Abstract
Riboswitches are non-protein coding RNA elements typically found in the 5’ untranslated region (5’-UTR) of mRNAs that utilize metabolite binding to control expression of their own transcript. The RNA-ligand interaction causes conformational changes in the RNA that direct the cotranscriptional folding of a downstream secondary structural switch that interfaces with the expression machinery. This review describes the structural themes common to the different RNA-metabolite complexes studied to date and conclusions that can be made regarding how these RNAs efficiently couple metabolite binding to gene regulation. Emphasis is placed on the temporal aspects of riboswitch regulation that are central to the function of these RNAs and the need to augment the wealth of data on metabolite receptor domains with further studies on the full regulatory element, particularly in the context of transcription.
Keywords: Riboswitch, metabolite binding, cotranscriptional folding, gene expression
1. Introduction
Bacteria continuously modulate gene expression in reaction to physical and chemical fluctuations in the environment, allowing them to tune their metabolism appropriately to avoid wasteful energy expenditure or inappropriate physiological responses. This formidable challenge employs numerous mechanisms of gene regulation including a large repository of non-protein coding RNAs with diverse biological functions. The regulatory capacity of RNA is readily illustrated by cis-acting elements found in 5’-UTRs of bacterial mRNAs that respond to physiological stimuli without the aid of proteins, thereby allowing mRNA to direct its own expression [1, 2]. These elements allow mRNAs to directly respond to many discrete environmental cues including changes in temperature, levels of uncharged tRNA, and the concentration of specific metal ions or small molecule metabolites [3]. The primary commonality of all of these regulatory mechanisms, along with some protein-assisted ones, is the presence of a sequence that adopts one of two mutually exclusive secondary structures that lead to expression or repression of the parent transcript [4]. This folding decision occurs cotranscriptionally, with ligand binding steering the RNA into one of two folding pathways that ultimately dictates its fate.
Metabolite sensing RNAs, better known as riboswitches, represent a fundamental mechanism of gene regulation that is widespread in the bacterial kingdom; they control ~4% genes in the Bacillus subtilis genome alone [5]. In bacteria, they are always found in the 5’-UTR, although examples have been identified in the introns or 3’-UTR in eukaryotic transcripts [6]. There are currently at least twenty classes of riboswitches that recognize a broad range of ligands including purine nucleobases [7, 8], amino acids [9, 10], vitamin cofactors [11, 12], aminosugars [13], metal ions [14], and second messenger molecules [15]. Many “orphan” classes have also been identified using phylogenetic analyses but their metabolites await identification [16, 17].
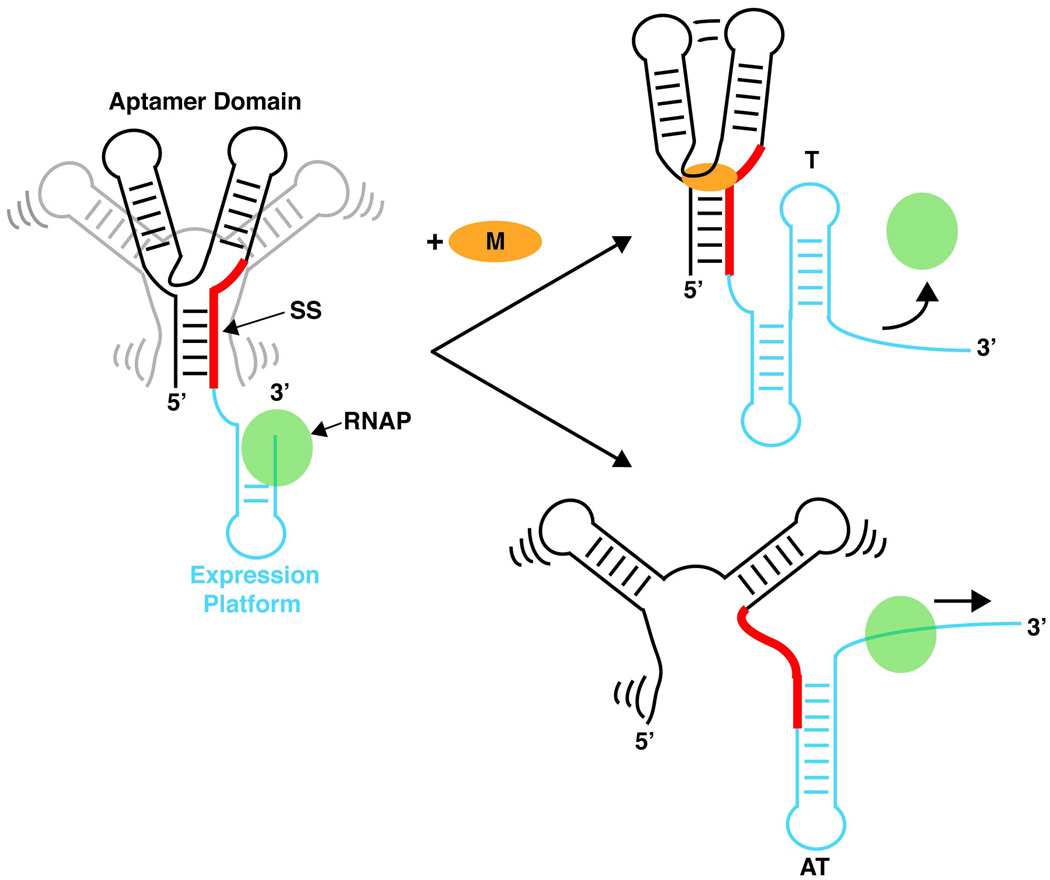
Riboswitches are typically composed of two distinct domains: a metabolite receptor known as the aptamer domain and an expression platform whose secondary structure signals the regulatory response (Figure 1). Due to the polarity of transcription, the aptamer domain is transcribed first, providing time for this receptor to sense the cellular environment before the expression platform is fully synthesized. Embedded within the aptamer domain is the switching sequence (Figure 1, red strand), a sequence shared between the aptamer domain and expression platform. The domain this sequence becomes incorporated into dictates which of the two secondary structures the expression domain adopts and thus, the expression fate of the mRNA. Though the term “switch” implies that these secondary structures can interconvert, there is significant evidence that they are typically heavily biased towards a default fold even in the presence of saturating ligand concentrations [18], suggesting that many riboswitches operate more like cotranscriptional fuses.
Figure 1. Riboswitch regulated transcription termination.
The aptamer domain (black/gray) forms prior to transcription of the expression platform (cyan), allowing this receptor to sense the concentration of metabolite (orange circle). Metabolite binding collapses an ensemble of the aptamer domain into a single conformation that sequesters the switching sequence (SS) away from pairing with the expression platform. This creates a folding pathway that leads to formation of a terminator stem (T) that causes transcription attenuation. In the event that ligand binding does not occur, particular elements of the aptamer domain remain loosely structured, leading to a conformational switch, that pairs the SS in the expression platform to form the anti-terminator stem (AT), allowing RNA polymerase (RNAP) to continue transcribing into the downstream open reading frame.
Although riboswitches provide a relatively simple means of control, these RNAs can be arranged in tandem to generate more complex regulatory responses [4, 19]. The simplest form of this involves a tandem arrangement consists of two riboswitches that respond to the same ligand. The gvcT operon for example is controlled by a glycine riboswitch with two aptamer domains that cooperatively bind glycine, a remarkable feature that had only previously been observed for proteins [9, 20]. This allows a more digital response to small changes in ligand concentration that is unattainable by single copy riboswitches. In other cases the expression response can be coordinated by distinct metabolites, such as that found in the 5’-UTR of the Bacillus clausii metE gene that contains both S-adenosylmethionine (SAM) and adenosylcobalamin (B12) responsive elements [21]. Each of these riboswitches can independently promote transcription termination upon ligand binding making this mRNA responsive to changes in the concentration of either metabolite. Riboswitches can also overlap with other posttranscriptional control mechanisms. This was recently described for the Enterococcus faecalis eut operon, whose expression allows ethanolamine utilization within the human gastrointestinal tract and is thought to affect the virulence of this organism [22]. The operon’s polycistronic transcript contains at least one B12 riboswitch that terminates transcription in the presence of coenzyme B12 and many copies of a conserved RNA stem loop that is bound by a response regulatory protein to promote antitermination [22]. In such cases, post-transcriptional regulation rivals the complexity of DNA binding transcription factors that can finely tune gene expression from a specific promoter.
The versatility of riboswitches is further demonstrated by their ability to regulate by a number of means. The most common mode is that of transcription attenuation whereby ligand binding guides formation of a rho-independent terminator in the expression platform resulting in transcriptional termination (Figure 1) [23]. However, there are also several examples of transcriptional “on” switches, such as the pbuE riboswitch in B. subtilis, that use ligand binding to direct formation of an antiterminator while the default folding pathway leads to termination, demonstrating the modularity of these RNAs [8]. In translational control, riboswitches utilize ligand binding to sequester the Shine-Dalgarno sequence within a helix, thereby impeding association with the 30S ribosomal subunit [12]. Additionally, riboswitches can alter the rate of mRNA degradation by directing cleavage or splicing events that promote degradation of the RNA. The glmS riboswitch takes a unique approach to this by acting as a ribozyme that uses glucosammine-6-phosphate (GN6P) as an enzymatic cofactor that participates directly in cleavage of the 5’-UTR resulting in destabilization of the transcript [13, 24, 25]. In contrast, eukaryotic thiamine pyrophosphate (TPP) riboswitches, found in the 3’-UTR or intergenic region of specific thiamine biosynthesis genes, can control alternative splicing of the mRNA by sequestering splice sites within the riboswitch upon ligand binding [6, 26].
The discovery of RNA switches in the 3’-UTR of other eukaryotic genes leads to speculation that this region may provide a fertile ground for the discovery of novel mechanisms of riboregulation in these organisms. Indeed, a recently discovered RNA switch in the 3’-UTR of the human VEGFA mRNA forms mutually exclusive structures that alter the production of this growth factor during oxidative stress in myeloid cells [27]. Although this regulatory element is not a true riboswitch, it does adopt mutually exclusive folds that each bind different proteins, each coordinating distinct expression responses [27]. This process is driven by inhibiting proteasomal degradation of one protein during oxidative stress that allows for its intracellular accumulation, thus tipping the balance towards one of the two conformations. Mechanistic details of this process await further investigation, but this study highlights the continuing expansion of our knowledge of the scope of riboregulation in eukaryotic organisms.
2. Structural themes of riboswitch aptamer domains
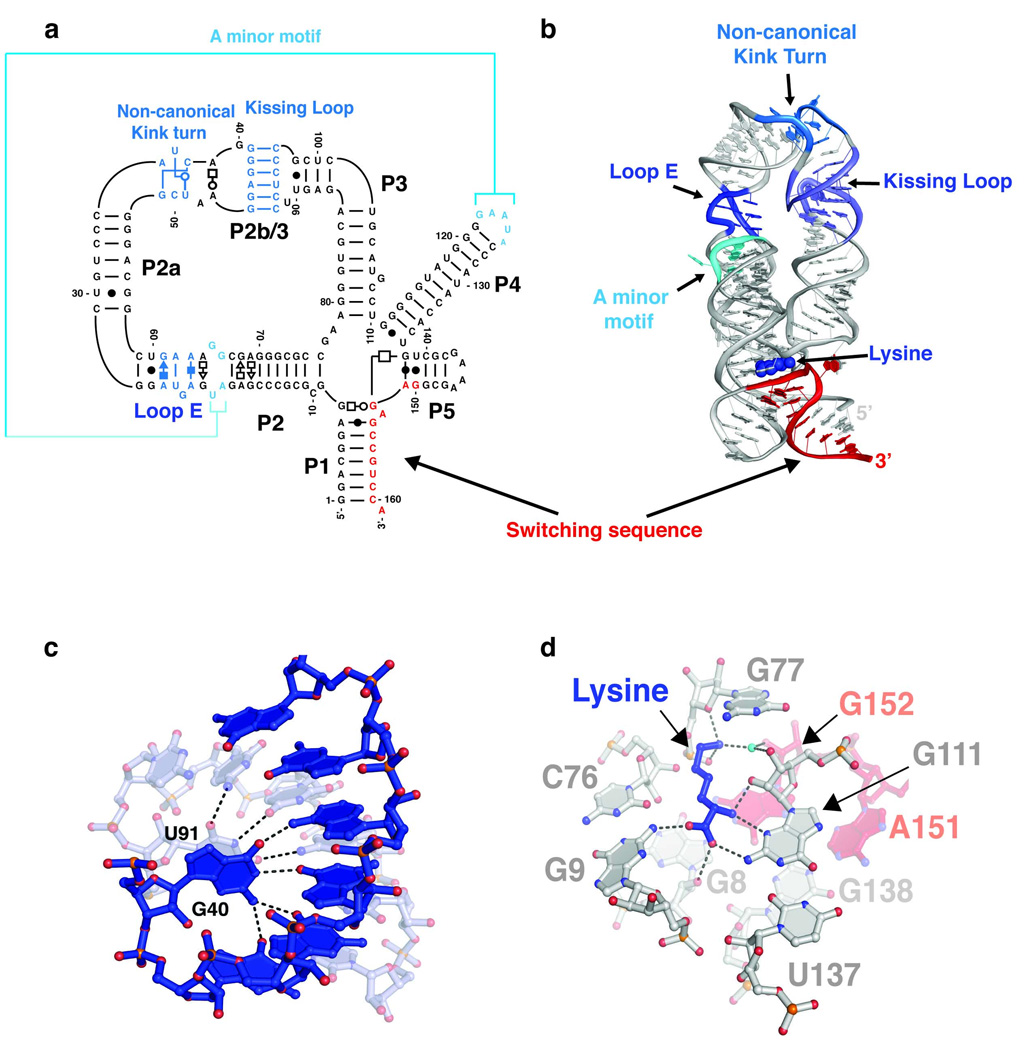
Many riboswitch regulated pathways are essential to either bacterial survival or virulence, prompting efforts to explore their utility as antibiotic targets [28]. In part, this has motivated efforts to obtain high resolution models of riboswitch-ligand complexes that explain the molecular details of ligand binding and provide clues into how these interactions are coupled to gene regulation. These studies have revealed a number of themes common to the various riboswitches—despite their obvious structural differences—and can be summarized by the recent crystal structure of a lysine-responsive riboswitch (L-box) from Thermotoga maritima (Figure 2) [29, 30].
Figure 2. Structural themes exemplified in the lysine riboswitch from the asd gene of T. maritima.
(a) Secondary structure of the L-box RNA. The non-canonical kink turn motif found in this RNA differs from the canonical K-turn consensus sequence found in other phylogenetic variants of this riboswitch class (L-box consensus). (b) A cartoon representation of the crystal structure of the lysine riboswitch (PDB ID 3D0U). Lysine is bound at the center of the five way junction, whose fold is facilitated by highly conserved tertiary structures that are highlighted in shades of blue; the mutually exclusive switching sequence is highlighted in red. (c) Close up view of the kissing loop complex formed between L2 and L3. U91 and G40 form a dinucleotide stack that coordinates perfectly with the major groove of this six base pair helix. (d) Hydrogen bonding contacts made between the ligand and the riboswitch illustrate the participation of the switching sequence (red) peripheral to the binding site (A151 and G152). Coloring is the same as panel (b).
First is the observation that riboswitch architectures are constructed from the same basic recurring tertiary motifs found in other biological RNAs, confirming early predictions that most of the RNA “erector set” has now been observed [31]. For instance, kink-turns, ribose zippers, and loop E motifs, all abundant in the ribosome and other large RNAs, introduce bending and long range tertiary interactions in many riboswitch aptamer domains, including the L-box (Figure 2a and 2b), that are essential to their regulatory functions. A classic GNRA tetraloop/tetraloop receptor which is critical to folding of group I introns [32, 33] is also conserved in particular sub-classes of the dicyclic GMP binding riboswitch [15]. The SAM-II and preQ1 riboswitches both conform to classic H-type pseudoknot folds when bound to their respective ligands [34–36], a fold adopted by many viral RNAs that promote frameshifting during translation [37] and a key functional element of telomerase RNA [38]. Surprisingly, even some of the more complex riboswitch architectural modules have been previously observed; the structure of an FMN binding riboswitch contains two symmetrical T-loop domains (P3–P5 and P2–P6) that align with the H19–H20 domain of the Haloarcula marismorti 23S rRNA with an r.m.s.d. of 1.48 Å, a striking similarity [39].
A second structural trend that encompasses more global aspects of riboswitch folding is the recurrence of helical bundles formed by coaxially stacked Waston-Crick paired helices held parallel to one other by the various motifs mentioned above. In the SAM-I riboswitch for example, two coaxial helical stacks pack together to yield an arrangement similar to the catalytic core of group I introns [40, 41]. Like SAM-I, the purine and TPP riboswitches also form two helix bundles, while the glmS, Mg2+, and lysine riboswitches all have a three helix bundle arrangement [2]. In the T. maritima L-box helical packing is facilitated by a non-canonical kink turn (Figure 2a and 2b) that bends the P2 helix by ~120° to position L2 properly for formation of a kissing loop complex with L3, similar to interaction that promotes genome dimerization in the human immunodeficiency virus [42]. The L-box kissing loop motif is further stabilized by a unique dinucleotide stack that interacts within the major groove of this six base pair L2–L3 helix, a feature that likely promotes thermostability in this RNA (Figure 2c). The third coaxial stack (P4/P5) is cemented to the P1/P2a stack through an A-minor interaction mediated by two highly conserved adenines in the L4 pentaloop (A123 and A124) docking into the minor groove of P2a adjacent to a canonical Loop E motif [43, 44]. Mutations that disrupt these motifs in the L-box compromise its affinity for lysine and ability to control transcription, demonstrating the linkage between tertiary structure formation and ligand binding [45, 46].
A third feature that provides a structural basis for the high affinity and selectivity of riboswitches is the use of binding pockets that typically envelop the metabolite, allowing recognition of nearly every functional group [47]. The L-box uses a five-way helical junction (Figure 2a) as a binding site that completely encapsulates lysine, coordinating binding through a network of hydrogen bonds to the main chain atoms and an electrostatic interaction with positively charged side chain (Figure 2d). The length and charge of the side chain account for lysine specificity over smaller amino acids that could easily be accommodated within this pocket. Ornithine, with a side chain that is a single carbon unit shorter than lysine, is unable to promote transcription termination in vitro at concentrations >10 mM [10]. Additionally, the L-box makes use of a metal ion (K+) in the binding pocket to help meditate RNA-ligand recognition [30], a strategy also employed by the TPP [48, 49] and FMN riboswitches [50].
Finally, almost all riboswitch structures have revealed that the switching sequence incorporated within the binding site or immediately adjacent, implying an intimate linkage between ligand binding and sequestration of this sequence within the aptamer domain. Indeed, in combination with chemical probing techniques, it is clear that ligand binding is accompanied by conformational changes around the binding pocket that often include the switching sequence. This concept was first uncovered through a structural analysis of the B. subtilis xpt-pbuX guanine riboswitch, a transcriptional regulator, in which ligand binding stabilizes formation of two base triples between residues in the three-way junction and the switching sequence [51]. A different solution is adopted by the SAM-II riboswitch where the SAM ligand makes contacts with the Shine-Dalgarno sequence, thereby directly obstructing ribosome binding [34]. Thus the aptamer domain and the expression platform are fully integrated into a single SAM binding pseudoknot. In the L-box, the switching sequence (Figure 2a, red) forms part of the binding pocket [29]. Chemical and in-line footprinting of the RNA indicate that lysine induces the G8•G152 and G138•A151 pairing interactions (Figure 2d) that stabilize incorporation of the switching sequence into the aptamer domain [10, 29, 30, 52]. Thus, in every riboswitch that has been structurally characterized to date, ligand binding is directly coupled to conformational changes that involve the switching sequence, which forms the basis for their regulatory activity.
3. Riboswitch free states and ligand induced folding
A comprehensive understanding of riboswitch regulation requires equal consideration of the free state structure of the aptamer domain. In its unliganded form, the RNA must maintain a conformation (or, more precisely, an ensemble of conformations), in which the binding site is accessible to the ligand. Second, comparison of free and bound state aptamer structures yields atomic-level details about the nature of the conformational changes associated with ligand binding and how these changes lead to regulatory control. Finally, the free state must be able to efficiently direct the RNA along the default folding pathway upon synthesis of the expression platform if ligand binding does not occur, which often requires dissembling part of the secondary structure of the aptamer domain (Figure 1).
In light of the large structural rearrangements usually depicted by cartoons and secondary structure representations, it may seem surprising that many aptamer domains are highly structured in the absence of ligand, undergoing only local conformational changes upon binding. Riboswitches that fit this description have been classified type I RNAs [2], with the purine riboswitch serving as the current model system. Studies using a variety of biochemical and structural techniques have shown that this RNA is globally organized in the absence of ligand by a conserved loop-loop interaction and only a small number of nucleotides in the highly conserved three way junction remain loosely structured [7, 53, 54]. Conformational ordering of these nucleotides occurs only after formation of an initial encounter complex with the ligand. The identity of many of the nucleotides in the binding pocket are crucial not for ligand recognition, but rather maintaining the site in an “open” conformation [18, 55]. Chemical probing and mutagenesis revealed that a number of nucleotides around the binding pocket, if altered, lead to a marked loss in the ability to efficiently recognize ligand, despite the fact that the structure of the bound complex is identical to wild type. These studies strongly argue that that there is considerable selective pressure on the sequence of the aptamer domain to maintain ligand-accessible conformations that do not trigger the inappropriate regulatory response [55].
Analyses of the L-box have revealed that this RNA is also a type I riboswitch. Small angle X-ray scattering (SAXS) data show that the L-box undergoes a large compaction upon addition of Mg2+ [29], consistent with the requirement of this ion for stabilizing key tertiary elements [56, 57]. The addition of lysine however, causes very little observable change in SAXS and native gel electrophoresis experiments, indicating that the ligand induced conformational change of this RNA is limited to local regions, much like the purine riboswitch. Chemical probing of the lysine riboswitch demonstrated this change involves a small set of nucleotides found in the five-way junction that form non-canonical pairing interactions in the core of the binding pocket [29] (Figure 2d). Notably, the crystal structure of the unliganded L-box has also been solved, and is nearly identical to its bound conformation [29, 30]. While this certainly does not represent the full conformational ensemble of the free-state RNA in solution, it suggests this conformation is populated to some degree in the absence of ligand. Since the binding pocket is inaccessible to lysine in this conformation, it must exchange with a set of open conformers in the absence of ligand, the rate of which likely has considerable implications on the rate of ligand binding and thereby the regulatory mechanism (vide infra).
Other riboswitch aptamers appear to be more unfolded in the free state, and experience global conformational change upon ligand binding, giving them the classification of type II RNAs [41]. These RNAs have bipartite binding pockets that span the coaxial stacks of riboswitches as illustrated by the thiamine pyrophosphate (TPP) riboswitch. In-line probing and limited nuclease digestion demonstrate that ligand binding is coupled to the formation of a GNRA like tetraloop docking interaction between loop 5 (L5) and the P3 helix of this RNA, suggesting that the free state does not adopt the helical packing observed by crystallography [12, 48, 49]. Fluorescence spectroscopy of RNA labeled site-specifically with 2-aminopurine showed that the L5-P3 interaction forms rapidly upon addition of TPP, serving to orient the P4/P5 coaxial stack in parallel with the P2/P3 stack, nucleating slower formation of the three way junction and P1 helix that contains the regulatory switching sequence [58]. Thus ligand binding is propagated into distal conformational changes that result in a regulatory response. Interestingly, it has been proposed that folding and binding can be decoupled in this RNA by mutations that retain ligand binding, but prevent folding of a crucial base quadruple in the three way junction [49].
4. Temporal aspects of riboswitch regulation
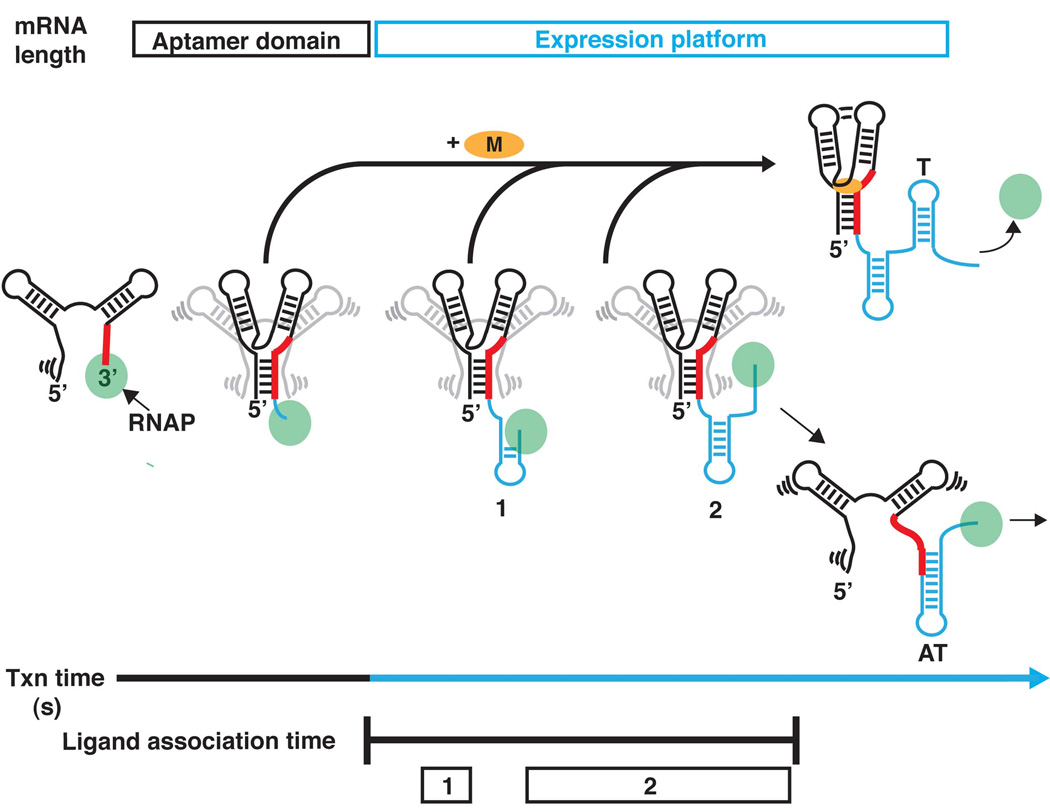
Since the early recognition that riboswitches are modular RNAs [12], most structural studies have focused on aptamer domains in isolation, providing only a snapshot in the life of a riboswitch. However, it is important to recognize mRNA folding in the context of transcription is central to the regulatory function of these RNAs. During each individual transcription event, the aptamer domain must first be able to rapidly fold into an active conformation with high fidelity (the “aptamer” time regime; black, Figure 3) and bind to its effector ligand or undergo a potential secondary structural rearrangement in the absence of ligand. These events are slated to occur in the time required to transcribe the full riboswitch (the “expression” time regime; blue, Figure 3). As bacterial polymerases have average elongation rates of ~50 nucleotides per second [59], it appears that riboswitches must have strict kinetic constraints imposed upon them. The degree to which this holds true may vary depending on the overall length and sequence composition of the expression platform of a given transcript.
Figure 3. A riboswitch timeline.
During the aptamer time regime (black), the sensor domain is synthesized and acquires secondary structure rapidly followed by slower formation of Mg2+-dependent tertiary structure. Upon formation of a competent aptamer domain the opportunity for ligand binding becomes possible. During the expression time regime (blue) the expression platform is synthesized, but the aptamer domain remains binding competent until the riboswitch has committed to forming the antiterminator element (“ligand association time”). During this time there are often two programmed transcriptional pauses (boxes 1 and 2) that stall the polymerase, and thereby influence the amount of time the riboswitch has for sensing its environment before committing to formation of one of the two mutually exclusive secondary structures in the expression platform.
To study the kinetics of folding during the “aptamer” time regime of an adenine binding riboswitch, a recent study by Block and coworkers applied single molecule force spectroscopy [60]. The rate of RNA refolding was monitored by fully extending the RNA under high force, followed by measuring the probability of obtaining fully refolded RNA as a function of time. This was done in the presence of adenine concentrations ranging from 1 to 200 µM to obtain true rate constants from the pseudo-first order constants determined at each concentration. This analysis yielded a rate constant of 0.4 s−1 for formation of a competent receptor, and kon and koff of values of 8×104 M−1 s−1 and 0.2 s−1 respectively for the ligand binding step. This translates to time constants (time required for ~37% of the reaction to go to completion) of approximately 3 seconds for folding, 4 seconds for ligand association (assuming a concentration of 3 µM adenine), and ~8 seconds for ligand dissociation. Thus, folding and ligand binding of the RNA aptamer occur on a timescale similar to transcription of the riboswitch (~4 seconds for transcription of 200 nucleotides without pausing; vide infra), supporting the hypothesis that the individual rate constants of folding and binding have a stronger influence on the outcome of gene expression than the equilibrium binding of ligand. It can also be reasoned that mutations that influence the rate of tertiary structure acquisition during the aptamer time regime (Figure 3; blue) would prevent the receptor from forming on a timescale relevant to transcription, which has been proposed as a possible mechanism of some naturally occurring mutations that disrupt regulation by the L-box but have relatively subtle effects on equilibrium lysine binding [29, 52].
A consequence of riboswitches being under “kinetic control” is that there are large differences between the affinity (the apparent equilibrium dissociation constant, KD) of isolated aptamer domains for their effector, and the concentration of ligand required to achieve half maximal regulatory response (the T50) in single round in vitro transcription assays. This phenomenon that has been observed for many riboswitch classes, and relates to the time required to transcribe the expression platform [10, 61, 62]. The most comprehensive study of the relationship of riboswitch function to the rate of transcription was conducted using the FMN riboswitch from the B. subtilis ribD operon [63]. The apparent KD of the FMN aptamer domain is 10 nM while the T50 is 500 nM in the absence of factors that promote transcriptional pausing [62]. However, slowing the speed of transcription by either reducing NTP concentrations, or by adding NusA (a protein that increases RNA polymerase (RNAP) pausing [64]), causes a ~2-fold decrease in the T50 to 200 nM. The speed of transcription and the efficiency of riboswitch regulation are therefore intimately coupled [62], underscoring the need to understand ligand recognition not in the context of isolated aptamers, but also as a part of transcription.
The induced fit binding mechanism of riboswitches, a feature shared by many RNA binding interactions, plays an important role in the regulatory mechanism of these RNAs [65, 66]. As alluded to above, ligand binding in the purine riboswitch is ~104 slower than diffusion limited processes. This is also true of the TPP and FMN riboswitches, implying that this is a general phenomenon in riboswitches [58, 60, 62, 63, 67]. These rates suggest that effector binding to these riboswitches is far too slow to be able to control transcription at concentrations close to the equilibrium dissociation constant. In part, this explains the higher ligand concentrations needed to effect half maximal transcriptional control. However, the ribD FMN riboswitch revealed the importance of intrinsic RNAP pausing at uridine-rich tracts encoded in the expression platform [63]. Two pauses were identified that occur immediately after the aptamer domain and after the 3’-sequence of the antiterminator (Figure 3), giving the RNA additional time (~1 and ~10 seconds respectively for the ribD FMN riboswitch) to interrogate the cellular environment before the conformational switch can happen. Despite this pausing phenomenon, the binding reaction still does not have ample time to reach equilibrium, though it does reduce the ligand concentration needed to control transcription.
It has been proposed that programmed pausing is a general strategy employed by non-coding RNA to efficiently fold during transcription. Ubiquitous housekeeping RNAs such as RNase P, SRP RNA, and tmRNA have adapted to the temporal pressures of transcription by forming “labile” intermediates during programmed pauses in the early stages of transcription [68]. These intermediates function to sequester 5’ sequences that form long-range helices (separated by >50 nt) in the native structure, and disruption of pausing that results in loss of intermediate formation leads to less efficient folding. A reasonable speculation is that the labile intermediates can be rapidly rearranged to their native state, thus providing a mechanism for preventing the RNA from adopting stabile but inactive folds. This is analogous to riboswitches whose long range P1 helix, which defines the 5’ and 3’ boundaries of the aptamer domain, is generally implicated in structural switching. This element would be expected to undergo rearrangement if the aptamer domain remains unbound, similar to the labile folding intermediates identified in the above RNAs.
5. Riboswitches in the cellular milieu
While structural and biochemical characterization of riboswitch-ligand interactions have illuminated many aspects of their regulatory mechanisms, these data cannot always be completely reconciled with their biological activity. This is exemplified in a recent study of the variability in the ligand responsiveness of 11 different SAM responsive transcriptional units in B. subtilis genome [69]. These riboswitches were demonstrated to have a ~250 fold range in KD and T50 in vitro. Additionally, in vivo Northern blotting, qRT-PCR, and lacZ reporter analyses revealed a a large degree of variability in the fold change of terminated transcription in the presence and absence of methionine: this ranged from a 1.2-fold increase in termination for the metK gene in the presence of methionine, to a 340-fold increase for the metE gene as measured by qRT-PCR. The function of the gene products is thought to be tied to this disparity. Genes involved directly in methionine biosynthesis experienced the tightest regulation, the greatest level of induction, and the longest delays before induction could be detected. Meanwhile riboswitches controlling methionine transport and genes of unknown function displayed lower levels of repression during growth in methionine supplemented media, and lower magnitudes of induction during methionine starvation. This study suggests that there are structural differences in the aptamer domain or expression platform of these SAM riboswitches may account for these disparities, motivating the need to correlate these observations to available structural and sequence based data. Furthermore, differences in the time of transcription (i.e. transcriptional pausing) at these different genetic loci may tune the response, requiring further characterization of this phenomenon for each transcript.
Interestingly, not all of these riboswitches behaved as expected. The cysH gene does not experience induction upon methionine starvation [69], which may be explained by previous reports that this operon is regulated primarily at the DNA level by a transcriptional repressor protein that responds to levels of the cystine precursor O-acetyl-L-serine [70]. The metK gene on the other hand experiences a transient rise in read through product at 0.5 hours, but falls back to basal levels after one hour [69]. This expression pattern suggests that this transcript may also be regulated at either the translational level or by the effects of mRNA degradation. A consequence of overlapping post-transcriptional regulation may therefore lead to differences in riboswitch response efficiency, further highlighting the need to correlate biochemical data with biological activity.
Another motivation for studying ligand interactions in their cellular context is to determine the potential of these RNAs as novel antimicrobial targets. Ribosomal RNA sets a precedence for RNA based strategies, as its known to be the target of aminoglycoside antibiotics, and many riboswitches have already been linked to antibiotic effects of naturally occurring compounds. For example, the L-box has long been recognized as a mutational hotspot in microbes resistant to the lysine analogue S-(2-aminoethyl)-L-cysteine [71, 72]. A recent study implicated role of the L-box in mediating the toxicity of this and other lysine analogues in B. subtilis [52]. However, shortly after this study was published, it was discovered that the primary target of these compounds in E. coli was one of the two lysyl-tRNA synthetase variants (LysRS) in this organism and the toxic effects are due to incorporation into nascent polypeptides during translation [73]. Mutations to the L-box cause inefficient repression of lysine biosynthesis genes, thus allowing lysine pools to become elevated and effectively compete for the charging of LysRS [73]. A similar study demonstrated that the TPP riboswitch serves as a mutational hotspot for relieving the toxic effects of the TPP analogue pyrithiamine pyrophosphate (PTPP) in B. subtilis, though many of the PTPP resistant E.coli mutants that were sequenced apparently acquired resistance by some other mechanism [74]. However, the toxic effects of roseoflavin, an FMN analogue, to B. subtilis are a clearly at least in part due to direct targeting of the riboswitch upstream of the ribD operon by the antimicrobial [75]. The homologous operon in Streptomyces davawensis—an organism that naturally produces this antibacterial agent—is also responsive to roseoflavin leaving the mechanism of this bacteria’s natural resistance to this antimicriobial compound unclear, highlighting the need to understand the biological roles of riboswitches in their cellular context.
6. Conclusions
With a wealth of structural information available (currently, at least 10 individual aptamer-ligand complexes have been solved by X-ray crystallography), a significant challenge remains to correlate these data with in vitro and in vivo studies that take into account the temporal aspects of riboswitch synthesis and function. In particular, new approaches need to be developed that can readily monitor cotranscriptional riboswitch folding with an eye towards its relationship to rates of ligand binding, the role of transcriptional pausing in the expression platform, and secondary structural rearrangements and their relationship to efficient genetic regulation. Determination of, response kinetics and regulatory capacity (i.e. fold induction or repression) in vivo may also reveal factors that overlap with riboswitch regulation to further tune expression; concepts that will need to be addressed to determine efficacy of targeting riboswitches for therapeutic purposes. An overarching goal of these studies will be to provide a clear link between structural and functional studies that takes into consideration the complicated folding process of a riboswitch in the cell.
Acknowledgments
The authors would like to thank P. Ceres, A.L. Edwards, J.J. Johnson, R.K. Montange, C.D. Stoddard, F.E. Reyes, and Q. Vicens for providing insightful discussion during the preparation of this manuscript. Funding for this work was provided by an NIH grant to R.T.B. (GM073850) and an NIH Molecular Biophysics Pre-doctoral Training Grant to A.D.G. (T32 GM065103).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Roth A, Breaker RR. The Structural and Functional Diversity of Metabolite-Binding Riboswitches. Annu Rev Biochem. 2009 doi: 10.1146/annurev.biochem.78.070507.135656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Montange RK, Batey RT. Riboswitches: emerging themes in RNA structure and function. Annual review of biophysics. 2008;37:117–133. doi: 10.1146/annurev.biophys.37.032807.130000. [DOI] [PubMed] [Google Scholar]
- 3.Irnov, Kertsburg A, Winkler WC. Genetic control by cis-acting regulatory RNAs in Bacillus subtilis: general principles and prospects for discovery. Cold Spring Harb Symp Quant Biol. 2006;71:239–249. doi: 10.1101/sqb.2006.71.021. [DOI] [PubMed] [Google Scholar]
- 4.Stoddard CD, Batey RT. Mix-and-match riboswitches. ACS Chem Biol. 2006;1:751–754. doi: 10.1021/cb600458w. [DOI] [PubMed] [Google Scholar]
- 5.Winkler WC, Breaker R. Regulation of bacterial gene expression by riboswitches. Annu Rev Microbiol. 2005;59:487–517. doi: 10.1146/annurev.micro.59.030804.121336. [DOI] [PubMed] [Google Scholar]
- 6.Wachter A, Tunc-Ozdemir M, Grove BC, Green PJ, Shintani DK, Breaker R. Riboswitch control of gene expression in plants by splicing and alternative 3' end processing of mRNAs. Plant Cell. 2007;19:3437–3450. doi: 10.1105/tpc.107.053645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mandal M, Boese B, Barrick J, Winkler WC, Breaker R. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell. 2003;113:577–586. doi: 10.1016/s0092-8674(03)00391-x. [DOI] [PubMed] [Google Scholar]
- 8.Mandal M, Breaker R. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat Struct Mol Biol. 2004;11:29–35. doi: 10.1038/nsmb710. [DOI] [PubMed] [Google Scholar]
- 9.Mandal M, Lee M, Barrick J, Weinberg Z, Emilsson GM, Ruzzo WL, Breaker R. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science. 2004;306:275–279. doi: 10.1126/science.1100829. [DOI] [PubMed] [Google Scholar]
- 10.Sudarsan N, Wickiser JK, Nakamura S, Ebert MS, Breaker R. An mRNA structure in bacteria that controls gene expression by binding lysine. Genes & Development. 2003;17:2688–2697. doi: 10.1101/gad.1140003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nahvi A, Sudarsan N, Ebert MS, Zou X, Brown KL, Breaker R. Genetic control by a metabolite binding mRNA. Chemistry & Biology. 2002;9:1043–1049. doi: 10.1016/s1074-5521(02)00224-7. [DOI] [PubMed] [Google Scholar]
- 12.Winkler W, Nahvi A, Breaker R. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- 13.Winkler WC, Nahvi A, Roth A, Collins JA, Breaker R. Control of gene expression by a natural metabolite-responsive ribozyme. Nature. 2004;428:281–286. doi: 10.1038/nature02362. [DOI] [PubMed] [Google Scholar]
- 14.Dann CE, Wakeman CA, Sieling CL, Baker SC, Irnov I, Winkler WC. Structure and mechanism of a metal-sensing regulatory RNA. Cell. 2007;130:878–892. doi: 10.1016/j.cell.2007.06.051. [DOI] [PubMed] [Google Scholar]
- 15.Sudarsan N, Lee ER, Weinberg Z, Moy RH, Kim JN, Link KH, Breaker RR. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science. 2008;321:411–413. doi: 10.1126/science.1159519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barrick J, Corbino KA, Winkler WC, Nahvi A, Mandal M, Collins J, Lee M, Roth A, Sudarsan N, Jona I, Wickiser JK, Breaker R. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc Natl Acad Sci USA. 2004;101:6421–6426. doi: 10.1073/pnas.0308014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weinberg Z, Barrick J, Yao Z, Roth A, Kim J, Gore J, Wang J, Lee E, Block K, Sudarsan N, Neph S, Tompa M, Ruzzo W, Breaker R. Identification of 22 candidate structured RNAs in bacteria using the CMfinder comparative genomics pipeline. Nucleic Acids Research. 2007;35:4809–4819. doi: 10.1093/nar/gkm487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mulhbacher J, Lafontaine D. Ligand recognition determinants of guanine riboswitches. Nucleic Acids Research. 2007;35:5568–5580. doi: 10.1093/nar/gkm572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tucker BJ, Breaker R. Riboswitches as versatile gene control elements. Current Opinion in Structural Biology. 2005;15:342–348. doi: 10.1016/j.sbi.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 20.Kwon M, Strobel SA. Chemical basis of glycine riboswitch cooperativity. RNA. 2008;14:25–34. doi: 10.1261/rna.771608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sudarsan N, Hammond M, Block K, Welz R, Barrick J, Roth A, Breaker R. Tandem Riboswitch Architectures Exhibit Complex Gene Control Functions. Science. 2006;314:300–304. doi: 10.1126/science.1130716. [DOI] [PubMed] [Google Scholar]
- 22.Fox KA, Ramesh A, Stearns JE, Bourgogne A, Reyes-Jara A, Winkler WC, Garsin DA. Multiple posttranscriptional regulatory mechanisms partner to control ethanolamine utilization in Enterococcus faecalis. Proc Natl Acad Sci U S A. 2009;106:4435–4440. doi: 10.1073/pnas.0812194106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barrick J, Breaker R. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol. 2007;8:R239. doi: 10.1186/gb-2007-8-11-r239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Collins JA, Irnov I, Baker S, Winkler WC. Mechanism of mRNA destabilization by the glmS ribozyme. Genes & Development. 2007;21:3356–3368. doi: 10.1101/gad.1605307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCarthy TJ, Plog MA, Floy SA, Jansen JA, Soukup JK, Soukup GA. Ligand requirements for glmS ribozyme self-cleavage. Chem Biol. 2005;12:1221–1226. doi: 10.1016/j.chembiol.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 26.Cheah MT, Wachter A, Sudarsan N, Breaker RR. Control of alternative RNA splicing and gene expression by eukaryotic riboswitches. Nature. 2007;447:497–500. doi: 10.1038/nature05769. [DOI] [PubMed] [Google Scholar]
- 27.Ray PS, Jia J, Yao P, Majumder M, Hatzoglou M, Fox PL. A stress-responsive RNA switch regulates VEGFA expression. Nature. 2009;457:915–919. doi: 10.1038/nature07598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blount K, Breaker R. Riboswitches as antibacterial drug targets. Nat Biotechnol. 2006;24:1558–1564. doi: 10.1038/nbt1268. [DOI] [PubMed] [Google Scholar]
- 29.Garst AD, Heroux A, Rambo RP, Batey RT. Crystal structure of the lysine riboswitch regulatory mRNA element. J Biol Chem. 2008;283:22347–22351. doi: 10.1074/jbc.C800120200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Serganov A, Huang L, Patel DJ. Structural insights into amino acid binding and gene control by a lysine riboswitch. Nature. 2008;455:1263–1267. doi: 10.1038/nature07326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moore PB. Structural motifs in RNA. Annu Rev Biochem. 1999;68:287–300. doi: 10.1146/annurev.biochem.68.1.287. [DOI] [PubMed] [Google Scholar]
- 32.Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Kundrot CE, Cech TR, Doudna JA. Crystal structure of a group I ribozyme domain: principles of RNA packing. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- 33.Costa M, Michel F. Frequent use of the same tertiary motif by self-folding RNAs. EMBO J. 1995;14:1276–1285. doi: 10.1002/j.1460-2075.1995.tb07111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gilbert SD, Rambo RP, Van Tyne D, Batey RT. Structure of the SAM-II riboswitch bound to S-adenosylmethionine. Nat Struct Mol Biol. 2008;15:177–182. doi: 10.1038/nsmb.1371. [DOI] [PubMed] [Google Scholar]
- 35.Kang M, Peterson R, Feigon J. Structural Insights into riboswitch control of the biosynthesis of queuosine, a modified nucleotide found in the anticodon of tRNA. Mol Cell. 2009;33:784–790. doi: 10.1016/j.molcel.2009.02.019. [DOI] [PubMed] [Google Scholar]
- 36.Klein DJ, Edwards TE, Ferré-D'Amaré AR. Cocrystal structure of a class I preQ(1) riboswitch reveals a pseudoknot recognizing an essential hypermodified nucleobase. Nat Struct Mol Biol. 2009 doi: 10.1038/nsmb.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brierley I, Pennell S, Gilbert RJ. Viral RNA pseudoknots: versatile motifs in gene expression and replication. Nat Rev Microbiol. 2007;5:598–610. doi: 10.1038/nrmicro1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Theimer CA, Feigon J. Structure and function of telomerase RNA. Curr Opin Struct Biol. 2006;16:307–318. doi: 10.1016/j.sbi.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 39.Krasilnikov AS, Mondragón A. On the occurrence of the T-loop RNA folding motif in large RNA molecules. RNA. 2003;9:640–643. doi: 10.1261/rna.2202703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adams PL, Stahley MR, Kosek AB, Wang J, Strobel SA. Crystal structure of a self-splicing group I intron with both exons. Nature. 2004;430:45–50. doi: 10.1038/nature02642. [DOI] [PubMed] [Google Scholar]
- 41.Montange RK, Batey RT. Structure of the S-adenosylmethionine riboswitch regulatory mRNA element. Nature. 2006;441:1172–1175. doi: 10.1038/nature04819. [DOI] [PubMed] [Google Scholar]
- 42.Ennifar E, Walter P, Ehresmann B, Ehresmann C, Dumas P. Crystal structures of coaxially stacked kissing complexes of the HIV-1 RNA dimerization initiation site. Nat Struct Biol. 2001;8:1064–1068. doi: 10.1038/nsb727. [DOI] [PubMed] [Google Scholar]
- 43.Leontis NB, Stombaugh J, Westhof E. Motif prediction in ribosomal RNAs Lessons and prospects for automated motif prediction in homologous RNA molecules. Biochimie. 2002;84:961–973. doi: 10.1016/s0300-9084(02)01463-3. [DOI] [PubMed] [Google Scholar]
- 44.Leontis NB, Westhof E. The 5S rRNA loop E: chemical probing and phylogenetic data versus crystal structure. RNA. 1998;4:1134–1153. doi: 10.1017/s1355838298980566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blouin S, Lafontaine DA. A loop loop interaction and a K-turn motif located in the lysine aptamer domain are important for the riboswitch gene regulation control. RNA. 2007;13:1256–1267. doi: 10.1261/rna.560307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grundy FJ, Lehman SC, Henkin TM. The L box regulon: lysine sensing by leader RNAs of bacterial lysine biosynthesis genes. Proc Natl Acad Sci U S A. 2003;100:12057–12062. doi: 10.1073/pnas.2133705100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Edwards TE, Klein DJ, Ferré-D'Amaré AR. Riboswitches: small-molecule recognition by gene regulatory RNAs. Current Opinion in Structural Biology. 2007;17:273–279. doi: 10.1016/j.sbi.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 48.Serganov A, Polonskaia A, Phan AT, Breaker R, Patel DJ. Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nature. 2006;441:1167–1171. doi: 10.1038/nature04740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thore S, Leibundgut M, Ban N. Structure of the eukaryotic thiamine pyrophosphate riboswitch with its regulatory ligand. Science. 2006;312:1208–1211. doi: 10.1126/science.1128451. [DOI] [PubMed] [Google Scholar]
- 50.Serganov A, Huang L, Patel DJ. Coenzyme recognition and gene regulation by a flavin mononucleotide riboswitch. Nature. 2009;458:233–237. doi: 10.1038/nature07642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Batey RT, Gilbert SD, Montange RK. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature. 2004;432:411–415. doi: 10.1038/nature03037. [DOI] [PubMed] [Google Scholar]
- 52.Blount KF, Wang JX, Lim J, Sudarsan N, Breaker R. Antibacterial lysine analogs that target lysine riboswitches. Nat Chem Biol. 2007;3:44–49. doi: 10.1038/nchembio842. [DOI] [PubMed] [Google Scholar]
- 53.Noeske J, Buck J, Furtig B, Nasiri H, Schwalbe H, Wohnert J. Interplay of 'induced fit' and preorganization in the ligand induced folding of the aptamer domain of the guanine binding riboswitch. Nucleic Acids Research. 2006;35:572–583. doi: 10.1093/nar/gkl1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Noeske J, Richter C, Grundl MA, Nasiri HR, Schwalbe H, Wöhnert J. An intermolecular base triple as the basis of ligand specificity and affinity in the guanine- and adenine-sensing riboswitch RNAs. Proc Natl Acad Sci USA. 2005;102:1372–1377. doi: 10.1073/pnas.0406347102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gilbert SD, Love CE, Edwards AL, Batey RT. Mutational analysis of the purine riboswitch aptamer domain. Biochemistry. 2007;46:13297–13309. doi: 10.1021/bi700410g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Draper DE, Grilley D, Soto AM. Ions and RNA folding. Annu Rev Biophys Biomol Struct. 2005;34:221–243. doi: 10.1146/annurev.biophys.34.040204.144511. [DOI] [PubMed] [Google Scholar]
- 57.Woodson SA. Metal ions and RNA folding: a highly charged topic with a dynamic future. Curr Opin Chem Biol. 2005;9:104–109. doi: 10.1016/j.cbpa.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 58.Lang K, Rieder R, Micura R. Ligand-induced folding of the thiM TPP riboswitch investigated by a structure-based fluorescence spectroscopic approach. Nucleic Acids Research. 2007;35:5370–5378. doi: 10.1093/nar/gkm580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ryals J, Little R, Bremer H. Temperature dependence of RNA synthesis parameters in Escherichia coli. J Bacteriol. 1982;151:879–887. doi: 10.1128/jb.151.2.879-887.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Greenleaf WJ, Frieda KL, Foster DA, Woodside MT, Block SM. Direct observation of hierarchical folding in single riboswitch aptamers. Science. 2008;319:630–633. doi: 10.1126/science.1151298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McDaniel BA, Grundy FJ, Artsimovitch I, Henkin TM. Transcription termination control of the S box system: direct measurement of S-adenosylmethionine by the leader RNA. Proc Natl Acad Sci U S A. 2003;100:3083–3088. doi: 10.1073/pnas.0630422100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wickiser JK, Cheah MT, Breaker R, Crothers DM. The kinetics of ligand binding by an adenine-sensing riboswitch. Biochemistry. 2005;44:13404–13414. doi: 10.1021/bi051008u. [DOI] [PubMed] [Google Scholar]
- 63.Wickiser J, Winkler W, Breaker R, Crothers D. The Speed of RNA Transcription and Metabolite Binding Kinetics Operate an FMN Riboswitch. Molecular Cell. 2005;18:49–60. doi: 10.1016/j.molcel.2005.02.032. [DOI] [PubMed] [Google Scholar]
- 64.Yakhnin AV, Babitzke P. NusA-stimulated RNA polymerase pausing and termination participates in the Bacillus subtilis trp operon attenuation mechanism invitro. Proc Natl Acad Sci U S A. 2002;99:11067–11072. doi: 10.1073/pnas.162373299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Leulliot N, Varani G. Current topics in RNA-protein recognition: control of specificity and biological function through induced fit and conformational capture. Biochemistry. 2001;40:7947–7956. doi: 10.1021/bi010680y. [DOI] [PubMed] [Google Scholar]
- 66.Williamson JR. Induced fit in RNA-protein recognition. Nat Struct Biol. 2000;7:834–837. doi: 10.1038/79575. [DOI] [PubMed] [Google Scholar]
- 67.Gilbert SD, Stoddard CD, Wise SJ, Batey RT. Thermodynamic and kinetic characterization of ligand binding to the purine riboswitch aptamer domain. J Mol Biol. 2006;359:754–768. doi: 10.1016/j.jmb.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 68.Wong TN, Sosnick TR, Pan T. Folding of noncoding RNAs during transcription facilitated by pausing-induced nonnative structures. Proc Natl Acad Sci USA. 2007;104:17995–18000. doi: 10.1073/pnas.0705038104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tomsic J, McDaniel BA, Grundy FJ, Henkin TM. Natural variability in S-adenosylmethionine (SAM)-dependent riboswitches: S-box elements in bacillus subtilis exhibit differential sensitivity to SAM In vivo and in vitro. Journal of Bacteriology. 2008;190:823–833. doi: 10.1128/JB.01034-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mansilla MC, Albanesi D, de Mendoza D. Transcriptional control of the sulfur-regulated cysH operon, containing genes involved in L-cysteine biosynthesis in Bacillus subtilis. Journal of Bacteriology. 2000;182:5885–5892. doi: 10.1128/jb.182.20.5885-5892.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lu Y, Shevtchenko TN, Paulus H. Fine-structure mapping of cis-acting control sites in the lysC operon of Bacillus subtilis. FEMS Microbiol Lett. 1992;71:23–27. doi: 10.1016/0378-1097(92)90536-w. [DOI] [PubMed] [Google Scholar]
- 72.Patte JC, Akrim M, Mejean V. The leader sequence of the Escherichia coli lysC gene is involved in the regulation of LysC synthesis. FEMS Microbiol Lett. 1998;169:165–170. doi: 10.1111/j.1574-6968.1998.tb13313.x. [DOI] [PubMed] [Google Scholar]
- 73.Ataide SF, Wilson SN, Dang S, Rogers TE, Roy B, Banerjee R, Henkin TM, Ibba M. Mechanisms of resistance to an amino acid antibiotic that targets translation. ACS Chem Biol. 2007;2:819–827. doi: 10.1021/cb7002253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sudarsan N, Cohen-Chalamish S, Nakamura S, Emilsson GM, Breaker R. Thiamine pyrophosphate riboswitches are targets for the antimicrobial compound pyrithiamine. Chemistry & Biology. 2005;12:1325–1335. doi: 10.1016/j.chembiol.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 75.Lee ER, Blount KF, Breaker RR. Roseoflavin is a natural antibacterial compound that binds to FMN riboswitches and regulates gene expression. RNA Biol. 2009;6 doi: 10.4161/rna.6.2.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]