Abstract
Metastasis is a critical event in the progression of head and neck squamous cell carcinoma (HNSCC). To identify microRNAs associated with HNSCC metastasis, 6 paired HNSCC cell lines with different metastatic potential were examined. Using microarrays, a panel of differentially expressed microRNAs was identified, including reduction of miR-138 in highly metastatic cells. Ectopic transfection of miR-138 suppressed cell invasion and led to cell cycle arrest and apoptosis. Knockdown of miR-138 enhanced cell invasion and suppressed apoptosis. Thus, our results suggested miR-138 acts as a tumor suppresser and may serve as a therapeutic target for HNSCC patients at risk of metastasis.
Keywords: HNSCC, microRNA, miR-138, metastasis, invasion
1. Introduction
Head and neck cancer, predominantly head and neck squamous cell carcinoma (HNSCC), is one of the most devastating diseases. The American Cancer Society estimated that more than 45,000 new cases of HNSCC were diagnosed in 2007, representing approximately 3% of all malignancies [1]. HNSCC exhibits frequent local/regional invasion and metastasis. Despite the improvements in surgery, radiotherapy and chemotherapy, the prognosis for HNSCC patients has not significantly improved for the past 3 decades. Improvement in patient survival requires better understanding of tumor invasion and metastasis so that aggressive tumors can be detected early in the disease process and targeted therapeutic interventions can be developed.
MicroRNAs (miRNAs) are a class of endogenous small non-coding RNAs that control the target gene expression at the post-transcriptional level. It is currently estimated that the human genome may have 800–1,000 miRNAs [2]. Although they account for only a minor fraction of the expressed genome, microRNAs are pivotal regulators of diverse cellular processes including proliferation, differentiation, apoptosis, survival, motility and morphogenesis. Several microRNAs have been functionally classified as proto-oncogenes or tumor suppressors and are aberrantly expressed in different cancer types including leukemia [3, 4], lymphoma [5], breast cancer [6, 7], colorectal cancer [8], lung cancer [9, 10], liver cancer [11, 12], and head and neck cancer [13–16]. Dysregulation (e.g., overexpression or loss of expression) of these “cancerous” microRNAs contributes to tumor initiation and progression by promoting uncontrolled proliferation, favoring survival, inhibiting differentiation and/or promoting invasive behavior [17, 18]. This study seeks to identify and validate the microRNA candidates that contribute to metastasis in HNSCC.
2. Materials and methods
2.1. Cell culture and transfection
Human HNSSC cell lines were used in this study (see Supplementary Table 1 for description on those cell lines). These cells were maintained in DMEM/F12 supplemented with 10% FBS, 100 U/mL penicillin and 100 μg/mL streptomycin (GIBCO) at 37°C in a humidified incubator containing 5% CO2. For functional analysis, miR-138 mimics, non-targeting miRNA mimics (Dharmacon), specific locked nucleic acid (LNA) inhibitor for miR-138 (anti-miR-138 LNA) and scrambled LNA probe (Exiqon) were used. The miR mimics and LNA inhibitors were transfected into the appropriate cells using DharmaFECT Transfection Reagent 1 (Dharmacon) according to the manufacturer’s instructions. In brief, cells were plated in 6 cm diameter cell culture dishes to 60% confluence. For each dish, 7.5 μl of miR-138 mimic (20 μM), or anti-miR-138 LNA (20 μM) and 6 μl of DharmaFECT Transfection Reagent were added into 750 μl of antibiotic-free opti-MEM medium (Invitrogen), separately, and then mixed together for forming the transfection complex. The transfection complex (100 nM) was added to cells and incubated for 24 h before replacing the medium.
2.2. Wound healing assay
Cell migration was measured using a wound healing assay as described previously [19]. In brief, cells were seeded in 12-well plates and cultured to confluence. Wounds of 1 mm width were created with a plastic scriber, and cells were washed and incubated in a serum-free medium. 24 hours after wounding, cultures were fixed and observed under a microscope. A minimum of 5 randomly chosen areas were measured.
2.3. In vitro cell invasion assay
The invasion assay was performed using a Cultrex 96-well membrane invasion assay kit (R&D Systems) as described previously [20]. Briefly, in day 1, 50 μl of 0.5 × BME coating solution was placed in each well of a top invasion chamber, and the cells were starved in serum-free medium. On day 2, the cells were harvested and seeded into the top chamber at 5 × 104 cells/well. 150 μl of serum-free medium were added to each well of the bottom invasion chamber. The device was assembled and incubated at 37°C in an incubator containing 5% CO2 for 24 h. On day 3, the cells in the top chamber were washed with PBS and transfected with desired miRNA reagents at a final concentration of 100 nM. Correspondingly, the medium in the bottom chamber was replaced with fresh DMEM/F12 medium containing 10% FBS. Forty-two hours after transfection, medium in both chambers were aspirated and each well was washed with 1× Washing Buffer. 150 μl of Cell Dissociation Solution/Calcein AM was added to the bottom chamber and incubated at 37 °C for 1 hour. Experiments were performed at least twice, separately, and run in quadruplicate. The top chamber was removed, and the bottom plate was measured at 485 nm excitation and 520 nm emission. The data was compared to the standard curve to determine the number of cells that have invaded. A separate standard curve was used for each cell type. The percentage of cell invasion was calculated as the number of the invaded cells divided by the number of the cells at the start of the assay.
2.4. MicroRNA microarray
Total RNA from HNSCC cell lines was isolated using a miRNeasy Mini Kit from Qiagen. The quality and quantity of the RNA samples were assessed by standard electrophoresis and spectrophotometer methods. Microarray analysis was performed by Genosensor Corporation (Tempe, AZ) based on the GenoExplorer microRNA Full Kit protocol. The GenoExplorer human miRNA array contains triplicated probesets representing approximately 900 microRNA, including both precursor microRNA and mature microRNA. Duplicated array assays were performed for each sample. Detectable probes were defined as probe signal intensity equal or above the signal threshold (array background + 2 × background standard deviation). Arrays were normalized based on global signal intensity. Differential miRNA expression was determined using a two-sided t-test on a single miRNA basis.
2.5. Real-time RT-PCR analysis
The relative expression level of miR-138 in HNSCC cell lines was determined based on a quantitative 2-step RT-PCR assay using mirVana™ qRT-PCR microRNA Detection Kit as per the manufacturer’s protocol (Ambion). The quantitative PCR was performed using iQ SYBR Green Supermix (Bio-Rad) in a BIO-RAD iCycler iQ real-time PCR detection system. Specific primer sets for miR-138 and U6 were obtained from Ambion. The relative expression level of miR-138 was determined using the 2−delta delta Ct analysis method [21], where U6 was used as an internal reference.
2.6. Flow cytometry-based apoptosis and cell cycle analysis
Cells were grown in 6-well plates to about 60% confluence and transiently transfected with the desired miRNA reagents at a final concentration of 100 nM. The cells were digested and collected after 48 h post-transfection, and washed with PBS twice. For cell apoptosis measurement, the cells were resuspended in 1× Binding Buffer, and 5 μl of Annexin FITC Conjugate, and 10 μl of Propidium Iodide Solution were added to each cell suspension, separately. For cell cycle analysis, the cells were resuspended in PBS and then fixed in ethanol at −20°C overnight. The cells were washed with PBS and resuspended in Staining Solution (50 μg/mL of propidium iodide, 1 mg/mL of RNase A, 0.1% Triton X-100 in PBS). The stained cells (1×105) were then analyzed with a flow cytometer (FACScalibur, Becton-Dickinson).
3. Results
3.1. microRNA profiling on HNSCC cell lines with different metastatic potential
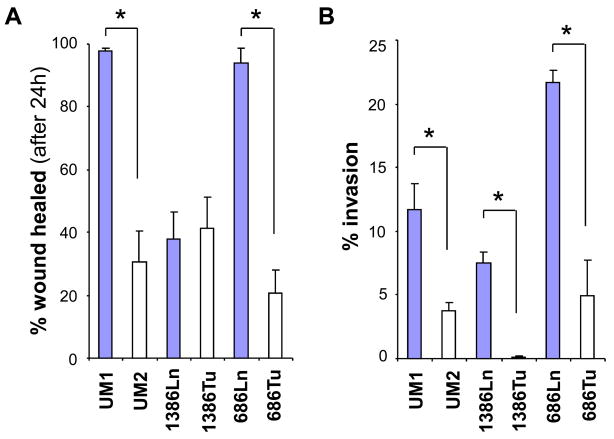
Six paired HNSCC cell lines (UM1/UM2, 1386Tu/1386Ln and 686Tu/686Ln) were assembled for microRNA profiling analysis. The UM1 and UM2 are paired cell lines with different metastatic potential that generated from a single patient with SCC of the tongue [19]. The 1386Tu/1386Ln and 686Tu/686Ln are paired cell lines generated from primary tumors and lymph node metastatic diseases from the HNSCC patients. To confirm the difference in metastatic potential of these paired cell lines, cell migration and cell invasion were measured with the wound healing assay and the transwell in vitro cell invasion assay, respectively. As showed in Figure 1A, UM1 and 686Ln exhibited faster wound healing ability when compared to their paired cell lines, UM2 and 686Tu, respectively, indicating enhanced cell migration. No statistical difference in wound healing was observed between 1386Ln and 1386Tu cells. As showed in Figure 1B, UM1, 1386Ln and 686Ln exhibited statistically significantly elevated invasions when compared to their paired cell lines, UM2, 1386Tu and 686Tu, respectively (p < 0.05).
Figure 1. Differential metastatic potentials in HNSCC cell lines.
The differences in cell migration and cell invasion of paired HNSCC cell lines (UM1/UM2, 1386Ln/Tu, and 686Ln/Tu) were evaluated with the wound healing assay (A) and the transwell in vitro cell invasion assay (B) as described in the Material and Methods section. The result represents 3 independent experiments with quadruplicates of each measurement. * indicates p < 0.05.
Microarray analysis was performed to identify the differentially expressed microRNA. As shown in Table 1, 10 miRNAs were downregulated, and 14 miRNAs were upregulated in at least two cell lines with more aggressive phenotype compared to their corresponding counterparts. Among those, reduced expression of miR-138 and enhanced expression of miR-566-pre were consistently observed in all 3 aggressive cell lines (UM1, 1386Ln and 686Ln).
Table 1.
Differentially expressed microRNA identified by microarray analysis *
| UM1 vs. UM2 | 1386Ln vs. 1386Tu | 686Ln vs. 686Tu | |||
|---|---|---|---|---|---|
| microRNA | fold diff. (UM1/UM2) | microRNA | fold diff. (1386Ln/Tu) | microRNA | fold diff. (686Ln/Tu) |
| miR-7 | 0.26 | miR-7 | 0.57 | miR-21 | 0.67 |
| miR-21 | 0.56 | miR-98 | 0.67 | miR-138 | 0.42 |
| miR-98 | 0.64 | miR-99b | 0.61 | miR-594-pre | 0.67 |
| miR-99b | 0.36 | miR-101 | 0.65 | ||
| miR-101 | 0.64 | miR-125a | 0.59 | ||
| miR-125a | 0.26 | miR-138 | 0.59 | ||
| miR-138 | 0.40 | miR-193 | 0.54 | ||
| miR-193 | 0.60 | miR-224 | 0.63 | ||
| miR-224 | 0.27 | ||||
| miR-594-pre | 0.39 | ||||
| miR-18 | 1.81 | let-7a | 1.48 | let-7a | 1.58 |
| miR-27a | 1.81 | let-7b | 1.53 | let-7b | 1.36 |
| miR-27b | 1.61 | let-7c | 1.47 | let-7c | 1.36 |
| miR-31 | 2.88 | miR-16 | 1.68 | miR-16 | 1.43 |
| miR-32 | 1.54 | miR-18 | 1.95 | miR-32 | 1.58 |
| miR-106b | 1.62 | miR-27a | 1.65 | miR-106b | 2.04 |
| miR-203 | ∝ ** | miR-27b | 1.63 | miR-200a | 1.48 |
| miR-566-pre | 4.77 | miR-31 | 1.46 | miR-203 | 2.11 |
| miR-574 | 1.81 | miR-200a | ∝ *** | miR-566-pre | 2.25 |
| miR-566-pre | 1.84 | ||||
| miR-574 | 2.72 | ||||
Differentially expressed in at least 2 pairs of cell lines.
miR-203 was not detected in UM2.
miR-200a was not detected in 1386Tu.
3.2. Differential expression of miR-138 in HNSCC cell lines
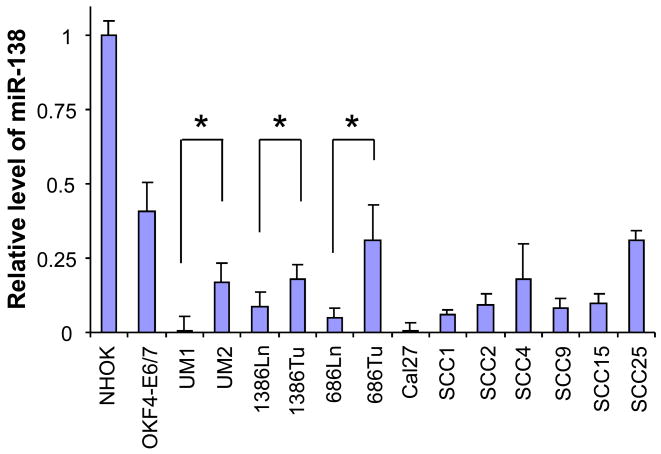
Real-time quantitative RT-PCR was performed to validate the expressional difference of miR-138 in these paired cell lines as well as 7 additional HNSCC cell lines, an immortalized non-tumorgenic cell line (OKF4-E6/7), and the normal oral keratinocyte cell culture (NHOK) (Figure 2). Compared to the non-tumorgenic cells (OKF4-E6/7 and NHOK), the expression level of miR-138 was reduced in all 13 HNSCC cell lines tested. Furthermore, for the paired HNSCC cell lines (UM1/UM2, 1386Tu/1386Ln and 686Tu/686Ln), relatively lower miR-138 levels were observed in all 3 highly invasive cell lines.
Figure 2. Reduced miR-138 level in HNSCC cell lines.
The qRT-PCR was performed as described in the Material and Methods section to assess the miR-138 level in a panel of HNSCC cell lines, and an immortalized non-tumorgenic cell lines (OKF4-E6/7), and normal oral keratinocyte cell culture (NHOK). Statistical differences in miR-138 levels were observed for the paired HNSCC cell lines (UM1/UM2, 1386Ln/Tu, and 686Ln/Tu). * indicates p < 0.05.
3.3. The effects of miR-138 on invasion, cell cycle and apoptosis
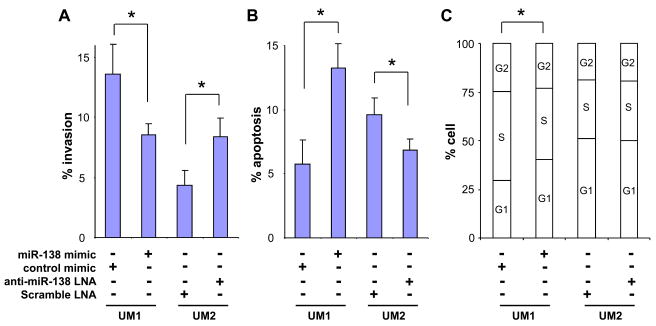
To validate the involvement of miR-138 dysregulation in HNSCC tumorigenesis and metastasis, functional analysis were performed to test the effects of miR-138 on cell invasion, apoptosis, and cell cycle. As shown in Figure 3A, ectopic transfection of miR-138 mimics led to significantly decreased invasion of UM1 cells when compared to cells tranfected with the negative control. Introducing anti-miR-138 LNA to the UM2 cells led to a significant increase in cell invasion. Apparent decrease in migration was also observed in UM1 cells transfected with miR-138, but the difference was not statistically significant (data not shown). No difference in migration was observed in UM2 cells transfected with anti-miR-138 LNA (data not shown). As shown in Figure 3B, significant increase in apoptosis was observed in UM1 cells transfected with the miR-138 mimic., whereas reduction in apoptosis was observed in UM2 cells that were transfected with anti-miR-138 LNA. As Ectopic expression of miR-138 in UM1 cells also led to changes in cell cycle, where significant reduction in S and accumulation in G1 were observed in UM1 cells transfected with miR-138 mimics. Given that sub-G1 DNA content is indicative of apoptosis, these data also supported our observations on apoptosis. No apparent difference in cell cycle was observed in UM2 cells that were transfected with anti-miR-138 LNA.
Figure 3. miR-138 influences invasion, cell cycle, and apoptosis in HNSCC cells.
The miR-138 mimic and negative control microRNA mimic were introduced into the UM1 cells. The anti-miR-138 LNA and scrambled LNA probe were introduced into the UM2 cells. Invasion was measured as described in our previous study [20] (A). Apoptosis (B) and cell cycle (C) were measured using flow cytometry as described in the Material and Methods section. Data represents at least 3 independent triplicated experiments with similar results. * indicates p < 0.05.
4. Discussion
Metastasis is the major hallmark of malignancy. Cancer cell invasion is one of the essential early events in metastasis. In this study, we aim to identify the microRNA alterations that correlate with enhanced invasive behavior in HNSCC. We used an established cell culture model for our study [20], which consists of 3 pairs of HNSCC cell lines that were isolated from the same patients. Among these HNSCC cell line pairs, UM1/UM2 and 686Ln/686Tu exhibit significant differences in migration and invasion; however, 1386Ln/1386Tu exhibit differences in cell invasion but not in migration. Nevertheless, the consistent differences in cell invasion for all 3 HNSCC pairs suggested that this cell culture model can provide us with a useful system to study cell invasion in HNSCC.
Among these identified microRNAs, many of them have been previously implicated in tumorigenesis and metastasis, including let-7 family members (including let-7a, let-7b and let-7c) [22], miR-7 [23], miR-16 [24, 25], miR-21 [26, 27], miR-27 family (including miR-27a and miR-27b) [28, 29], miR-98 [16], miR-99b [30], miR-101 [31, 32], miR-106b [33, 34], miR-125a [35], miR-138 [13, 36], miR-193 [37], miR-200a [38], miR-203 [39], miR-224 [12]. However, for a number of identified candidate microRNAs, their functional involvements in cancer are not clear, including those of miR-18, miR-31, miR-32, miR574. The role of miR-31 in tumorigenesis is not entirely clear. While up-regulation of miR-31 has also been observed in HNSCC [13, 14], colorectal cancer [40, 41], and hepatocellular carcinoma [42], reduced expression of miR-31 was observed in breast cancer [43], and frequent homozygous deletion of miR-31 gene was reported in urothelial carcinomas [44]. Similarly, the role of miR-32 in tumorigenesis is also not clear. While miR-32 up-regulation has been observed in multiple myeloma [45] and prostate cancer [46], decreased miR-32 was observed in bronchial squamous cell carcinoma [47]. Also, the function of miR-18 and miR-574 in tumorigenesis is currently not known. Further studies are needed to fully explore the functional relevance of these microRNAs in tumorigenesis and metastasis of HNSCC.
Among the identified microRNA dysregulations, reduced expression of miR-138 was consistently observed in all 3 highly invasive cell lines (UM1, 1386Ln and 686Ln). Down-regulation of miR-138 has been previously observed in SCC of the tongue [13] and thyroid carcinoma [36]. Two putative genes for miR-138 precursors, termed pre-miR-138-1 and pre-miR-138-2, have been predicted in mouse genome recently [48]. Their human homologs have been located on chromosome 3p21.33 and 16q13, respectively. Interestingly, loss of heterozygosity (LOH) at both chromosome loci have been frequently detected in HNSCC and appears to correlate with tumor progression (i.e., cervical lymph node metastasis) [49–51]. With our new observations presented in this study, it is logical to suggest that miR-138-1 and miR-138-2 may be the tumor suppresser genes that reside in these 2 LOH regions. More functional analyses will be needed to fully explore the potential tumor suppressor role(s) of miR-138 in HNSCC. It is worth knowing that down-regulation of miR-138 has been previously associated with over-expression of hTERT in human anaplastic thyroid carcinoma cell lines [36]. However, we did not observe any apparent change in hTERT expression after ectopic transfection of miR-138 in our cell lines (data not shown). Nevertheless, our results, together with previous observations suggest that miR-138 is a potential tumor suppresser, and may serve as a novel therapeutic target for HNSCC patients at risk of metastatic disease.
Supplementary Material
Acknowledgments
This work was supported in part by NIH PHS grants K22DE014847, RO1CA139596, RO3CA135992, and a grant from Prevent Cancer Foundation (to X.Z.). X.L. is supported in part by grants from the National Natural Science Foundation (30700952). MDA 686Tu, MDA 686Ln, MDA 1386Tu, MDA 1386Ln cells were gifts from Dr. PG. Sacks of the New York University. We thank Ms. Katherine Long for her editorial assistance.
Footnotes
Conflicts of Interest:
None declared.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pickle LW, Hao Y, Jemal A, Zou Z, Tiwari RC, Ward E, Hachey M, Howe HL, Feuer EJ. A new method of estimating United States and state-level cancer incidence counts for the current calendar year. CA Cancer J Clin. 2007;57:30–42. doi: 10.3322/canjclin.57.1.30. [DOI] [PubMed] [Google Scholar]
- 2.Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 3.Calin GA, Liu CG, Sevignani C, Ferracin M, Felli N, Dumitru CD, Shimizu M, Cimmino A, Zupo S, Dono M, et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc Natl Acad Sci U S A. 2004;101:11755–11760. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Metzler M, Wilda M, Busch K, Viehmann S, Borkhardt A. High expression of precursor microRNA-155/BIC RNA in children with Burkitt lymphoma. Genes Chromosomes Cancer. 2004;39:167–169. doi: 10.1002/gcc.10316. [DOI] [PubMed] [Google Scholar]
- 6.Kondo N, Toyama T, Sugiura H, Fujii Y, Yamashita H. miR-206 Expression is down-regulated in estrogen receptor alpha-positive human breast cancer. Cancer Res. 2008;68:5004–5008. doi: 10.1158/0008-5472.CAN-08-0180. [DOI] [PubMed] [Google Scholar]
- 7.Bhaumik D, Scott GK, Schokrpur S, Patil CK, Campisi J, Benz CC. Expression of microRNA-146 suppresses NF-kappaB activity with reduction of metastatic potential in breast cancer cells. Oncogene. 2008 doi: 10.1038/onc.2008.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Michael MZ, SM OC, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1:882–891. [PubMed] [Google Scholar]
- 9.Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 10.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 11.Murakami Y, Yasuda T, Saigo K, Urashima T, Toyoda H, Okanoue T, Shimotohno K. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25:2537–2545. doi: 10.1038/sj.onc.1209283. [DOI] [PubMed] [Google Scholar]
- 12.Wang Y, Lee AT, Ma JZ, Wang J, Ren J, Yang Y, Tantoso E, Li KB, Ooi LL, Tan P, et al. Profiling microRNA expression in hepatocellular carcinoma reveals microRNA-224 up-regulation and apoptosis inhibitor-5 as a microRNA-224-specific target. J Biol Chem. 2008;283:13205–13215. doi: 10.1074/jbc.M707629200. [DOI] [PubMed] [Google Scholar]
- 13.Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP, Wei WI. Mature miR-184 as Potential Oncogenic microRNA of Squamous Cell Carcinoma of Tongue. Clin Cancer Res. 2008;14:2588–2592. doi: 10.1158/1078-0432.CCR-07-0666. [DOI] [PubMed] [Google Scholar]
- 14.Kozaki K, Imoto I, Mogi S, Omura K, Inazawa J. Exploration of tumor-suppressive microRNAs silenced by DNA hypermethylation in oral cancer. Cancer Res. 2008;68:2094–2105. doi: 10.1158/0008-5472.CAN-07-5194. [DOI] [PubMed] [Google Scholar]
- 15.Tran N, McLean T, Zhang X, Zhao CJ, Thomson JM, O’Brien C, Rose B. MicroRNA expression profiles in head and neck cancer cell lines. Biochem Biophys Res Commun. 2007;358:12–17. doi: 10.1016/j.bbrc.2007.03.201. [DOI] [PubMed] [Google Scholar]
- 16.Hebert C, Norris K, Scheper MA, Nikitakis N, Sauk JJ. High mobility group A2 is a target for miRNA-98 in head and neck squamous cell carcinoma. Mol Cancer. 2007;6:5. doi: 10.1186/1476-4598-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 18.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 19.Nakayama S, Sasaki A, Mese H, Alcalde RE, Matsumura T. Establishment of high and low metastasis cell lines derived from a human tongue squamous cell carcinoma. Invasion Metastasis. 1998;18:219–228. doi: 10.1159/000024515. [DOI] [PubMed] [Google Scholar]
- 20.Ye H, Wang A, Lee BS, Yu T, Sheng S, Peng T, Hu S, Crowe DL, Zhou X. Proteomic Based Identification of Manganese Superoxide Dismutase 2 (SOD2) as a Metastasis Marker for Oral Squamous Cell Carcinoma. Cancer Genomics Proteomics. 2008;5:85–94. [PMC free article] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18:505–516. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 23.Webster RJ, Giles KM, Price KJ, Zhang PM, Mattick JS, Leedman PJ. Regulation of epidermal growth factor receptor signaling in human cancer cells by microRNA-7. J Biol Chem. 2009;284:5731–5741. doi: 10.1074/jbc.M804280200. [DOI] [PubMed] [Google Scholar]
- 24.Kaddar T, Chien WW, Bertrand Y, Pages MP, Rouault JP, Salles G, Ffrench M, Magaud JP. Prognostic value of miR-16 expression in childhood acute lymphoblastic leukemia relationships to normal and malignant lymphocyte proliferation. Leuk Res. 2009 doi: 10.1016/j.leukres.2008.12.015. [DOI] [PubMed] [Google Scholar]
- 25.Kaddar T, Rouault JP, Chien WW, Chebel A, Gadoux M, Salles G, Ffrench M, Magaud JP. Two new miR-16 targets: Caprin-1 and HMGA1, proteins implicated in cell proliferation. Biol Cell. 2009 doi: 10.1042/BC20080213. [DOI] [PubMed] [Google Scholar]
- 26.Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26:2799–2803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 27.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 28.Mertens-Talcott SU, Chintharlapalli S, Li X, Safe S. The oncogenic microRNA-27a targets genes that regulate specificity protein transcription factors and the G2-M checkpoint in MDA-MB-231 breast cancer cells. Cancer Res. 2007;67:11001–11011. doi: 10.1158/0008-5472.CAN-07-2416. [DOI] [PubMed] [Google Scholar]
- 29.Tsuchiya Y, Nakajima M, Takagi S, Taniya T, Yokoi T. MicroRNA regulates the expression of human cytochrome P450 1B1. Cancer Res. 2006;66:9090–9098. doi: 10.1158/0008-5472.CAN-06-1403. [DOI] [PubMed] [Google Scholar]
- 30.Wu W, Lin Z, Zhuang Z, Liang X. Expression profile of mammalian microRNAs in endometrioid adenocarcinoma. Eur J Cancer Prev. 2009;18:50–55. doi: 10.1097/CEJ.0b013e328305a07a. [DOI] [PubMed] [Google Scholar]
- 31.Friedman JM, Liang G, Liu CC, Wolff EM, Tsai YC, Ye W, Zhou X, Jones PA. The putative tumor suppressor microRNA-101 modulates the cancer epigenome by repressing the polycomb group protein EZH2. Cancer Res. 2009;69:2623–2629. doi: 10.1158/0008-5472.CAN-08-3114. [DOI] [PubMed] [Google Scholar]
- 32.Su H, Yang JR, Xu T, Huang J, Xu L, Yuan Y, Zhuang SM. MicroRNA-101, down-regulated in hepatocellular carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer Res. 2009;69:1135–1142. doi: 10.1158/0008-5472.CAN-08-2886. [DOI] [PubMed] [Google Scholar]
- 33.Ivanovska I, Ball AS, Diaz RL, Magnus JF, Kibukawa M, Schelter JM, Kobayashi SV, Lim L, Burchard J, Jackson AL, et al. MicroRNAs in the miR-106b family regulate p21/CDKN1A and promote cell cycle progression. Mol Cell Biol. 2008;28:2167–2174. doi: 10.1128/MCB.01977-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, de Martino I, Iliopoulos D, Pilozzi E, Liu CG, Negrini M, et al. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13:272–286. doi: 10.1016/j.ccr.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 35.Scott GK, Goga A, Bhaumik D, Berger CE, Sullivan CS, Benz CC. Coordinate suppression of ERBB2 and ERBB3 by enforced expression of micro-RNA miR-125a or miR-125b. J Biol Chem. 2007;282:1479–1486. doi: 10.1074/jbc.M609383200. [DOI] [PubMed] [Google Scholar]
- 36.Mitomo S, Maesawa C, Ogasawara S, Iwaya T, Shibazaki M, Yashima-Abo A, Kotani K, Oikawa H, Sakurai E, Izutsu N, et al. Downregulation of miR-138 is associated with overexpression of human telomerase reverse transcriptase protein in human anaplastic thyroid carcinoma cell lines. Cancer Sci. 2008;99:280–286. doi: 10.1111/j.1349-7006.2007.00666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ovcharenko D, Kelnar K, Johnson C, Leng N, Brown D. Genome-scale microRNA and small interfering RNA screens identify small RNA modulators of TRAIL-induced apoptosis pathway. Cancer Res. 2007;67:10782–10788. doi: 10.1158/0008-5472.CAN-07-1484. [DOI] [PubMed] [Google Scholar]
- 38.Bracken CP, Gregory PA, Kolesnikoff N, Bert AG, Wang J, Shannon MF, Goodall GJ. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 2008;68:7846–7854. doi: 10.1158/0008-5472.CAN-08-1942. [DOI] [PubMed] [Google Scholar]
- 39.Lena AM, Shalom-Feuerstein R, di Val Cervo PR, Aberdam D, Knight RA, Melino G, Candi E. miR-203 represses ‘stemness’ by repressing DeltaNp63. Cell Death Differ. 2008;15:1187–1195. doi: 10.1038/cdd.2008.69. [DOI] [PubMed] [Google Scholar]
- 40.Bandres E, Cubedo E, Agirre X, Malumbres R, Zarate R, Ramirez N, Abajo A, Navarro A, Moreno I, Monzo M, et al. Identification by Real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol Cancer. 2006;5:29. doi: 10.1186/1476-4598-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Slaby O, Svoboda M, Fabian P, Smerdova T, Knoflickova D, Bednarikova M, Nenutil R, Vyzula R. Altered expression of miR-21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology. 2007;72:397–402. doi: 10.1159/000113489. [DOI] [PubMed] [Google Scholar]
- 42.Wong QW, Lung RW, Law PT, Lai PB, Chan KY, To KF, Wong N. MicroRNA-223 is commonly repressed in hepatocellular carcinoma and potentiates expression of Stathmin1. Gastroenterology. 2008;135:257–269. doi: 10.1053/j.gastro.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 43.Yan LX, Huang XF, Shao Q, Huang MY, Deng L, Wu QL, Zeng YX, Shao JY. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. Rna. 2008;14:2348–2360. doi: 10.1261/rna.1034808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Veerla S, Lindgren D, Kvist A, Frigyesi A, Staaf J, Persson H, Liedberg F, Chebil G, Gudjonsson S, Borg A, et al. MiRNA expression in urothelial carcinomas: Important roles of miR-10a, miR-222, miR-125b, miR-7 and miR-452 for tumor stage and metastasis, and frequent homozygous losses of miR-31. Int J Cancer. 2008 doi: 10.1002/ijc.24183. [DOI] [PubMed] [Google Scholar]
- 45.Pichiorri F, Suh SS, Ladetto M, Kuehl M, Palumbo T, Drandi D, Taccioli C, Zanesi N, Alder H, Hagan JP, et al. MicroRNAs regulate critical genes associated with multiple myeloma pathogenesis. Proc Natl Acad Sci U S A. 2008;105:12885–12890. doi: 10.1073/pnas.0806202105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ambs S, Prueitt RL, Yi M, Hudson RS, Howe TM, Petrocca F, Wallace TA, Liu CG, Volinia S, Calin GA, et al. Genomic profiling of microRNA and messenger RNA reveals deregulated microRNA expression in prostate cancer. Cancer Res. 2008;68:6162–6170. doi: 10.1158/0008-5472.CAN-08-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mascaux C, Laes JF, Anthoine G, Haller A, Ninane V, Burny A, Sculier JP. Evolution of microRNA expression during human bronchial squamous carcinogenesis. Eur Respir J. 2009;33:352–359. doi: 10.1183/09031936.00084108. [DOI] [PubMed] [Google Scholar]
- 48.Obernosterer G, Leuschner PJ, Alenius M, Martinez J. Post-transcriptional regulation of microRNA expression. Rna. 2006;12:1161–1167. doi: 10.1261/rna.2322506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Piccinin S, Gasparotto D, Vukosavljevic T, Barzan L, Sulfaro S, Maestro R, Boiocchi M. Microsatellite instability in squamous cell carcinomas of the head and neck related to field cancerization phenomena. Br J Cancer. 1998;78:1147–1151. doi: 10.1038/bjc.1998.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hogg RP, Honorio S, Martinez A, Agathanggelou A, Dallol A, Fullwood P, Weichselbaum R, Kuo MJ, Maher ER, Latif F. Frequent 3p allele loss and epigenetic inactivation of the RASSF1A tumour suppressor gene from region 3p21.3 in head and neck squamous cell carcinoma. Eur J Cancer. 2002;38:1585–1592. doi: 10.1016/s0959-8049(01)00422-1. [DOI] [PubMed] [Google Scholar]
- 51.Wang X, Gleich L, Pavelic ZP, Li YQ, Gale N, Hunt S, Gluckman JL, Stambrook PJ. Cervical metastases of head and neck squamous cell carcinoma correlate with loss of heterozygosity on chromosome 16q. Int J Oncol. 1999;14:557–561. doi: 10.3892/ijo.14.3.557. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.