Abstract
Objective
Our goal was to identify phosphorylation sites that regulate serum response factor (SRF) activity to gain a better understanding of the signaling mechanisms that regulate SRF’s involvement in smooth muscle cell (SMC)-specific and early response gene expression.
Methods and Results
By screening phosphorylation deficient and mimetic mutations in SRF −/− ES cells, we identified T159 as a phosphorylation site that significantly inhibits SMC-specific gene expression in an ES cell model of SMC differentiation. This residue conforms to a highly conserved consensus cAMP-dependent protein kinase (PKA) site, and in vitro and in vivo labeling studies demonstrated that it was phosphorylated by PKA. Results from gel shift and chromatin immunoprecipitation assays demonstrated that T159 phosphorylation inhibited SRF binding to SMC-specific CArG elements. Interestingly, the myocardin factors could at least partially rescue the effects of the T159D mutation under some conditions, but this response was promoter specific. Finally, PKA signaling had much less of an effect on c-fos promoter activity and SRF binding to the c-fos CArG.
Conclusions
Our results indicate that phosphorylation of SRF by PKA inhibits SMC-specific transcription suggesting a novel signaling mechanism for the control of SMC phenotype.
Keywords: smooth muscle, serum response factor, protein kinase A
Introduction
Smooth muscle cells (SMC) phenotypic modulation plays an important role in the progression of several prominent cardiovascular diseases, including atherosclerosis, hypertension, and restenosis 1. While it is well known that this process is controlled by local environmental cues (see 2 for review), the precise signaling mechanisms that regulate this process are unclear. Extensive evidence indicates that serum response factor (SRF), is a critical transcriptional regulator of SMC differentiation maker gene expression 3-8. However, because SRF is a ubiquitously expressed gene that also regulates early response and cardiac- and skeletal muscle-specific gene expression (see 9 for review), it is clear that additional mechanisms are involved.
The identification of the myocardin family of transcription factors (myocardin and the Myocardin-Related-Transcription Factors, MRTF-A and MRTF-B) was a particularly important advance because these SRF co-factors strongly transactivate SMC-specific gene expression (see 10 for review). Importantly, knock-out studies in mice have demonstrated that myocardin and MRTF-B are required for the differentiation of specific SMC sub-types while MRTF-A is required for the expression of SM α-actin that occurs in mammary epithelial cells during lactation. 10 Early studies also demonstrated that SRF is phosphorylated at multiple residues just N-terminal to the DNA binding domain by several kinases including casein kinase II, CaM kinase IV, and MAPKAP kinase 2 11-17. Phosphorylation of these sites (especially Ser 103) increased SRF affinity for the c-fos CArG. However, because SRF phosphorylation had only marginal effects on SRF’s ability to stimulate c-fos expression and because SRF phosphorylation was not altered by serum stimulation 18, the physiologic significance of these findings were unclear.
A more recent study indicates that SRF phosphorylation may regulate SMC differentiation marker gene expression. Iyer et.al demonstrated that SRF was phosphorylated at S162 by PKC-α and that this modification attenuated SRF-dependent cardiac and SM α-actin promoter activity by inhibiting SRF binding to CArG elements 19. Interestingly, SRF binding to the c-fos CArG was not significantly affected by this mechanism because of the stabilizing effects of Elk-1 within the ternary complex.
To better examine the role of SRF phosphorylation on SMC differentiation marker gene expression we have screened a variety of SRF phosphorylation mutants in an SRF −/− ES cell model of SMC differentiation. Our results suggest that SRF is phosphorylated at T159 by cAMP-dependent kinase (PKA), and that this phosphorylation inhibits SMC-specific transcription by inhibiting SRF binding to the CArG elements within the SMC-specific promoters.
Materials and Methods
Plasmids and SRF mutations
SRF mutations (see figure 1) were generated using the QuikChange mutagenesis kit (Stratagene) or sequence overlap extension. All SRF constructs were subcloned into flag-tagged pcDNA3.1 for mammalian expression or pGEX-4T1 (Amersham) for the generation of GST-fusions. Please see supplemental methods for more detailed protocols.
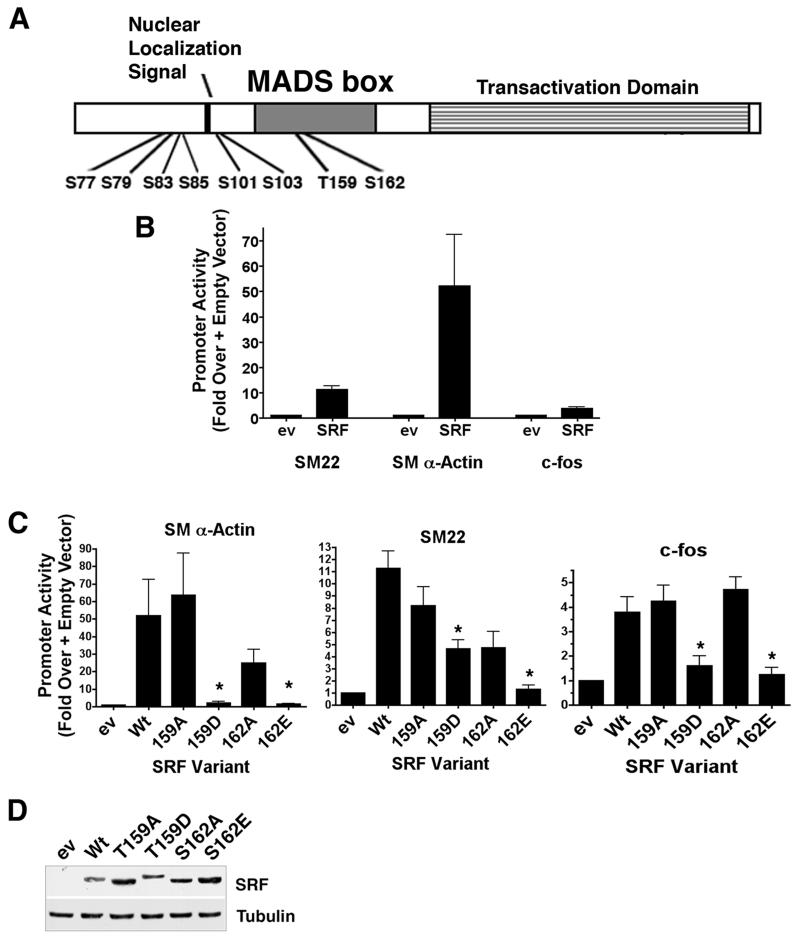
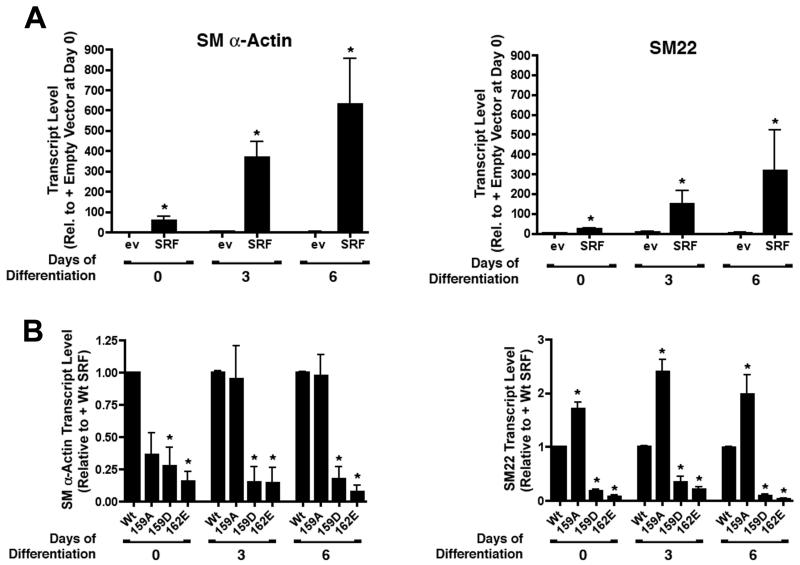
Figure 1. The phosphomimetic T159D mutation negatively regulated SRF-dependent SMC-specific transcription.
A) Schematic of SRF functional domains and phosphorylation sites tested in these studies. B) SRF was transfected into SRF −/− ES cells along with SM22, SM α-actin, or c-fos promoter/luciferase constructs. Luciferase activity was measured 24h post-transfection and expressed relative to that measured in the presence of an equal amount of empty expression vector. C) The indicated SRF variants and promoter-reporter constructs were co-transfected into ES cells. Luciferase activity after 24h was measured and is expressed relative to empty expression vector set to 1. *p<0.05 versus Wt SRF D) Western blot showing expression levels of the SRF variants in ES cells 24h post-transfection.
Cell culture, transfections, and promoter assays
The SRF −/− embryonic stem cells were a generous gift from Alfred Nordheim; (Tubingen University, Germany) and their culture and differentiation have been previously described 20-22. SM α-actin and SM22 message levels were measured by quantitative RT-PCR from RNA isolated at day 0, 3, and 6 of the differentiation protocol. The primary rat aortic SMC cell cultures, transient transfections, and promoter luciferase assays have been previously described 23. Statistical comparisons between groups were made using the 2-tailed Student’s t test with statistical significance accepted at p<0.05.
Gel shift analyses
Flag-tagged SRF and myocardin factors were translated in vitro using the TnT kit (Promega). Binding reactions contained 1μL SRF, 2μL of myocardin factor, 20,000 cpms of 32P-labeled oligonucleotide probe, and 0.20μg dIdC in binding buffer (10mM Tris, pH 7.5, 50mM NaCl, 100mM KCl, 1mM DTT, 1mM EDTA, 5% glycerol). For supershifts, 1μl of anti-flag antibody (Sigma) was added after the first 20 min of incubation.
Phosphorylation assays
GST-SRF fusion proteins were purified from BL21 bacterial lysates using glutathione sepharose (Amersham Biosciences). For in vitro kinase reactions 0.08 μg of purified constitutively active PKA (New England BioLabs) or 8-pCPT-cGMP-stimulated PKG-Iα (Promega) was incubated with 2μg GST-SRF in 25μL kinase buffer (50mM Tris, 10mM MgCl2, 200μM ATP, and 10μCi γ32P-ATP). For in vivo labeling, full length Wt or T159A flag-tagged SRF variants were expressed in Cos-7 cells. Following washing in phosphate-free media, cells were incubated with 1mCi ortho 32P for 2h and then treated with 20μM forskolin for 2.5h or 100μM 8-pCPT-cGMP. SRF was immunoprecipitated from RIPA lysates using anti-flag agarose beads (Sigma). Phosphorylation assays in SMC were mostly similar except that endogenous SRF was immunoprecipiatated using an anti-SRF Ab (Santa Cruz).
Chromatin immunoprecipitation assays
ChIP assays were performed according to the X-ChIP protocol (Abcam). In brief, cells were fixed for 10 min in 1% formaldehyde, placed in lysis buffer (50mM TrisHCl (pH 8.0), 10mM EDTA, 1% SDS), and then chromatin was sheared into 500 bp to 1000 bp fragments by sonication. Equal amounts of sample were immunoprecipitated with 2.5μg of anti-SRF (Santa Cruz), or anti-flag (Sigma) Ab. ChIP PCRs were performed using Red Taq ReadyMix (Sigma) using primers that were previously described 24. To control for non-specific binding, immunoprecipitations were also performed in the absence of primary Ab (see supplemental figure 1 for results).
Results
Identification of T159 as a potential phosphorylation site that inhibits SRF activity
Because little is known about the effects of SRF phosphorylation on SMC-specific transcription, we decided to screen SRF phosphorylation mutations for their ability to alter SMC-specific promoter activity when expressed in SRF −/− ES cells. SRF −/− ES cells have been shown to be stable when grown in LIF containing media and to have similar proliferation rates to Wt ES cells 25. However, they have an impaired immediate early growth response and do not express SRF-dependent muscle specific markers such as cardiac-, skeletal-, and SM α-actin 21,22. As expected, the SM22 and SM α-actin promoters were virtually inactive in SRF −/− ES cells but were strongly stimulated (by 12 and 50 fold, respectively) by co-transfection of Wt SRF (figure 1b).
Based upon previous reports on SRF phosphorylation, we generated phosphorylation deficient (T/S to A) and phospho-mimetic (T/S to D/E) mutations to the residues shown in figure 1a and expressed these SRF variants in SRF −/− ES cells. In excellent agreement with a previous study by Iyer et.al. 19, the S162E phosphomimetic dramatically inhibited the activity of both SMC-specific promoters (fig 1c) providing significant validation of our screening approach. The phosphomimetic mutation T159D also completely inhibited SRF’s effects on the SM α-actin promoter and reduced SM22 promoter by about 60%. The c-fos promoter exhibited relatively high basal activity in the SRF −/− ES cells and was only stimulated 4 fold by expression of Wt SRF suggesting that its activity was less dependent upon SRF. Nevertheless, SRF-dependent activation of the c-fos promoter was attenuated by the T159D and S162E mutations. Except for the T103A mutation, which inhibited SRF-dependent activation of the SM22 and SM α-actin promoters by 40–50%, none of the other SRF phosphorylation variants tested had major effects on SRF activity in this model that were not accounted for by differences in SRF expression levels (data not shown).
T159 was phosphorylated by protein kinase A
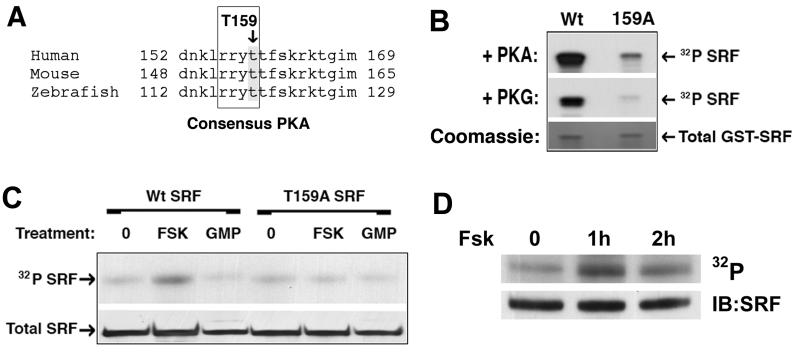
Given the novelty and efficacy of the T159D SRF mutation, we wanted to delineate the molecular mechanisms involved in this effect. Sequence analysis revealed that T159 conforms to a consensus phosphorylation site for cAMP and cGMP-dependent protein kinases, RRXS/T (fig 2a). As a first step to determine whether PKA or PKG could phosphorylate T159, we performed in vitro kinases assays. A GST fusion protein containing the N-terminal third of SRF (AA1–203) was incubated with purified active PKG or PKA. As shown in figure 2b, both of these kinases strongly phosphorylated the Wt construct and this phosphorylation was dramatically inhibited by the T159A mutation. To test whether these kinases could phosphorylate SRF in vivo, we transfected full length flag-tagged SRF into Cos-7 cells and activated PKA or PKG by treatment with forskolin or 8-pCPT-cGMP, respectively. Since Cos-7 cells express very low levels of PKG, the cells to be treated with 8-pCPT-cGMP were co-transfected with PKG. As shown in figure 2c, treatment of cells with forskolin resulted in a significant increase in 32P incorporation in SRF immunoprecipitants and this increase was completely abrogated by the T159A mutation. Activation of PKG failed to significantly increase SRF phosphorylation even though these conditions lead to substantial phosphorylation of the PKG substrate VASP (data not shown). To test whether SRF was phosphorylated by PKA in SMC, we treated 32P labeled primary rat SMC cultures with forskolin and immunoprecipitated endogenous SRF. Although SRF was phosphorylated to some extent under basal conditions, activation of PKA increased this signal (fig 2d). Taken together these results indicate that T159 is a target for PKA in SMC.
Figure 2. PKA phosphorylated SRF T159 in vitro and in vivo.
A) SRF sequence conservation near the consensus PKA/PKG phosphorylation site at T159 B) GST-SRF fusions (AA 1–203) containing Wt or T159A protein sequence were incubated with γ32P-ATP in the presence of active PKA or PKG for 15 min. Following removal of unincorporated label, samples were separated on an SDS-Page gel and exposed to film. C) Cos-7 cells expressing flag-tagged Wt and T159A SRF were incubated with 1mCi ortho 32P for 2h and then stimulated with 20μM forskolin or 100μM 8-pCPT-cGMP for 2h. SRF was then immunoprecipitated from RIPA lysates using anti-flag agarose. Following washing immunoprecipitants were run on an SDS-Page gel, transferred to nitrocellulose, and exposed to film. D) Endogenous SRF was immunoprecipitated from primary SMCs labeled with ortho 32P and then treated with FSK for the indicated times. Immunoprecipitants were run on an SDS-Page gel, transferred to nitrocellulose, and exposed to film.
SRF T159D had promoter-specific effects on myocardin factor transactivation
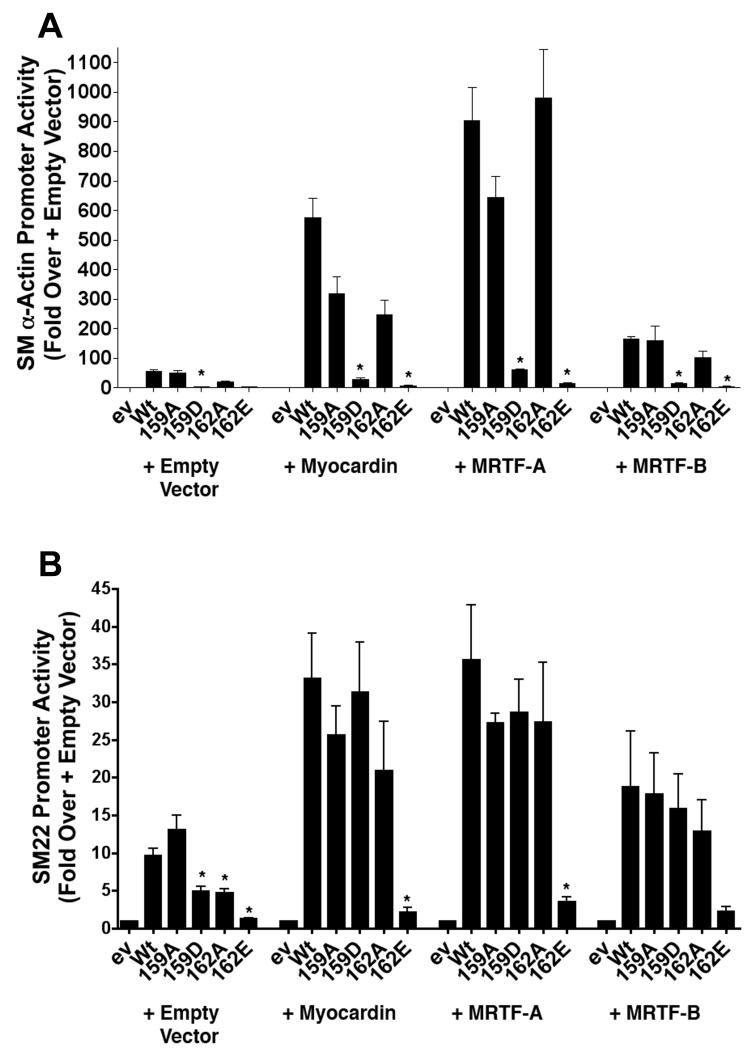
Since SMC differentiation marker gene expression is regulated mainly by SRF’s interaction with the myocardin family of cofactors, we next tested the functional significance of the SRF T159 phosphomimetic on SM α-actin and SM22 transactivation by these factors. As shown in figure 3a, expression of myocardin factors in SRF −/− ES cells strongly increased SM α-actin promoter activity only in the presence of Wt SRF and the T159D mutation almost completely inhibited this response. Interestingly, as shown in 3b, the T159D mutation had no effect on SM22 promoter activity when co-expressed with the myocardin factors suggesting that the effects of this mutation could be rescued under some circumstances. The S162E mutation strongly inhibited both promoters under these conditions.
Figure 3. Myocardin factor over-expression rescued the inhibitory effects of the T159 mutation in a promoter-specific fashion.
SM α-actin (A) and SM22 (B) promoter luciferase constructs were co-transfected into SRF −/− ES cells along with the indicated SRF variant and myocardin factor. Luciferase activity was measured at 24h. *p<0.05 versus Wt SRF.
Phosphorylation of T159 inhibited SRF-CArG binding
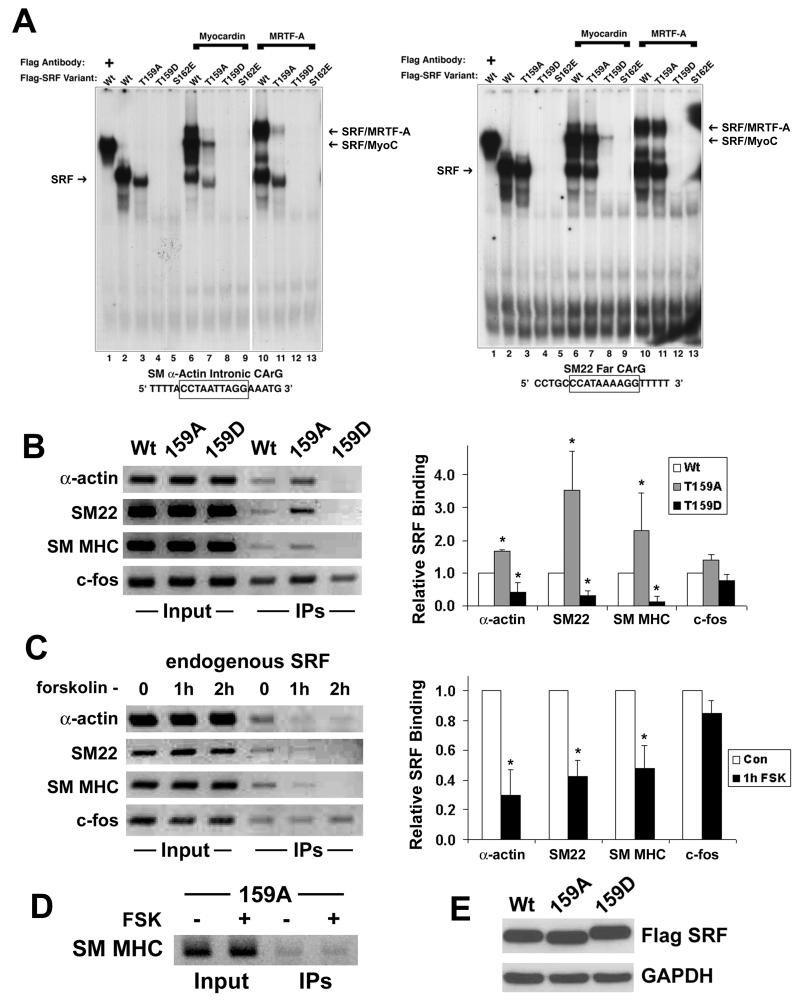
T159 is located within the amphipathic αI helix of the SRF MADS box that interacts with the CArG element. It lies on the same helical face as S162, and like S162, has been shown to interact with nucleotides flanking the CArG sequence 26,27. Given that S162 phosphorylation inhibited SRF-CArG binding 19, we hypothesized that the inhibitory effect of the T159D mutation on SMC-specific transcription was due to a similar mechanism. To test this, we measured the binding of Wt SRF and SRF phosphorylation variants to CArG elements using gel shift assays. As shown in the left panel of figure 4a, incubation of in vitro translated Wt flag-tagged SRF with the SM α-actin intronic CArG resulted in a prominent complex that was supershifted by the addition of anti-flag Ab. SRF binding was slightly reduced by the T159A mutation but was completely inhibited by both the T159D and S162E mutations (lanes 4 And 5). As shown in lanes 6–13 the addition of myocardin or ΔNMRTF-A resulted in the formation of a ternary complex with Wt SRF, but not with the T159D or S162E SRF variants.
Figure 4. Phosphorylation of T159 inhibited SRF binding to the SMC-specific CArGs.
A) Radiolabeled oligonucleotide probes containing either the SM α-actin intronic CArG (left panel) or the SM22 far CArG (right panel) were incubated with the indicated SRF variant in the absence or presence of myocardin or ΔNMRTF-A. All proteins were in vitro translated and the total amount of TnT protein in each reaction was maintained by the addition of unprogrammed lysate. After 30 min binding reactions were run on a 5% non-denaturing gel, transferred to filter paper, and visualized by autoradiography. For supershifts (lane 1), 1μL of anti-flag antibody was added after 20 minutes of incubation. B) 10T1/2 cells were transfected with the Wt, T159A, or T159D SRF variant. After 48h cells were processed for ChIP assays by standard protocols (see methods for more details). PCR reactions were performed on immunoprecipitants using primers spanning the SM α-actin intronic CArG, the SM22 far CArG, a SM MHC 5′ CArG, and the c-fos CArG. Quantification of 3 separate ChIp assays is shown in the right panel. C) SMCs grown in 10% FBS were treated with 20μM forskolin for the indicated time before processing for ChIP assays. * p<0.05 versus vehicle treated. D) 10T1/2 cells expressing the T159 SRF variant were treated with forskolin for 1h before processing for ChIP assays. E) Western blot of SRF variant expression in 10T1/2 cells.
Interestingly, the results of gel shift assays with the SM22 far CArG were slightly different (Fig 4a, right panel). First, the T159A mutation had much less of an effect on SRF binding and myocardin factor complex formation. More importantly, the presence of myocardin could partially restore binding of the T159D variant to the SM22 far CArG (lane 8). The presence of ΔNMRTF-A also resulted in a ternary complex (lane 12) that was more clearly visible after slightly longer exposure times. These data support previous studies from our lab indicating that differences in CArG and/or CArG flanking sequence have important effects on CArG/SRF/myocardin factor ternary complex formation 28 and indicate that SRF phosphorylation may have preferential effects on certain CArG-dependent genes. In addition, these data may explain the ability of myocardin factor over-expression to rescue the effects of the T159D mutation on SM22 promoter activity.
To test whether T159 phosphorylation affected SRF binding in vivo, we performed chromatin immunoprecipitation (ChIP) assays in 10T1/2 cells over-expressing Wt, T159A, and T159D SRF variants. In this assay, if SRF is present at a CArG-containing promoter region upon fixation, then these promoter fragments would be detected by PCR in SRF immunoprecipitates. As shown in figure 4b (and quantified at right), the binding of the T159A variant to the CArG-containing regions of the SM α-actin, SM22, and SM MHC promoters was significantly stronger than that observed with Wt SRF even though these proteins were expressed at identical levels (Fig 4E). Consistent with our gel shift data, binding of the T159D SRF variant to all of the SMC-specific promoters was significantly weaker than Wt. Binding of the T159A SRF variant to the SM MHC promoter in 10T1/2 cells was resistant to the effects of forskolin (Fig 4d), directly implicating T159 phosphorylation by PKA in this mechanism. Importantly, we also used ChIP assays in primary SMC to demonstrate that the interaction of endogenous SRF with the SMC-specific promoters was inhibited by forskolin treatment (figure 4c). Interestingly, the phosphorylation mutations or forskolin treatment had no significant effect on SRF binding to the CArG-containing region of the c-fos promoter.
The T159D mutation inhibited SMC differentiation of ES Cells and SMC
The results presented so far indicate that SRF is phosphorylated by PKA at T159 and that this phosphorylation can inhibit SMC-specific promoter activity. To further test this regulatory mechanism on SMC differentiation, we used a previously described monolayer ES cell model in which endogenous SMC differentiation marker gene expression is induced by removal of LIF and addition of retinoic acid 20-22. As expected, SMC differentiation marker gene mRNAs were not detected in SRF −/− ES cells transfected with empty expression vector at any time-point of the differentiation protocol. In contrast, transfection of Wt SRF into SRF −/− ES cells induced endogenous SM α-actin and SM22 message levels by 55 and 20 fold, respectively, and these levels were further increased at day 3 and day 6 of the differentiation protocol (figure 5a). The T159D and S162E mutations significantly reduced SM α-actin and SM22 message levels at day zero and prevented any further increase in message levels at the later time-points (figure 5b). Interestingly, the T159A mutation actually increased SM22 message levels suggesting at least some basal SRF phosphorylation under these conditions.
Figure 5. The T159D mutation inhibited endogenous SMC marker gene expression in an ES cell model of SMC differentiation.
A) SRF −/− ES cells were transfected with Wt SRF. After 24 h cells were placed in differentiation media (minus LIF, plus 500nM retinoic acid) for 0, 3, and 6 days. SM α-actin and SM22 message levels were measured by quantitative PCR. * p<0.05 versus plus empty expression vector. B) SRF −/− ES cells expressing the indicated SRF variants were subjected to the SMC differentiation protocol. SM α-actin and SM22 mRNA levels measured at 0, 3, and 6 d are expressed relative to Wt SRF set to 1. * p<0.05 versus Wt SRF
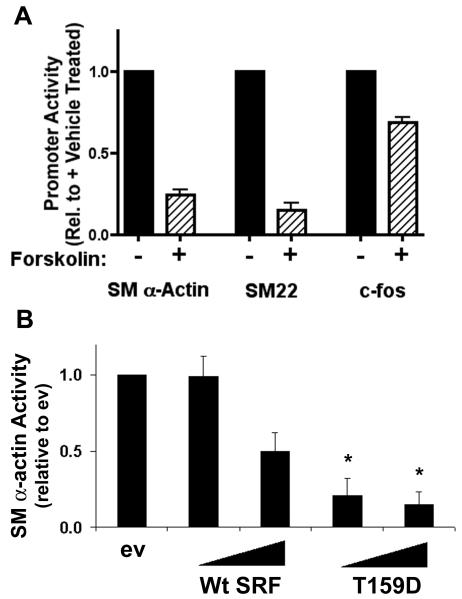
Finally, we performed several experiments to test whether PKA-dependent phosphorylation of SRF affected SMC-specific transcription in mature SMC. As shown in figure 6b, forskolin dramatically inhibited SM α-actin and SM22 promoter activity in primary SMC cultures but had only minor effects on c-fos. We also tested whether the T159D functioned as a dominant negative in SMC. As previously demonstrated by us and others, expression of Wt SRF in SMC led to a dose-dependent inhibition of SM α-actin promoter activity most likely due to the squelching of transcription factors or co-factors required for promoter activity. Importantly the inhibitory effects of the T159D mutation were significantly greater than Wt providing additional evidence that this signaling mechanism plays a role in the regulation of SMC phenotype.
Figure 6. PKA activation inhibited SM α-actin and SM22 promoter activity in primary SMC.
A) Primary SMCs were transfected with SM α-actin, SM22, or c-fos promoter luciferase constructs. Luciferase activity was measured after treatment with 20μM forskolin for 9h. * p<0.05 versus vehicle. B) Primary SMC cultures were co-transfected with SM α-actin-luciferase and increasing concentrations of Wt or T159D SRF. * p<0.05 versus same concentration of Wt SRF.
Discussion
Extensive evidence indicates that SRF plays a critical role in the regulation of SMC-specific and early response growth gene expression, and a major goal of these studies was to identify signaling mechanisms that regulate SRF’s involvement in these disparate gene programs. Although many kinases have been identified that phosphorylate SRF, the significance of these modifications in regard to the regulation of SMC-specific gene expression are somewhat unclear. Thus, in the present study we used a relatively unbiased approach to identify phosphorylation sites on SRF that regulate its ability to drive SMC-specific expression. By expressing phosphorylation mutants in SRF −/− ES cells, we identified T159 as a phosphorylation site that negatively regulates SRF activity. T159 was phosphorylated by PKA in vitro and in vivo, and gel shift and ChIP assays strongly suggest that this phosphorylation regulates SRF’s interaction with SMC-specific CArG elements.
Several studies have characterized SRF’s interaction with CArG-containing oligonucleotides, and the major contacts between SRF and DNA are between the basic residues within the SRF αI helix and the core CArG sequence 26,27,29. However, as shown in the original crystal structure of the core SRF dimer bound to DNA 26, T159 and S162 of one SRF αI helix contact a residue on the 5′ flank of the CArG element 26. Interestingly, Mo et al. demonstrated that the presence of the SRF accessory protein, SAP-1, within the ternary complex significantly increased SRF contacts with the CArG flanking regions of the c-fos SRE including additional interactions between S162/T159 on the second αI helix with a thymine residue to the 3′ side of the CArG box 27. Thus, the inhibitory effects of the T159 and S162 phosphorylation observed in the present studies are consistent with the importance of these residues for SRFs interaction with DNA. Interestingly, a similar crystal structure study on the SAP-1-containing ternary complex by Hassler et. al. did not detect an interaction between T159 and CArG flanking sequence 29. Although the precise nature of this discrepancy is somewhat unclear, the CArG-containing oligonucleotide used by this group was substantially different suggesting that sequence variations within or flanking the CArG element may play a role in determining SRF conformation in the ternary complex. Our demonstration that the myocardin factors could specifically rescue the effects of the T159D mutation on SRF binding to and activation of the SM22 promoter indicates that the myocardin factors alter SRF-CArG interactions and supports the idea that sequence variation within the CArG or CArG flanking regions plays an important role in this mechanism.
The ability of the T159A mutation to enhance SRF binding in ChIP assays and the observation that T159A binding in this assay was resistant to forskolin treatment provide additional support for a direct role for PKA-mediated phosphorylation in the regulation of SRF binding. The observation that the T159A mutation did not consistently increase SMC-specific transcription in functional assays suggests that basal levels of T159 phosphorylation are relatively low or that SRF binding is not rate limiting under the conditions tested. Interestingly, the T159A had negative effects on transcriptional activity in some experiments and on DNA binding in gel shift assays. Similar effects were seen with the 162A mutation (by us and by Iyer et.al. 19) perhaps suggesting that these S/T residues in their non-phosphorylated form may be important for SRF function.
Activation of PKA has many affects in the cell and its ability to inhibit SRF binding to SMC-specific CArG elements may only partially explain its effects on SMC-specific transcription. Indeed, it is well known that PKA antagonizes RhoA-dependent signaling in a variety of cell types (including SMC) resulting in inhibition of actin stress fiber formation 30,31. Since we have shown that actin polymerization positively regulates SMC-specific transcription by enhancing the nuclear localization of the MRTFs 23,28,32, it PKA may also inhibit SMC-specific transcription by this mechanism. Interestingly, PKA and RhoA signaling also have antagonistic effects on SMC contractility, and we postulate that these pathways may act in combination to couple agonist-induced changes in SMC contractility to subsequent changes in contractile protein gene expression.
Several lines of evidence from the current study indicated that T159 phosphorylation has less of an effect on c-fos promoter activity. First, the overall activity of the c-fos promoter was less dependent upon the presence of SRF. Second, SRF binding to the c-fos SRE in ChIP assays was not affected by the T159D mutation or forskolin treatment. These results likely reflect the stabilization of SRF binding to the c-fos SRE by the TCFs 19. Finally, PKA treatment of SMC resulted in a relatively small reduction in c-fos promoter activity. Because the c-fos promoter is also regulated by a cAMP response element, the positive effects of PKA on CREB may limit the negative effects of PKA on SRF. While most previous studies suggest that PKA activation inhibits SMC proliferation (see 33 for review), our data suggest that it may be more important for down regulating the SMC differentiation gene program. Clearly, SMC growth and differentiation are not mutually exclusive 2 and PKA signaling may have independent effects on these important SMC parameters.
Our results also have important implications on control of SMC phenotype by environmental cues that regulate cAMP levels. A previous study by Davis et. al. demonstrated that activation of PKA in rat aortic SMC inhibited while dominant negative PKA enhanced SRF-dependent transcription 34. Subsequently, this group showed that activation of PKA by the Gαs-coupled agonists, isoproterenol and ATP, resulted in a decrease in endogenous SMC differentiation marker gene expression 35. In contrast, Fetalvero et al. demonstrated in human aortic SMC that the prostacyclin analog, iloprost, enhanced SMC differentiation marker gene expression by a mechanism that was dependent upon PKA 36. Although the discrepancy between these studies could be explained by species differences, it is also possible that agonist-specific differences in the level, timing, or localization of PKA activation by these agonists may also play a role. Of interest, genetic deletion of the inhibitory RIα subunit of PKA in the mouse (which led to constitutive PKA activity) resulted in defective mesoderm formation 37, a phenotype very similar to that observed in SRF knock out mice 22,25.
Our findings are somewhat contrary to an earlier study demonstrating that a T159 phosphomimetic enhanced cardiac α-actin promoter activity in CV-1 cells 38. In the original report of these results, the T159D mutation did not affect SRF binding, and the positive effects of the T159D variant were attributed to increased association of SRF with additional transcription factors such as GATA-4 and Nkx2.5. However, during the completion on of the current study these authors published an erratum to this manuscript that was prompted by the discovery of a sequencing error in their T159D construct (Biochemistry. 2008 Feb 5;47(5):1464). Although they now conclude that the T159D mutation inhibits SRF binding, they confirmed that this mutation increases cardiac α-actin-promoter activity in CV-1 cells. ES cells grown in monolayer culture do not express Nkx2.5 and Gata-4 39,40 and the lack of this (or some other) positive transcription factor interaction could help explain this discrepancy. It is also important to note that these authors demonstrated that T159 was a possible substrate for myotonic dystrophy protein kinase (DMPK) and PKCα. In the current study, we focused on the role of PKA because it had previously been shown to have negative effects on SMC-specific transcription. While, we cannot rule out that DMPK and/or PKCα phosphorylate SRF at T159, our data indicate that such a modification would inhibit SMC-specific transcription.
In summary, the results from the present study indicate that phosphorylation of SRF at T159 by PKA inhibits SMC-specific transcription by inhibiting SRF’s interaction with CArG elements. The promoter specific effects of this modification in the presence of the myocardin factors suggest that it may be an important mechanism by which SRF differentially regulates the many genes that are CArG-dependent. Thus, it will be critical to further investigate the role of PKA signaling on SMC phenotype and to identify the consequences of this modification on SMC function.
Supplementary Material
Acknowledgements
a) Sources of funding - This work was supported by NIH Grants HL-070953 (CPM) and HL-071054 (JMT).
b) Acknowledgements - none
c) Disclosures - none
References
- 1.Ross R. The pathogenesis of atherosclerosis: a perspective for the 1990s. Nature. 1993;362:801–9. doi: 10.1038/362801a0. [DOI] [PubMed] [Google Scholar]
- 2.Owens GK, Kumar MS, Wamhoff BR. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol Rev. 2004;84:767–801. doi: 10.1152/physrev.00041.2003. [DOI] [PubMed] [Google Scholar]
- 3.Mack CP, Owens GK. Regulation of smooth muscle alpha-actin expression in vivo is dependent on CArG elements within the 5′ and first intron promoter regions. Circ Res. 1999;84:852–61. doi: 10.1161/01.res.84.7.852. [DOI] [PubMed] [Google Scholar]
- 4.Mack CP, Thompson MM, Lawrenz-Smith S, Owens GK. Smooth muscle alpha-actin CArG elements coordinate formation of a smooth muscle cell-selective, serum response factor-containing activation complex. Circ Res. 2000;86:221–32. doi: 10.1161/01.res.86.2.221. [DOI] [PubMed] [Google Scholar]
- 5.Madsen CS, Hershey JC, Hautmann MB, White SL, Owens GK. Expression of the smooth muscle myosin heavy chain gene is regulated by a negative-acting GC-rich element located between two positive-acting serum response factor-binding elements. J Biol Chem. 1997;272:6332–40. doi: 10.1074/jbc.272.10.6332. [DOI] [PubMed] [Google Scholar]
- 6.Miano JM, Carlson MJ, Spencer JA, Misra RP. Serum response factor-dependent regulation of the smooth muscle calponin gene [In Process Citation] J Biol Chem. 2000;275:9814–22. doi: 10.1074/jbc.275.13.9814. [DOI] [PubMed] [Google Scholar]
- 7.Li L, Liu Z, Mercer B, Overbeek P, Olson EN. Evidence for serum response factor-mediated regulatory networks governing SM22alpha transcription in smooth, skeletal, and cardiac muscle cells. Dev Biol. 1997;187:311–21. doi: 10.1006/dbio.1997.8621. [DOI] [PubMed] [Google Scholar]
- 8.Herring BP, Smith AF. Telokin expression in A10 smooth muscle cells requires serum response factor. Am J Physiol. 1997;272:C1394–404. doi: 10.1152/ajpcell.1997.272.4.C1394. [DOI] [PubMed] [Google Scholar]
- 9.Miano JM, Long X, Fujiwara K. Serum response factor: master regulator of the actin cytoskeleton and contractile apparatus. Am J Physiol Cell Physiol. 2007;292:C70–81. doi: 10.1152/ajpcell.00386.2006. [DOI] [PubMed] [Google Scholar]
- 10.Pipes GC, Creemers EE, Olson EN. The myocardin family of transcriptional coactivators: versatile regulators of cell growth, migration, and myogenesis. Genes Dev. 2006;20:1545–56. doi: 10.1101/gad.1428006. [DOI] [PubMed] [Google Scholar]
- 11.Misra RP, Rivera VM, Wang JM, Fan PD, Greenberg ME. The serum response factor is extensively modified by phosphorylation following its synthesis in serum-stimulated fibroblasts. Mol Cell Biol. 1991;11:4545–54. doi: 10.1128/mcb.11.9.4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heidenreich O, Neininger A, Schratt G, Zinck R, Cahill MA, Engel K, Kotlyarov A, Kraft R, Kostka S, Gaestel M, Nordheim A. MAPKAP kinase 2 phosphorylates serum response factor in vitro and in vivo. J Biol Chem. 1999;274:14434–43. doi: 10.1074/jbc.274.20.14434. [DOI] [PubMed] [Google Scholar]
- 13.Manak JR, de Bisschop N, Kris RM, Prywes R. Casein kinase II enhances the DNA binding activity of serum response factor. Genes Dev. 1990;4:955–67. doi: 10.1101/gad.4.6.955. [DOI] [PubMed] [Google Scholar]
- 14.Prywes R, Dutta A, Cromlish JA, Roeder RG. Phosphorylation of serum response factor, a factor that binds to the serum response element of the c-FOS enhancer. Proc Natl Acad Sci U S A. 1988;85:7206–10. doi: 10.1073/pnas.85.19.7206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gauthier-Rouviere C, Vandromme M, Lautredou N, Cai QQ, Girard F, Fernandez A, Lamb N. The serum response factor nuclear localization signal: general implications for cyclic AMP-dependent protein kinase activity in control of nuclear translocation. Mol Cell Biol. 1995;15:433–44. doi: 10.1128/mcb.15.1.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janknecht R, Hipskind RA, Houthaeve T, Nordheim A, Stunnenberg HG. Identification of multiple SRF N-terminal phosphorylation sites affecting DNA binding properties. Embo J. 1992;11:1045–54. doi: 10.1002/j.1460-2075.1992.tb05143.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marais RM, Hsuan JJ, McGuigan C, Wynne J, Treisman R. Casein kinase II phosphorylation increases the rate of serum response factor-binding site exchange. Embo J. 1992;11:97–105. doi: 10.1002/j.1460-2075.1992.tb05032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manak JR, Prywes R. Phosphorylation of serum response factor by casein kinase II: evidence against a role in growth factor regulation of fos expression. Oncogene. 1993;8:703–11. [PubMed] [Google Scholar]
- 19.Iyer D, Chang D, Marx J, Wei L, Olson EN, Parmacek MS, Balasubramanyam A, Schwartz RJ. Serum response factor MADS box serine-162 phosphorylation switches proliferation and myogenic gene programs. Proc Natl Acad Sci U S A. 2006;103:4516–21. doi: 10.1073/pnas.0505338103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Philippar U, Schratt G, Dieterich C, Muller JM, Galgoczy P, Engel FB, Keating MT, Gertler F, Schule R, Vingron M, Nordheim A. The SRF Target Gene Fhl2 Antagonizes RhoA/MAL-Dependent Activation of SRF. Mol Cell. 2004;16:867–80. doi: 10.1016/j.molcel.2004.11.039. [DOI] [PubMed] [Google Scholar]
- 21.Schratt G, Weinhold B, Lundberg AS, Schuck S, Berger J, Schwarz H, Weinberg RA, Ruther U, Nordheim A. Serum response factor is required for immediate-early gene activation yet is dispensable for proliferation of embryonic stem cells. Mol Cell Biol. 2001;21:2933–43. doi: 10.1128/MCB.21.8.2933-2943.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weinhold B, Schratt G, Arsenian S, Berger J, Kamino K, Schwarz H, Ruther U, Nordheim A. Srf(−/−) ES cells display non-cell-autonomous impairment in mesodermal differentiation. Embo J. 2000;19:5835–44. doi: 10.1093/emboj/19.21.5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lockman K, Hinson JS, Medlin MD, Morris D, Taylor JM, Mack CP. Sphingosine 1-Phosphate Stimulates Smooth Muscle Cell Differentiation and Proliferation by Activating Separate Serum Response Factor Co-factors. J Biol Chem. 2004;279:42422–30. doi: 10.1074/jbc.M405432200. [DOI] [PubMed] [Google Scholar]
- 24.Lockman K, Taylor JM, Mack CP. The histone demethylase, Jmjd1a, interacts with the myocardin factors to regulate SMC differentiation marker gene expression. Circ Res. 2007;101:e115–23. doi: 10.1161/CIRCRESAHA.107.164178. [DOI] [PubMed] [Google Scholar]
- 25.Arsenian S, Weinhold B, Oelgeschlager M, Ruther U, Nordheim A. Serum response factor is essential for mesoderm formation during mouse embryogenesis. Embo J. 1998;17:6289–99. doi: 10.1093/emboj/17.21.6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pellegrini L, Tan S, Richmond TJ. Structure of serum response factor core bound to DNA. Nature. 1995;376:490–8. doi: 10.1038/376490a0. see comments. [DOI] [PubMed] [Google Scholar]
- 27.Mo Y, Ho W, Johnston K, Marmorstein R. Crystal structure of a ternary SAP-1/SRF/c-fos SRE DNA complex. J Mol Biol. 2001;314:495–506. doi: 10.1006/jmbi.2001.5138. [DOI] [PubMed] [Google Scholar]
- 28.Hinson JS, Medlin MD, Lockman K, Taylor JM, Mack CP. Smooth muscle cell-specific transcription is regulated by nuclear localization of the myocardin-related transcription factors. Am J Physiol Heart Circ Physiol. 2007;292:H1170–80. doi: 10.1152/ajpheart.00864.2006. [DOI] [PubMed] [Google Scholar]
- 29.Hassler M, Richmond TJ. The B-box dominates SAP-1-SRF interactions in the structure of the ternary complex. Embo J. 2001;20:3018–28. doi: 10.1093/emboj/20.12.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hirshman CA, Zhu D, Panettieri RA, Emala CW. Actin depolymerization via the beta-adrenoceptor in airway smooth muscle cells: a novel PKA-independent pathway. Am J Physiol Cell Physiol. 2001;281:C1468–76. doi: 10.1152/ajpcell.2001.281.5.C1468. [DOI] [PubMed] [Google Scholar]
- 31.Nguyen GH, French R, Radhakrishna H. Protein kinase A inhibits lysophosphatidic acid induction of serum response factor via alterations in the actin cytoskeleton. Cell Signal. 2004;16:1141–51. doi: 10.1016/j.cellsig.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 32.Staus DP, Blaker AL, Taylor JM, Mack CP. Diaphanous 1 and 2 regulate smooth muscle cell differentiation by activating the myocardin-related transcription factors. Arterioscler Thromb Vasc Biol. 2007;27:478–86. doi: 10.1161/01.ATV.0000255559.77687.c1. [DOI] [PubMed] [Google Scholar]
- 33.Bornfeldt KE, Krebs EG. Crosstalk between protein kinase A and growth factor receptor signaling pathways in arterial smooth muscle. Cell Signal. 1999;11:465–77. doi: 10.1016/s0898-6568(99)00020-0. [DOI] [PubMed] [Google Scholar]
- 34.Davis A, Hogarth K, Fernandes D, Solway J, Niu J, Kolenko V, Browning D, Miano JM, Orlov SN, Dulin NO. Functional significance of protein kinase A activation by endothelin-1 and ATP: negative regulation of SRF-dependent gene expression by PKA. Cell Signal. 2003;15:597–604. doi: 10.1016/s0898-6568(02)00148-1. [DOI] [PubMed] [Google Scholar]
- 35.Hogarth DK, Sandbo N, Taurin S, Kolenko V, Miano JM, Dulin NO. Dual role of PKA in phenotypic modulation of vascular smooth muscle cells by extracellular ATP. Am J Physiol Cell Physiol. 2004;287:C449–56. doi: 10.1152/ajpcell.00547.2003. [DOI] [PubMed] [Google Scholar]
- 36.Fetalvero KM, Shyu M, Nomikos AP, Chiu YF, Wagner RJ, Powell RJ, Hwa J, Martin KA. The prostacyclin receptor induces human vascular smooth muscle cell differentiation via the protein kinase A pathway. Am J Physiol Heart Circ Physiol. 2006;290:H1337–46. doi: 10.1152/ajpheart.00936.2005. [DOI] [PubMed] [Google Scholar]
- 37.Amieux PS, Howe DG, Knickerbocker H, Lee DC, Su T, Laszlo GS, Idzerda RL, McKnight GS. Increased basal cAMP-dependent protein kinase activity inhibits the formation of mesoderm-derived structures in the developing mouse embryo. J Biol Chem. 2002;277:27294–304. doi: 10.1074/jbc.M200302200. [DOI] [PubMed] [Google Scholar]
- 38.Iyer D, Belaguli N, Fluck M, Rowan BG, Wei L, Weigel NL, Booth FW, Epstein HF, Schwartz RJ, Balasubramanyam A. Novel phosphorylation target in the serum response factor MADS box regulates alpha-actin transcription. Biochemistry. 2003;42:7477–86. doi: 10.1021/bi030045n. [DOI] [PubMed] [Google Scholar]
- 39.Capo-Chichi CD, Rula ME, Smedberg JL, Vanderveer L, Parmacek MS, Morrisey EE, Godwin AK, Xu XX. Perception of differentiation cues by GATA factors in primitive endoderm lineage determination of mouse embryonic stem cells. Dev Biol. 2005;286:574–86. doi: 10.1016/j.ydbio.2005.07.037. [DOI] [PubMed] [Google Scholar]
- 40.Christoforou N, Miller RA, Hill CM, Jie CC, McCallion AS, Gearhart JD. Mouse ES cell-derived cardiac precursor cells are multipotent and facilitate identification of novel cardiac genes. J Clin Invest. 2008;118:894–903. doi: 10.1172/JCI33942. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.