Abstract
The innate antiviral factor APOBEC3G (A3G) possesses RNA binding activity and deaminates HIV-1 DNA. High-molecular-mass forms of A3G can be isolated from a variety of cell types, but exhibit limited deaminase activity relative to low-molecular-mass species prepared under RNA-depleted conditions. To investigate the fundamental oligomeric state and shape of A3G, we conducted sedimentation velocity analyses of the pure enzyme under RNA-deficient conditions. The results reveal a predominant dimer in equilibrium with minor monomeric and tetrameric species. Hydrodynamic modeling of the dimer supports an extended cylindrical shape that assembles into an elongated tetramer. Overall, the results provide physical restraints for the A3G quaternary structure that have implications for modulating antiviral function.
A3G is an enzyme expressed in CD4+ T cells, the primary target of HIV-1 infection (1). The protein belongs to the APOBEC family of cytidine deaminases, which are distinguished by a Zn2+-dependent hydrolytic reaction that converts cytidine to uridine in the context of DNA or RNA substrates (Figure 1A) (2). Interest in the A3G mechanism of action has been fueled by the discovery of its antiviral properties that can block infectivity by producing G-to-A hypermutations in the coding strand of proviral DNA (3, 4). The HIV-1 protein Vif can counteract A3G by recruiting it to a cellular Cullin-RING ligase that polyubiquitinates A3G leading to its proteasomal degradation (5–7). Packaging of A3G into viral particles is important for antiviral activity and is dependent on its interaction with HIV-1 Gag (8, 9) Thus, the A3G subunit organization has implications for understanding its stability and interplay with viral factors.
Figure 1.
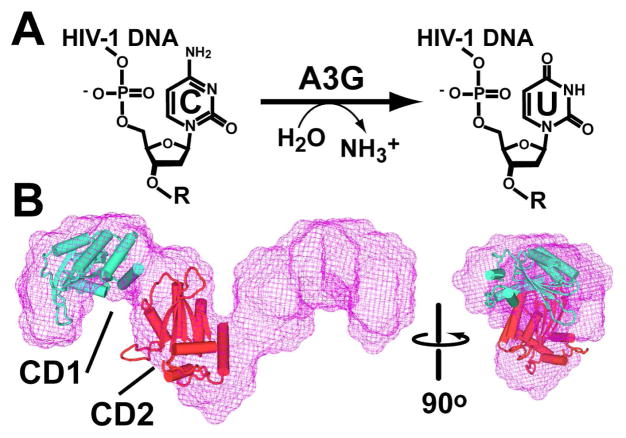
The reaction of A3G and its molecular envelope from SAXS. (A) The hydrolytic deamination reaction in the context of viral DNA. (B) The most probable molecular envelope (mesh) comprises a dimer with P2 symmetry (13). Two C-terminal A3G deaminase domains (each comprising residues 197 to 380 from ref. (16)) were rigid-body modeled as described in the Supporting Information. The orientation of each domain (CD1 and CD2) in a subunit is hypothetical, but demonstrates the SAXS envelope can accommodate four CD domains, equivalent to a dimer.
Prior analyses of A3G’s molecular mass demonstrated that MDa-sized ribonucleoprotein particles of the enzyme develop in infected cells (10). These large assemblies have severely diminished deaminase function, but can be treated with RNase to restore A3G activity in vitro (2, 10). This observation suggests that bridging RNA leads to higher-order assemblies with impaired antiviral function (2). Although low-molecular-mass forms of A3G are necessary for deaminase activity, reports of the fundamental quaternary structure have varied widely and suggest stable monomeric (11, 12), dimeric (13, 14), and tetrameric (15) species. These disparate observations have implications for deaminase function, homology modeling, as well as the accessibility of A3G to viral proteins, such as Gag or Vif.
Among APOBEC family members, the subunit organization of A3G is unusual because it comprises tandem deaminase domains, CD1 and CD2, on a single polypeptide chain. The three-dimensional structure of A3G has not been reported, although the C-terminal deaminase domain has been determined by crystallography and NMR (16–18). Previously, our lab reported a small angle X-ray scattering (SAXS) analysis of the intact enzyme, which revealed a novel, elongated shape with a maximum length (DMax) of 14 nm (Figure 1B). Rigid-body modeling of a fundamental deaminase domain, as well as volumetric considerations, supported a dimer of A3G subunits in solution (13). A subsequent crystal structure of the distant paralogue APOBEC2 (A2) exhibited an elongated shape (DMax = 12.7 nm) that affirmed the feasibility of an extended A3G dimer (19). However, at 21.3 kDa each A2 subunit is less than half the mass of an A3G subunit of 46.4 kDa. This disparity represents an impediment to homology modeling of the intact molecule. Moreover, absence of a clear A3G quaternary structure has caused this aspect of the enzyme’s organization to be largely neglected despite the importance of surface accessibility in Vif or Gag binding.
The hydrodynamic properties of full-length A3G have not been documented at present. As such, we describe herein a sedimentation velocity analysis of the pure, human enzyme by use of analytical ultracentrifugation (AUC). Data were analyzed by two methods: (i) the time-derivative of the concentration profile, g(s*) (20, 21), and (ii) direct boundary modeling based on numerical solutions to the Lamm equations (22). The results reveal the number of A3G species in solution, the enzyme’s self-associative properties, and the sedimentation coefficients of discrete species. These parameters place physical restraints on the A3G quaternary structure that allow hydrodynamic modeling, which is discussed in light of existing SAXS, high-resolution structural, and biochemical data.
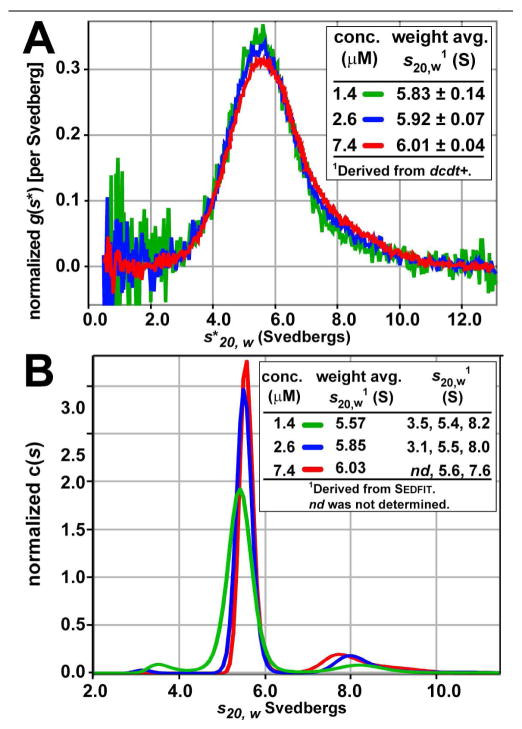
A3G with a C-terminal four-His tag was purified from baculovirus-infected insect cells as described (ref. (13) and the Supporting Information). As a preface to AUC, the protein was purified by gel filtration revealing a dominant peak consistent with the mass of an A3G dimer (Figure S1). Sample purity was estimated to be >99% based on SDS PAGE analysis. Intact A3G is notoriously difficult to produce in large quantities (13), which limited the concentration range attainable for AUC studies. Sufficient quantities of protein were obtained to subject three concentrations of A3G to sedimentation velocity analysis (Supporting Information). Initial processing of the data utilized the model-independent sedimentation coefficient distribution, g(s*), approach implemented in dcdt+ (21). An overlay of the normalized g(s*) distributions for each concentration is depicted in Figure 2A. The respective plots do not superimpose perfectly, which is evidence for one or more reversible interactions. The self-association reaction is also evident from the shift of the weight-average sedimentation coefficient to higher values with increasing protein concentration (Figure 2A, inset).
Figure 2.
Sedimentation velocity analysis of A3G. (A) Model-independent sedimentation coefficient distribution, g(s*), analysis used to derive the weight-average sedimentation coefficients shown in the inset. The protein concentrations were obtained by integration of the g(s*) plot. Values of s have been corrected to standard conditions, s20,w, at 20 °C in water. (B) The continuous sedimentation coefficient distribution, c(s) of A3G. Both weight average and individual sedimentation coefficients were calculated.
To extend the hydrodynamic investigation of A3G, we performed a differential distribution analysis of Lamm equation solutions using the program Sedfit (22). This approach yields relatively higher resolution information and is more sensitive to discrete species than the g(s*) approach. Plots of the normalized, continuous sedimentation coefficient distributions, c(s), are depicted in Figure 2B. Calculation of the weight-average s20,w for each concentration corroborated the results from the g(s*) approach (Figure 2B, inset). Specifically, the weight-average s20,w increases as a function of A3G concentration, which is further evidence for a self-associating system.
Importantly, the c(s) analysis of A3G indicates the presence of three species. Based on the known molecular mass of a protein subunit, the observed distributions are consistent with a monomer-dimer-tetramer reacting system in the concentration range examined. The dominant species exhibits an s20,w value of ~5.6 S that is consistent with a 94 kDa dimer of subunits. The ratio of the experimental frictional coefficient to the frictional coefficient of a sphere of the same mass (f/f0) was calculated to be ~1.45. This value reports on the relative geometric shape asymmetry (23) and suggests dimeric A3G is an elongated particle in solution. A monomeric species of A3G was observed over the range of our analysis, but was the smallest population, even at the lowest protein concentration of 1.4 μM (Figure 2B). Values of s20,w for the monomer were variable between 3.5 and 3.1 S. Poor signal for the monomer at the highest concentration, 7.4 μM, precluded a reliable measurement of the sedimentation coefficient.
A tetrameric species of A3G was observed as a minor species at all concentrations with a range of 8.2 to 7.6 S (Figure 2B, inset). Although efforts were made to fit the monomer-dimer-tetramer data to a self-association scheme, no result was obtained due to limitations in the available concentration range. However, fit of the data to a monomer-dimer self-association scheme gave a Kd of ~100–200 nM using Sedphat (24). Systematic deviations of the fit were present due to neglect of the dimer-tetramer equilibrium, although the amount of dimer is ~10-fold greater than monomer at 1.4 μM (Figure 2B) in accord with the Kd estimate.
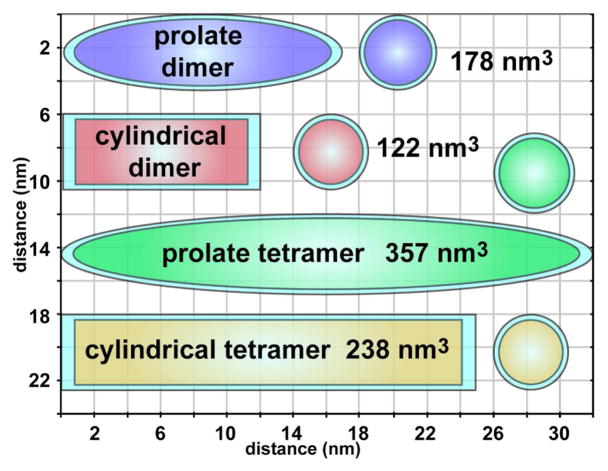
To elucidate the volume and shape properties of A3G, we explored simple hydrodynamic models for the AUC data at 7.4 μM by use of Sednterp (25), which considers subunit mass and the observed sedimentation coefficient. The ~5.6 S dimeric species could be modeled as a prolate ellipsoid (major axis 17 nm and a minor axes of 4.4 nm), a solid cylinder (12 nm × 4.4 nm) or an oblate ellipsoid (11 nm × 3 nm). Because the flat disk from the oblate calculation was thinner than the known A3G C-terminal domain (4.6 nm × 4.4 nm × 4.1 nm) (16), it was not considered a viable shape for the dimer or tetramer. As such, only the shapes and volumes for the prolate and cylindrical dimers were considered (Figure 3). We then evaluated these models in the context of the A3G C-terminal domain (16), which has a calculated hydrated volume of 30.8 nm3 (Supporting Information).
Figure 3.
Hydrodynamic modeling and volumes of A3G based on s20,w values from Figure 2, inset at 7.4 μM. The light-blue shell around each model represents the hydration expansion (~14%). Spheres represent the model viewed down an object’s major axis.
Due to the high sequence homology (53%) between A3G CD2 (22.1 kDa) and CD1 (24.3 kDa), it is reasonable to assume that each domain has a similar tertiary fold, and that the dimeric volume of A3G is nearly equal to four volumes of the C-terminal crystal structure or 123 nm3. However, due to the 2.2 kDa difference between CD1 and CD2, we conducted a calculation based on A3G’s partial specific volume (Supporting Information) and amino acid sequence. The resulting volume was 114 nm3 for a dimer, which agrees well with our coordinate-based calculation.
Each volume is consistent with the value of 122 nm3 calculated for the hydrodynamically modeled A3G cylindrical dimer. In contrast, the volume obtained from the prolate dimer is considerably larger at 178 nm3. Additional support for a cylindrical A3G dimer comes from its 12 nm length, which agrees with the extended shape of the A2 tetramer (DMax is 12.7 nm), as well as the solution SAXS analysis of intact A3G (DMax of 14 ± 1.4 nm) in ref. (13).
We then explored shapes for the A3G tetramer. Whereas the cylindrical tetramer yielded dimensions of 25 nm × 4.3 nm and a volume of 238 nm3, a prolate ellipsoid had dimensions of 32 nm × 4.7 nm and a volume of 357 nm3 (Figure 3). Volumes based on the A3G sequence and C-terminal domain, respectively, suggested the tetramer displaces 228 to 246 nm3. Again the results support a cylindrical model over a prolate one. If the tetramer were comprised of two end-to-end dimers, DMax would be 24 nm, which concurs with a 25 nm cylindrical tetramer (Figure 3).
In summary, A3G is predominantly a dimer in solution that exists in equilibrium with minor tetrameric and monomeric species. This observation accounts for the various reported oligomeric states. Hydrodynamic modeling suggests the dimer is elongated with a cylindrical quaternary structure. This model, as well the observed s20,w range of 5.4 to 5.6 S and the f/f0 of ~1.45 reported herein, is consistent with the SAXS analysis that reported the extended dimer in Figure 1B (13), which exhibits a calculated s20,w of 5.5 S and an f/f0 of 1.55 (Supporting Information). The s20,w for a tetrameric A3G shape is compatible with a cylinder as well, possibly comprised of tandem end-to-end cylindrical dimers. Previously, A3G was proposed to form a tetrameric filament (15), which agrees with our observations. Although exact dimensions and the spatial orientations of subunits in the quaternary structure will require higher-resolution analyses, our results imply that low-molecular mass, functional forms of A3G comprise an equilibrium mixture of monomeric, dimeric and tetrameric subunits. Self-association leading to inactive high-molecular mass assemblies results from RNA bridging between the fundamental forms. Knowledge that A3G has self-associative properties implies that high-molecular mass forms of the enzyme achieve some stability via protein-protein interactions, which has implications for dissolution of such aggregates to restore antiviral activity.
Supplementary Material
Acknowledgments
We are grateful to Jeffrey W. Lary at the University of Connecticut Analytical Ultracentrifuge Facility for data collection and analysis.
Footnotes
Support from the National Institutes of Health (NIH) grant AI076085 to JEW is acknowledged. JDS was supported by training grant NIH T32 AI49815.
SUPPORTING INFORMATION AVAILABLE
Supporting methods and figures are available for AUC and hydrodynamic modeling. Materials are available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Sheehy AM, Gaddis NC, Choi JD, Malim MH. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 2.Chiu YL, Greene WC. Annu Rev Immunol. 2008;26:317–353. doi: 10.1146/annurev.immunol.26.021607.090350. [DOI] [PubMed] [Google Scholar]
- 3.Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, Neuberger MS, Malim MH. Cell. 2003;113:803–809. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- 4.Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. Nature. 2003;424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- 5.Yu X, Yu Y, Liu B, Luo K, Kong W, Mao P, Yu XF. Science. 2003;302:1056–1060. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- 6.Conticello SG, Harris RS, Neuberger MS. Curr Biol. 2003;13:2009–2013. doi: 10.1016/j.cub.2003.10.034. [DOI] [PubMed] [Google Scholar]
- 7.Marin M, Rose KM, Kozak SL, Kabat D. Nat Med. 2003;9:1398–1403. doi: 10.1038/nm946. [DOI] [PubMed] [Google Scholar]
- 8.Luo K, Liu B, Xiao Z, Yu Y, Yu X, Gorelick R, Yu XF. J Virol. 2004;78:11841–11852. doi: 10.1128/JVI.78.21.11841-11852.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zennou V, Perez-Caballero D, Gottlinger H, Bieniasz PD. J Virol. 2004;78:12058–12061. doi: 10.1128/JVI.78.21.12058-12061.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiu YL, Soros VB, Kreisberg JF, Stopak K, Yonemoto W, Greene WC. Nature. 2005;435:108–114. doi: 10.1038/nature03493. [DOI] [PubMed] [Google Scholar]
- 11.Chen KM, Martemyanova N, Lu Y, Shindo K, Matsuo H, Harris RS. FEBS Lett. 2007;581:4761–4766. doi: 10.1016/j.febslet.2007.08.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friew YN, Boyko V, Hu WS, Pathak VK. Retrovirology. 2009;6:56. doi: 10.1186/1742-4690-6-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wedekind JE, Gillilan R, Janda A, Krucinska J, Salter JD, Bennett RP, Raina J, Smith HC. J Biol Chem. 2006;281:38122–38126. doi: 10.1074/jbc.C600253200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bennett RP, Salter JD, Liu X, Wedekind JE, Smith HC. J Biol Chem. 2008;283:33329–33336. doi: 10.1074/jbc.M803726200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chelico L, Sacho EJ, Erie DA, Goodman MF. J Biol Chem. 2008;283:13780–13791. doi: 10.1074/jbc.M801004200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holden LG, Prochnow C, Chang YP, Bransteitter R, Chelico L, Sen U, Stevens RC, Goodman MF, Chen XS. Nature. 2008;456:121–124. doi: 10.1038/nature07357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furukawa A, Nagata T, Matsugami A, Habu Y, Sugiyama R, Hayashi F, Kobayashi N, Yokoyama S, Takaku H, Katahira M. Embo J. 2009;28:440–451. doi: 10.1038/emboj.2008.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harjes E, Gross PJ, Chen KM, Lu Y, Shindo K, Nowarski R, Gross JD, Kotler M, Harris RS, Matsuo H. J Mol Biol. 2009;389:819–832. doi: 10.1016/j.jmb.2009.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prochnow C, Bransteitter R, Klein MG, Goodman MF, Chen XS. Nature. 2007;445:447–451. doi: 10.1038/nature05492. [DOI] [PubMed] [Google Scholar]
- 20.Philo JS. Anal Biochem. 2000;279:151–163. doi: 10.1006/abio.2000.4480. [DOI] [PubMed] [Google Scholar]
- 21.Philo JS. Anal Biochem. 2006;354:238–246. doi: 10.1016/j.ab.2006.04.053. [DOI] [PubMed] [Google Scholar]
- 22.Schuck P. Biophys J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lebowitz J, Lewis MS, Schuck P. Protein Sci. 2002;11:2067–2079. doi: 10.1110/ps.0207702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schuck P. Anal Biochem. 2003;320:104–124. doi: 10.1016/s0003-2697(03)00289-6. [DOI] [PubMed] [Google Scholar]
- 25.Hayes DB, Laue TM, Philo J. Sedimentation Interpretation Program. 1.09. University of New Hampshire; 2006. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.