) were searched for; however, these structures were not found, likely due to their low abundances and low ionization efficiencies.
Analysis of Combined Data
Data from all three levels of analysis, i.e. intact protein, released glycan and glycopeptide, were next combined to provide a detailed characterization of r-αhCG and to verify data consistency. This combination increased the information available from the profiling results of the intact glycoforms. In this analysis, it was assumed that structures identified for the released glycans were the only species that contributed to the glycoforms of the protein. In order to confirm compositions of suggested glycoforms, structures of all observed glycans, including those below 1%, were used to generate a theoretical list of potential protein glycoforms. Assuming that both glycosylation sites are occupied and that the same glycan could be attached to both sites, the total number of combinations was calculated. A perl script was written to enumerate all combinations of glycans and calculate the masses and total glycan compositions corresponding to all theoretical glycoforms. A total of 528 combinations of the glycans were calculated. If only one site were occupied, 32 additional glycoforms or a total of 560 were assumed to be possible. . However, out of all 560 glycoforms, only 286 glycoforms would have a unique glycan composition that could be distinguished by mass alone. Table S3 lists all theoretical glycoform compositions and their masses.
Next, the calculated total glycan compositions were compared with the structures of all 60 glycoforms derived from the CE-MS analysis of the intact glycoproteins (Table S3). The experimentally determined compositions were compared to the list of 560 theoretical structures derived from observed glycans. It was found that the majority of high intensity protein glycoforms could indeed be matched to theoretical glycan compositions. For example, the intact protein glycoform labeled d in Figure 2A, with an average mass of 15,265 Da and total glycan composition of HexNAc9Hex11NeuAc5, could be associated with a single combination of glycans HexNAc4Hex5NeuAc2 and HexNAc5Hex6NeuAc3 (Table S1). However, it is important to note that for many of the intact glycoforms, more than one combination of glycan structures could to match the total glycan composition. For example, the intact protein glycoform with a mass of 16,556 Da, peak q in Figure 2A, could be associated with 8 different combinations of observed glycans with the same total glycan composition of HexNAc12Hex17NeuAc4. This result is in part due to the complexity of r-αhCG glycosylation in the murine cell line. Finally, not all observed 60 intact glycoforms could be matched to a combination of glycans.
In the hope of determining which combination(s) of glycans actually corresponded to observed glycoforms, when there was more than one possibility, we combined the data obtained from the CE-MS analysis of the intact r-αhCG forms with site specific glycan information from the glycopeptide analysis. For example, the r-αhCG glycoform with a glycan mass composition HexNAc9Hex14NeuAc2 (Figure 2A, band a, mass 14,877 Da) could be matched to two combinations of glycans: a) HexNAc4Hex6NeuAc and HexNAc5Hex8NeuAc and b) HexNAc5Hex7NeuAc2 and HexNAc4Hex7. Since the intact glycoform with a molecular mass of 14,877 Da was one of the most abundant forms, see Table 1, it is expected that the glycans and glycopeptides associated with this glycoform should be also highly abundant. On the other hand, since the total relative amount of glycan with the composition HexNAc4Hex7 is below one percent (Table S1), this glycan was not considered to contribute to the highly abundant glycoform a in Figure 2A. As a result, a single combination of glycans was considered for the site specific assignment in this case. For example, the glycan HexNAc4Hex6NeuAc is likely present at site N102 because there is a 22% abundance compared to 4.5% on the other site (Table S2). Similarly, HexNAc5Hex8NeuAc is likely present at site N76 because its relative abundance is 30% compared to only 1.7% on site N102. Unfortunately, such site specific glycosylation assignment could only be performed for a limited number of glycoforms because most of the high abundant forms were associated with more than one combination of glycans. Nevertheless, the determination of the glycan and glycopeptide compositions provided useful additional information to the CE/MS profiling of the intact protein glycoforms.
We also briefly examined glycan compositions of low abundant intact protein glycoforms migrating at longer times in the CE run, suggesting the presence of large numbers of sialic acids (up to 9) in the intact glycoproteins, see Figure 2B (also Table S3). With only two glycosylation sites in r-αhCG, one site must, presumably, contain at least 5 sialic acids, if the total for the the glycoform were 9 sialic acids. Importantly, several structures with 5 sialic acids were observed in the analysis of released glycans (Table S1). More detailed studies would be needed to assign additional glycoform structures for this very complex recombinant protein.
Analysis of r-αhCG Expressed in CHO Cell Culture
Having examined the glycoforms of r-αhCG from a murine cell line, we briefly applied CE/LTQ-FT-MS to characterize intact glycoforms of the same protein expressed in CHO cells. It was expected that different cell lines and culture conditions could significantly alter the glycosylation of r-αhCG. Recombinant-αhCG obtained from CHO cells was supplied in a formulated form containing high concentrations of phosphoric acid and sucrose, substances that are incompatible with CE analysis. To overcome this sample matrix effect, the protein was purified from its excipients using molecular weight cutoff filters (see Experimental Section).
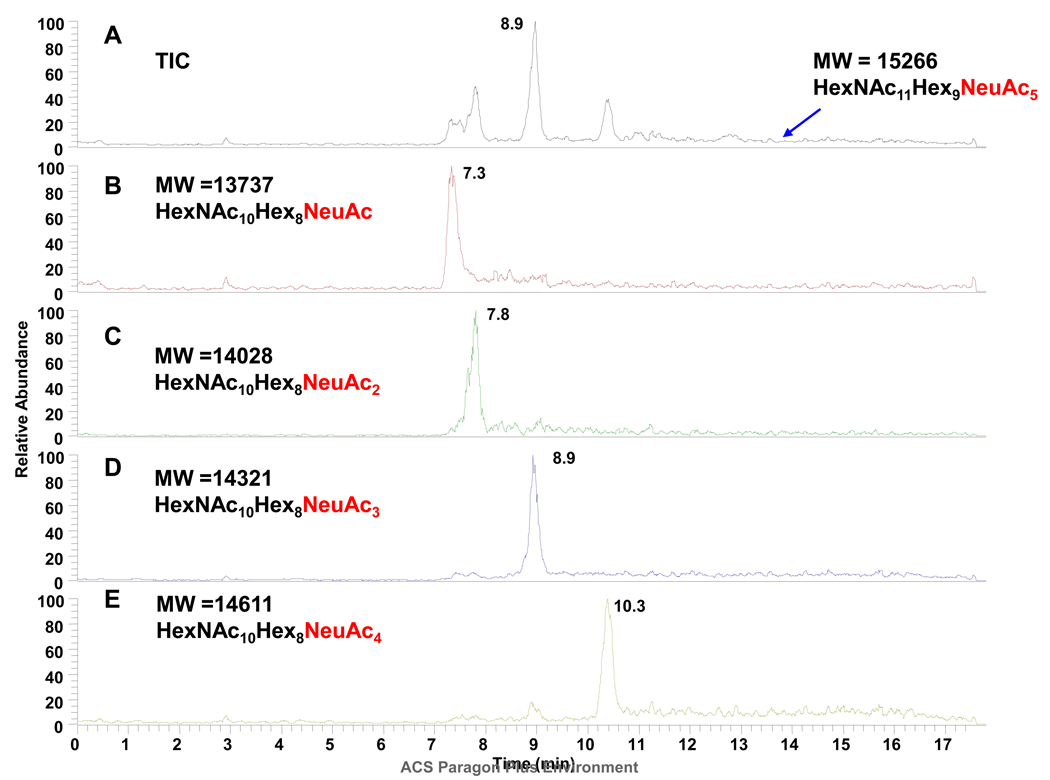
The intact forms of the desalted CHO sample were separated and analyzed by CE-MS (Figure 3). We found that the glycosylation of intact r-αhCG produced in a CHO cell line was indeed far less complex (Figure 3A) than the protein expressed in mouse cells (Figure 1A), as seen in the smaller number of electrophoretic peaks observed. Using the high mass accuracy of the FT-MS, compositions of major intact protein glycoforms were determined, see Table 2. The glycosylation was found to be limited to only several forms primarily differing in the number of sialic acids. Based on the separation pattern and high mass accuracy, glycoforms with up to 5 sialic acids were derived from the CHO-derived product, in contrast to glycoforms from the mouse cell line which contained up to 9 sialic acids. The analysis further revealed that CHO cells derived r-αhCG contained mainly bi- and triantennary glycans, with up to three sialic acids.
Figure 3. CE/MS analysis of r-αhCG produced in CHO cells.
A) Total ion electropherogram. (B)–(E) extracted ion electropherograms for individual high level glycoforms. Other conditions as in Figure 2.
Table 2.
Major glycoforms observed by CE/MS analysis of r-αhCG produced in a CHO cell line.
| Glycoprotein MW, Da |
Glycan mass | HexNAc | Hex | NeuAc | Abundance |
|---|---|---|---|---|---|
| 12,404 | 2208 | 4 | 5 | 2 | 10.2% |
| 13,446 | 3250 | 8 | 10 | 0 | 2.5% |
| 13,737 | 3541 | 8 | 10 | 1 | 11.5% |
| 14,028 | 3832 | 8 | 10 | 2 | 25.1% |
| 14,320 | 4124 | 8 | 10 | 3 | 32.1% |
| 14,611 | 4415 | 8 | 10 | 4 | 10.1% |
| 14,685 | 4489 | 9 | 11 | 3 | 2.6% |
| 14,976 | 4780 | 9 | 11 | 4 | 3.8% |
| 15,266 | 5070 | 9 | 11 | 5 | 2.1% |
Comparison of the CE-MS profiling for CHO cell derived r-αhCG with those for murine derived r-αhCG indicates that there are dramatic differences between the two cell lines, emphasizing the importance of selection of the proper cell line (and conditions) to achieve the desired glycosylation pattern. Interestingly, based on calculated compositions, one of the high abundant glycoforms (molecular mass of 12,404 Da) from the CHO cell line could be matched to the glycan composition HexNAc4Hex5NeuAc2, likely representing a single N-linked glycan with the other site on the protein unoccupied. This assignment is consistent with the migration position of this glycoform, as it migrates between the form with two glycans with one sialic acid (13,373 Da, HexNAc10Hex8NeuAc) and the form with two glycans with two sialic acids (14,028 Da, HexNAc10Hex8NeuAc2). The results illustrate that intact glycoprotein analysis by CE-MS can provide a rapid means for profiling changes in glycosylation. In summary, it was found, as expected, CHO derived proteins have much simpler glycosylation patterns than those derived from the murine cell culture.
Conclusions
We have described in this paper the use of high resolution CE/LTQ-FT-MS for the profiling of the intact glycoforms of r-αhCG produced in a murine cell line, followed by a brief comparison to the product produced in a CHO cell line. The studies demonstrate that the high resolution CE/MS method can be rapid and information rich, although the direct assignments of glycosylation sites might be challenging due to a high number of possible isomeric structures. Nevertheless, the studies suggest that CE/MS can be an important tool for rapid assessment of the recombinant product quality either for product release or for in-process control. In future studies, additional glycoproteins can be examined using this approach with the FT-MS or an alternative high resolution MS instrument such as the Orbitrap or newer high resolution qTOFs. Nevertheless, in agreement with other studies7, the effectiveness of the CE/MS method in its role in glycoprotein characterization has been demonstrated. Finally, high resolution techniques, as described in this paper, can also play a role in assessing comparability of a glycoprotein therapeutic a biosimilar to an innovator product22. Unlike analysis using cIEF, commonly employed for assessment of intact glycoform abundance, CZE can separate glycoforms not only based on charge but also on size and shape. In addition, while not observed in this study, CE/MS has the potential to determine glycosylation along with modifications such as oxidation or protein backbone truncation.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Ian Parsons for his contributions in scientific discussions. This research was support in part by NIH GM15847 (BLK). Contribution No. 941 from the Barnett Institute.
References
- 1.Krull IS, Kazmi S, Zhong H, Santora LC. Methods Mol Biol. 2003;213:197–218. doi: 10.1385/1-59259-294-5:197. [DOI] [PubMed] [Google Scholar]
- 2.Smith RD, Udseth H. Nature. 1988;331:639–640. doi: 10.1038/331639a0. [DOI] [PubMed] [Google Scholar]
- 3.Kelly JF, Locke SJ, Ramaley L, Thibault P. J Chromatogr A. 1996;720:409–427. doi: 10.1016/0021-9673(94)01197-4. [DOI] [PubMed] [Google Scholar]
- 4.Yeung B, Porter TJ, Vath JE. Anal Chem. 1997;69:2510–2516. doi: 10.1021/ac9611172. [DOI] [PubMed] [Google Scholar]
- 5.Demelbauer UM, Plematl A, Kremser L, Allmaier G, Josic D, Rizzi A. Electrophoresis. 2004;25:2026–2032. doi: 10.1002/elps.200305936. [DOI] [PubMed] [Google Scholar]
- 6.Amon S, Zamfir A, Rizzi A. Electrophoresis. 2008;12:2485–2507. doi: 10.1002/elps.200800105. [DOI] [PubMed] [Google Scholar]
- 7.Neususs C, Demelbauer U, Pelzing M. Electrophoresis. 2005;26:1442–1450. doi: 10.1002/elps.200410269. [DOI] [PubMed] [Google Scholar]
- 8.Balaguer E, Demelbauer U, Pelzing M, Sanz-Nebot V, Barbosa J, Neusub C. Electrophoresis. 2006;27:2638–2650. doi: 10.1002/elps.200600075. [DOI] [PubMed] [Google Scholar]
- 9.Balaguer E, Neususs C. Anal Chem. 2006;78:5384–5393. doi: 10.1021/ac060376g. [DOI] [PubMed] [Google Scholar]
- 10.Sanz-Nebot V, Balaguer E, Benavente F, Neususs C, Barbosa J. Electrophoresis. 2007;28:1949–1957. doi: 10.1002/elps.200600648. [DOI] [PubMed] [Google Scholar]
- 11.Domann PJ, Pardos-Pardos AC, Fernandes DL, Spencer DI, Radcliffe CM, Royle L, Dwek RA, Rudd PM. Proteomics. 2007;7 Suppl 1:70–76. doi: 10.1002/pmic.200700640. [DOI] [PubMed] [Google Scholar]
- 12.Guttman A. Nature. 1996;380:461–462. doi: 10.1038/380461a0. [DOI] [PubMed] [Google Scholar]
- 13.Edge CJ, Rademacher TW, Wormald MR, Parekh RB, Butters TD, Wing DR, Dwek RA. Proc Natl Acad Sci USA. 1992;89:6338–6342. doi: 10.1073/pnas.89.14.6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim YG, Kim SY, Hur YM, Joo HS, Chung J, Lee DS, Royle L, Rudd PM, Dwek RA, Harvey DJ, Kim BG. Proteomics. 2006;6:1133–1142. doi: 10.1002/pmic.200500275. [DOI] [PubMed] [Google Scholar]
- 15.Ninonuevo M, An H, Yin H, Killeen K, Grimm R, Ward R, German B, Lebrilla C. Electrophoresis. 2005;26:3641–3649. doi: 10.1002/elps.200500246. [DOI] [PubMed] [Google Scholar]
- 16.Harvey DJ. J Mass Spectrom. 2005;40:642–653. doi: 10.1002/jms.836. [DOI] [PubMed] [Google Scholar]
- 17.Ciucanu I, Costello CE. J Am Chem Soc. 2003;125:16213–16219. doi: 10.1021/ja035660t. [DOI] [PubMed] [Google Scholar]
- 18.Ashline DJ, Lapadula AJ, Liu YH, Lin M, Grace M, Pramanik B, Reinhold VN. Anal Chem. 2007;79:3830–3842. doi: 10.1021/ac062383a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wuhrer M, Catalina MI, Deelder AM, Hokke CH. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;849:115–128. doi: 10.1016/j.jchromb.2006.09.041. [DOI] [PubMed] [Google Scholar]
- 20.Stenman UH, Tiitinen A, Alfthan H, Valmu L. Hum Reprod Update. 2006;12:769–784. doi: 10.1093/humupd/dml029. [DOI] [PubMed] [Google Scholar]
- 21. http://prospector.ucsf.edu/
- 22.Dove A. Nature Biotechnology. 2001;19:117–120. doi: 10.1038/84365. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.